Abstract
Despite the increasing importance of long noncoding RNA in physiology and disease, their role in endothelial biology remains poorly understood. Growing evidence has highlighted them to be essential regulators of human embryonic stem cell differentiation. SENCR, a vascular-enriched long noncoding RNA, overlaps the Friend Leukemia Integration virus 1 (FLI1) gene, a regulator of endothelial development. Therefore, we wanted to test the hypothesis that SENCR may contribute to mesodermal and endothelial commitment as well as in endothelial function. We thus developed new differentiation protocols allowing generation of endothelial cells from human embryonic stem cells using both directed and hemogenic routes. The expression of SENCR was markedly regulated during endothelial commitment using both protocols. SENCR did not control the pluripotency of pluripotent cells; however its overexpression significantly potentiated early mesodermal and endothelial commitment. In human umbilical endothelial cell (HUVEC), SENCR induced proliferation, migration, and angiogenesis. SENCR expression was altered in vascular tissue and cells derived from patients with critical limb ischemia and premature coronary artery disease compared to controls. Here, we showed that SENCR contributes to the regulation of endothelial differentiation from pluripotent cells and controls the angiogenic capacity of HUVEC. These data give novel insight into the regulatory processes involved in endothelial development and function.
Introduction
Proangiogenic therapy to promote regeneration of damaged tissue is a challenge for medicine. Human embryonic stem cells (hESC) offer a broad perspective to regenerative medicine due to their ability of self-renewal and differentiation into the three germ layers including mesoderm.1 Mesodermal commitment is dependent on epithelial to mesenchymal transition (EMT) described by the loss of epithelial marker CD326 and the acquisition of the neural cell adhesion molecule CD56.2 The mesodermal specification is an important step in vascular development in controlling the de novo emergence of primordial endothelial cells (EC).3 The potential for hESC to form endothelium has been studied using 2D monolayer-directed differentiation or 3D embryoid body (EB) differentiation culture.4 However, a specific developmental stimulus sufficient to support the specification of large numbers of functional EC from hESC (hESC-EC) is poorly understood. Accordingly, the comprehension of the molecular mechanisms governing hESC fate is a crucial step for their clinical use in cell-based therapies.
Noncoding RNAs (ncRNAs) have emerged to be essential regulators of cell function and identity. NcRNAs are classified by size and include long ncRNAs (lncRNA, >200 nucleotides) and small ncRNAs (<200 nucleotides) such as microRNA (miRNA). MiRNA play an important role in maintaining pluripotent state and stimulating differentiation of hESC including EC commitment.5 LncRNA participate in a variety of biological processes, such as chromosome imprinting, epigenetic regulation, and reprogramming of induced pluripotent stem cells.6 Recently, functional lncRNA have emerged as novel regulators of hESC pluripotency and differentiation.7 However, the role of lncRNA in mesodermal and EC commitment and function is poorly understood. Recently, FENDRR and BRAVHEART have been described as mesodermal-specific lncRNA and shown to be important for heart development.8,9 TIE1-AS was the first lncRNA described as an essential modulator of EC development and altered in patients with vascular malformation.10 ALIEN and PUNISHER have been identified to be orchestrators of cardiovascular commitment and EC function, respectively.11 Overall, these reports highlight the importance of lncRNA in the control of cell lineage specification during embryonic development.
SENCR is a human vascular-enriched lncRNA located on the chromosome 11 and existing as 2 isoforms including a full length (SENCR V1) and an alternative spliced variant (SENCR V2)12 (Supplementary Figure S1). SENCR has been described to stabilize the differentiated state of human vascular smooth muscle cells (VSMC) and is transcribed antisense from the first intron of the Friend Leukemia Integration virus 1 (FLI1) gene.12 FLI1 is a member of E26 Transformation-specific (ETS) family expressed in the first step of EC development and is one of the earliest expressed transcription factors (TF) involved in EC specification.13
Since SENCR was found to be most abundant in EC and to correlate with expression of FLI1,12 we hypothesized that it may play a role during EC differentiation and in EC function. To investigate this, we developed two novel protocols allowing efficient EC differentiation from hESC using direct and haemogenic routes. We then profiled the dynamic regulation of SENCR during endothelial differentiation. We further characterized its role in controlling the mesodermal and endothelial commitment of hESC. Finally, we revealed an alteration of SENCR in vascular tissue and EC derived from patients with critical limb ischemia and premature coronary artery disease.
Results
Differentiation protocols using direct routing or hemogenic endothelium formation allow generation of EC from hESC
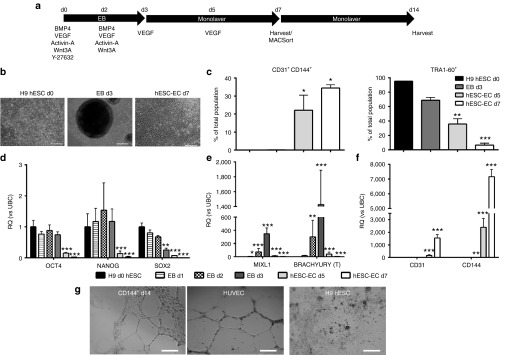
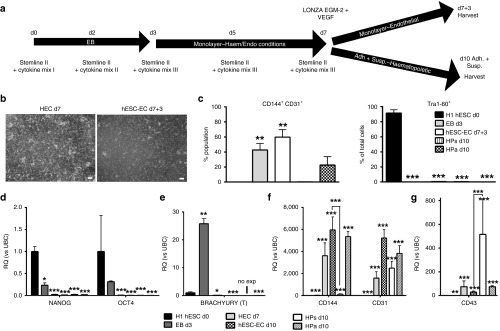
To investigate the role of lncRNA during EC development, we established two novel protocols for the derivation of hESC-EC; direct and indirect, the latter routed through hemogenic endothelium. For direct hESC-EC differentiation, H9 d0 hESC were forced into EB, plated out on d3 and cultured for 4 days in EGM-2 (Figure 1a,b). At d7, EC markers CD144 and CD31 were increased, while pluripotency-marker TRA1-60 was decreased (Figure 1c, Supplementary Figure S2a top). Pluripotency-associated genes OCT4, NANOG, and SOX2 were decreased from d3 (Figure 1d), mesodermal genes BRACHYURY and MIXL1 peaked at d3 (Figure 1e), and EC genes CD31 and CD144 were upregulated from d3 (Figure 1f). Similar results were obtained using H1 hESC line (Supplementary Figure S2a bottom, Supplementary Figure S2b). D7 hESC-EC CD144+ and CD144- were purified and expanded for 7 days, then expression of EC markers CD144 and CD31 was analyzed (Supplementary Figure S3). After 7 days of expansion, CD144+ hESC-EC, coexpressing CD31, were able to form tube-like structures on matrigel (Figure 1g) supporting their functionality. Hemogenic endothelial cells (HEC) are progenitors, hypothesised to give rise to EC and hematopoietic progenitor cells (HP) during development,14 previously identified as CD144+CD73−CD235a−CD43−CD117intermediate.15 EC generation through HEC was developed using the first 7 days of an HP differentiation protocol (Supplementary Methods, Figure 2a). By d7, we observed the formation of suspension HP (HPs), which bud off adherent HP (HPa). Within HPa cells, we identified a CD31+CD144+CD235A−CD117−CD43mixedCD73mixed population with cells expressing either CD43 or CD73 (Supplementary Figure S4), similar to previously described for HEC.15 EC capacity of d7-9 HPa was examined by culturing d7, 8 or d9 HPa cells in EGM-2 medium containing FBS for 3, 2 or 1 day until d10 (hESC-EC d7+3, d8+2, d9+1 respectively). Generation of hESC-EC was efficient using 3 days in EC culture media (hESC-EC d7+3) (Supplementary Figure S5). HESC-EC d7+3 formed confluent monolayers, with a reduction in budding and suspension cells (Figure 2b). HEC were cultured in HP and EC conditions to compare their EC and hematopoietic capacities (Figure 2a). The percentage of CD144+CD31+ cells was significantly higher in the hESC-EC d7+3 compared to d7 HEC and d10 HPs/a, and a decrease in TRA1-60+ cells was observed throughout differentiation (Figure 2c). Moreover, pluripotent genes NANOG and OCT4 were downregulated from d3, and BRACHYURY expression peaked at d3 (Figure 2d,e). The EC-associated gene, CD144 was upregulated in d7 HEC, d10 HPa, in hESC-EC d7+3, and was observed only at very low levels in d10 HPs (Figure 2f). CD31 was upregulated in all populations after d3, and its expression has been associated with both EC and HP cells (Figure 2f). CD43, a HP-associated gene, was more highly expressed in d10 HPs compared to d7 HEC and hESC-EC d7+3 (Figure 2g).
Figure 1.
Direct differentiation of human embryonic stem cells (hESC) to endothelial cell (EC). (a) Schematic representation of direct hESC-EC differentiation. (b) Morphological analysis of H9 hESC, d3 EB, and d7 hESC-EC (scale bars 250 µm). (c) Fluorescence-activated cells sorting quantification for CD144/CD31 (left) and TRA1-60 (right) in H9 hESC, d3 EB, and d5-7 hESC-EC (n = 3, error bars = standard error of the mean). (d–f) Quantitative reverse transcription-PCR analysis of pluripotency (d), mesodermal (e), and endothelial (f) genes (n = 4, error bars = RQmax). (c–f) Repeated measures analysis of variance, Tukey's post-hoc comparisons, *P < 0.05, **P < 0.01, ***P < 0.001 compared to d0. (g) Tubule formation analysis of d14 CD144+CD31+ hESC-EC. d0 H9 hESC was used as a negative control and human umbilical endothelial cell as a positive control. Images taken at 10× magnification (scale bars represent 100 µm). n = 3; images are representative for sample group.
Figure 2.
Indirect endothelial cell (EC) differentiation formation of embryonic stem cells (HEC). (a) Schematic representation of indirect hESC-EC differentiation. (b) Morphological analysis of d7 HEC and d7+3 hESC-EC (scale bars 250 µm). (c) Quantification of fluorescence-activated cells sorting for both CD144+CD31+ (left) and TRA1-60+ (right) in H1 hESC, d3 EB, d7 HEC, d7+3 hESC-EC, d10 HPs, and d10 HPa. (n = 3, error bars = SEM). (d–g) Quantitative analysis of pluripotent (d), mesodermal (e), endothelial (f), and hematopoietic (g) genes (n = 3, error bars = RQmax). (c–i) Repeated measures analysis of variance, Tukey's post-hoc comparisons, *P < 0.05, **P < 0.01, ***P < 0.001 compared to d0 H1 hESC pluripotent control.
CD326lowCD56high mesoderm progenitor populations during hESC-EC differentiation
Using direct and HEC routes, a population of mesodermal progenitor (MP) appeared transiently at d3 and were identified as CD326lowCD56high as previously described2 (Supplementary Figure S6). CD326lowCD56high d3 population was isolated using fluorescence-activated cells sorting after direct EC differentiation (Supplementary Figure S7a) and gene profile was analysed (Supplementary Figure S7b). This data supports emergence of early MP identified as CD326lowCD56high. These data suggest that these two protocols were able to efficiently generate CD144+CD31+ hESC-EC with a consistent EC phenotype allowing us to interrogate the role of SENCR in EC development.
SENCR is dynamically regulated in endothelial and erythroid derivatives of hESC
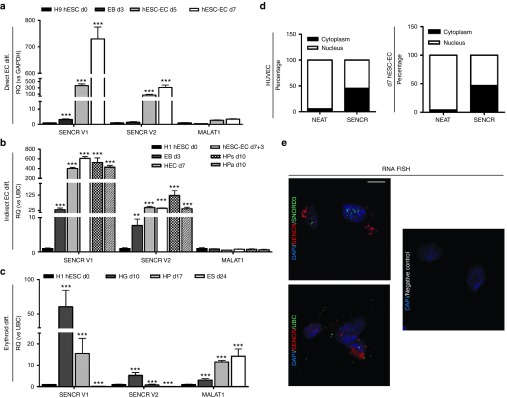
Because SENCR is enriched in EC and correlates with expression of FLI1,12 a regulator of EC development,13 we first quantified SENCR expression during EC differentiation using our protocol. We also profiled MALAT1 expression which is known to regulate EC function.16 During direct hESC-EC differentiation (Figure 3a), SENCR V1 but not V2 was upregulated at d3 and sustained in hESC-EC at d5 and 7. SENCR V2 and MALAT1 were upregulated at d5 and increased at d7. SENCR V1 was consistently induced to a higher level than SENCR V2 and MALAT1. During generation of EC through HEC (Figure 3b), SENCR V1 and V2 were induced from d3 and upregulated in d7 HEC, d7+3 hESC-EC, and d10 HPa/s. SENCR V1 was still the more highly induced variant. MALAT1 was not modulated. During erythroid differentiation (Figure 3c), SENCR V1 was the higher expressed lncRNA in d10 haemangioblast. In d17 HP, SENCR V1 and V2 were silenced while MALAT1 remained constantly upregulated. In d24 erythroid cells, SENCR V1 and V2 were switched off when MALAT1 was stably expressed. We concluded that SENCR V1 was consistently and highly upregulated in early endothelial progenitor and remained expressed in hESC-EC. As SENCR V1 (hereafter called SENCR) was the more abundant variant in hESC-EC, we focused on its role in differentiation. Since FLI1 is the antisense gene of SENCR, we analysed any correlation between FLI1 and SENCR during differentiation. Consistent with previous findings in mature vascular cells,12 expression of SENCR and FLI1 was positively correlated during EC and HP differentiation (Supplementary Figure S8). Time course expression demonstrated that FLI1 and SENCR were activated at d1 and were then constantly upregulated during EC differentiation and in human umbilical endothelial cell (HUVEC)(Supplementary Figure S9a). Expression of ETV2, an ETS factor upstream of FLI1,17 peaked at d5 then was silenced in d7 hESC-EC and in HUVEC (Supplementary Figure S9a). However, ETV2 was more highly expressed compared to SENCR and FLI1 in H9 hESC and in d1-2 EB (Supplementary Figure S9b). As cellular localisation is crucial to understand the potential function of lncRNA, we investigated expression of SENCR in intracellular compartment. We found an equal distribution of SENCR between both nucleus and cytoplasm whereas NEAT exhibited a high selectivity for nuclear localisation in HUVEC and hESC-EC (Figure 3d). RNA FISH experiments confirmed the equal distribution of SENCR in nucleus and cytoplasm in HUVEC compared to UBC and nuclear SNORD3 (Figure 3e)
Figure 3.
Regulation of SENCR in human embryonic stem cells (hESC) derivatives. (a–c) Histograms represent expression of SENCR V1, SENCR V2, and MALAT1 during: (a) Direct EC differentiation versus H9 hESC compared to d3 EB, d5-7 hESC-EC; (b) Indirect differentiation of hESC into endothelial cells (EC) versus H1 hESC compared to d3 EB, d7 HEC, d7+3 hESC-EC, d10 HPa/s; (c) Erythroid differentiation versus H1 hESC compared to d10 haemangioblast (HG), d17 HP, d24 erythroid cells (ES). (a–c) Repeated measures analysis of variance, Tukey's post-hoc comparisons, n = 3, *P < 0.05, **P < 0.01, ***P < 0.005 compared to d0 (all error bars = RQmax). (d) Subcellular localization of SENCR and NEAT1 in human umbilical endothelial cell (HUVEC) (left) and d7 hESC-EC (right) using RNA fractionation. The equal volumes of nuclear and cytoplasmic RNA, corresponding to equivalent numbers of cells were used for cDNA synthesis. The abundance of transcript in nuclear and cytoplasmic fraction was analysed by quantitative reverse transcription-PCR and normalized using UBC. (e) RNA FISH analysis targeting SENCR (red) versus UBC (green) and nuclear SNORD3 (green) in HUVEC (scale bars 25 µm).
SENCR overexpression does not impact on pluripotency
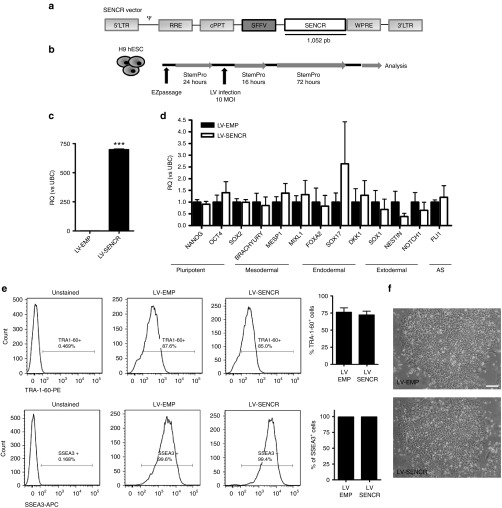
To assess whether SENCR may impact on hESC pluripotency, we used self-inactivating vesicular stomatitis virus-pseudotyped lentivirus (LV) vectors harbouring the SENCR sequence under the SFFV promoter (Figure 4a). SENCR overexpression by LV particles (Figure 4b,c) did not alter: the pluripotent and germ-layer gene expression profile of hESC (Figure 4d); the expression of pluripotency-associated surface markers TRA-1-60 and SSEA3 (Figure 4e); and the colony shape of hESC (Figure 4f). These data revealed that SENCR overexpression was not sufficient to drive hESC to exit their pluripotent state.
Figure 4.
SENCR overexpression does not impact on human embryonic stem cells (hESC) pluripotency. (a) Schematic representation of constructed recombinant lentivirus (LV) vectors harbouring SENCR sequence under SFFV promoter. LTR, long-terminal-repeat; Ψ = Psi, packaging signal; RRE, Rev response element; cPPT, central polypurine tract; SFFV, promoter of spleen focus-forming virus; wPRE, Woodchuck hepatitis virus posttranscriptional regulatory element. (b) Experimental design used to transduce pluripotent H9 hESC. (c–f) All of measures were performed 3 days postinfection and LV-SENCR-infected group is compared to LV-control infected group. (c,d) Gene analysis for SENCR expression (c) and pluripotent/germ-layer genes (d) by quantitative reverse transcription-PCR (all error bars = RQmax). (e) Representative fluorescence-activated cells sorting and quantification of immunophenotype profile analysis for TRA-1-60 and SSEA-3 (all error bars = SEM). (f) Image representing morphology of H9 hESC colony (scale bar 125 μm). (a–f) n = 3, Student's t-test. ***P < 0.005.
SENCR overexpression potentiates the mesodermal and endothelial commitment of hESC
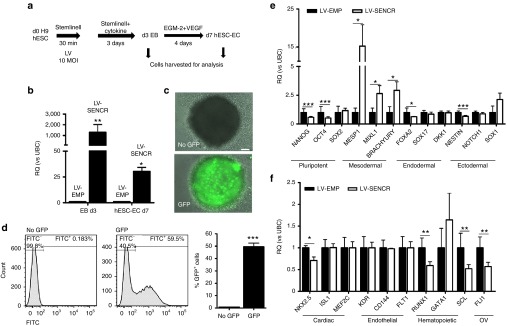
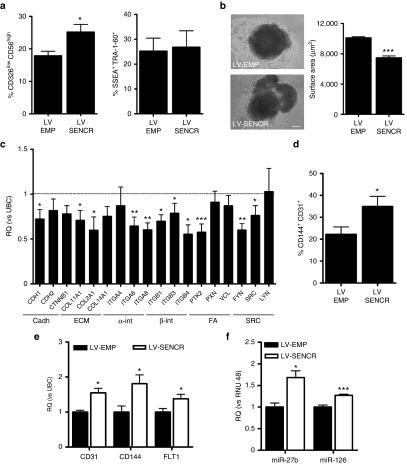
To understand whether the modulation of SENCR may regulate hESC-EC differentiation, we developed a transduction protocol allowing SENCR overexpression during EB development (Figure 5a). Toxicity of LV infection was evaluated by flow cytometry (Supplementary Figure S10). SENCR was overexpressed 3 days postinfection and was also stably overexpressed 7 days after transduction (Figure 5b). LV infection using vectors carrying the green fluorescence protein (GFP) under control of the SFFV promoter demonstrated an efficient GFP expression in EB 3 days postinfection (Figure 5c,d). SENCR overexpression reduced the expression of pluripotent (NANOG, OCT4), endodermal (FOXA2), and ectodermal genes (NESTIN) but enhanced the expression of mesodermal genes (MESP1, MIXL1, BRACHYURY) (Figure 5e). Specific cardiac (NKX2.5), hematopoietic (RUNX1, SCL), and FLI genes were reduced while EC genes were unchanged (Figure 5f). Since exogenous SENCR enhanced mesodermal gene expression, we then investigated whether SENCR drives MP commitment. An increase in CD326lowCD56high MP was observed 3 days after SENCR overexpression while pluripotency-associated markers SSEA-3 and TRA1-60 were unchanged (Figure 6a) (Supplementary Figure S11a,b). EB overexpressing SENCR displayed a less compact and more irregular shape as well as a reduction in surface area (Figure 6b). EMT, characterized by a loss of epithelial gene such as CDH1, is a process governing the emergence of MP18 and we therefore wanted to understand the mechanism involved in this phenotype. A custom TaqMan Low Density Array identified stem cell adhesion molecules that were downregulated in EB overexpressing SENCR including CDH1 (Figure 6c). We found also that SENCR overexpression after 7 days, stimulated generation of hESC-EC expressing CD144+CD31+ (Figure 6d) (Supplementary Figure S11c) as well as increased expression of EC-genes (CD31, CD144, FLT1) and EC-miRNA (miR-27b, miR-126) (Figure 6e,f). Collectively, these data revealed the potential role of SENCR in controlling the EC commitment from hESC through MP.
Figure 5.
SENCR overexpression during EB formation induces mesodermal genes expression. (a) Experimental design allowing SENCR overexpression during EB formation. (b) SENCR expression analysis by quantitative reverse transcription-PCR 3 and 7 days post-transduction (all error bars = RQmax). (c–f) All of measures were performed 3 days postinfection. (c,d) Fluorescence microscopy (c, scale bar 125 µm) and quantification by fluorescence-activated cells sorting of GFP+ cells (d) in EB infected with or without LV-GFP (all error bars = SEM). (e,f) quantitative reverse transcription-PCR analysis showing the expression of pluripotent, mesodermal, endodermal, ectodermal (e), cardiac, endothelial, hematopoietic and FLI1 overlapping gene (OV) (f) in LV-SENCR group compared to LV-control (all error bars = RQmax). (a–f) n = 5, Student's t-test, *P < 0.05, **P < 0.01, ***P < 0.005.
Figure 6.
SENCR overexpression potentiates the mesodermal and endothelial commitment of human embryonic stem cells. (a–c) All of measures were performed 3 days postinfection and LV-SENCR group is compared to LV-control group. (a) Histograms represent fluorescence-activated cells sorting analysis for MP CD326lowCD56high (left) and pluripotent marker TRA1-60+SSEA3+ (right). (b) Measure of area of EB by ImageJ expressed in µm2 (scale bar 125µm) (all error bars = SEM). (c) Custom Taqman low-density array analysis of stem cells adhesion genes expression including cadherins (cadh), extracellular matrix (ECM), α-integrins (α-int), β-integrins (β-int), focal adhesion (FA), and Src-Kinase (SRC) (n = 5, all error bars = RQmax). (d–f) All measures were performed 7 days postinfection and LV-SENCR group is compared to LV-control group. (d) Histogram representation of immunophenotype profiles of cells expressing endothelial cell markers CD144 and CD31 (n = 3, all error bars = SEM). (e,f) Endothelial genes (e) and miRNA (f) expressions analysed by quantitative reverse transcription-PCR (n = 3, all error bars = RQmax). (a–f) Student's t-test, *P < 0.05, **P < 0.01, ***P < 0.005.
SENCR regulates proliferation, migration, and tube-like formation of HUVEC
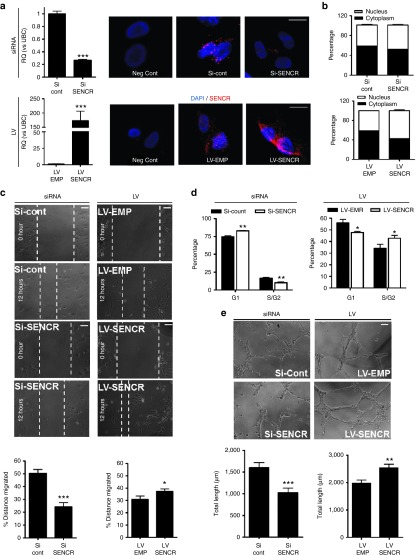
Because SENCR expression remained elevated in hESC-EC, we investigated the role of SENCR in HUVEC. We first elucidated the response of SENCR expression in HUVEC subjected to treatment with vascular endothelial growth factor (VEGF), a cytokine involved in biological activity of EC.19 VEGF stimulated HUVEC proliferation (Supplementary Figure S12a) and induced SENCR expression transiently and in a time dependant manner with a peak 12 hours after stimulation (Supplementary Figure S12b) demonstrating its role in physiology of EC. To understand the role of SENCR in EC, loss- and gain-of-function studies were performed in HUVEC transfected with siRNA-Pool or infected with LV, respectively. SENCR knockdown and overexpression were confirmed by RNA FISH and quantitative reverse transcription-PCR (Figure 7a). Moreover siRNA and LV did not impact on cellular localization of SENCR (Figure 7b). We examined the ability of SENCR to affect cell motility using the scratch “wound” healing assays in HUVEC. Silencing and overexpression of SENCR reduced and stimulated, respectively, the covering of wounded area after 12 hours suggesting that SENCR promotes EC migration (Figure 7c). Cell cycle was next analysed to understand whether SENCR modulation may cause cell cycle arrest. SENCR silencing led to an increase of cells in the G1 phase supported by a decrease of cells in the S/G2 phase whereas its overexpression had the opposite effect (Figure 7d). Since migration and proliferation are important in angiogenesis, we focused on the impact of SENCR modulation in this process. Knockdown and overexpression of SENCR impaired and stimulated, respectively, capillary-like structures on matrigel (Figure 7e).
Figure 7.
SENCR controls proliferation, migration and tube like formation in human umbilical endothelial cell (HUVEC). (a) SENCR expression analysis by quantitative reverse transcription-PCR and RNA FISH (scale bar 25 µm) in HUVEC subjected to siRNA-Pool transfection targeted or not against SENCR (top) and to LV infection (bottom) (all error bars = RQmax). (b) Subcellular localization using RNA fractionation of SENCR in HUVEC subjected to siRNA transfection (top) and LV infection (bottom). (c) Following siRNA transfection (left) or LV infection (right) HUVEC were starved for 12 hours and mechanically disrupted with a sterile 200 μl tip, then photographed immediately at 0 and 12 hours latter. Migrated distance was measured as percentage by ImageJ and representative images are shown (scale bar 125 µm). (d) Percentage of cells in G0/G1 and S/G2 measured by flow cytometry following siRNA transfection (left) and LV infection (right). (e) Following siRNA transfection (left) or LV infection (right) HUVEC were starved for 12 hours and were allowed to sprout in a three-dimensional matrix for 12 hours. Cumulative sprout length was quantified using angiogenesis analyser for ImageJ and representative images are shown (scale bar 125 µm) (all error bars = SEM). (a–d) n = 3, Student's t-test, *P < 0.05, **P < 0.01, ***P < 0.005.
SENCR controls the expression of angiogenic-related genes in HUVEC
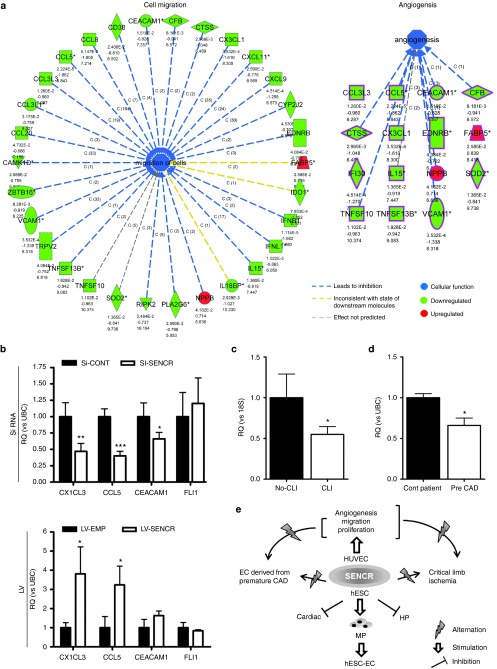
To understand the targeted gene modulated by silencing of SENCR, we next carried out a microarray analysis using siRNA-treated cells. A schematic presentation (Figure 8a) shows that known migratory and angiogenic genes were downregulated after silencing of SENCR. We focused our study on CCL5, CEACAM1 and CX3CL1 described as proangiogenic gene.20,21,22,23 We confirmed that knockdown of SENCR resulted in a decrease of these genes and its overexpression induced an upregulation of CCL5 and CX3CL1 (Figure 8b). As shown by Bell et al.,12 modulation of SENCR did not alter the expression of FLI1 in this scenario demonstrating that SENCR regulated EC function independently of FLI1 (Figure 8b). These data revealed SENCR as a regulator of angiogenesis-related function in EC.
Figure 8.
SENCR controls the expression of angiogenic-related gene and is altered in human sample derived from ischemic condition. (a) These figures illustrate genes with expressional changes after SENCR silencing belong to the cell migration (left) and angiogenesis (right). Gene expression profiles were uploaded to the Ingenuity Analysis Software and based on the differential expression of these genes, the most relevant biochemical network with functional links was assessed. (b) Quantitative reverse transcription-PCR was used to analyse expression of CX1CL3, CCL5, CEACAM1 as well as FLI1 in human umbilical endothelial cell subjected to siRNA transfection (top) and LV infection (bottom) (all error bars = RQmax), n = 3, Student's t-test, *P < 0.05, **P < 0.01, ***P < 0.005. (c) Comparison of SENCR expression for the critical limb ischemia group (CLI, n = 10, *P < 0.05) versus control group (No-CLI, n = 9). (d) Comparison of SENCR expression for superficial forearm vein endothelial cells derived from premature coronary artery disease (CAD) group (Pre-CAD, n = 8, *P < 0.05) versus control group (Cont patient, n = 8) (all error bars = RQmax). Student's t-test. (e) Schematic representation of the role of SENCR in hESC-EC differentiation and endothelial cell function.
SENCR dysregulation is associated with premature coronary artery disease and critical limb ischemia
Since SENCR regulates angiogenic-related function in HUVEC, we interrogated whether SENCR alteration may be associated with angiogenesis dysfunction in human samples. SENCR level was reduced, versus control limb muscle sample, in human critical limb ischemia (CLI) (Figure 8c), an occlusive peripheral arterial disease resulting in inadequate perfusion of the limb.24 Alterations in EC function are common to a variety of pathologies including CLI and coronary artery disease (CAD).25 Particularly, EC dysfunction has been associated with the premature onset of CAD.26 EC derived from the superficial forearm veins were previously isolated from pre-CAD patients and healthy control subjects. These cells have a high expression of mature endothelial cell antigens vWF, CD31, CD146, and KDR, and lack expression of the haematopoietic antigen, CD45.27 This study demonstrated impaired proliferation, adhesion, and migration of vessel wall EC derived from the superficial forearm veins in these patients compared to matched control subjects.27 Consistent with these observations, we found that SENCR expression was significantly reduced in vessel wall EC isolated from patients with premature CAD compared to control subjects (Figure 8d). Collectively, these data showed that SENCR expression is down regulated in patients with EC dysfunction and atherosclerotic vascular disease.
Discussion
We aimed to investigate the role of SENCR in EC differentiation and function by using two novel differentiation protocols allowing efficient hESC-EC generation, through direct and haemogenic routes. We demonstrated that SENCR was regulated during hESC-EC commitment, and through gain-of-function study, we revealed SENCR as an early induced lncRNA capable of promoting mesodermal and EC commitment. Therefore, loss- and gain-of-function experiments identified SENCR as a regulator of proliferation, migration, and angiogenesis in EC. Finally, altered SENCR expression was found in CLI tissues and in EC derived from premature CAD (Figure 8e).
SENCR is vascular-enriched lncRNA overlapping FLI1 gene,12 a member of ETS family, which has been described as an early regulator of hemato-endothelial development.13,17,28 ETS factors are involved in developmental processes and ETS-binding sites are not specific for EC-expressed gene loci making difficult the comprehension of mechanisms involved in ETS factor-regulating EC specification. ETS factors controlling-EC gene depend on complex processes including binding partners, post-transcriptional modification and flanking sequence content.29 The functional role of ETS factors in EC fate has been, for example, explained by a probable cooperative action with EC TF such as GATA-2.13 With tissue-specific expression patterns during development, lncRNA may be orchestrators of this process and some reports have already related their role in specification of germ-layer9,11 and adult cells.9,10,11 Here, we showed the dynamic regulation of a lncRNA, SENCR, and an overlapping ETS gene, FLI1, during hESC-EC differentiation in precursor and differentiated hESC-EC. These data support a previous report associating induction of FLI1 to EC development28 and revealed SENCR as an early mesodermal lncRNA remaining expressed in hESC-EC. Addition of exogenous SENCR during EB formation increased the generation of MP expressing CD326lowCD56high and induced expression of mesodermal genes including BRACHYURY, MIXL1, and MESP1. EMT, controlling generation of MP, is mediated by TF such as MESP1 and characterized by the loss of epithelial genes such as CDH1.2 MESP1, through EMT, drives generation of cardiovascular MP including EC and VSMC whilst inhibiting the formation of other MP lineages such as HP.30 MESP1 was upregulated when CDH1 was downregulated after SENCR overexpression suggesting that SENCR can positively manipulate cardiovascular MP specification. This correlated with the fact that EB overexpressing SENCR failed to form compact structures and with a report which described CDH1 as a factor of EB aggregation.31 Exogenous SENCR also induced a downregulation of extracellular matrix molecules such as COL2A1 and of β-integrins genes such as ITGB1, both pathways linked to cardiac specification.32 Moreover, a previous study related the size of EB to efficiency of EC differentiation33 suggesting that EB morphology impacts on specification of vascular MP. Similarly, addition of exogenous SENCR resulted in a reduction in the size of EB. These demonstrated that SENCR may impact on the generation of vascular MP through molecular and morphological changes. NKX2.5, involved in cardiac development,34 as well as SCL and RUNX1, both involved in HP specification,35,36 were downregulated after SENCR overexpression while EC genes were unchanged reaffirming the hypothesis that SENCR is part of a molecular axis, driving vascular fate while inhibiting the formation of other mesodermal lineages.
Our observations revealed that exogenous SENCR impacts on vascular mesodermal specification, essential for EC development,3,6 is in concordance with the findings that SENCR overexpression led to an increase in CD31+CD144+ hESC-EC at d7 of differentiation. This was accompanied with an enhancement in EC-specific genes and miRNA including CD144, CD31, FLT1, miR-126 and miR-27b. SENCR, and FLI1 shared the same expression profile throughout EC and erythroid differentiation highlighting their identical transcriptional regulation during development. To act at transcriptional level, ETS factors bind to a core “GGAA/T” binding element,37 and FLI1 itself is activated by ETV2 via an ETS binding site located -192 upstream its start site.17 Therefore, analysis of genomic sequences revealed an ETS binding site -149 upstream SENCR start site, and transcription of both FLI1 and SENCR gene was activated from d1 of differentiation. Moreover, studies supported common TF activating both protein coding, and antisense lncRNA.38 Upon vascular development, ETV2 expression turns off and FLI controls its own expression through an auto-regulatory loop.17 Consistent with these statements, expression of ETV2 was consistently increased on the first day of differentiation and silenced in fully differentiated hESC-EC and in HUVEC. Moreover, FLI1 and SENCR expression was not dependant on ETV2 silencing at the end of differentiation. This asks the question of how SENCR remained expressed as well in EC. The influence of SENCR modulation on FLI1 expression and vice versa has not yet been described12 eliminating cis-acting effect on each other. Our gain- and loss-of-function experiment in HUVEC confirmed this fact; however, we found that expression of exogenous SENCR in EB led to a reduction of FLI1 transcription. This may be explained by the downregulation of SCL, induced by exogenous SENCR. Indeed SCL and FLI1 form a regulatory circuit controlling specification of HP by maintaining expression of each other.39 Nevertheless, despite the reduction of FLI1 in EB, its level seems to be sufficient to drive EC differentiation through MP in concert with exogenous SENCR transcript. Active transcription of SENCR during EC differentiation may be explained as FLI1 by a positive auto-regulatory loop similarly to the lncRNA HULC, in hepatocellular carcinoma which exists as part of an intricate auto-regulatory network resulting in an increase of its own expression.40 Another explanation of the high levels of SENCR expression in hESC-EC and cultured EC may be due to the stability of the RNA after post-transcriptional modification similar to ZFAS1 lncRNA which is extremely stable and highly expressed in mouse neuroblastoma cells.41
SENCR was initially discovered as a vascular enriched lncRNA and described to inhibit migration and stimulate contractile gene expression of VSMC.12 The function of SENCR in adult EC has not yet been studied; although its high expression in EC12 reflected its potential importance. Along this line, silencing and overexpression of SENCR impaired and enhanced, respectively, sprouting of cultured EC as well as the expression of proangiogenic genes including CCL5, CEACAM-1, and CX1CL3. Angiogenesis is a multistep process including a first step where EC migrate into the extracellular space, proliferate, and form capillary sprouts and tubular structures. CEACAM-1 is a transmembrane protein expressed on cell surface of capillary EC and has been described to stimulate angiogenesis through promigratory process.23 In correlation with these observations, the migratory capacity of EC was inhibited and stimulated after SENCR silencing and overexpression, respectively. Bell et al.12 described SENCR as a suppressor of promigratory phenotype in VSMC highlighting, its probable function in disease processes such as atherosclerosis where VSMC migration contributes to the pathogenesis of neo-intima formation. Currently, treatments for CAD use drug-eluting stents allowing the delivery of a nonspecific antiproliferative agent acting on both VSMC and EC leading to a delay of re-endothelialisation and restenosis.42 Ideally, treatments would inhibit the VSMC proliferation and migration, whilst promoting EC function. In this direction, SENCR is able to inhibit promigratory phenotype of VSMC while stimulating angiogenesis of EC similar to miR-126 which inhibits VSMC migration while restoring EC function in animal models of vascular balloon injury.43 The proangiogenic and promigratory effect induced by SENCR on cultured EC were also correlated in limb ischemia, pathology induced by vascular injury, where SENCR expression was altered. Consistent with that, CX3CL1, downregulated after SENCR silencing, has been demonstrated to significantly reverse limb ischaemia in a rat model via the induction of effective revascularization.21 Amongst other genes, CX3CL1 simulates tissue neovascularization through the enhancement of EC proliferative capacity. Our findings showed that SENCR also impacts on cell cycle progression by stimulating cells to enter S/G2 phase. SENCR was also impaired in vessel wall EC, derived from patient with premature CAD, displaying defects in proliferative and migratory processes. The purpose of this study was to define a novel molecular mechanism that may have an application in the development of effective hESC-EC therapies for ischemic disease. Only a few reports have described the role of lncRNA in vascular development and pathology.29 For example, MALAT1 has been described to be significantly upregulated during hypoxia and controlling sprouting and migratory capacity of EC;44 however loss-of-function studies in mice revealed that it is not essential for development.7 TIE1-AS is the only lncRNA described to be essential for EC development and altered in human vascular injury.10 Here, we described for the first time an ETS-related lncRNA involved in EC development and function as well as its association with human vascular pathology.
Materials and Methods
Cell culture. The hESC lines H1 and H9 (WiCell Research Institute, Madison, WI, http://wicell.org) were cultured in a feeder-free culture system on recombinant human vitronectin and using StemPro hESC serum-free medium (Life Technologies, CA) supplemented with 20 ng/ml basic fibroblast growth factor (Peprotech, NJ). Cells were passaged mechanically using StemPro EZPassage mechanical stem cell passaging tool (Life Technologies Europe, Bleiswijk, Netherlands). Protocols allowing generation of EC via direct and haemogenic routes are described in the Supplementary Method.
Study population. Tissue samples and vessel-wall derived EC were obtained from patients with critical limb ischemia (patients with (n = 9) or without (n = 10) critical limb ischemia from leftover limb muscle) and premature coronary artery disease (EC derived from the superficial forearm veins, n = 8) respectively, following written informed consent, and in accordance with the Declaration of Helsinki. Patient characteristics and protocols for muscle biopsy and isolation of EC are described in the Supplementary Method (Supplementary Tables S5 and S6).
Reagents and other detailed methods are described in the Supplementary Method, Supplementary Tables S1 to S4 and Supplementary References.
SUPPLEMENTARY MATERIAL Figure S1. Schematic representation of genomic localization of both version of SENCR and FLI1. Figure S2. hESC-EC direct differentiation using the H9 and H1 hESC line. Figure S3. MACSorting of d7 hESC-EC from direct differentiation. Figure S4. Identification of a HEC population existing on d7 of an established hematopoietic differentiation protocol. Figure S5. Optimisation of an indirect hESC-EC differentiation protocol. Figure S6. Identification of CD326lowCD56high MP population during direct and indirect hESC-EC differentiation. Figure S7. Purification and characterisation of a CD326lowCD56high MP population in hESC-EC differentiation. Figure S8. Expression of FLI1 during differentiation of hESC into EC and erythroid cells. Figure S9. Time course expression of ETV2, FLI1 and SENCR during hESC-EC differentiation and in HUVEC. Figure S10. Assessment of toxicity after LV infection using Zombie aqua staining. Figure S11. SENCR enhances differentiation of CD326lowCD56high and CD144+CD31+ hESC-EC. Figure S12. SENCR expression analysis after VEGF treatment in HUVEC. Table S1. Information for specific antibodies used in FACS analysis and sorting. Table S2. Information for isotype control antibodies used in FACS analysis and sorting. Table S3. Information for probes used in TaqMan-based quantitative real-time PCR assays. Table S4. Primer-pairs used for SYBR-Green-based quantitative real-time PCR assays. Table S5. Clinical characteristics of patients with or without critical limb ischaemia, from whom leftover limb muscle samples from cardiovascular surgery were analysed. Table S6. Clinical characteristics of patients with or without premature coronary artery diseases. Methods.
Acknowledgments
We acknowledge Gregor Aitchison and Nicola Britton for the technical assistance. This work has been funded by the British Heart Foundation grant SP/10/005/28298, British Heart Foundation Centre for Vascular Reparation grant (RM/13/1/30158), and Scottish Universities Life Science Alliance. Disclosures: None.
Supplementary Material
References
- Thomson, JA, Itskovitz-Eldor, J, Shapiro, SS, Waknitz, MA, Swiergiel, JJ, Marshall, VS et al. (1998). Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147. [DOI] [PubMed] [Google Scholar]
- Evseenko, D, Zhu, Y, Schenke-Layland, K, Kuo, J, Latour, B, Ge, S et al. (2010). Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc Natl Acad Sci USA 107: 13742–13747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamme, I, Frölich, T and Risau, W (1997). Molecular mechanisms of vasculogenesis and embryonic angiogenesis. J Cell Physiol 173: 206–210. [DOI] [PubMed] [Google Scholar]
- Descamps, B and Emanueli, C (2012). Vascular differentiation from embryonic stem cells: novel technologies and therapeutic promises. Vascul Pharmacol 56: 267–279. [DOI] [PubMed] [Google Scholar]
- Kane, NM, Thrasher, AJ, Angelini, GD and Emanueli, C (2014). Concise review: MicroRNAs as modulators of stem cells and angiogenesis. Stem Cells 32: 1059–1066. [DOI] [PubMed] [Google Scholar]
- Wapinski, O and Chang, HY (2011). Long noncoding RNAs and human disease. Trends Cell Biol 21: 354–361. [DOI] [PubMed] [Google Scholar]
- Fatica, A and Bozzoni, I (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15: 7–21. [DOI] [PubMed] [Google Scholar]
- Grote, P, Wittler, L, Hendrix, D, Koch, F, Währisch, S, Beisaw, A et al. (2013). The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24: 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff, CA, Scheuermann, JC, Surface, LE, Bradley, RK, Fields, PA, Steinhauser, ML et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152: 570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, K, Blum, Y, Verma, A, Liu, Z, Pramanik, K, Leigh, NR et al. (2010). A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood 115: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian, L, Aguirre, A, Sancho-Martinez, I, Benner, C, Hishida, T, Nguyen, TB et al. (2015). Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation 131: 1278–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, RD, Long, X, Lin, M, Bergmann, JH, Nanda, V, Cowan, SL et al. (2014). Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol 34: 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val, S and Black, BL (2009). Transcriptional control of endothelial cell development. Dev Cell 16: 180–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrón-Barthe, L, Temiño, S, Villa del Campo, C, Carramolino, L, Isern, J and Torres, M (2014). Clonal analysis identifies hemogenic endothelium as the source of the blood-endothelial common lineage in the mouse embryo. Blood 124: 2523–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, KD, Vodyanik, MA, Togarrati, PP, Suknuntha, K, Kumar, A, Samarjeet, F et al. (2012). Identification of the hemogenic endothelial progenitor and its direct precursor in human pluripotent stem cell differentiation cultures. Cell Rep 2: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalik, KM, You, X, Manavski, Y, Doddaballapur, A, Zörnig, M, Braun, T et al. (2014). Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 114: 1389–1397. [DOI] [PubMed] [Google Scholar]
- Abedin, MJ, Nguyen, A, Jiang, N, Perry, CE, Shelton, JM, Watson, DK et al. (2014). Fli1 acts downstream of Etv2 to govern cell survival and vascular homeostasis via positive autoregulation. Circ Res 114: 1690–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, SJ and Robertson, EJ (2009). Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol 10: 91–103. [DOI] [PubMed] [Google Scholar]
- Carmeliet P and Jain RK (2011). Molecular mechanisms and clinical applications of angiogenesis. Nature 473: 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suffee, N, Hlawaty, H, Meddahi-Pelle, A, Maillard, L, Louedec, L, Haddad, O et al. (2012). RANTES/CCL5-induced pro-angiogenic effects depend on CCR1, CCR5 and glycosaminoglycans. Angiogenesis 15: 727–744. [DOI] [PubMed] [Google Scholar]
- Ryu, J, Lee, CW, Hong, KH, Shin, JA, Lim, SH, Park, CS et al. (2008). Activation of fractalkine/CX3CR1 by vascular endothelial cells induces angiogenesis through VEGF-A/KDR and reverses hindlimb ischaemia. Cardiovasc Res 78: 333–340. [DOI] [PubMed] [Google Scholar]
- Horst, AK, Ito, WD, Dabelstein, J, Schumacher, U, Sander, H, Turbide, C et al. (2006). Carcinoembryonic antigen-related cell adhesion molecule 1 modulates vascular remodeling in vitro and in vivo. J Clin Invest 116: 1596–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, MM, Singer, BB, Klaile, E, Obrink, B and Lucka, L (2005). Transmembrane CEACAM1 affects integrin-dependent signaling and regulates extracellular matrix protein-specific morphology and migration of endothelial cells. Blood 105: 3925–3934. [DOI] [PubMed] [Google Scholar]
- Varu, VN, Hogg, ME and Kibbe, MR (2010). Critical limb ischemia. J Vasc Surg 51: 230–241. [DOI] [PubMed] [Google Scholar]
- Khazaei, M, Moien-Afshari, F and Laher, I (2008). Vascular endothelial function in health and diseases. Pathophysiology 15: 49–67. [DOI] [PubMed] [Google Scholar]
- Choi, BJ, Prasad, A, Gulati, R, Best, PJ, Lennon, RJ, Barsness, GW et al. (2013). Coronary endothelial dysfunction in patients with early coronary artery disease is associated with the increase in intravascular lipid core plaque. Eur Heart J 34: 2047–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittan, M, Hunter, A, Boulberdaa, M, Fujisawa, T, Skinner, EM, Shah, AS et al. (2015). Impaired vascular function and repair in patients with premature coronary artery disease. Eur J Prev Cardiol 22: 1557–1566. [DOI] [PubMed] [Google Scholar]
- Liu, F, Walmsley, M, Rodaway, A and Patient, R (2008). Fli1 acts at the top of the transcriptional network driving blood and endothelial development. Curr Biol 18: 1234–1240. [DOI] [PubMed] [Google Scholar]
- Miano, JM and Long, X (2015). The short and long of noncoding sequences in the control of vascular cell phenotypes. Cell Mol Life Sci 72: 3457–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley, RC, Gill, JG, Murphy, TL, Langer, EM, Cai, M, Mashayekhi, M et al. (2008). Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell 3: 55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, R, Cai, KQ, Escudero, DO and Xu, XX (2009). Cell adhesive affinity does not dictate primitive endoderm segregation and positioning during murine embryoid body formation. Genesis 47: 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, D, Ou, DB, Wei, T, Ding, L, Liu, XT, Hu, XL et al. (2013). Collagen/β(1) integrin interaction is required for embryoid body formation during cardiogenesis from murine induced pluripotent stem cells. BMC Cell Biol 14: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, YS, Chung, BG, Ortmann, D, Hattori, N, Moeller, HC and Khademhosseini, A (2009). Microwell-mediated control of embryoid body size regulates embryonic stem cell fate via differential expression of WNT5a and WNT11. Proc Natl Acad Sci USA 106: 16978–16983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, SM, Yelon, D, Conlon, FL and Kirby, ML (2010). Myocardial lineage development. Circ Res 107: 1428–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real, PJ, Ligero, G, Ayllon, V, Ramos-Mejia, V, Bueno, C, Gutierrez-Aranda, I et al. (2012). SCL/TAL1 regulates hematopoietic specification from human embryonic stem cells. Mol Ther 20: 1443–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancrin, C, Sroczynska, P, Stephenson, C, Allen, T, Kouskoff, V and Lacaud, G (2009). The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature 457: 892–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettgen, P (2006). Regulation of vascular inflammation and remodeling by ETS factors. Circ Res 99: 1159–1166. [DOI] [PubMed] [Google Scholar]
- Wu, Z, Liu, X, Liu, L, Deng, H, Zhang, J, Xu, Q et al. (2014). Regulation of lncRNA expression. Cell Mol Biol Lett 19: 561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimanda, JE, Ottersbach, K, Knezevic, K, Kinston, S, Chan, WY, Wilson, NK et al. (2007). Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc Natl Acad Sci USA 104: 17692–17697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J, Liu, X, Wu, H, et al. (2010). CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 38: 5366–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, MB, Johnston, RL, Inostroza-Ponta, M, Fox, AH, Fortini, E, Moscato, P et al. (2012). Genome-wide analysis of long noncoding RNA stability. Genome Res 22: 885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka, F, Finn, AV, Yazdani, SK, Nakano, M, Kolodgie, FD and Virmani, R (2012). The importance of the endothelium in atherothrombosis and coronary stenting. Nat Rev Cardiol 9: 439–453. [DOI] [PubMed] [Google Scholar]
- Santulli, G, Wronska, A, Uryu, K, Diacovo, TG, Gao, M, Marx, SO et al. (2014). A selective microRNA-based strategy inhibits restenosis while preserving endothelial function. J Clin Invest 124: 4102–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X, Shan, D, Chen, J and Jing, Q (2014). miRNAs and lncRNAs in vascular injury and remodeling. Sci China Life Sci 57: 826–835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.