Abstract
Introduction
Pharmacokinetic outcomes of transporter-mediated drug-drug interactions (TMDDIs) are increasingly being evaluated clinically. The goal of our study was to determine the effects of selective inhibition of multi-drug and toxin extrusion protein 1 (MATE1) using famotidine, on the pharmacokinetics and pharmacodynamics of metformin in healthy volunteers.
Methods
Volunteers received metformin alone or with famotidine in a crossover design. As a positive control, the longitudinal effects of famotidine on the plasma levels of creatinine (an endogenous substrate of MATE1) were quantified in parallel. Famotidine unbound concentrations in plasma reached 1 μM, thus exceeding the in vitro concentrations that inhibit MATE1 (IC50 0.25 μM). Based on current regulatory guidance, these concentrations are expected to inhibit MATE1 clinically (i.e., Cmax,u/IC50 >0.1).
Results
Consistent with MATE1 inhibition, famotidine administration significantly altered creatinine plasma and urine levels in opposing directions (P<0.005). Interestingly, famotidine increased the estimated bioavailability of metformin (Ae∞/Dose, P < 0.005) without affecting its systemic exposure (AUC or Cmax) as a result of a counteracting increase in metformin renal clearance. Moreover, metformin-famotidine co-therapy caused a transient effect on oral glucose tolerance tests (AUCglu,0.5, P < 0.005).
Conclusions
These results suggest that famotidine may improve the bioavailability and enhance the renal clearance of metformin.
1 Introduction
The MATE transporters (i.e., MATE1 [SLC47A1] and MATE2 [SLC47A2]) mediate the excretion of endogenous and exogenous compounds into urine and bile and play important roles in the clinical pharmacokinetics and pharmacodynamics of numerous drugs[1]. Recently, the International Transporter Consortium (ITC) recommended that evaluation of MATE mediated drug-drug interactions (DDIs) be incorporated into drug development programs. Particularly, the ITC recommended that: a clinical DDI study should be considered for new molecular entities that 1) are predicted to cause inhibition of MATEs at clinically relevant concentrations, and 2) are identified as MATE substrates and for which renal secretion accounts for at least 25% of their total elimination. In the first case, for new molecular entities that inhibit MATE1 or MATE2 in in vitro assays, a sensitive substrate for both MATEs such as metformin was recommended for use as the victim drug in the clinical study. In the second case, for new drugs that are substrates of MATE1, the ITC recommended the use of a clinically validated MATE probe inhibitor (i.e., >10-fold more potent for MATEs compared to organic cation transporter 2 [OCT2]) such as cimetidine or pyrimethamine in the clinical study [1]. However, at clinical concentrations, cimetidine and pyrimethamine inhibit both MATE1 and MATE2. Therefore, the effect of a selective MATE1 inhibitor (>10-fold more potent for MATE1 compared to MATE2 and OCT2) on pharmacokinetics, particularly renal drug secretion, is not known.
In the kidney, MATE1 and MATE2 are highly expressed on the apical membrane of the renal epithelium and have somewhat overlapping substrate specificities (e.g., metformin, procainamide and thiamine but not oxaliplatin or cephalexin) [2–8]. Interestingly MATE1 transporters are ubiquitously expressed in both the kidney proximal and distal tubules, as well as in intestine tissue and on the canalicular membranes of hepatocytes, whereas MATE2 transporters are localized specifically in the kidney proximal tubules in humans and are absent in mice[8–10]. Creatinine is an endogenous metabolite eliminated unchanged in the urine by glomerular filtration and tubular secretion (10–40%) [11]. While transporter-mediated tubular secretion of creatinine is reported to involve OCT2/3, organic anion transporter 2 (OAT2, uptake into the renal cell) and subsequently the MATE1/2 transporters (efflux from the renal cell into the urine) [12,13], inhibition of the efflux transporter MATE1, in particular, has been suggested as the mechanism for the decrease in renal clearance of creatinine observed in multiple clinical studies [12].
Determination of metformin pharmacokinetics is currently the gold standard for investigating clinical TMDDIs involving renal organic cation transporter inhibition. Metformin, an anti-hyperglycemic agent, is commonly recommended as first-line pharmacotherapy for treating type 2 diabetes mellitus (T2DM). A small cationic drug at physiologic pH (>99% ionized in blood and in urine), metformin interacts with organic cation transporters (OCTs and MATEs) to traverse membranes for access to target tissues, such as the liver, and for elimination from the body, (i.e., through the kidney) [14,15]. The importance of MATEs in the clinical pharmacokinetics of metformin has been clearly documented, yet MATE1 specific evidence in humans is currently limited to genetic studies [16–22]. Evidence in rodents show that deletion of the murine MATE1 gene, (Mate1 [−/−]) can significantly reduce metformin urinary excretion and significantly increase metformin plasma, kidney and liver concentrations compared to wildtype mice (Mate1 [+/+]) [16,17]. Since mice do not express the MATE2 transporter, it is not surprising that non-selective chemical MATE inhibitors (e.g., cimetidine and pyrimethamine) effectively mimic rodent knockout strains by reducing metformin renal clearance and increasing plasma exposure in mice, as well as in healthy human volunteers [23–25]. Although clinically validated selective MATE1 inhibitors are not currently available, genetic studies investigating single nucleotide polymorphisms (SNPs) in the human MATE1 gene provide insight regarding the selective importance of MATE1 in humans [18–22]. Particularly, a SNP located in the intronic region of the MATE1 gene (G>A, SNP rs2289669) is associated with better response (greater glycated hemoglobin (A1C) reduction) to metformin therapy in T2DM patients [18–21], although the mechanism for this effect is unknown. Likewise, a common mutation in the promoter region of the MATE1 gene (T>C, SNP rs2252281) is associated with improved response to metformin in T2DM patients (greater A1C reduction) as well as in healthy volunteers (greater glucose-lowering effect)[22]. Functional studies demonstrate that the T>C mutation in the promoter region of the MATE1 gene alters the binding affinity for MATE1 transcription factors (i.e., activator AP-1 and repressor AP-2rep), which reduces MATE1 mRNA expression levels in in vitro assays [26]. Although an effect of genetic variants in MATE1 on human pharmacokinetics has not been observed, these studies suggest that MATE1 plays an important role in the distribution of metformin to target tissues as well as in the glycemic response to metformin.
The goal of this study was to determine the effect of selective MATE1 inhibition on the pharmacokinetics and pharmacodynamics of metformin, as well as on creatinine levels as a positive control, in healthy volunteers. Recently, in a large prescription drug screening study, Wittwer et al. [27] identified 84 MATE1 inhibitors, including four compounds predicted to selectively inhibit MATE1 at clinical concentrations (i.e., famotidine, imatinib, indinavir and ritonavir). For this study, we selected to evaluate famotidine as an in vitro and in vivo inhibitor of metformin transport by MATE1 in human cells and in healthy volunteers, respectively. Famotidine, an H2-receptor antagonist used to reduce gastric acid secretion, is marginally bound to plasma proteins (approximately 20%) and is eliminated by renal (65–70%) and by metabolic (30–35%) routes [28–32]. We hypothesized that selective inhibition of MATE1 by famotidine would phenocopy the effects of the reduced expression MATE1 promoter variant (T> C, SNP rs2252281), resulting in improved glycemic response to metformin. In addition, we hypothesized that in the presence of a selective MATE1 inhibitor, the renal clearance of metformin and creatinine would decrease.
2 Methods
2.1 Clinical Study Procedures
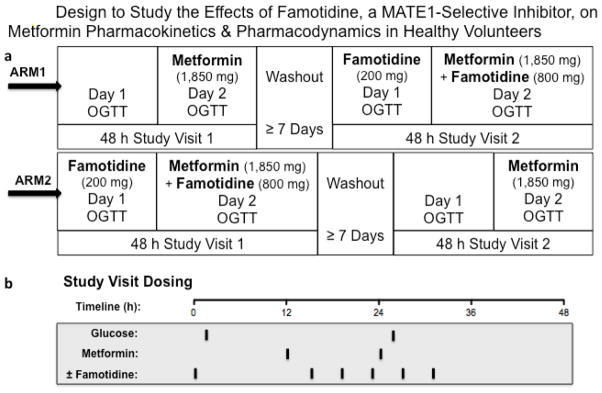
Healthy volunteers were recruited as described in the Supplemental Material. In this open-label, two-phase crossover study, an equal number of volunteers completed the metformin alone followed by metformin plus famotidine treatment sequence (n=6), as completed the same treatments but in reverse order, i.e. metformin plus famotidine followed by metformin alone (n=6) (See Clinical Study Design, Fig. 1). The interval between treatment phases was set to a minimum of 7days to allow for complete elimination of the study drugs. Study day procedures are described below.
Fig. 1.
a) Volunteers in Arm 1 received metformin alone (1,850 mg in two divided doses), followed by a 7-day washout, then metformin + famotidine (1,850 mg in two-divided doses and 1,000 mg in six divided doses, respectively) during two separate study treatment visits. Volunteers in Arm 2 received the same treatments but in reverse order, i.e., metformin + famotidine, washout, then metformin alone. b) During each 48h treatment visit, Oral Glucose Tolerance Tests (OGTTs) were performed after a 10h overnight fasts on Day 1 and on Day 2. Black bars indicate time of drug administration
2.2 Metformin Alone Phase
Volunteers reported to Clinical Research Center (CRC) at San Francisco General Hospital (SFGH) after a three-day carbohydrate controlled diet (200–250 g/day) and an overnight fast (10h), where they remained for the duration of the visit (48h). Vital signs were obtained, inclusion/exclusion criteria were confirmed, and a 3h oral glucose tolerance test (OGTT) (75 g) was performed in the morning of Day 1. Subjects were administered a 1,000 mg immediate release tablet (IRT) of metformin in the evening of Day 1, followed by an 850 mg IRT 12h later on Day 2. On Day 2, after fasting (10h), a second OGTT was administered 2h after the second dose of metformin (Fig. 1b). A normal meal schedule (standardized carbohydrate-controlled meal) was resumed on both study days after completion of the OGTT. For 12h following the second dose of metformin, volunteers were asked to drink 8 oz of water every 2h to maintain urine flow and pH. A series of venous blood samples were drawn at 0, 0.5, 1, 2h after the first metformin dose and 0, 0.5, 1, 2, 2.25, 2.5, 2.75, 3, 3.5, 4, 5, 6, 8, 10, 12, and 24h after the second metformin dose for pharmacokinetic analysis of metformin. Blood samples were collected at 0, 15, 30, 45, 60, 90, 120, and 180 minutes after each glucose administration for pharmacodynamic analysis (glucose concentrations). Each blood sample was collected into heparinized tubes and kept on ice until centrifugation (3000 rpm for 10 min at 4°C). Urine samples were collected at the following time intervals: pre-dose, 0–12h after the first metformin dose and 0–2, 2–4, 4–6, 6–8, 8–12 and 12–24h after the second metformin dose. Urine volume and pH were recorded for each interval. Aliquots of all plasma and urine specimens were stored at −80°C pending bioanalysis.
2.3 Metformin Plus Famotidine Phase
Procedures for the Metformin Plus Famotidine Phase were repeated as described above, with the addition of six oral doses of famotidine (1,000 mg in total) administered over the course of the 48h visit. A single dose of famotidine (200 mg) was administered 1h prior to the OGTT on Day 1. Multiple dosing of famotidine (160 mg given at 4 hourly intervals) beginning 9 hours before the second dose of metformin in the morning of Day 2 (See Study Design, Fig. 1b). Eight additional blood draws (5 mL each) were taken at 0, 0.5, 1, 1.5, 2, 2.5, 3 and 4h on Day 1 for single dose pharmacokinetic analysis of famotidine.
2.4 Analytical Methods
Metformin and famotidine concentrations in plasma and urine were concomitantly quantified using liquid chromatography– tandem mass spectrometry (LC/MS/MS). The assay included modifications from a previously reported method [22,33] as follows: each aliquot of clinical sample (plasma or urine) was mixed with 3 volumes of acetonitrile containing internal standards (i.e., metformin-d6 for metformin and phenformin for famotidine). The mobile phase consisted of 80% acetonitrile, 20% double-distilled water, 0.1% 10 mM ammonium formate, and 0.02% 5 mM formic acid (v/v). The assay was validated and the range of the calibration curve in plasma was 5–2000 ng/mL and in urine was 40–4000 ng/mL for both famotidine and metformin. Both the intra-day and inter-day assay coefficients of variation were less than 10% for both analytes. Glucose concentrations in plasma and creatinine concentrations in plasma and urine were determined by the CLIA certified Clinical Laboratory at SFGH. For in vitro methodology see Supplemental Material.
2.5 Clinical Pharmacokinetics
The pharmacokinetic parameters of metformin (0–24h after the second dose, 1,850 mg in total) and famotidine (0–4h and 12h after a single 200 mg dose) were evaluated using non-compartmental analysis, as previously described (Phoenix WinNonlin, 6.3.0; Pharsight, a Certara Company, St. Louis, MO, USA) [33]. Participants and treatment periods with missing or inaccurate urine volumes were excluded from the statistical analysis. Since inhibition of MATE1 is known to alter creatinine concentrations, and famotidine does not affect glomerular filtration [34,35], the CKD-EPI creatinine equation was used to calculate creatinine clearance (CLCR) and estimate glomerular filtration rate (GFR) [36]. Metformin renal secretion clearance (CLRS) was calculated based on that estimated GFR using the equation CLRS = CLR − fu · GFR, where fu is the fraction of unbound metformin in plasma. Metformin total renal clearance was calculated using the total amount of metformin excreted in urine (Ae36) divided by the total area under the curve (AUC36) after two doses. The plasma AUC36 was determined by a population approach using population pharmacokinetic parameters from a model previously developed in healthy volunteers under similar study conditions together with integration techniques in NONMEM [37]. Bioavailability was calculated as the total amount of metformin excreted in the urine estimated to infinity divided by the total dose of metformin (1,850 mg), assuming that metformin is eliminated entirely by the kidney (fe=1) [38]. For a description of the pharmacometric modeling and simulation methodologies for famotidine and creatinine, see Supplemental Material.
2.6 Statistical Analysis
The size of the study population (n=12) was calculated to allow a detection of a true difference of more than 30% in AUC3 of glucose with a power of 80% and a significance level of 5%. Data are presented as mean ± SEM unless otherwise specified. Paired nonparametric Student’s t-tests were used to analyze the differences in metformin pharmacokinetic and pharmacodynamic parameters during metformin alone versus metformin plus famotidine, as well as for between genotypes, using GraphPad Prism 5.0 (GraphPad Software Inc., San Diego, CA, USA). Geometric mean ratios (GMR) and 90% confidence interval (CI) about the GMR were calculated using Stata 12.1 (StataCorp LP, College Station, Texas, USA) to estimate clinically relevant differences in metformin pharmacokinetic parameters with standard bioequivalence boundaries of 80–125 %.
3 Results
3.1 Famotidine In Vitro Assays
The inhibitory effect of famotidine on 14C-labeled metformin transport was evaluated in HEK293 cells stably overexpressing organic cation transporters (Supplemental Fig. 1). The mean (95% CI) IC50 values for famotidine against metformin transport were 0.25 μM (0.20–0.32 μM), 2.5 μM (1.5–4.0 μM), 19 μM (10–35 μM), and 66 μM (48–90 μM) in hMATE1, hMATE2 hOCT1 and hOCT2 cells, respectively. In this manuscript the term “selective” MATE1 inhibitor refers to the selectivity of famotidine for MATE1 in comparison to these transporters.
3.2 Healthy Volunteers
A total of 14 healthy male and female volunteers were recruited from the Studies of Pharmacogenetics in Ethnically Diverse Populations (SOPHIE) cohort. Two volunteers experienced nausea after metformin dosing and voluntarily withdrew from the study (See Study Design, Fig. 1). Twelve subjects, all of European ancestry, completed both phases of the study and are included in the pharmacokinetic and pharmacodynamic analyses. Demographic characteristics are found in Table 1.
Table 1.
Demographic Characteristics of Twelve Healthy Volunteers
| Continuous Covariate | Mean | SEM | Median | Range |
|---|---|---|---|---|
| Age (years) | 28 | 1.6 | 28 | 21 – 40 |
| BMI (kg/m2) | 27 | 1.7 | 25 | 21 – 37 |
| CLCR (mL/min/1.73m2) | 113 | 3.0 | 114 | 99 – 129 |
| Categorical Covariate Sex [n (%)] |
Value | |||
| Male | 8 (67) | |||
| Female | 4 (33) | |||
BMI, body mass index; CLCR, creatinine clearance estimated from CKD-EPI (2009) equation http://www.kidney.org/professionals/KDOQI/gfr_calculator
3.3 Clinical Pharmacokinetics of Famotidine
The maximal concentration of unbound famotidine in plasma ([I]) exceeded approximately 4-fold the in vitro hMATE1 IC50 ([I]=1.0 ± 0.064 μM, Supplemental Fig. 1). At such concentrations, famotidine was well tolerated during co-administration with metformin in this cohort of twelve healthy volunteers. Using the Cmax,u of famotidine as [I], the corresponding [I]/IC50 values for metformin transporters hMATE1, hMATE2, hOCT1 and hOCT2, were 4.0, 0.40, 0.053 and 0.015, which indicates that famotidine reached plasma levels predictive of highly selective inhibition of metformin transport by MATE1 in the kidney, without appreciably inhibiting metformin transport by OCT1 or OCT2 and with only minimal inhibition of MATE2 (Supplemental Fig. 2). Therefore, to our knowledge, famotidine is the first clinically validated highly selective MATE1 inhibitor using oral doses of 160 to 200 mg. For population pharmacokinetic parameter estimates of famotidine see Supplemental Material.
3.4 Clinical Pharmacokinetics of Metformin
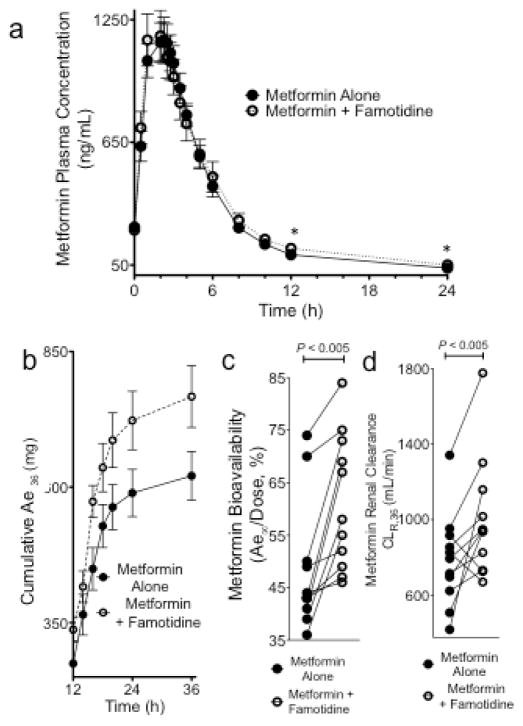
Mean metformin plasma concentration-time curves during metformin alone and during metformin with famotidine, are shown in Fig. 2a. The pharmacokinetic parameters of metformin are comparable to previous reports in healthy volunteers [22,33,39,40]. Famotidine did not significantly affect the Cmax, AUC or total clearance (CL/F) of metformin (Table 2). However, famotidine increased the cumulative amount of metformin excreted in the urine at total time of 36h and at time estimated to infinity (P<0.005, Fig. 2b). Famotidine significantly increased the estimated metformin bioavailability (F, Ae∞/Dose) on average by 24% (Fig. 2c, P<0.005, GMR (90%CI) 1.24 (1.12–1.38). When the treatment groups were compared in terms of total metformin renal clearance (1,850 mg in total over 36h) using integration techniques in NONMEM (Table 2), there was a 28% increase in metformin renal clearance (P<0.05, GMR [90%CI] 1.28 [1.06–1.55]) and 36% increase in metformin secretory clearance (P<0.05, GMR [90%CI] 1.36 [1.07–1.73]) (Fig. 2d, Table 2). Interestingly, there was an increase in metformin fractional renal clearance 0–12h post metformin dosing (Supplemental Fig. 3). Although no difference in urine flow rate between treatment phases was documented, famotidine significantly decreased the urine pH between 0 and 12h after metformin dosing (P<0.005) (Table 2).
Fig. 2.
The effect of famotidine on metformin pharmacokinetics in healthy volunteers (n=12). a) The mean metformin plasma concentration-time curves from 0–24h following metformin dosing (1,850 mg in total) alone (closed circles) and during co-administration with famotidine (1,000 mg over 48h) in healthy volunteers (n=12). The asterisks indicate significant differences, P < 0.01. b) Cumulative urinary excretion of metformin after two doses of metformin (1,850 mg in total) alone (solid circles) or during co-therapy with famotidine (open circles). Values are expressed as mean ± SEM. c) Estimated metformin bioavailability [Ae∞/Dose]. d) Metformin renal clearance (CLR) for 36h, computed using NONMEM, during metformin treatment alone (square) and during co-administration with famotidine (triangle).
Table 2.
Effect of Famotidine, a MATE1 Selective Inhibitor, on Metformin Pharmacokinetics and Pharmacodynamics in Twelve Healthy Volunteers
| Parameter | Units | Metformin | Metformin + Famotidine | P | GMR (90%CI) | ||
|---|---|---|---|---|---|---|---|
| Mean ± SEM | Median | Mean± SEM | Median | ||||
| Metformin Pharmacokinetics | |||||||
| Cmax | ng/mL | 1,260 ± 97 | 1,255 | 1,290 ± 120 | 1,200 | 0.97 | 1.01 (0.83–1.22) |
| Tmax | h | 2.0 ± 0.24 | 2.0 | 1.6 ± 0.23 | 1.0 | 0.23 | 0.80 (0.60–1.06) |
| T 1/2 | h | 7.6 ± 0.53 | 7.9 | 9.1 ± 1.1 | 8.4 | 0.077 | 1.17 (1.01–1.34) |
| CL/F | mL/min | 1,540 ± 112 | 1,510 | 1,500 ± 149 | 1,510 | 0.99 | 0.95 (0.82–1.11) |
| V/F | L | 1,020 ± 105 | 1,060 | 1,210 ± 193 | 1,070 | 0.34 | 1.11 (0.92–1.35) |
| AUC24 | ng* h/mL | 6,980 ± 466 | 6,780 | 7,480 ± 664 | 6,760 | 0.99 | 1.05 (0.90–1.23) |
| AUC36 | ng* h/mL | 14,200 ± 1,040 | 13,400 | 14,300 ± 1,390 | 12,600 | 0.99 | 0.98 (0.82–1.17) |
| AUC∞ | ng* h/mL | 14,600 ± 1,040 | 14,100 | 15,000 ± 1,400 | 13,300 | 0.99 | 1.00 (0.84–1.20) |
| Ae36 | mg | 621 ± 45 | 576 | 800 ± 51 | 729 | 0.0010 | 1.22 (1.11–1.35) |
| Ae∞ | mg | 629 ± 48 | 730 | 817 ± 52 | 754 | 0.0010 | 1.24 (1.12–1.37) |
| F(Ae∞/Dose*) | % | 47 ± 3.6 | 43 | 61 ± 3.9 | 58 | 0.0038 | 1.24 (1.12–1.38) |
| CLR,met,36 | mL/min | 767 ± 70 | 754 | 1,000 ± 96 | 942 | 0.042 | 1.28 (1.06–1.55) |
| CLRS,met,36 | mL/min | 643 ± 71 | 637 | 877 ± 93 | 809 | 0.032 | 1.36 (1.07–1.73) |
| Metformin Pharmacodynamics: | |||||||
| Parameter | Units | Baseline (No Drug) | Metformin | P | GMR (90%CI) | ||
| Mean ± SEM | Median | Mean ± SEM | Median | ||||
| AUCglu,0.5 | mg* h/dL | 58 ± 1.9 | 57 | 55 ± 1.5 | 55 | 0.012 | 0.94 (0.90–0.98) |
| AUCglu,3 | mg* h/dL | 360 ± 16 | 373 | 356 ± 14 | 343 | 0.79 | 0.99 (0.94–1.04) |
| Parameter | Units | Metformin | Metformin + Famotidine | P | GMR (90%CI) | ||
| Mean ± SEM | Median | Mean ± SEM | Median | ||||
| AUCglu,0.5 | mg* h/dL | 55 ± 1.5 | 55 | 52 ± 1.4 | 52 | 0.021 | 0.95 (0.92–0.99) |
| AUC glu,3 | mg* h/dL | 356 ± 14 | 343 | 355 ± 11 | 354 | 0.79 | 1.00 (0.94–1.07) |
| Other Plasma and Urinary Measurements | |||||||
| Urine Flow | mL/min | 3.4 ± 0.32 | 3.6 | 3.4 ± 0.29 | 3.1 | 0.99 | 1.00 (0.87–1.14) |
| Urine pH | - | 6.34 ± 0.064 | 6.4 | 6.11 ± 0.070 | 6.1 | 0.0049 | 0.96 (0.95–0.98) |
| CLR,creat,0 | mL/min | 124 ± 5 | 126 | 125 ± 6 | 127 | 0.72 | 1.00 (0.98–1.02) |
| CLR,creat,12 | mL/min | 121 ± 6.1 | 121 | 111 ± 7 | 108 | 0.014 | 0.91 (0.86–0.97) |
| CLR,creat,36 | mL/min | 123 ± 7.0 | 130 | 108 ± 6 | 106 | 0.028 | 0.88 (0.82–0.95) |
| CLR,creat,48 | mL/min | 123 ± 6 | 121 | 113 ± 6 | 106 | 0.0037 | 0.92 (0.88–0.96) |
Blood and urine collection was complete for all participants, except in one individual with one incomplete urine collection (0–2h after metformin therapy during the metformin plus famotidine phase). The individual with the incomplete urine collection was excluded when calculating Ae, F and CLR. Total amount of metformin excreted in the urine through time infinity with and without famotidine was estimated by fitting the mean amount excreted in the urine versus time to a Hill equation where Ae∞ represented the plateau value. Data are expressed as mean ± SEM unless stated otherwise. P values of less than 0.05 are considered significant.
Dose refers to metformin-free base (1,332 mg).
GMR, Geometric Mean Ratio; CI, confidence interval; Cmax, maximal plasma concentration; Tmax, time to the maximal plasma concentration; T1/2, plasma terminal elimination half-life; CL/F, apparent systemic clearance; V/F, apparent volume of distribution; AUC24/36/∞, area under the metformin plasma concentration–time curve from 12 to 36h or 0 to 36h or 0 to infinity, respectively, where 0 is the time of first metformin dose administration; F, estimated bioavailability; Ae36/∞, amount of metformin excreted in the urine from 0 to 36h after initial dose or to infinity, respectively; CLR,met,36 metformin renal clearance from 0 to 36h; CLRS,met,36 metformin secretory clearance from 0 to 36h, calculated as CLRS = CLR − fu · GFR where CLR, fu, and GFR are metformin renal clearance, fraction of unbound metformin in plasma, and glomerular filtration rate, respectively, assuming negligible metformin reabsorption. The value of fu was set at 1, and GFR was calculated using the CKD-EPI (2009) equation, without adjustment for body surface area to express GFR in mL/min (http://www.kidney.org/professionals/KDOQI/gfr_calculator); AUCglu,0.5/3 area under the glucose blood concentration time–curve from 0 to 0.5h or 3h, respectively, where 0 is the time of glucose administration; CLR,creat,0/12/36/48 creatinine renal clearance at time 0,12, 36 or 48h after the first famotidine dose administration, respectively.
3.5 Clinical Pharmacodynamics of Metformin
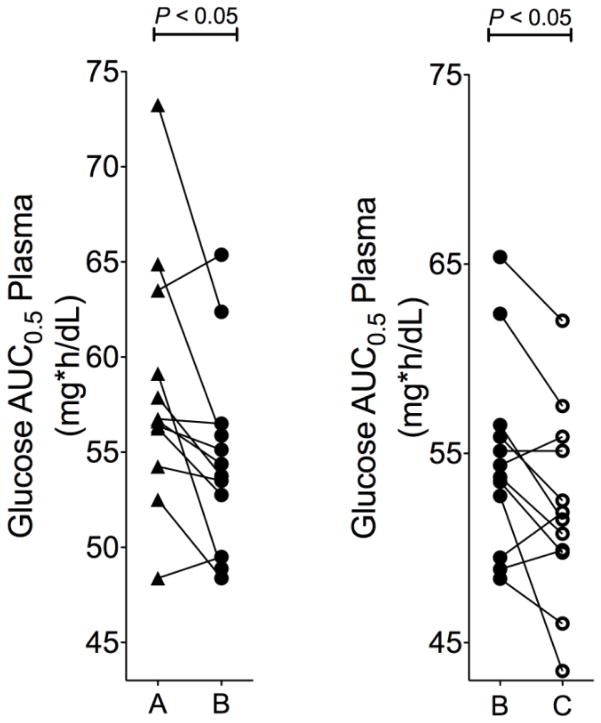
Healthy volunteers (n=12) underwent 3h OGTTs to evaluate the effect of famotidine on the glucose-lowering response to metformin. Baseline plasma glucose concentrations (before glucose ingestion) were similar during each treatment phase. Glucose AUCs before and after metformin therapy alone and after metformin-famotidine co-therapy, as a comparison of metformin glucose-lowering response in healthy volunteers, are shown in Fig. 3 and in Table 2. The AUC of glucose during the first 30 minutes after glucose ingestion was significantly reduced after metformin alone (P<0.05, GMR [90% CI] 0.94 [0.90–0.98]) and was further reduced after metformin-famotidine co-therapy compared to metformin therapy alone (P<0.05, GMR [90%CI] 0.95 [0.92–0.99]) (Fig. 3). Famotidine alone produced no significant effect on glucose AUC (data not shown). Thus, the ability of metformin to reduce the AUC of glucose during the first 30 minutes after glucose ingestion was significantly improved by famotidine. However, the improvement in glucose AUC reduction was not sustained beyond 30 minutes.
Fig. 3.
The difference in area under the glucose concentration-time curve from 0–0.5h (AUCglu,0.5) a) before (A, triangles) and after metformin (B, solid circles) and b) after metformin (B, solid circles) compared to after metformin plus famotidine (C, open circles)
4 Discussion
In this study, using famotidine, we determined the effect of selective MATE1 inhibition on the pharmacokinetics and pharmacodynamics of metformin and creatinine in healthy volunteers. Our major findings were: i) famotidine increases the estimated bioavailability (Ae∞/Dose) of metformin, ii) selective MATE1 inhibition does not result in reduced renal clearance or net clearance by secretion of metformin, iii) selective MATE1 inhibition is sufficient to alter creatinine levels in opposing directions, iv) famotidine’s effects on metformin pharmacokinetics differ from those of traditional non-selective MATE1/MATE2 inhibitors and v) famotidine transiently improves the glucose lowering effects of metformin. The data supporting these findings and their implications to understanding the mechanisms of metformin pharmacokinetics and pharmacodynamics are discussed below.
Previous studies demonstrate that the rate of metformin elimination is much faster than its rate of absorption, which indicates flip-flop kinetics, that is, the terminal linear portion of the plasma concentration-time curve actually reflects absorption instead of elimination [38,41]. In this study, although the Cmax and AUC of metformin did not significantly increase with famotidine co-therapy, metformin concentrations at later time-points (i.e., 12 and 24h post-metformin dosing) were significantly higher. Additionally, the time to Cmax (i.e., Tmax) decreased 20% (GMR 90%CI 0.80 (0.60–1.06) with famotidine (P<0.01), however the terminal half-life did not significantly change (Table 2). Importantly, the bioavailability and cumulative amount of metformin excreted in the urine significantly increased following famotidine co-administration (Fig. 2b,c). Since metformin does not undergo appreciable metabolism or biliary excretion, interactions of famotidine with metformin transporters involved in its absorption may explain the apparent increase in bioavailability[42–45]. MATE1 is expressed in human intestine, liver and it was localized to cell membrane as well as intracellular organelles that may play a role in metformin transport [2–7,9–10]. In addition, tissue distribution studies in mice treated with the MATE inhibitor pyrimethamine indicate that liver and kidney levels of metformin are significantly increased even without a change in plasma concentrations [16,17,24,46,47]. These reports, together with our results, suggest that famotidine may increase the rate and extent of metformin absorption or alter hepatic distribution resulting in less intestinal elimination and improved absorption. Moreover, famotidine has clear effects in increasing gastric pH, which would result in a greater fraction of the drug in the unionized state, facilitating passive absorption of the drug [31]. Further studies are clearly needed to determine the mechanisms by which famotidine enhances metformin’s estimated bioavailability.
Famotidine did not reduce metformin renal clearance as we had hypothesized. Instead, we observed increased metformin renal clearance and renal clearance by secretion during famotidine co-administration after 1,850 mg in total (Fig. 2d). The famotidine-induced increases in both metformin absorption and renal clearance likely had opposing effects, which resulted in no change in metformin plasma exposure (Cmax or AUC) and no change in metformin clearance (CL/F) (Table 2). These results differ from previous findings with the non-selective MATE inhibitors pyrimethamine and cimetidine. In particular, administration of pyrimethamine or cimetidine result in reduced renal clearance and secretory clearance of MATE substrates, including metformin [23,25,40,47–50].
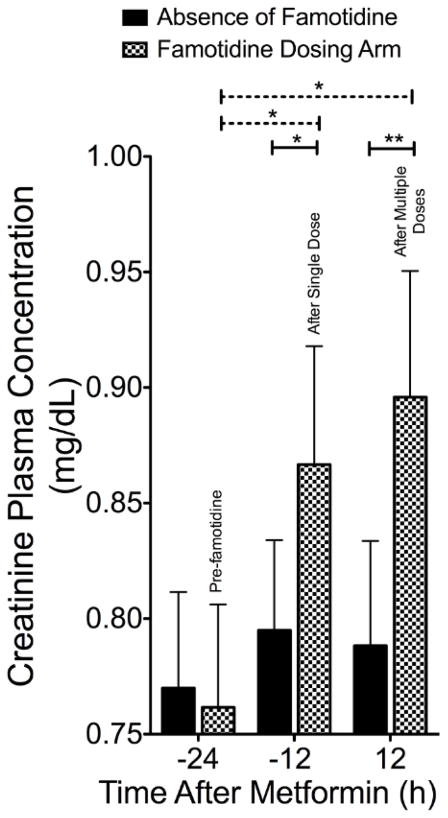
Several possible mechanisms may explain the differences between our results with famotidine versus those obtained previously with pyrimethamine and cimetidine. First, it is possible that the drug simply did not interact with MATE1 during our study, even though it achieved four times higher plasma levels compared with its IC50 for inhibition of MATE1. Perhaps famotidine concentrations in the vicinity of the transporter (i.e., intracellularly on the apical surface of the renal tubule) may not have been sufficiently high to inhibit MATE1. However, famotidine was determined to be an unequivocal substrate of the transporters MATE1, MATE2, OCT1 and OCT2 in overexpressing cell lines (Supplemental Fig. 4) with renal clearance of 290 ± 32 mL/min, indicating some tubular secretion. Therefore, unbound concentrations of famotidine in the proximal tubule would be expected to be similar to or even higher than those in the plasma. Moreover, our data suggest that famotidine did inhibit MATE1. That is, in this study, creatinine concentrations were significantly altered in a manner that is consistent with previous reports of drugs that are potent inhibitors of MATE1 [12,51]; urinary creatinine clearance was significantly decreased (Table 1) and creatinine concentrations in plasma were significantly higher 12h after famotidine (200 mg) administration alone compared to pre-famotidine concentrations, as well as compared to matched controls in the absence of famotidine at 12h and 36h (Fig. 4). A model was developed to quantify the impact of famotidine on creatinine plasma levels in healthy volunteers while separating out diurnal rhythms in the data (Supplemental material, Supplemental Fig. 5,6, Supplemental Table 3,4,5). The model described the data well, both for the mean curve and the variability between patients, and the effect of famotidine on creatinine turnover could be quantified with good precision and proved to be significant (P< 0.001). It is possible that MATE1 may be a more sensitive transporter of creatinine than metformin, which would need to be determined experimentally, however our data suggest that famotidine did inhibit MATE1, but did not reduce metformin secretory clearance. The second explanation for the apparent lack of significant reduction of metformin renal clearance, despite alteration in creatinine concentrations, is that, metformin renal clearance may be blood flow limited, [52], whereas creatinine clearance is less sensitive to changes in renal blood flow. It is possible that famotidine may have increased renal blood blow in our study resulting in enhanced metformin renal clearance though to our knowledge changes in renal hemodynamics with famotidine have not been described. Third, selective MATE1 inhibition may not be sufficient to cause a significantly observable reduction in metformin secretory clearance. That is, unlike creatinine, for metformin non-selective inhibition of both apical transporters MATE1 and MATE2 may be required. Lastly, inhibition of basolateral transporters (e.g., OCT2) may be more important to metformin pharmacokinetics than apical membrane transporters (e.g., MATEs). Famotidine inhibition of metformin transport is over 200 fold more selective for MATE1 than for OCT2 (Supplemental Fig. 1), whereas cimetidine appears to be a considerably more potent inhibitor of OCT2 [53]. Thus, cimetidine may be a particularly nonselective inhibitor of metformin renal secretory transporters, OCT2, MATE1 and MATE2, in comparison to famotidine, which is highly selective for MATE1.
Fig. 4.
The effect of famotidine on plasma creatinine concentrations in healthy volunteers (n=12). Creatinine plasma concentrations at 0, 12 and 36h post the first dose (200 mg) of famotidine (checked bars) and at the same times during metformin alone (solid bars). Dashed lines indicate comparisons within the same study visit, solid lines indicate comparisons between study visits with *P < 0.05 and ** P <0.01. Data represent mean ± SEM
The increase in total metformin renal clearance and the trend towards an increased fractional renal secretory clearance of metformin are difficult to explain. It is possible that famotidine inhibits transporter-mediated reabsorption of metformin, which may be mediated through MATE1 or another transporter. For example, famotidine was found to be a potent inhibitor of metformin transport by plasma membrane monoamine transporter (PMAT, SLC29A4), which has been implicated in facilitating reabsorption of organic cations in the kidney [54] (Supplemental Fig. 7). Furthermore, the decrease in urinary pH associated with famotidine administration may have resulted in altered proton dependent transport. Though speculative, it’s possible that famotidine increased metformin renal clearance by decreasing its rate of passive reabsorption in renal tubule. Alternatively famotidine may have inhibited an as yet unidentified transporter involved in metformin reabsorption in the kidney. Finally, as noted, famotidine may have affected renal blood flow. This result clearly needs further study.
In terms of pharmacodynamics, the effects of famotidine on metformin-induced decreases in OGTT were in the direction that we hypothesized. That is, we observed a significant effect of famotidine in enhancing metformin’s response to OGTT in the first 30 minutes, when famotidine concentrations were the highest. These results are consistent with results obtained previously for the reduced function variant of MATE1 (rs2252281) [22]. Inhibition of MATE1 efflux would result in higher metformin concentrations in the liver and kidney and improved efficacy. However, in this study the effects were small and cannot be extrapolated to diabetic patients.
5 Conclusion
In conclusion, this study demonstrates that famotidine, a selective inhibitor of MATE1, significantly increases the estimated bioavailability of metformin and produces a transient increase in the pharmacologic response to metformin. Rather than a significant reduction in metformin secretory clearance, we observed an enhanced renal clearance of metformin with famotidine, which is in contrast to nonselective MATE inhibitors [23,25,40] that reduce metformin renal clearance. Taken together, the study suggests that improved bioavailability of metformin as well as enhanced renal clearance can be obtained with famotidine. Our studies highlight the importance of conducting clinical drug-drug interaction studies because findings may differ from predictions based on in vitro and animal studies.
Supplementary Material
Table 3.
Population Pharmacokinetic Parameter Estimates for Famotidine in Twelve Healthy Volunteers
| Population Parameter Estimate | Typical Values (R.S.E) |
|---|---|
| CL/F (L/h) | 116 (7.2%) |
| V/F (L) | 715 (6.0%) |
| ka (h−1) | 1.14 (11%) |
| Bioavailability (F) (%) | 1 |
| Lag time (h) | 0.27 (32%) |
|
| |
| Between Subject Variability (BSV) | |
|
| |
| BSV-CL (%) | 6.4% (25%) |
| BSV-F (%) | 26% (53%) |
| BSV-Lagtime (%) | 77% (99%) |
|
| |
| Residual Error | |
|
| |
| Additive Error (μg/L) | 0.26 (8.6%) |
| Proportional Error (%) | 1.4 (26%) |
Table 4.
Final Parameter Estimates for the Population Pharmacokinetic Describing the Effect of Famotidine on Creatinine Plasma Levels in Twelve Healthy Volunteers
| Population Parameters | Typical Values Without Famotidine (R.S.E.) | Typical Value With Famotidine (R.S.E.) |
|---|---|---|
| Baseline (mg/L) | 7.62 (3.7%) | 8.07 (5.5%) |
| Amplitude 1 (mg/L) | 1.11 (50%) | 1.15 (49%) |
| Phase Shift 1 (mg/L) | 2.41 (3.3%) | 2.37 (2.6%) |
| Amplitude 2 (mg/L) | 1.14 (49%) | |
| Phase Shift 2 (mg/L) | −0.79 (8.3%) | |
|
| ||
| Between Subject Variability (BSV) | ||
|
| ||
| BSV-Baseline (%) | 19 (27%) | 18 (19%) |
|
| ||
| BSV-Phase Shift (%) | 1.9 (89%) | 2.7 (89%) |
|
| ||
| Residual Error | ||
|
| ||
| Proportional Error (%) | 0.049 (5.7%) | 0.059 (120%) |
Key Points.
The study highlights an effect of famotidine on metformin urinary excretion and pharmacologic action suggesting that the bioavailability and response to metformin can be improved with a MATE1 inhibitor.
MATE1 inhibition by oral famotidine, at doses of 160 to 200 mg, is sufficient to increase creatinine plasma levels and decrease creatinine urine levels. A population pharmacokinetic model was developed to quantify the influence of famotidine on creatinine concentrations in plasma.
During drug development, clinical studies evaluating selective transporter-mediated drug-interactions may be less important than nonselective interactions for compounds such as metformin.
Acknowledgments
The authors would like to thank the SOPHIE Cohort and Study #6112 participants for contributing their time. The authors also thank Hector Vizoso, Nurse Manager, and the staff of the CRC, Chav Doherty, Research Study Coordinator, and the staff of the Clinical Laboratory at San Francisco General Hospital for their excellent service and assistance with our clinical study, as well as Dr. Yong Huang, Dr. Howard Horng and Mr. Nick Massenkoff at the UCSF Drugs Services Unit for access to their bioanalytical facilities and for their technical support.
Funding
This project was funded by the National Institutes of Health (NIH) grant GM61390. This work was also funded by NIH/National Center for Research Resources (NIH/NCRR) University of California, San Francisco (UCSF)-CTSI grant UL1 RR024131. J.E.H. was funded by the National Research Service Award T32 GM07546 from the NIH and by the Department of Clinical Pharmacy at UCSF. M.B.W. was funded by the Swiss National Science Foundation’s grant for prospective researchers PBBSP3-133384.
Glossary
- IC50
Concentration of drug producing 50% inhibition
- Cmax,u
Maximum unbound plasma drug concentration
- Ae∞
Cumulative amount of unchanged drug excreted in urine from time zero to infinity
- AUC
Area under the plasma concentration-time curve
- Cmax
maximum concentration in plasma
- AUCglu,0.5
area under the glucose plasma concentration-time curve between time zero and 0.5 hours
Footnotes
The authors are solely responsible for the contents and do not necessarily represent the official views of the NIH.
Conflict of interest
KMG, SWY, XZ, YH have declared the following conflicts of interest that might be relevant to the content of this manuscript: KMG and SWY are co-founders of Apricity Therapeutics, which develops drugs that exploit membrane transporters to enhance their pharmacologic action. KMG receives funds from several pharmaceutical companies (AstraZeneca, Pfizer, Sanofi Aventis and GlaxoSmithKline) for research in her laboratory. XZ and YH are employees of Optivia Biotechnology Inc., a transporter CRO company.
JEH, AAZ, RAC, MBW, RJK, SG, SLS, CMB and RMS, have no conflicts of interest that might be relevant to the content of this manuscript.
Compliance and Ethical Standards
Ethical Approval
This analysis was approved by the Committee on Human Research (CHR), which is the Institutional Review Board (IRB) at UCSF, approval # 10-02578.
References
- 1.Hillgren KM, Keppler D, Zur AA, Giacomini KM, Stieger B, Cass CE, et al. Emerging transporters of clinical importance: an update from the International Transporter Consortium. Clin Pharmacol Ther. 2013;94:52–63. doi: 10.1038/clpt.2013.74. [DOI] [PubMed] [Google Scholar]
- 2.Lu M, Symersky J, Radchenko M, Koide A, Guo Y, Nie R, et al. Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc Natl Acad Sci. 2013;110:2099–104. doi: 10.1073/pnas.1219901110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu M, Radchenko M, Symersky J, Nie R, Guo Y. Structural insights into H+-coupled multidrug extrusion by a MATE transporter. Nat Struct Mol Biol. 2013;20:1310–7. doi: 10.1038/nsmb.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damme K, Nies AT, Schaeffeler E, Schwab M. Mammalian MATE (SLC47A) transport proteins: impact on efflux of endogenous substrates and xenobiotics. Drug Metab Rev. 2011;43:499–523. doi: 10.3109/03602532.2011.602687. [DOI] [PubMed] [Google Scholar]
- 5.Motohashi H, Inui K. Multidrug and toxin extrusion family SLC47: Physiological, pharmacokinetic and toxicokinetic importance of MATE1 and MATE2-K. Mol Aspects Med. 2013;34:661–8. doi: 10.1016/j.mam.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Yonezawa A, Inui K. Importance of the multidrug and toxin extrusion MATE/SLC47A family to pharmacokinetics, pharmacodynamics/toxicodynamics and pharmacogenomics. Br J Pharmacol. 2011;164:1817–25. doi: 10.1111/j.1476-5381.2011.01394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanihara Y, Masuda S, Sato T, Katsura T, Ogawa O, Inui K. Substrate specificity of MATE1 and MATE2-K, human multidrug and toxin extrusions/H+-organic cation antiporters. Biochem Pharmacol. 2007;74:359–71. doi: 10.1016/j.bcp.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Masuda S, Terada T, Yonezawa A, Tanihara Y, Kishimoto K, Katsura T, et al. Identification and functional characterization of a new human kidney-specific H+/organic cation antiporter, kidney-specific multidrug and toxin extrusion 2. J Am Soc Nephrol. 2006;17:2127–35. doi: 10.1681/ASN.2006030205. [DOI] [PubMed] [Google Scholar]
- 9.Otsuka M, Matsumoto T, Morimoto R, Arioka S, Omote H, Moriyama Y. A human transporter protein that mediates the final excretion step for toxic organic cations. Proc Natl Acad Sci. 2005;102:1–6. doi: 10.1073/pnas.0506483102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JH, Lee JE, Kim Y, Lee H, Jun H, Lee S. Multidrug and toxic compound extrusion protein-1 (MATE1/SLC47A1) is a novel flavonoid transporter. J Agric Food Chem. 2014;62:9690–8. doi: 10.1021/jf500916d. [DOI] [PubMed] [Google Scholar]
- 11.Levey AS, Perrone RD, Madias NE. Serum creatinine and renal function. Annu Rev Med. 1988;39:465–90. doi: 10.1146/annurev.me.39.020188.002341. [DOI] [PubMed] [Google Scholar]
- 12.Lepist E-I, Zhang X, Hao J, Huang J, Kosaka A, Birkus G, et al. Contribution of the organic anion transporter OAT2 to the renal active tubular secretion of creatinine and mechanism for serum creatinine elevations caused by cobicistat. Kidney Int. 2014:1–8. doi: 10.1038/ki.2014.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imamura Y, Murayama N, Okudaira N, Kurihara A, Okazaki O, Izumi T, et al. Prediction of Fluoroquinolone-Induced Elevation in Serum Creatinine Levels: A Case of Drug–Endogenous Substance Interaction Involving the Inhibition of Renal Secretion. Clin Pharmacol Ther. 2009;89:81–8. doi: 10.1038/clpt.2010.232. [DOI] [PubMed] [Google Scholar]
- 14.Gong L, Goswami S, Giacomini KM, Altman RB, Klein TE. Metformin pathways. Pharmacogenet Genomics. 2012;22:820–7. doi: 10.1097/FPC.0b013e3283559b22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham GG, Punt J, Arora M, Day RO, Doogue MP, Duong JK, et al. Clinical pharmacokinetics of metformin. Clin Pharmacokinet. 2011:81–98. doi: 10.2165/11534750-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 16.Tsuda M, Terada T, Mizuno T, Katsura T, Shimakura J, Inui Ki. Targeted disruption of the multidrug and toxin extrusion 1 (mate1) gene in mice reduces renal secretion of metformin. Mol Pharmacol. 2009;75:1280–6. doi: 10.1124/mol.109.056242. [DOI] [PubMed] [Google Scholar]
- 17.Toyama K, Yonezawa A, Masuda S, Osawa R, Hosokawa M, Fujimoto S, et al. Loss of multidrug and toxin extrusion 1 (MATE1) is associated with metformin-induced lactic acidosis. Br J Pharmacol. 2012;166:1183–91. doi: 10.1111/j.1476-5381.2012.01853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker ML, Visser LE, van Schaik RHN, Hofman A. Genetic variation in the multidrug and toxin extrusion 1 transporter protein influences the glucose-lowering effect of metformin in patients with diabetes: A preliminary study. Diabetes. 2009;58:745–49. doi: 10.2337/db08-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tkáč I, Klimčáková L, Javorský M, Fabianová M, Schroner Z, Hermanová H, et al. Pharmacogenomic association between a variant in SLC47A1 gene and therapeutic response to metformin in type 2 diabetes. Diabetes Obes Metab. 2013;15:189–91. doi: 10.1111/j.1463-1326.2012.01691.x. [DOI] [PubMed] [Google Scholar]
- 20.Becker ML, Visser LE, van Schaik RHN, Hofman A, Uitterlinden AG, Stricker BHC. Interaction between polymorphisms in the OCT1 and MATE1 transporter and metformin response. Pharmacogenet Genomics. 2010;20:38–44. doi: 10.1097/FPC.0b013e328333bb11. [DOI] [PubMed] [Google Scholar]
- 21.Jablonski KA, McAteer JB, de Bakker PIW, Franks PW, Pollin TI, Hanson RL, et al. Common Variants in 40 Genes Assessed for Diabetes Incidence and Response to Metformin and Lifestyle Intervention in the Diabetes Prevention Program. Diabetes. 2010;59:2672–81. doi: 10.2337/db10-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stocker SL, Morrissey KM, Yee SW, Castro RA, Xu L, Dahlin a, et al. The Effect of Novel Promoter Variants in MATE1 and MATE2 on the Pharmacokinetics and Pharmacodynamics of Metformin. Clin Pharmacol Ther. 2012;93:186–94. doi: 10.1038/clpt.2012.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusuhara H, Ito S, Kumagai Y, Jiang M, Shiroshita T, Moriyama Y, et al. Effects of a MATE Protein Inhibitor, Pyrimethamine, on the Renal Elimination of Metformin at Oral Microdose and at Therapeutic Dose in Healthy Subjects. Clin Pharmacol Ther. 2011;89:837–44. doi: 10.1038/clpt.2011.36. [DOI] [PubMed] [Google Scholar]
- 24.Ito S, Kusuhara H, Kuroiwa Y, Wu C, Moriyama Y, Inoue K, et al. Potent and Specific Inhibition of mMate1-Mediated Efflux of Type I Organic Cations in the Liver and Kidney by Pyrimethamine. J Pharmacol Exp Ther. 2010;333:341–50. doi: 10.1124/jpet.109.163642. [DOI] [PubMed] [Google Scholar]
- 25.Somogyi A, Stockley C, Keal J, Rolan P, Bochner F. Reduction of metformin renal tubular secretion by cimetidine in man. Br J Clin Pharmacol. 1987;23:545–51. doi: 10.1111/j.1365-2125.1987.tb03090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ha Choi J, Wah Yee S, Kim MJ, Nguyen L, Ho Lee J, Kang J-O, et al. Identification and characterization of novel polymorphisms in the basal promoter of the human transporter, MATE1. Pharmacogenet Genomics. 2009;19:770–80. doi: 10.1097/FPC.0b013e328330eeca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wittwer MB, Zur AA, Khuri N, Kido Y, Kosaka A, Zhang X, et al. Discovery of Potent, Selective Multidrug and Toxin Extrusion Transporter 1 (MATE1, SLC47A1) Inhibitors Through Prescription Drug Profiling and Computational Modeling. J Med Chem. 2013;56:781–95. doi: 10.1021/jm301302s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pepcid label. Merck & Co., Inc; 2011. [Google Scholar]
- 29.Yeh KC, Chremos AN, Lin JH, Constanzer ML, Kanovsky SM, Hucker HB, et al. Single-dose pharmacokinetics and bioavailability of famotidine in man. Results of multicenter collaborative studies. Biopharm Drug Dispos. 1987;8:549–60. doi: 10.1002/bdd.2510080606. [DOI] [PubMed] [Google Scholar]
- 30.Dowling TC, Frye RF, Fraley DS, Matzke GR. Characterization of tubular functional capacity in humans using para-aminohippurate and famotidine. Kidney Int. 2001;59:295–303. doi: 10.1046/j.1523-1755.2001.00491.x. [DOI] [PubMed] [Google Scholar]
- 31.Chremos AN. Clinical pharmacology of famotidine: a summary. J Clin Gastroenterol. 1987;9(Suppl 2):7–12. doi: 10.1097/00004836-198707002-00003. [DOI] [PubMed] [Google Scholar]
- 32.Kroemer H, Klotz U. Pharmacokinetics of famotidine in man. Int J Clin Pharmacol Ther Toxicol. 1987;25:458–63. [PubMed] [Google Scholar]
- 33.Shu Y, Brown C, Castro RA, Shi RJ, Lin ET, Owen RP, et al. Effect of Genetic Variation in the Organic Cation Transporter 1, OCT1, on Metformin Pharmacokinetics. Clin Pharmacol Ther. 2007;83:273–80. doi: 10.1038/sj.clpt.6100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin JH, Los LE, Ulm EH, Duggan DE. Kinetic studies on the competition between famotidine and cimetidine in rats. Evidence of multiple renal secretory systems for organic cations. Drug Metab Dispos. 1988;16:52–6. [PubMed] [Google Scholar]
- 35.Dowling T, Frye R, Fraley D, Matzke G. Characterization of tubular functional capacity in humans using para-aminohippurate and famotidine. Kidney Int. 2001;59:295–303. doi: 10.1046/j.1523-1755.2001.00491.x. [DOI] [PubMed] [Google Scholar]
- 36.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF, Feldman HI, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–12. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goswami S, Yee SW, Stocker S, Mosley JD, Kubo M, Castro R, et al. Genetic Variants in Transcription Factors Are Associated With the Pharmacokinetics and Pharmacodynamics of Metformin. Clin Pharmacol Ther. 2014;96:370–9. doi: 10.1038/clpt.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pentikäinen PJ, Neuvonen PJ, Penttilä A. Pharmacokinetics of metformin after intravenous and oral administration to man. Eur J Clin Pharmacol. 1979;16:195–202. doi: 10.1007/BF00562061. [DOI] [PubMed] [Google Scholar]
- 39.Sambol NC, Chiang J, O’Conner M, Liu CY, Lin ET, Goodman AM, et al. Pharmacokinetics and Pharmacodynamics of Metformin in Healthy Subjects and Patients with Noninsulin-Dependent Diabetes Mellitus. J Clin Pharmacol. 1996;36:1012–21. doi: 10.1177/009127009603601105. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z-J, Yin OQP, Tomlinson B, Chow MSS. OCT2 polymorphisms and in-vivo renal functional consequence: studies with metformin and cimetidine. Pharmacogenet Genomics. 2008;18:637–45. doi: 10.1097/FPC.0b013e328302cd41. [DOI] [PubMed] [Google Scholar]
- 41.Stepensky D, Friedman M, Srour W, Raz I, Hoffman A. Preclinical evaluation of pharmacokinetic–pharmacodynamic rationale for oral CR metformin formulation. J Control release. 2001;71:107–15. doi: 10.1016/s0168-3659(00)00374-6. [DOI] [PubMed] [Google Scholar]
- 42.Zamek-Gliszczynski MJ, Bao JQ, Day JS, Higgins JW. Metformin sinusoidal efflux from the liver is consistent with negligible biliary excretion and absence of enterohepatic cycling. Drug Metab Dispos. 2013;41:1967–71. doi: 10.1124/dmd.113.053025. [DOI] [PubMed] [Google Scholar]
- 43.Han TK, Proctor WR, Costales CL, Cai H, Everett RS, Thakker DR. Four Cation-Selective Transporters Contribute to Apical Uptake and Accumulation of Metformin in Caco-2 Cell Monolayers. J Pharmacol Exp Ther. 2015;352:519–28. doi: 10.1124/jpet.114.220350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamichi N, Shima H, Asano S, Ishimoto T, Sugiura T, Matsubara K, et al. Involvement of carnitine/organic cation transporter OCTN1/SLC22A4 in gastrointestinal absorption of metformin. J Pharm Sci. 2013;102:3407–17. doi: 10.1002/jps.23595. [DOI] [PubMed] [Google Scholar]
- 45.Zhou M, Xia L, Wang J. Metformin Transport by a Newly Cloned Proton-Stimulated Organic Cation Transporter (Plasma Membrane Monoamine Transporter) Expressed in Human Intestine. Drug Metab Dispos. 2007;35:1956–62. doi: 10.1124/dmd.107.015495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hume WE, Shingaki T, Takashima T, Hashizume Y, Okauchi T, Katayama Y, et al. The synthesis and biodistribution of [(11)C]metformin as a PET probe to study hepatobiliary transport mediated by the multi-drug and toxin extrusion transporter 1 (MATE1) in vivo. Bioorg Med Chem. 2013;21:7584–90. doi: 10.1016/j.bmc.2013.10.041. [DOI] [PubMed] [Google Scholar]
- 47.Li Q, Guo D, Dong Z, Zhang W, Zhang L, Huang S-M, et al. Ondansetron can enhance cisplatin-induced nephrotoxicity via inhibition of multiple toxin and extrusion proteins (MATEs) Toxicol Appl Pharmacol. 2013;273:100–9. doi: 10.1016/j.taap.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Somogyi a, McLean a, Heinzow B. Cimetidine-procainamide pharmacokinetic interaction in man: Evidence of competition for tubular secretion of basic drugs. Eur J Clin Pharmacol. 1983;25:339–45. doi: 10.1007/BF01037945. [DOI] [PubMed] [Google Scholar]
- 49.Van Crugten J, Bochner F, Keal J, Somogyi A, Pharmacology OF. Selectivity of the cimetidine-induced alterations in the renal handling of organic substrates in humans. Studies with anionic, cationic and zwitterionic drugs. J Pharmacol Exp Ther American Society for Pharmacology and Experimental Therapeutics. 1986;236:481–7. [PubMed] [Google Scholar]
- 50.Yasui-Furukori N, Uno T, Sugawara K, Tateishi T. Different effects of three transporting inhibitors, verapamil, cimetidine, and probenecid, on fexofenadine pharmacokinetics. Clin Pharmacol Ther. 2005;77:17–23. doi: 10.1016/j.clpt.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 51.Ray AS. Creat Ren Transp. San Diego, CA, USA: AAPS Annual Meeting and Exposition; 2014. Creatinine and Renal Transporters. Endog. Biomarkers Mother Nature’s Guid. to Predict. Drug-Drug Interact. [Google Scholar]
- 52.Wang D-S, Jonker JW, Kato Y, Kusuhara H, Schinkel AH, Sugiyama Y. Involvement of organic cation transporter 1 in hepatic and intestinal distribution of metformin. J Pharmacol Exp Ther. 2002;302:510–5. doi: 10.1124/jpet.102.034140. [DOI] [PubMed] [Google Scholar]
- 53.Motohashi H, Uwai Y, Hiramoto K, Okuda M, Inui K-I. Different transport properties between famotidine and cimetidine by human renal organic ion transporters (SLC22A) Eur J Pharmacol. 2004;503:25–30. doi: 10.1016/j.ejphar.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 54.Xia L, Engel K, Zhou M, Wang J. Membrane localization and pH-dependent transport of a newly cloned organic cation transporter (PMAT) in kidney cells. Am J Physiol Renal Physiol. 2007;292:F682–90. doi: 10.1152/ajprenal.00302.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.