Abstract
Bacillus cereus is an important cause of foodborne infectious disease and food poisoning. However, B. cereus has also been used as a probiotic in human medicine and livestock production, with low standards of safety assessment. In this study, we evaluated the safety of 15 commercial probiotic B. cereus preparations from China in terms of mislabeling, toxin production, and transferable antimicrobial resistance. Most preparations were incorrectly labeled, as they contained additional bacterial species; one product did not contain viable B. cereus at all. In total, 18 B. cereus group strains—specifically B. cereus and Bacillus thuringiensis—were isolated. Enterotoxin genes nhe, hbl, and cytK1, as well as the ces-gene were assessed by PCR. Enterotoxin production and cytotoxicity were confirmed by ELISA and cell culture assays, respectively. All isolated B. cereus group strains produced the enterotoxin Nhe; 15 strains additionally produced Hbl. Antimicrobial resistance was assessed by microdilution; resistance genes were detected by PCR and further characterized by sequencing, transformation and conjugation assays. Nearly half of the strains harbored the antimicrobial resistance gene tet(45). In one strain, tet(45) was situated on a mobile genetic element—encoding a site-specific recombination mechanism—and was transferable to Staphylococcus aureus and Bacillus subtilis by electro-transformation. In view of the wide and uncontrolled use of these products, stricter regulations for safety assessment, including determination of virulence factors and transferable antimicrobial resistance genes, are urgently needed.
Keywords: Bacillus cereus, probiotic, enterotoxin, antimicrobial resistance, China, tetracycline resistance gene tet(45), site-specific recombination, dif site
Introduction
Bacillus cereus is a common cause of foodborne infectious disease: in a 10-year US survey B. cereus was ranked the fifth most common bacterial species causing foodborne outbreaks (http://www.cdc.gov/foodsafety/pdfs/fdoss-98-08-appendix-contributing-factors-508c.pdf; for detailed outbreak analysis see Bennett et al., 2013). Worldwide, Chinese food is frequently associated with foodborne B. cereus outbreaks (Notermans and Batt, 1998). Accordingly, B. cereus accounted for 13.4% of 1082 bacterial foodborne outbreaks between 1994 and 2005 in China; B. cereus was ranked as the second after Salmonella spp. amongst the most frequent bacterial causes of foodborne disease in Chinese inland provinces (Wang et al., 2007). This might be partly attributed to consumption habits, since B. cereus outbreaks are regularly associated with the consumption of rice (Ankolekar et al., 2009). In addition to rice (and pasta), meat, and dairy products are also important sources of foodborne B. cereus outbreaks in humans (Ruusunen et al., 2013; López et al., 2015). There are two different types of foodborne disease related to B. cereus: intoxication due to pre-formed cereulide or infection, followed by intra-intestinal production of enterotoxins such as Nhe and Hbl (Stenfors Arnesen et al., 2008). Foodborne B. cereus disease is generally self-limiting, although severe cases and even deaths have been reported (Bottone, 2010).
Though, being known as an important foodborne pathogen, B. cereus is licensed as a probiotic for men and livestock. In China, the safety of B. cereus probiotics is assessed and regulated; however, these regulations are partly rendered obsolete by recent research on B. cereus, particularly the identification of new virulence factors (Kim et al., 2015; Omer et al., 2015; Kilcullen et al., 2016). Thus, there is a lack of appropriate safety assessment for these probiotics in China—and also in other parts of the world—although, probiotics are even used to treat bacterial infections (Isolauri, 2003). In legal terms, probiotics are usually considered as supplements or foods—a fact which is critically discussed (Arnold, 2013). Apart from one pseudo-outbreak (Kniehl et al., 2003), no case of foodborne B. cereus infection due to the use of probiotics has theretofore been reported. However, a fatal infection following the application of a probiotic Bacillus subtilis preparation was reported in an immune-compromised patient (Oggioni et al., 1998).
Probiotic strains are applied to livestock as well. Thus, the use of B. cereus probiotics in livestock production might provide an additional hazard for human health. In China, animal wastes are traditionally used as feed in fish ponds; this creates a melting pot for bacteria of human, animal, and environmental origin and thereby facilitates horizontal gene transfer (Allen et al., 2010). Thus, besides the lack of detectable toxicity, future probiotic safety regulations should take into account the potential of transferring antibiotic resistance genes.
We hypothesize that toxin production, transferable antimicrobial resistance, and contamination with other bacteria are important safety aspects of probiotics, but insufficiently assessed up to now. Therefore, we intended to re-evaluate these important safety parameters in commercial probiotic B. cereus products.
Materials and methods
Isolation and identification of bacteria
Products were collected between 2013 and 2015 from nine provinces in China (Table 1). This investigation included all four B. cereus preparations licensed for human use. From the original packaging, the capsules with the dried probiotic preparations were removed with a tweezer, dipped into 80% ethanol, flame-treated and soaked in sterile distilled water until the capsules where dissolved. Subsequently, the content was suspended and 0.1 ml of the suspension was plated on Columbia agar with 5% sheep blood (Oxoid, Wesel, Germany). Single colonies of each different phenotype were passaged on new agar plates. Matrix assisted laser desorption/ionization (MALDI-TOF) mass spectrometry was used for bacterial identification (Schulthess et al., 2014). Phenotypic identification by means of biochemical tests with the API 50 CH and 20 E kits (BioMerieux) were used to further confirm and/or characterize the B. cereus group bacteria. The total DNA of each isolate was extracted from overnight cultures in casein hydrolysate-yeast medium plus 1% glucose (CGY) using the DNeasy blood and tissue kit (Qiagen, Germany), according to the protocol for Gram-positive bacteria. An extended multiplex PCR assay with five pairs of universal primers was used to identify the cry-group genes in the Bacillus thuringiensis strains (Ben-Dov et al., 1997).
Table 1.
Characterization of commercial B. cereus containing probiotic products in China.
| Product No. | Strain No. | Origin | Label | Other species | Intended for use in | Species | Toxin type |
|---|---|---|---|---|---|---|---|
| 1 | 1 | Jiangsu | mono | none | Human | B. cereus | Nhe, Hbl |
| 2 | 2a | Jiangsu | mono | none# | B. cereus | Nhe, Hbl | |
| 2b | B. cereus | Nhe | |||||
| 3 | 3 | Henan | mono | none | B. cereus | Nhe, Hbl | |
| 4. | 4 | Zhejiang | mixed | not determined | B. cereus | Nhe, Hbl | |
| 5 | 5c | Jiangsu | mono | other Bacillus spp. | Animal | B. cereus | Nhe |
| 6 | 6f | Hebei | mono | other Bacillus spp. Acinetobacter sp. Enterococcus sp. no ID. |
B. cereus | Nhe, Hbl | |
| 7 | 7d | Hebei | mono | other Bacillus spp. | B. thuringiensis | Nhe, Hbl, Cry | |
| 8 | 8h | Hebei | mixed | other Bacillus spp. | B. cereus | Nhe, Hbl | |
| 9 | 9b | Hebei | mixed | other Bacillus spp. Staphylococcus sp. Pseudomonas sp. |
B. thuringiensis | Nhe, Hbl, Cry | |
| 9i | B. cereus | Nhe | |||||
| 10 | Hebei | mixed | other Bacillus spp. Enterococcus sp. Cronobacter sp. Acinetobacter sp. |
none | |||
| 11 | 11 | Shandong | mono | none | B. cereus | Nhe, Hbl | |
| 12 | 12c | Jiangxi | mixed | other Bacillus spp. | B. cereus | Nhe, Hbl | |
| 13 | 13d | Guangdong | mono | other Bacillus spp. | B. cereus | Nhe, Hbl | |
| 14 | 14e | Shanxi | mixed | other Bacillus spp. | Plant | B. cereus | Nhe, Hbl |
| 14f | B. thuringiensis | Nhe, Hbl, Cry | |||||
| 15 | 15a | Hubei | mixed | other Bacillus spp. | B. thuringiensis | Nhe, Hbl, Cry | |
| 15d | B. cereus | Nhe, Hbl |
“Toxin type” refers to the detection of the corresponding gene in the PCR-assay plus positive ELISA-reaction at the level of the reference strains plus cytotoxicity for Vero-cells.
two different B. cereus strains.
Detection of enterotoxins and emetic toxin
The four main toxin genes of B. cereus, ces, nhe, hbl, and cytK1, were detected as previously described (Wehrle et al., 2009). Enzyme-linked immunosorbent assays were used to detect the production of B. cereus toxins, based on monoclonal antibodies specifically recognizing different epitopes of each toxin component (Zhu K. et al., 2013). Cytotoxicity assays were performed on Vero and HEp-2 cells to evaluate the toxic potential of the strains due to the production of enterotoxins (Nhe, Hbl, and CytK1) and/or emetic toxin (cereulide), respectively (Wehrle et al., 2009). B. cereus NVH 75/95, DSMZ 4384, and MHI 165 were used as reference strains for Nhe, Hbl, and cereulide, respectively (Wehrle et al., 2009; Lindbäck et al., 2010). B. cereus NVH 391/98 served as reference strain for CytK1, with a variant nhe-gene and no expression of the Nhe complex in the bacterial culture medium (Fagerlund et al., 2007).
Assessment of phenotypic antimicrobial resistance
Antimicrobial resistance was assessed by a standardized microdilution assay (ISO 20776-1:2006) in cation adjusted Mueller Hinton broth (CAMHB). The assay included ampicillin, amoxicillin + clavulanate, cefaclor, cefoxitin, cefuroxime, ciprofloxacin, clindamycin, colistin, daptomycin, doxycycline, enrofloxacin, ertapenem, erythromycin, florfenicol, fosfomycin, gentamicin, imipenem, linezolid, meropenem, oxacillin, penicillin, quinupristin + dalfopristin, rifampicin, streptomycin, teicoplanin, telithromycin, tobramycin, and vancomycin.
Detection of antimicrobial resistance genes
The presence of the tetracycline resistance genes tet(A), tet(B), tet(C), tet(D), tet(K), tet(L)/tet(45), tet(M), and tet(O) was assessed by end-point PCR as described elsewhere (Smith et al., 2004; Srinivasan et al., 2008; You et al., 2012); this selection included the most commonly detected tet-genes in Bacillus cereus (Agersø et al., 2002), but also four genes—tet(A), tet(B), tet(C), tet(D)—that were not yet found in the genus Bacillus (http://faculty.washington.edu/marilynr/tetweb3.pdf). PCR products were purified and submitted for sequencing (Sequiserve, Vaterstetten, Germany). The obtained sequences were compared with public databases (NCBI, EMBL-EBI) to verify the PCR products and to distinguish between tet(L) and tet(45) (You et al., 2013).
Transferability of antimicrobial resistance
Transformation assay
Electro-transformation
The spread of antimicrobial resistance genes was determined in a modified transferability test as described elsewhere (Zhu W. et al., 2013). Plasmids of the B. cereus strain 9i were extracted by QIAfilter Plasmid Midi Kit (QIAGEN®). Subsequently, 500 ng of the purified plasmids were transformed into Staphylococcus aureus RN4220 electrocompetent cells, at 200 Ω, 25 μ Fd, 2500 V. After incubation at 37°C for 2 h, the resulting transformants were spread on brain heart infusion (BHI) agar plates containing 16 mg/L tetracycline, and incubated at 37°C for another 24 to 48 h. Isolated single colonies were tested for the presences of tet(45) by PCR assay, and the obtained PCR products were submitted for sequence analysis. Electro-transformation of B. subtilis ATCC 6051, Escherichia faecalis JH2-2, Bacillus licheniformis ATCC 14580 and Bacillus amyloliquefaciens ATCC 23842 followed the same protocol.
Chemical transformation
For the preparation of competent cells of B. subtilis ATCC 6051, B. subtilis was cultured on LB agar plates at 37°C for 24 h; a single colony was inoculated in 5 mL GM1 medium and shaken at 100 rpm (37°C) for 16 h. Subsequently, 2 mL of B. subtilis culture was transferred to 18 mL GM1 medium and shaken at 100 rpm (37°C) for another 3 h. Ten milliliters of the culture was subsequently transferred into 90 mL GM2 medium and shaken at 100 rpm (37°C) for 2 h. Lastly, all the cells were collected after centrifugation at 4000 rpm (4°C) for 10 min and re-suspended by 10 mL GM2 medium with the addition of 5 mL 30% glycerol to prepare the competent cells of B. subtilis. Chemical transformation was performed with Escherichia coli (DH5α) and B. subtilis competent cells as recipients using 0.5–1 μg plasmid DNA extracted from the donor strain B. cereus 9i, according to the manufacturer's instruction (TaKaRa).
Conjugation assay
Filter matings were performed with E. coli (EC600, rifampin resistant), S. aureus (RN4220), E. faecalis (JH2-2, rifampin resistant), and B. subtilis (ATCC 6051) as the recipient strains, following the method described elsewhere (Huys et al., 2004). Overnight cultures of the donor (B. cereus 9i) or recipient strains were diluted to 108 CFU/mL in brain heart infusion (BHI) broth (Land Bridge Technology). Five microliters of each dilution was added into 1 mL BHI broth and incubated at 37°C for 4 h. The cultures of donor (20 μL) and recipients (60 μL) were mixed and filtered through a sterile 0.2 μm membrane filter (Millipore), and placed at 37°C for overnight incubation. After mating, the colonies were washed with 1 mL BHI broth and plated on BHI agars, containing 8 μg/mL TET and further incubated at 37°C for 24 h. For the selection of potential transconjugants, the agar plates contained 8 μg TET /mL; for E. coli EC600 and E. faecalis JH2-2, 25 μg rifampin /mL was added. Potential transformants grown on the selective agar plates were identified by 16S rRNA-gene sequences and tested for the presences of tet(45) by PCR assay.
Genetic environment of tet(45)
The DNA of B. cereus 9i was extracted (Wizard Genomic DNA Purification Kit, Promega) and partially identified by high-throughput sequencing (Illumina Hiseq 2500, performed at BerryGenomics Inc., Beijing China). A preliminary draft assembly of the genomic fragments was conducted using CLC Genomics Workbench 5 (CLC Bio, Aarhus, Denmark).
Once the surrounding of tet(45) was known, we designed primer pairs targeting the upstream- and downstream-environment of tet(45): UP-tet(45) (fw: TGGTTGAAGAGGCAC TCTATGG, rv: GGCGTAAATTTGACTGTGAATGA), tet(45)-INT1 (fw: TCATCGTCA TTAGCTGGTTGGT, rv: AAGTTGCAGAGGACATGGAAAAAC), INT1-INT2 (fw: TTT TCGGGGTTGATGGGCAA, rv: CGATGAGGAAAAACATCGAGAATCA), INT2-HTH (fw: GCGTGGCTTGTCCTTTCTAAC, rv: AGAACCCCAACAAGACTCCC), in order to verify the environment of tet(45) in representatives of each different variant of tet(45). All PCR-reactions were performed with an initial denaturation of 5 min at 94°C, 35 cycles of 60 s at 94°C, 60 s at 54°C, 90 s at 72°C, and a final elongation of 5 min at 72°C. B. cereus 9i served as positive control.
Results
Presence of multiple bacterial species
A total of 66 morphologically different strains were isolated from 15 typical samples labeled as products containing B. cereus in mono-preparation (Product No. 1–3, 5–7, 11, and 13) or mixed preparation (Product No. 4, 8–10, 12, 14, and 15). Based on bacterial morphology and biochemical testing results, as well as cry-gene based multiplex-PCR and MALDI-TOF mass spectrometry, these 66 isolates were classified as belonging to more than 10 different bacterial species; 18 strains were identified as belonging to the B. cereus group (Table 1). Four of these strains were further identified as B. thuringiensis due to the presence of the plasmid-borne cry-genes encoding insecticidal crystal toxins (cry1+cry2+cry7/8 in strains 9b, 14f, and 15a; cry2 in strain 7d). One sample (No. 10) did not contain viable B. cereus, but viable B. subtilis spores, instead. In total, this product contained nine different viable bacterial strains, including Cronobacter sakazakii and Acinetobacter baumannii.
Detection of toxins
The PCR-assay detected nhe in all isolated B. cereus group strains (Table 1); hbl was detected in 15 isolates; accordingly, all strains were cytotoxic for Vero-cells. None of the isolates harbored ces-genes which are a hallmark of emetic strains, or cytK1. Enzyme-linked immunosorbent assays confirmed that non-hemolytic enterotoxin (Nhe) and hemolysin BL (Hbl) were expressed by the nhe-/hbl-positive strains at the levels of the respective positive controls.
Antimicrobial resistance
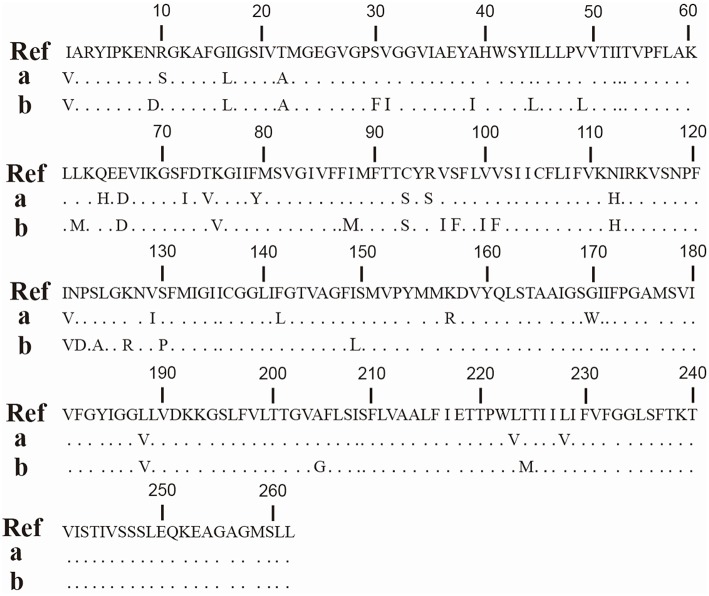
Microdilution according to ISO-20776 indicated natural (species-specific) insusceptibility to penicillins (ampicillin, oxacillin, penicillin), cephalosporins (cefaclor, cefoxitin, cefuroxime), and colistin, as well as varying susceptibility for daptomycin, amoxicillin+clavulanate, and fosfomycin (Table 2). Microdilution further revealed eight isolates (44%) with elevated MIC-values for doxycycline (1–4 mg/L) whereas all other isolates had MIC-values ≤ 0.125–0.25 mg doxycycline /L. The same eight strains were positive for tet(45). Sequence analysis revealed that seven tet(45) amplicons were identical among each other, but the amplicon of the eighth strain, 9i, showed a different sequence (Figure 1).
Table 2.
Minimum inhibitory concentrations (MICs, mg/l) of 22 antimicrobials and presence /absence of tet(45) in probiotic Bacillus cereus / Bacillus thuringiensis strains.
| No. | Intended for use in | Species | tet(45) | AMC | CIP | CLI | DPT | DOX | ENR | ERT | ERY | FLL | FOS | GEN | IMP | LEV | LIZ | MER | QDA | RIF | STR | TEI | TEL | TOB | VAN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Human | B. cereus | >64 | 0.125 | 4 | 2 | 0.25 | 0.125 | ≤0.25 | 2 | 2 | 16 | 0.5 | 1 | 0.125 | 2 | ≤0.25 | 0.5 | 0.5 | 8 | 0.5 | 0.063 | 0.5 | 1 | |
| 2a | B. cereus | >64 | 0.125 | 2 | >4 | ≤0.125 | 0.125 | ≤0.25 | 1 | 1 | 16 | 1 | 1 | 0.125 | 1 | 1 | 1 | 0.5 | 8 | 0.5 | 0.063 | ≤0.25 | 1 | ||
| 2b | B. cereus | 8 | 0.25 | 1 | 0.5 | ≤0.125 | 0.125 | ≤0.25 | 0.5 | 1 | 16 | 1 | 0.25 | 0.125 | 1 | ≤0.25 | 0.5 | 0.5 | 8 | 0.25 | ≤0.031 | 1 | 0.5 | ||
| 3 | B. cereus | > 64 | 0.125 | 2 | 2 | ≤0.125 | 0.125 | ≤0.25 | 1 | 1 | 16 | 1 | 1 | 1 | 0.5 | 0.5 | 0.5 | 8 | 0.5 | 0.063 | 0.5 | 1 | |||
| 5c | Animal | B. cereus | 4 | 0.25 | 1 | >4 | ≤0.125 | 0.125 | ≤0.25 | 8 | 1 | 16 | 0.5 | 0.25 | 0.25 | 2 | ≤0.25 | 1 | 0.5 | 8 | 0.25 | 0.5 | ≤0.25 | 1 | |
| 6f | B. cereus | + | >64 | 0.25 | 4 | 4 | 2 | 0.25 | 0.5 | 1 | 1 | 16 | 0.5 | 1 | 0.125 | 1 | 0.5 | 1 | 0.5 | 8 | 0.5 | 0.063 | 1 | 1 | |
| 7d | B. thuringiensis | + | >64 | 0.25 | 2 | 4 | 2 | 0.125 | ≤0.25 | 1 | 1 | 16 | 1 | 1 | 0.25 | 1 | 1 | 1 | 0.5 | 8 | 0.5 | ≤0.031 | 0.5 | 1 | |
| 8h | B. cereus | + | >64 | 0.125 | 2 | 4 | 2 | 0.125 | ≤0.25 | 1 | 1 | 16 | 1 | 1 | 0.125 | 1 | ≤0.25 | 1 | 0.5 | 8 | 0.5 | 0.063 | 0.5 | 1 | |
| 9b | B. thuringiensis | >64 | 0.125 | 1 | 4 | ≤0.125 | 0.125 | ≤0.25 | 0.5 | ≤0.5 | 64 | 2 | 1 | 0.125 | 1 | 1 | 1 | 0.5 | 8 | 0.25 | ≤0.031 | 1 | 1 | ||
| 9i | B. cereus | + | 64 | 0.5 | 2 | >4 | 4 | 0.25 | ≤0.25 | 1 | 1 | 64 | 0.5 | 0.25 | 0.25 | 2 | ≤0.25 | 1 | 1 | 8 | 1 | ≤0.031 | 0.5 | 2 | |
| 11 | B. cereus | + | >64 | 0.25 | 2 | >4 | 2 | 0.125 | ≤0.25 | 1 | 1 | 16 | 0.5 | 1 | 0.125 | 1 | ≤0.25 | 1 | 0.5 | 8 | 0.5 | 0.063 | 0.5 | 1 | |
| 12c | B. cereus | 64 | 0.125 | 2 | 2 | ≤0.125 | 0.125 | 0.5 | 1 | 1 | 16 | 0.5 | 2 | 0.125 | 1 | 0.5 | 0.5 | 0.5 | 8 | 0.5 | 0.125 | ≤0.25 | 1 | ||
| 13d | B. cereus | + | >64 | 0.125 | 4 | 4 | 2 | 0.25 | ≤0.25 | 1 | 1 | 16 | 0.5 | 1 | 0.125 | 1 | 0.5 | 1 | 0.5 | 8 | 0.5 | ≤0.031 | 1 | 1 | |
| 14e | Plant | B. cereus | + | 32 | 0.25 | 2 | 4 | 1 | 0.125 | ≤0.25 | 1 | 1 | 16 | 0.5 | 0.5 | 0.125 | 1 | ≤0.25 | 1 | 0.5 | 8 | 0.5 | ≤0.031 | 0.5 | 1 |
| 14f | B. thuringiensis | >64 | 0.125 | 1 | >4 | ≤0.125 | 0.125 | ≤0.25 | 0.5 | 1 | 32 | 1 | 1 | 0.125 | 1 | 0.5 | 0.5 | 0.5 | 8 | 0.25 | ≤0.031 | 1 | 1 | ||
| 15a | B. thuringiensis | >64 | 0.125 | 1 | 4 | ≤0.125 | 0.125 | ≤0.25 | 0.5 | 1 | 128 | 1 | 1 | 0.125 | 1 | ≤0.25 | 1 | 0.5 | 8 | 0.25 | ≤0.031 | 1 | 1 | ||
| 15d | B. cereus | + | 64 | 0.125 | 2 | 4 | 1 | 0.125 | ≤0.25 | 1 | 1 | 16 | 1 | 1 | 0.125 | 1 | ≤0.25 | 1 | 0.5 | 8 | 0.5 | ≤0.031 | 1 | 1 | |
| B. cereus ATCC 10876 | n.d. | 32 | 0.25 | 1 | n.d. | 0.5 | 0.125 | ≤0.125 | 0.5 | 1 | n.d. | 0.5 | 4 | 0.125 | 1 | ≤0.063 | 0.5 | n.d. | n.d. | n.d. | 0.063 | n.d. | 1 | ||
MICs were determined according to ISO-20776. All isolates where negative for tet(M), tet(K), tet(O), tet(A), tet(B), tet(C), tet(D). All tet(L)/tet(45)-amplicons revealed higher sequence identity with tet(45) than with tet(L).
AMC, amoxicillin + clavulanate; CIP, ciprofloxacin; CLI clindamycin, DPT, daptomycin; DOX, doxycycline; ENR, enrofloxacin; ERT, ertapenem;
ERY, erythromycin; FLL, florfenicol; FOS, fosfomycin; GEN, gentamicin; IMP, imipenem; LEV, levofloxacin; LIZ, linezolid; MER, meropenem;
QDA, qinupristin + dalfopristin; RIF, rifampicin; STR, streptomycin; TEI, teicoplain; TEL, telithromycin; TOB, tobramycin; VAN, vancomycin.
Figure 1.
Partial amino acid sequence of Tet(45). Amino acid sequence alignment of tet(45)-amplicons (a, strain No. 6f, 7d, 8h, 11, 13d, 14e and 15d) and the mobile genetic element- tet(45)-amplicon (b, strain No. 9i) with Tet(45) from Bhargavaea spp. (Ref, You et al., 2013; blastx, https://npsa-prabi.ibcp.fr).
Plasmid extraction and horizontal transfer
Further investigations demonstrated that B. cereus 9i contained two plasmids, one of ~10 kB and one of > 50 kB in size (Figure 2). Both plasmids were assumedly low copy plasmids, generating reproducible, but weak bands on agarose gel (Figure 2). The tet(45)-gene of 9i was successfully transferred into S. aureus RN4220 and B. subtilis ATCC6051 (but not E. coli) by electro-transformation. Transformed S. aureus and B. subtilis increased their doxycycline MIC-values from ≤ 0.25 to 2 mg/L. After electro-transformation, the tet(45)-gene was amplified in the DNA-extracts of the putative transformants. However, neither chemical transformation nor conjugation assays resulted in detectable transfer of tet(45).
Figure 2.
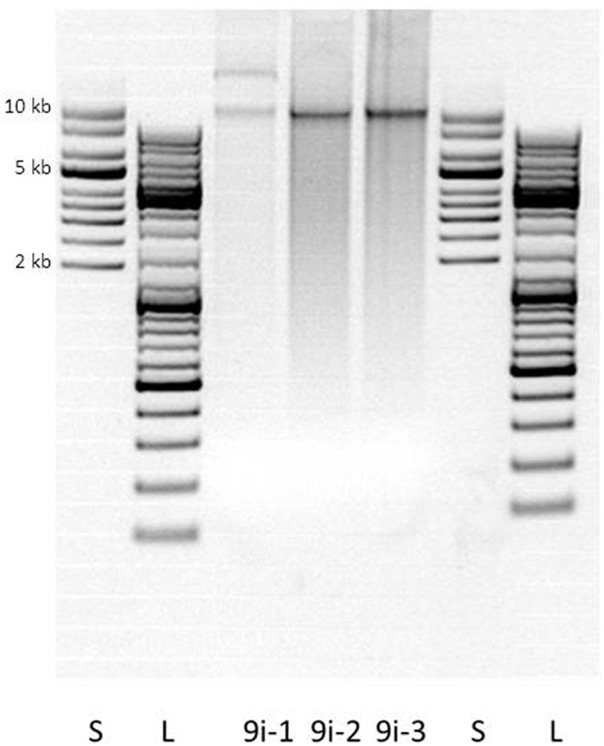
Agarose gel electrophoresis after plasmid extraction of B. cereus 9i. 1–3: three independent replicates. S supercoiled ladder, L linear ladder. Note: two putative plasmid bands were visible in a second agarose gel electrophoresis of extract 9i-3, at the same height as in extract 9i-1.
Genetic environment of tet(45)
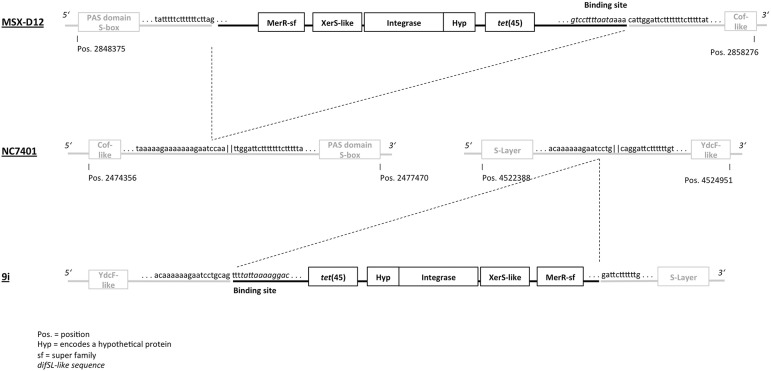
Partial sequence analysis of the DNA extracts is visualized in Figure 3. The tet(45)-gene is located on a putatively mobile genetic element (MGE) of 6789 bp (Genbank accession number KX091844). Upstream, the MGE starts with a dif SL-like binding motif (Figure 3) situated on a 413 bp sequence of unknown function, which precedes tet(45). Downstream, tet(45) is followed by a gene encoding a protein of unknown function. The insertion element further contains two different integrase genes (2181/1347 bp)—at least one of the integrases with significant similarity to a site-specific tyrosine recombinase, XerS—and a 552 bp gene encoding a MerR superfamily protein (putative excisionase) with a helix-turn-helix domain.
Figure 3.
Genetic environment of tet(45) in B. cereus 9i. Surrounding sequences of tet(45) in B. cereus 9i (this study) and B. cereus MSX-D12 (Timmery et al., 2011) are shown relatively to B. cereus NC7401 (AP007209.1). Note: most Genbank entries for Bacillus cereus show the same 5'-3'-orientation of the YdcF-like-/ S-layer protein encoding sequences and the Pas domain S-box-/Cof-like protein encoding sequences as 9i and MSX-D12, respectively, not as NC7401. NC7401 was chosen as reference sequence since the palindromic sequence between the Pas domain S-box-/Cof-like protein encoding sequences is conserved here, but not in the Bacillus cereus reference genomes.
With DNA-extracts of two representatives of the second tet(45)-variant, no amplicons were generated when we used primer pairs UP-tet(45), tet(45)-INT1, INT1-INT2, or INT2-HTH, while amplicons of the expected size were found for B. cereus 9i (positive control) in each PCR-reaction.
Sequences of the mobile genetic element and of the second tet(45) variant (partial sequence) were deposited in Genbank (accession numbers KX091844 and KX091845).
Discussion
Mislabeled probiotic products
Five products labeled as B. cereus-monopreparations contained bacterial species which did not belong to the B. cereus group. Mislabeling of probiotic Bacillus strains is a common problem also observed in Europe, where Bactisubtil has been firstly labeled to contain B. subtilis spores instead of B. cereus spores (Huys et al., 2013). Vice versa, in our study one preparation for use in livestock did not contain any viable B. cereus group strain, but B. subtilis spores, instead. In addition, the product contained two opportunistic human pathogens, A. baumannii and C. sakazakii; the latter outnumbered the B. subtilis spores (data not shown).
Toxins
Due to the presence of plasmid-borne cry-group-genes, which encode insecticidal toxins, four strains were identified as B. thuringiensis. Two of them were isolated from preparations that were not labeled to contain B. thuringiensis. B. cereus is closely related to B. thuringiensis: the 16S rRNA-gene sequences share more than 99% similarity (Stenfors Arnesen et al., 2008). The insecticidal toxins of B. thuringinesis are not harmful for mammals; however, the isolates also produced enterotoxins (Hbl and Nhe), as did all of the B. cereus group strains (Table 1). Hbl and Nhe are pore-forming toxins, each composed of three individual proteins, which show a specific order of binding to mammalian cells. Although the mode of action of these toxins is not fully understood, assembly of the components seems to occur partly in solution and is completed on the cell surface resulting in pore formation and cell lysis (Lindbäck et al., 2010; Sastalla et al., 2013). For both toxins, the presence of all three compounds is necessary to evoke toxicity, and only strains expressing complete Hbl and/or Nhe can be regarded as diarrheic (Stenfors Arnesen et al., 2008). This was the case for all isolated B. cereus group strains, the expression level of Nhe was even similar to the reference strain isolated from a food poisoning case in Norway (Lund and Granum, 1996; data not shown). A probiotic B. cereus strain licensed in the European Union has also been shown to produce biologically active enterotoxins (Nhe, Hbl; Duc et al., 2004). Interestingly, a preparation of this strain was withdrawn for use in animals, while still included in a licensed—but originally mislabeled—preparation for human use (Huys et al., 2013; Hong and Cutting, 2005).
Enteropathogenic B. cereus
The infectious dose of enteropathogenic B. cereus was proposed to be 1 × 105 to 1 × 108 CFU or spores/g (Granum and Lund, 1997). Variations in this infectious dose are attributed to strain traits, since strains differ both in the amount of toxins produced (Dommel et al., 2011; Jessberger et al., 2015; Lücking et al., 2015), and also in their capability for intestinal persistence. Probiotic strains are applied in a dose of 107–109CFU or spores/g; they have also been selected for their ability to colonize the intestinal tract (Duc et al., 2004). Thus, given their high likelihood to establish colonies in the intestine, unsafe probiotic strains might provide a major health risk, compared to strains casually ingested with food. Such health risks have been demonstrated in case studies. For example, a fatal infection with a probiotic Bacillus preparation in an immune-compromised patient has been reported (Oggioni et al., 1998).
Transferable antimicrobial resistance gene
Besides the lack of toxicity, the presence of transferable resistance genes or not is another major safety parameter for probiotic strains. At least one of the strains investigated in the current study carried a tetracycline resistance gene, tet(45), on a mobile genetic element. In Bacillus spp., some corresponding entries in Genbank are labeled as “tetracycline resistance MFS efflux pump,” which overcomes the problem that the protein encoded by tet(45) in Bacillus spp. is similar to the product of tet(L) so that partial sequences might not allow unambiguous distinction. However, the complete sequence presented here has higher similarity to the reference sequences of tet(45) than to the reference sequences of tet(L) (http://faculty.washington.edu/marilynr/tetweb4.pdf).
The mobile element contained a dif SL-like binding site, the tet(45)-gene, one ORF encoding a hypothetical protein, two integrase genes (one of the XerS superfamily, but without significant similarity to the XerS-encoding gene of Streptococcus suis), two non-coding-sequences of unknown function and a gene encoding a MerC-superfamily protein (putative excisionase). The coherent mobile element as found here was not described in Bacillus spp. before, but NCBI-BLAST revealed 93% identity to an inverse sequence of B. cereus MSX-D12 (pos. 2856985–2851281), isolated 2005 in the Antarctic Concordia Station by Timmery et al. (2011). At 5′, the putative MGE in 9i begins with an adenin-rich-motif (caaaaaagaatcc). This motif was identified as the “left-hand” part of a putative Xer-binding/cleavage site, since the 3′ (“right-half”)-site (tattaaaaggacag) downstream the motif had remarkably high identity to dif SL (Leroux et al., 2011; bold = consensus), whereas the sequence had much lower similarity to known Xer-binding sites of Bacillus spp. (e.g., cer, dif, dif BS). MSX-D12 presented a complementary dif SL-like sequence at 3′ (Figure 3).
The sequence situated upstream of the putative MGE in 9i contains one single caaaaaagaatcctg-cag-motif; an inverted repeat of the motif, gattcttttttg (missing the cag-sequence which is adhered at the 5′ cleavage site), follows immediately downstream of the 3′ end of the MGE. In MSX-D12, the same—but undisrupted—sequence is situated ~2,100,000 bp upstream of the putative mobile element described here, forming a perfect 30 bp palindrome (caaaaaagaatcctg||caggattcttttttg). Interestingly, the both opposed ends of the palindrome are identical to the opposed ends of the “left-hand”-part of dif SL (Leroux et al., 2011).
Identical upstream and downstream genes—encoding a YdcF-like protein and an S-layer protein—are found in MSX-D12 (not shown), NC7401 and 9i (Figure 3, respectively) next to both ends of the palindromic cleavage site. This raises strong suspicion that, in 9i, tet(45) was inserted into this sequence by help of (and together with) the site-specific recombinases encoded downstream. In MSX-D12, the MGE was inserted at another palindromic site (Figure 3), was inversely orientated, and its insertion was accompanied by a crossover-event so that the palindromic sequence at both ends of the MGE was directly mirrored (tatttttctttttttcttag||gattcttttttctttttat) instead of being complimentary.
Remarkably, while the binding site had high homology with the dif SL-site identified in Streptococcus and Lactococcus, the XerS-like protein had no significant similarity with the XerS-proteins of Streptococcus or Lactococcus, and two different integrase genes were combined in the MGE, hinting toward a XerC/XerD-like dimeric process of site-specific recombination instead of a “stand-alone” mechanism as proposed for XerS (Leroux et al., 2011).
B. cereus 9i contained two plasmids. However, in the plasmid extracts we could neither identify any gene associated with conjugation, nor did we succeed in transferring tet(45) by means of conjugation assays. The YdcF-like-protein and the S-layer-protein which are connected by the putative insertion site are chromosomally encoded in Bacillus spp. This does not necessarily mean that tet(45) is chromosomally integrated in 9i, since chromosomal genes could also be transferred to plasmids, as vice versa. Very recently (2016-03-21) there was a new Genbank entry of a B. thuringiensis plasmid (pBT1850294, CP014284.1) which encodes a significantly similar pair of integrase genes (66% nucleotide identity). However, we assume that the transfer of tet(45) to S. aureus and B. subtilis was not necessarily associated with its presence on one of the two plasmids found in 9i, but was associated with a small mobile genetic element, instead. Such elements could be easily integrated into naturally competent hosts after natural transformation, especially if insertion relies on conserved integrases like the Xer-superfamily (Midonet and Barre, 2014), as indicated here. The dif SL-like “right-hand”-motif is present in the genome of a variety of other bacteria, e.g., Lawsonia intracellularis (e.g., CP004029.1), Mycoplasma spp. (e.g., CP014346.1), Clostridia spp. (e.g., CP011803.1), A. baumannii (e.g., CP014528.1), Staphylococcus spp. (e.g., KT780704), and other Bacillus spp. (e.g., CP012600.1). While it is unknown whether these bacteria express recombinase enzymes that bind there, at least streptococci and lactococci are known to do so. Moreover, Aneurinibacillus soli (AP017312.1) and B. mycoides (CP007626.1) encode a pair of integrases (and also a MerC-superfamily protein) with significant similarity to the recombinase enzymes described here, suggesting a further possibility for interspecific and even intergeneric transfer of the MGE.
The detection of a mobile tet(45)-gene in probiotics is certainly undesirable. Due to the presence of a tetracycline resistance gene located on a mobile genetic element, together with insufficient data on toxicity, the probiotic B. cereus strain of Esporafeed Plus has been considered unsafe for use as a feed additive for calves and pigs in Europe in 1999 (http://ec.europa.eu/food/fs/sc/scan/out39_en.pdf, accessed March 31, 2016); more recently, Bacillus toyonensis was considered unsafe for use in chickens, pigs, cattle and rabbits for the same reason (http://www.efsa.europa.eu/en/efsajournal/pub/3766, accessed May 12, 2016). The tet(45)-gene has been originally detected in an environmental bacterium, Bhargavaea spp., from which it was successfully transferred to E. coli (You et al., 2012). Many antibiotic resistance genes (ARGs) of common human pathogenic bacteria are assumed to originate from environmental bacterial communities (Martínez, 2008; Forsberg et al., 2012). As a spore-forming bacterium and original soil inhabitant, B. cereus has a distinct ability to persist in and (re-)adapt to the environment (Carlin et al., 2010). Therefore, it cannot be ruled out that mobile resistance determinants may be further transferred to other soil bacteria, but also to other animal or human pathogens after excretion of such probiotic B. cereus strains. The detection of tet(M)-carrying B. cereus in soil was directly related to manure application in the past (Agersø et al., 2002; Roberts, 2012). In view of the wide and uncontrolled use of probiotics, strict regulations are urgently needed to decrease the probability of transfer of ARGs and safety assessment of these products must include the determination of antimicrobial resistance profiles.
Here, we summarize the possible health hazards of commercial probiotic products advertised to contain B. cereus in China due to mislabeling, toxin production and the presence of transferable antimicrobial resistance. The growing presence of pharmaceutical and biotechnological companies has contributed to an increase of B. cereus related products, but these products are often of poor quality and incorrectly designated. It must be emphasized that the safety of these commercially available probiotics is not a limited local public health issue in China, but may also be relevant worldwide due to global trade and international travel.
Author contributions
KZ, EM, and SD conceived the project. KZ, CH, YC, RM, YW, AD, RD, RB, and SD conceived and performed experiments and/or data analysis. CH, KZ, EM, and SD wrote the manuscript, RB conceived the graphical design. KZ and CH contributed equally to this work.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank G. Acar, S. Eberhardt, S. Strassner, and B. Fritz for expert technical assistance. We thank S. Ulrich, Chair of Food Safety, and G. Wolf, Chair of Bacteriology, LMU Munich, for performing the MALDI-TOF-analysis. This study is supported by the National Basic Research Program of China (2013CB127200), Special Fund for Agro-Scientific Research in the Public Interest (201203069-2) and Fund of Modern Agriculture Industry System Innovation in Beijing City Team (1051-25012090).
References
- Agersø Y., Jensen L. B., Givskov M., Roberts M. C. (2002). The identification of a tetracycline resistance gene tet(M), on a Tn916-like transposon, in the Bacillus cereus group. FEMS Microb. Lett. 214, 251–256. 10.1016/S0378-1097(02)00883-2 [DOI] [PubMed] [Google Scholar]
- Allen H., Donato J., Wang H., Cloud-Hansen K., Davies J., Handelsman J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. 10.1038/nrmicro2312 [DOI] [PubMed] [Google Scholar]
- Ankolekar C., Rahmati T., Labbé R. (2009). Detection of toxigenic Bacillus cereus and Bacillus thuringiensis spores in US rice. Int. J. Food Microbiol. 128, 460–466. 10.1016/j.ijfoodmicro.2008.10.006 [DOI] [PubMed] [Google Scholar]
- Arnold C. (2013). The pros and cons of probiotics. Lancet Infect. Dis. 7, 571–572. 10.1016/S1473-3099(13)70172-5 [DOI] [PubMed] [Google Scholar]
- Ben-Dov E., Zaritsky A., Dahan E., Barak Z., Sinai R., Manasherob R., et al. (1997). Extended screening by PCR for seven cry-group genes from field-collected strains of Bacillus thuringiensis. Appl. Environ. Microbiol. 63, 4883–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett S., Walsh K., Gould H. (2013). Foodborne disease outbreaks caused by Bacillus cereus, Clostridium perfringens, and Staphylococcus aureus–United States, 1998-2008. Clin. Infect. Dis. 57, 425–433. 10.1093/cid/cit244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottone E. (2010). Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 23, 382–398. 10.1128/CMR.00073-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin F., Brillard J., Broussole V., Clavel T., Duport C., Jobin M., et al. (2010). Adaptation of Bacillus cereus, an ubiquitous worldwide-distributed foodborne pathogen, to a changing environment. Food Res. Int. 43, 1885–1894. 10.1016/j.foodres.2009.10.024 [DOI] [Google Scholar]
- Dommel M., Lücking G., Scherer S., Ehling-Schulz M. (2011). Transcriptional kinetic analyses of cereulide synthetase genes with respect to growth, sporulation and emetic toxin production in Bacillus cereus. Food Microbiol. 28, 284–290. 10.1016/j.fm.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Duc L., Hong H., Barbosa T., Henriques A., Cutting S. (2004). Characterization of Bacillus probiotics available for human use. Appl. Environ. Microbiol. 70, 2161–2171. 10.1128/AEM.70.4.2161-2171.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerlund A., Brillard J., Fürst R., Guinebretière M. H., Granum P. E. (2007). Toxin production in a rare and genetically remote cluster of strains of the Bacillus cereus group. BMC Microbiol. 7:43. 10.1186/1471-2180-7-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg K., Reyes A., Wang B., Selleck E., Sommer M., Dantas G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111. 10.1126/science.1220761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granum P., Lund T. (1997). Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 157, 223–228. 10.1111/j.1574-6968.1997.tb12776.x [DOI] [PubMed] [Google Scholar]
- Hong H., Cutting S. (2005). The use of bacterial spore formers as probiotics. FEMS Microbiol. Rev. 29, 813–835. 10.1016/j.femsre.2004.12.001 [DOI] [PubMed] [Google Scholar]
- Huys G., Botteldoorn N., Delvigne F., De Vuyst L., Heyndrickx M., Pot B., et al. (2013). Microbial characterization of probiotics–advisory report of the Working Group “8651 Probiotics” of the Belgian Superior Health Council (SHC). Mol. Nutr. Food Res. 57, 1479–1504. 10.1002/mnfr.201300065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huys G., D'Haene K., Collard J., Swings J. (2004). Prevalence and molecular characterization of tetracycline resistance in Enterococcus isolates from food. Appl. Environ. Microbiol. 70, 1555–1562. 10.1128/AEM.70.3.1555-1562.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isolauri E. (2003). Probiotics for treating infectious diarrhoea. Gut 52, 436–437. 10.1136/gut.52.3.436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger N., Krey V., Rademacher C., Böhm M., Mohr A., Ehling-Schulz M., et al. (2015). From genome to toxicity: a combinatory approach highlights the complexity of enterotoxin production in Bacillus cereus. Front. Microbiol 6:560. 10.3389/fmicb.2015.00560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilcullen K., Teunis A., Popova T. G., Popov S. G. (2016). Cytotoxic potential of Bacillus cereus strains ATCC 11778 and 14579 against human lung epithelial cells under microaerobic growth conditions. Front. Microbiol. 7:69. 10.3389/fmicb.2016.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Han J., Park J., Lee J., Lee S., Cho J., et al. (2015). Various enterotoxin and other virulence factor genes widespread among Bacillus cereus and Bacillus thuringiensis strains. J. Microbiol. Biotechnol. 25, 872–879. 10.4014/jmb.1502.02003 [DOI] [PubMed] [Google Scholar]
- Kniehl E., Becker A., Forster D. (2003). Pseudo-outbreak of toxigenic Bacillus cereus isolated from stools of three patients with diarrhoea after oral administration of a probiotic medication. J. Hosp. Infect. 55, 33–38. 10.1016/S0195-6701(03)00133-6 [DOI] [PubMed] [Google Scholar]
- Leroux M., Jia F., Szatmari G. (2011). Characterization of the Streptococcus suis XerS recombinase and its unconventional cleavage of the difSL site. FEMS Microbiol. Lett. 324, 135–141. 10.1111/j.1574-6968.2011.02398.x [DOI] [PubMed] [Google Scholar]
- Lindbäck T., Hardy S. P., Dietrich R., Sodring M., Didier A., Moravek M., et al. (2010). Cytotoxicity of the Bacillus cereus Nhe enterotoxin requires specific binding order of its three exoprotein components. Infect. Immun. 78, 3813–3821. 10.1128/IAI.00247-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López A., Minnaard J., Pérez P., Alippi A. (2015). A case of intoxication due to a highly cytotoxic Bacillus cereus strain isolated from cooked chicken. Food Microbiol. 46, 195–199. 10.1016/j.fm.2014.08.005 [DOI] [PubMed] [Google Scholar]
- Lücking G., Frenzel E., Rütschle A., Marxen S., Stark T. D., Hofmann T., et al. (2015). Ces locus embedded proteins control the non-ribosomal synthesis of the cereulide toxin in emetic Bacillus cereus on multiple levels. Front. Microbiol. 6:1101. 10.3389/fmicb.2015.01101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund T., Granum P. (1996). Characterisation of a non-haemolytic enterotoxin complex from Bacillus cereus isolated after a foodborne outbreak. FEMS Microbiol. Lett. 141, 151–156. [DOI] [PubMed] [Google Scholar]
- Martínez J. L. (2008). Antibiotics and antibiotic resistance genes in natural environments. Science 321, 365–367. 10.1126/science.1159483 [DOI] [PubMed] [Google Scholar]
- Midonet C., Barre F. X. (2014). Xer site-specific recombination: promoting vertical and horizontal transmission of genetic information. Microbiol. Spectr. 2. 10.1128/microbiolspec.MDNA3-0056-2014 [DOI] [PubMed] [Google Scholar]
- Notermans S., Batt C. (1998). A risk assessment approach for food-borne Bacillus cereus and its toxins. Symp. Ser. Soc. Appl. Microbiol. 27, 51S–61S. 10.1046/j.1365-2672.1998.0840s151s.x [DOI] [PubMed] [Google Scholar]
- Oggioni M., Pozzi G., Valensin P., Galieni P., Bigazzi C. (1998). Recurrent septicemia in an immunocompromised patient due to probiotic strains of Bacillus subtilis. J. Clin. Microbiol. 36, 325–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer H., Alpha-Bazin B., Brunet J.-L., Armengaud J., Duport C. (2015). Proteomics identifies Bacillus cereus EntD as a pivotal protein for the production of numerous virulence factors. Front. Microbiol. 6:1004. 10.3389/fmicb.2015.01004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. C. (2012). Mechanisms of bacterial antibiotic resistance and lessons learned from environmental tetracycline-resistant bacteria, in Antimicrobial Resistance in the Environment, 1st Edn., eds Keen P., Montforts M. H. H. M. (Hoboken, NJ: John Wiley & Sons; ), 93–122. [Google Scholar]
- Ruusunen M., Salonen M., Pulkkinen H., Huuskonen M., Hellström S., Revez J., et al. (2013). Pathogenic bacteria in Finnish bulk tank milk. Foodborne Pathog. Dis. 10, 99–106. 10.1089/fpd.2012.1284 [DOI] [PubMed] [Google Scholar]
- Sastalla I., Fattah R., Coppage N., Nandy P., Crown D., Pomerantsev A. P., et al. (2013). The Bacillus cereus Hbl and Nhe tripartite enterotoxin components assemble sequentially on the surface of target cells and are not interchangeable. PLoS ONE 8:e76955. 10.1371/journal.pone.0076955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulthess B., Bloemberg G., Zbinden R., Böttger E., Hombach M. (2014). Evaluation of the Bruker MALDI Biotyper for identification of Gram-positive rods–development of a diagnostic algorithm for the clinical laboratory. J. Clin. Microbiol. 52, 1089–1097. 10.1128/JCM.02399-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M., Yang R., Knapp C., Niu Y., Peak N., Hanfelt M., et al. (2004). Quantification of tetracycline resistance genes in feedlot lagoons by real-time PCR. Appl. Environ. Microbiol. 70, 7372–7377. 10.1128/AEM.70.12.7372-7377.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan V., Nam H., Sawant A., Headrick S., Nguyen L., Oliver S. (2008). Distribution of tetracycline and streptomycin resistance genes and class 1 integrons in Enterobacteriaceae isolated from dairy and nondairy farm soils. Microb. Ecol. 55, 184–193. 10.1007/s00248-007-9266-6 [DOI] [PubMed] [Google Scholar]
- Stenfors Arnesen L., Fagerlund A., Granum P. (2008). From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32, 579–606. 10.1111/j.1574-6976.2008.00112.x [DOI] [PubMed] [Google Scholar]
- Timmery S., Hu X., Mahillon J. (2011). Characterization of Bacilli isolated from the confined environments of the Antarctic Concordia station and the International Space Station. Astrobiol 11, 323–334. 10.1089/ast.2010.0573 [DOI] [PubMed] [Google Scholar]
- Wang S., Duan H., Zhang W., Li J. (2007). Analysis of bacterial foodborne disease outbreaks in China between 1994 and 2005. FEMS Immunol. Med. Microbiol. 51, 8–13. 10.1111/j.1574-695X.2007.00305.x [DOI] [PubMed] [Google Scholar]
- Wehrle E., Moravek M., Dietrich R., Bürk C., Didier A., Märtlbauer E. (2009). Comparison of multiplex PCR, enzyme immunoassay and cell culture methods for the detection of enterotoxinogenic Bacillus cereus. J. Microbiol. Methods 78, 265–270. 10.1016/j.mimet.2009.06.013 [DOI] [PubMed] [Google Scholar]
- You Y., Hilpert M., Ward M. (2012). Detection of a common and persistent tet(L)-carrying plasmid in chicken-waste-impacted farm soil. Appl. Environ. Microbiol. 78, 3203–3213. 10.1128/AEM.07763-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y., Hilpert M., Ward M. (2013). Identification of Tet45, a tetracycline efflux pump, from a poultry-litter-exposed soil isolate and persistence of tet(45) in the soil. J. Antimicrob. Chemother. 68, 1962–1969. 10.1093/jac/dkt127 [DOI] [PubMed] [Google Scholar]
- Zhu K., Dietrich R., Didier A., Acar G., Märtlbauer E. (2013). Versatile antibody-sensing Boolean logic for the simultaneous detection of multiple bacterial toxins. Chem. Commun. 49, 9314–9316. 10.1039/c3cc45370g [DOI] [PubMed] [Google Scholar]
- Zhu W., Clark N., Patel J. (2013). pSK41-like plasmid is necessary for Inc18-like vanA plasmid transfer from Enterococcus faecalis to Staphylococcus aureus in vitro. Antimicrob. Agents Chemother. 57, 212–219. 10.1128/AAC.01587-12 [DOI] [PMC free article] [PubMed] [Google Scholar]