Abstract
Many changes in development, growth, hormone activity and environmental stimuli responses are mediated by mitogen-activated protein kinase (MAPK) cascades. However, in plants, studies on MAPKs have mainly focused on MPK3, MPK4 and MPK6. Here, a novel group B MAPK gene, GhMPK11, was isolated from cotton (Gossypium hirsutum L.) and characterized. Both promoter and expression pattern analyses revealed that GhMPK11 is involved in defense responses and signaling pathways. GhMPK11 overexpression in Nicotiana benthamiana plants could increase gibberellin 3 (GA3) content through the regulation of GA-related genes. Interestingly, either GhMPK11 overexpression or exogenous GA3 treatment in N. benthamiana plants could enhance the susceptibility of these plants to the infectious pathogens Ralstonia solanacearum and Rhizoctonia solani. Moreover, reactive oxygen species (ROS) accumulation was increased after pathogen infiltration due to the increased expression of ROS-related gene respiratory burst oxidative homologs (RbohB) and the decreased expression or activity of ROS detoxification enzymes regulated by GA3, such as superoxide dismutases (SODs), peroxidases (PODs), catalase (CAT) and glutathione S-transferase (GST). Taken together, these results suggest that GhMPK11 overexpression could enhance the susceptibility of tobacco to pathogen infection through the GA3 signaling pathway via down-regulation of ROS detoxification enzymes.
Keywords: Gossypium hirsutum L., GhMPK11, disease resistance, GA3, ROS
Introduction
In contrast with animals, when confronted with various environmental challenges, plants cannot escape danger because they are sessile. To adapt to different stresses, plants have evolved sophisticated signaling pathways to transduce biotic or abiotic stimuli into proper cellular responses (Xie et al., 2014). Among those pathways, mitogen-activated protein kinase (MAPK) cascades that are conserved in all eukaryotes function as a mechanism to sense invading pathogens and then transduce internal or external signals to downstream effectors, such as transcription factors.
Typical MAPK cascades contain MAPK kinase kinases (MAPKKKs), MAPK kinases (MAPKKs) and MAPKs. MAPKKKs can activate MAPKKs by phosphorylating a dual-specificity Ser/Thr or Tyr residue (Jonak et al., 2002; Rodriguez et al., 2010). Then, activated MAPKKs can phosphorylate Thr and Tyr residues in the activation loop (A-loop) of MAPKs (Xu et al., 2014). Finally, the activated MAPKs interact with downstream transcription factors and transduce signals into cellular responses typically by altering gene expression (Widmann et al., 1999; Ichimura et al., 2002). In Arabidopsis, 20 MAPKs have been identified. Based on their sequence homology and the conserved phosphorylation motifs, MAPKs can be divided into four groups: A, B, C, and D (MAPK Group, 2002). Group A-C MAPKs contain a TEY motif in their conserved domains, while group D MAPKs contain a TDY motif (Rodriguez et al., 2010; Meng and Zhang, 2013). MAPKs in groups A and B also have a conserved common docking (CD) domain (Tanoue et al., 2000).
Plant MAPK cascades are induced by numerous environmental stresses, such as wounding, drought, salinity, cold and diverse pathogens (Pitzschke et al., 2009; Rodriguez et al., 2010; Sinha et al., 2011; Rasmussen et al., 2012; Šamajová et al., 2013). Group A MAPKs have been found to be involved in environmental and hormonal responses. GhMPK6a (Gossypium hirsutum) can reduce tolerance to osmotic stress and bacterial infection and plays an important role in development (Li et al., 2013). In Arabidopsis, AtMPK3 and AtMPK6 participate in pathogen resistance and abiotic stress (Rodriguez et al., 2010; Meng and Zhang, 2013). Of the group C MAPKs, the expression level of PsMPK2 (Pisum sativum L.) is induced by wounding, JA, abscisic acid (ABA), and H2O2 (Ortiz-Masia et al., 2008). AtMPK1 and AtMPK2 have been reported to be involved in pathogen signaling (Dóczi et al., 2007). In addition, GhMPK7 might participate in SA-regulated broad-spectrum resistance to pathogen infection (Shi et al., 2010). AtMPK9, a group D MAPK, is involved in ABA signaling through ROS homeostasis and calcium and anion channels (Jammes et al., 2009). A recent study revealed that GhMPK17 might be involved in plant responses to high salinity, osmotic stresses and ABA signaling (Zhang et al., 2014). AtMPK4, a group B MAPK, regulates the levels of plant hormones and negatively regulates systemic acquired resistance and innate immunity in plants (Petersen et al., 2000; Brodersen et al., 2006; Brader et al., 2007; Gao et al., 2008). However, studies on other group B members are limited.
Mitogen-activated protein kinases exert their functions in association with diverse phytohormones. SA, JA and ET are well-established phytohormones involved in disease responses (Yang D.L. et al., 2008). Development-related hormones such as brassinosteroid (Nakashita et al., 2003), cytokinin (Siemens et al., 2006), auxin (Zhang et al., 2007), and ABA (Adie et al., 2007) are associated with disease resistance. The functions of these hormones in response to diseases vary, suggesting that plants might use diverse signaling pathways to manage defense responses during pathogen infection. GA signaling also functions as a stress signal similar to other phytohormones, such as ABA or SA, under adverse conditions (Achard et al., 2006; Willige et al., 2011; Gallego-Bartolomé et al., 2012; Golldack et al., 2013; Colebrook et al., 2014). However, previous studies have mainly focused on GA’s function in plant resistance to drought, salinity and other abiotic stresses, and its function in disease responses has been neglected (Achard et al., 2006; Shan et al., 2007; Li et al., 2012).
Cotton (G. hirsutum) is one of the most important economic crops and serves as an important source of food, fiber, oil and biofuel (Zhang et al., 2002; Sunilkumar et al., 2006; Chen et al., 2007). Furthermore, G. hirsutum produces more than 95% of the annual cotton crop worldwide, and its research value for genome size evolution studies cannot be ignored (Grover et al., 2004). In this study, a novel group B MAPK gene, GhMPK11 from G. hirsutum, was isolated and characterized. The transcription level of GhMPK11 could be induced by various stresses, and overexpression of GhMPK11 led to a higher level of GA3 than that in control plants. Furthermore, GhMPK11 overexpression and exogenous GA3 treatment could reduce plants resistance to pathogens. All of these results suggest that GhMPK11 may increase plants susceptibility to pathogens due to a higher level of GA3 in vivo. This study increases our understanding of MAPK signaling in cotton and indicates that GA signaling may have a role in disease responses.
Materials and Methods
Plant Materials and Treatments
Cotton (G. hirsutum L. cv lumian 22) seeds were grown in an incubator at 25 ± 1°C with a 16 h light/8 h dark cycle (light intensity of 200 μmol m-2 s-1; relative humidity of 60–75%). The 7-day-old cotton seedlings were sprayed with 10 mM H2O2, 100 μM ABA and 100 μM GA3. For pathogen treatment, 7-day-old cotton seedlings were inoculated with bacterial suspensions of Ralstonia solanacearum (OD600 = 0.6–0.8) and conidial suspensions of Rhizoctonia solani (105 conidia/mL) using the root dip method. The treated cotyledons were harvested at the appropriate times as indicated, frozen in liquid nitrogen and stored at -70°C for RNA extraction. Each treatment was repeated at least twice. N. benthamiana seeds were surface-sterilized, planted in soil, and maintained under a 16 h light/8 h dark photoperiod at 25°C. Three to four-leaf-stage N. benthamiana seedlings were transplanted into pots with soil and maintained under glasshouse conditions.
Cloning of the Full Length cDNA, Genomic Sequences and 5′ Flanking Region of GhMPK11
The cDNA and genomic DNA were isolated as described previously (Yu et al., 2012). Total RNA was extracted from the leaves of cotton seedlings using the CTAB method (Hu and Yu, 2007). To obtain the internal conserved fragment of GhMPK11, primers MF and MR were designed based on the nucleotide sequences and amino acids that are conserved among TcMPK4 (Theobroma cacao), AtMPK11 and AtMPK4 (Arabidopsis thaliana) and NaMPK4 (Nicotiana attenuate). Then, RT-PCR was performed to clone an internal fragment of GhMPK11. Next, primers 5N, 5W, 3N and 3W were designed based on the GhMPK11 fragment, and then TAIL PCR was performed to amplify the 5′ flanking region according to Liu and Chen (2007). The primers used in this study are shown in Supplementary Table S1. All products were purified, cloned into the pEASY-T3 vector (TransGen Biotech, China), transformed into the Escherichia coli strain DH5α and then sequenced. All sequencing was performed by BioSune using an ABI 3730 XL sequencer.
RNA Extraction
Total RNA was extracted from cotton seedlings according to the CTAB method described by Hu and Yu (2007). Total RNA was extracted from N. benthamiana seedlings using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) and digested with RNase-free DNaseI (Promega, Madison, WI, USA). The first strand cDNA was synthesized using the EasyScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China).
Expression Pattern of GhMPK11 Using Quantitative PCR
Total RNA was extracted from all experimental samples, and the first strand cDNA was synthesized as described above. Quantitative PCR (qRT-PCR) was performed as previously described (Wang et al., 2014). All primers used are listed in Supplementary Table S1. G. hirsutum polyubiquitin (UBI) or N. benthamiana β-actin genes were used as the standard control. The relative expression level of GhMPK11 was analyzed using the comparative CT method (2-ΔΔCT method), and three replicates of each sample were analyzed. Prism 5 software (GraphPad Software, Inc.) was used to determine significant differences.
Subcellular Localization of GhMPK11
The ORF of GhMPK11 was inserted upstream of the N-terminus of the GFP gene following the CaMV35S promoter. The recombinant vector was transformed into onion epidermal cells using the particle bombardment method as described by Yu et al. (2012). Then, the tissues were incubated in the dark at 25°C for 12 h. Nuclei were stained with 100 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) (Solarbio, Beijing, China) in phosphate-buffered saline for 10 min. The 35S::GFP construct was used as a control. In addition, the two recombinant plasmids were transferred into Agrobacterium tumefaciens strain GV3101. Agrobacterium cells were harvested by centrifugation, resuspended in infiltration buffer (10 mM MES, pH 5.7, 10 mM MgCl2, and 150 mM acetosyringone) and adjusted to a final OD600 of 1.0. After the Agrobacterium mixture was incubated for 3 h at room temperature in the dark, it was infiltrated into the leaves of 6-week-old N. benthamiana plants with a syringe. The 35S-GFP plasmid was used as a control. Fluorescence was observed 3 to 4 days after infiltration using a confocal laser scanning microscope (LSM 510 META, ZEISS, Germany).
Vector Construction and Genetic Transformation
The vector construction and genetic transformation were performed as previously described by Yu et al. (2012). Plants transformed with pBI121-GFP were used as controls. Transformation of the tobacco plants was confirmed by PCR. The transgenic T2 lines and vector plants were used for further experiments.
GA Content Measurement
Leaves were detached from plants OE GhMPK11 or the control were grounded in liquid nitrogen, and then soaked in 95% chromatographic methanol at 4°C overnight. The solution was filtered through a 0.45 μm membrane, and the GA3 content was measured via Agilent 1200 rapid resolution liquid chromatography (Agilent Technologies, Waldbronn, Germany). The mobile phase was 0.4 % (v/v) acetic acid + chromatographic methanol. Chromatography was performed at 254 nm at 30°C with a flow rate of 1.0 cm3 min-1. The marker for GA3 was accurately weighed and dissolved in chromatographic methanol to provide serial concentrations ranging from 0.0133 to 13.3 mg/mL. The standard curve was analyzed using the peak area, and the GA3 content was calculated.
DAB and NBT Staining Assays
For DAB staining, leaves were soaked in DAB solution (1 mg/mL, pH 3.8) in the dark at 25°C for 12 h. Then, the leaves were incubated in 95% ethanol overnight to remove the chlorophyll. For NBT staining, the leaves were soaked in NBT solution (0.1 mg/mL) in the dark for 12 h at 25°C. Then, the leaves were incubated in 95% ethanol overnight to remove the chlorophyll. Seedlings treated with water were used as controls.
GUS Histochemical Staining Assay
Transgenic Arabidopsis plants harboring the ProGhMPK11::GUS construct were generated using the floral dip method. T2 progeny were used for promoter activity analyses and stained with the GUS histochemical staining buffers as previously described (Jefferson, 1987).
Disease Resistance of the Transgenic Plants
For bacterial infection, the detached leaves of 8-week-old seedlings were inoculated with suspensions of R. solanacearum, a Gram-negative plant pathogenic bacterium (OD600 = 0.6–0.8). The bacteria were cultured in Luria–Bertani (LB) broth overnight at 37°C, harvested by centrifugation, and resuspended in sterile tap water. For fungal infection, R. solani was cultured on potato dextrose agar (PDA) medium for 2 weeks at 28°C, and the spores were then suspended in sterile tap water. R. solani spore suspensions (105 spores/mL) were infiltrated into leaves detached from 8-week-old T2 transgenic and control seedlings using a needleless syringe. At least three independent experiments were performed for each pathogen.
Pathogen Growth Assays
Total RNA was extracted from each sample using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The abundance of the fungus (R. solani) was estimated by the ITS gene copy number through qPCR using the primer pair ITS1/ITS4 (White et al., 1990). The bacterial (R. solanacearum) abundance was estimated by the 16S rRNA gene copy number through qPCR using the primer pair Eub338 and Eub518 (Rasche et al., 2011). Amplification reactions were carried out with SYBR Premix Ex Taq (TaKaRa, Japan) in a total volume of 20 μL. Standard curves were obtained using serial dilutions of a known copy number of plasmids containing an ITS or 16S rRNA gene fragment, and these curves were linear from 9.77 × 103 to 9.77 × 108 gene copies/μL (R2 = 0.998; ITS gene) and gene copies/μL (R2 = 0.998; 16S rRNA gene). All samples were analyzed in triplicate.
Enzyme Activity Assays and Oxidative Stress Experiments
For the enzyme activity assays, the leaves of transgenic and control plants were inoculated with R. solanacearum or R. solani and then tested for SOD, POD and CAT activity as previously described (Yang L. et al., 2008). Oxidative stress experiments were performed according to Lu et al. (2013) with a modification of the MV concentration.
Results
Isolation and Sequence Analysis of GhMPK11
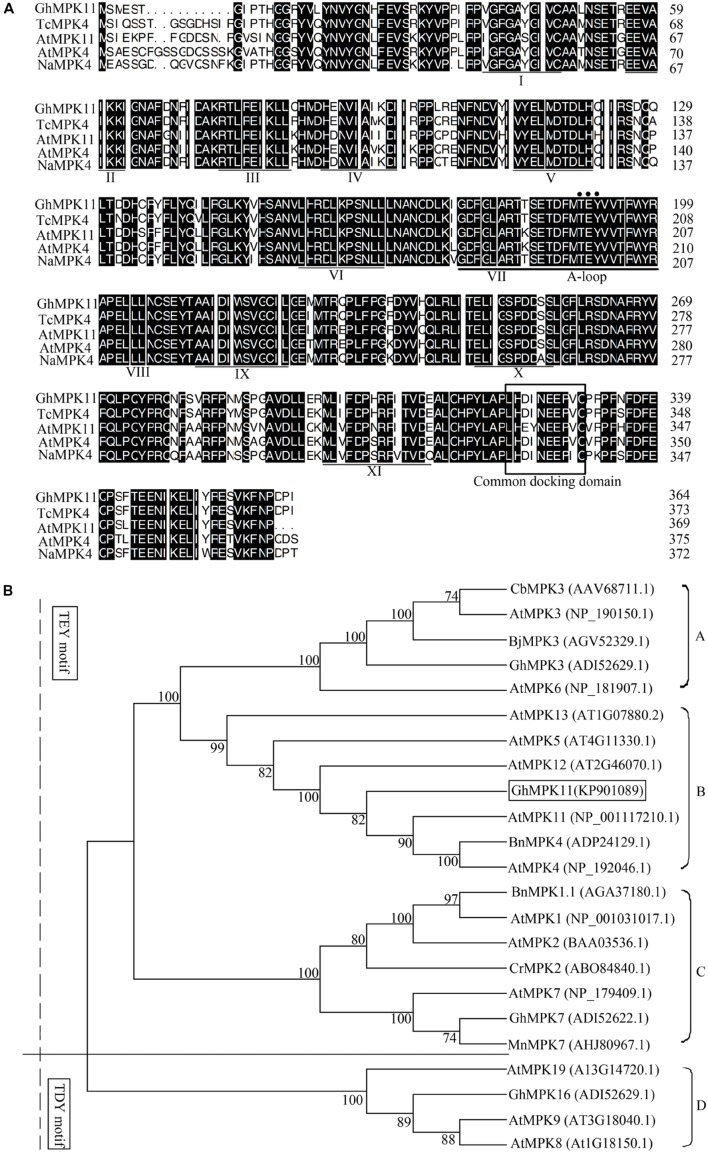
The full-length cDNA of GhMPK11 (KP901089) contained a 95-bp 5′ untranslated region (UTR), 40-bp 3′ UTR and 1098-bp ORF that encoded a 365-amino-acid protein. Multiple alignments indicated that GhMPK11 possesses a conserved TEY motif in the activation loop, 11 conserved subdomains, and a CD domain (Figure 1A). Multiple alignments also demonstrated high identities (84.18–92.78%) with homologous sequences, including TcMPK4 from T. cacao, AtMPK11 and AtMPK4 from A. thaliana and NaMPK4 from N. attenuata. To investigate the evolutionary relationships among MAPK proteins from different species, a phylogenetic analysis based on the amino acid sequences was performed using the neighbor-joining method and MEGA software version 4.1. The results demonstrated that GhMPK11 belongs to the group B MAPK family (Figure 1B).
FIGURE 1.
Sequence and phylogenetic analyses of GhMPK11 (GenBank accession no. KP901089). (A) Multiple alignments of the GhMPK11 protein with TcMPK4 (EOX95887.1), AtMPK11 (NP_001117210.1), AtMPK4 (NP_192046.1) and NaMPK11 (ADT91692.1). Identical amino acids are shaded in black. The conserved subdomains are indicated by numerals (I–XI) at the bottom of the sequences. The activation loop (A-loop) is underlined and contains the phosphorylation motif (TEY motif) marked by a circle. The common docking domain, which is the binding site of MAPKKs and GhMPK11, is boxed. (B) Phylogenetic analysis of MAPK proteins from different species. GhMPK11 is boxed. The GenBank accession numbers are indicated in parentheses. The GenBank IDs follow the protein names.
Subcellular Localization of GhMPK11
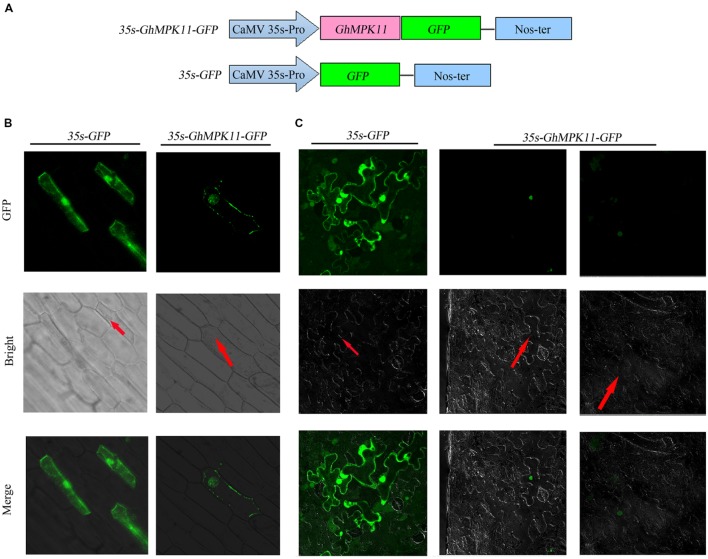
The Plant-mPLoc program was used to predict the subcelluar localization of GhMPK11 and indicated that GhMPK11 localizes in the nucleus. However, CELL version 2 predicted that GhMPK11 primarily localizes in the cytoplasm. To investigate the localization of GhMPK11, a biolistic transformation system was used for a transient assay. Two constructs, 35S-GhMPK11::GFP and 35S-GFP (Figure 2A), were introduced individually into onion epidermal cells. As shown in Figure 2B, fluorescent signals were found in the nucleus, and DAPI staining was also detectable in the nucleus (Supplementary Figure S1). Additionally, the 35S-GhMPK11::GFP and 35S-GFP constructs were transformed individually into tobacco cells, and fluorescence signals were found in the nucleus (Figure 2C). These results suggested that GhMPK11 localizes in the nucleus.
FIGURE 2.
The subcellular localization of GhMPK11 protein transiently expressed in onion epidermal cells and tobacco cells. (A) Schematic diagram of the 35S-GhMPK11-GFP fusion construct and the control 35S-GFP construct. GFP was fused to the C terminus of GhMPK11. Transient expression of the 35S-GhMPK11-GFP and 35S-GFP constructs in onion epidermal cells (B) and tobacco cells (C). Red arrows indicate the location of the nucleus. Green fluorescence was observed using a confocal microscope. Bar = 200 mm.
Expression Patterns of GhMPK11
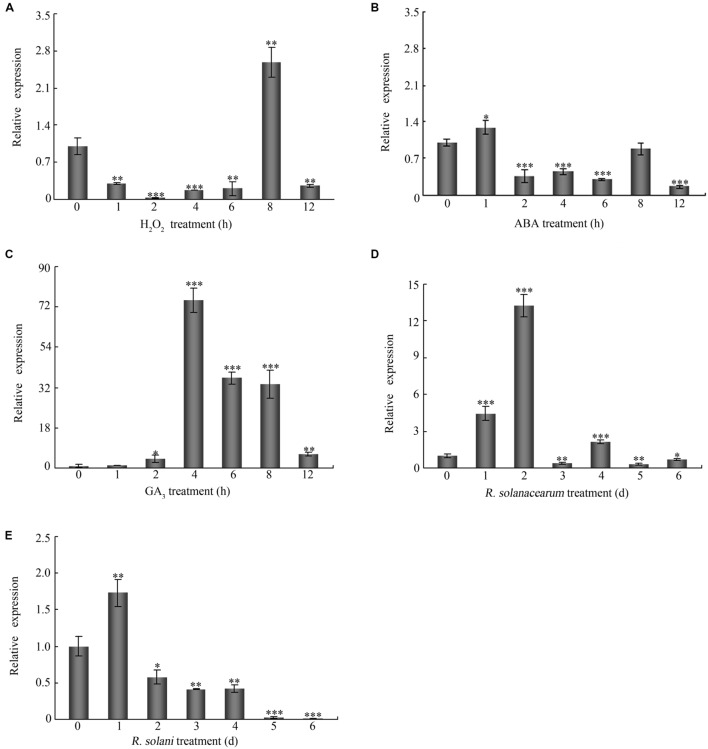
To study the effects of signaling molecules and biotic stresses on the expression of GhMPK11, 7-day-old cotton seedlings were treated with H2O2, ABA, GA3, R. solanacearum and R. solani. GhMPK11 expression increased strongly at 8 h under H2O2 treatment (Figure 3A). ABA treatment led to a slight increase in GhMPK11 transcription levels at 1 h after treatment (Figure 3B), while GA3 treatment sharply enhanced the expression level of GhMPK11 at 4 h after treatment (Figure 3C). Following R. solanacearum treatment, the expression level of GhMPK11 was rapidly induced within 2 h (Figure 3D). R. solani treatment also induced the transcription of GhMPK11 at 1 h, and then a gradual reduction was observed (Figure 3E). These results indicated that GhMPK11 could be induced by pathogens and signaling molecules.
FIGURE 3.
The expression patterns of GhMPK11. Seven-day-old cotton seedlings obtained from hydroponic culture were treated with 10 mM H2O2 (A), 100 μM ABA (B), 100 μM GA3 (C), R. solanacearum (D) and R. solani (E). The G. hirsutum polyubiquitin (UBI) gene was employed as an internal control. The data are the means ± SD from three independent experiments. The asterisks indicate statistically significant differences according to Student’s t-test (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001).
Promoter Analysis of GhMPK11
To characterize the underlying mechanism by which GhMPK11 responds to multiple stresses, a 1309-bp fragment was cloned upstream of the transcription start site (KP901088). The PlantCARE database was used to analyze the cis-acting regulatory elements. Some putative cis-acting regulatory elements, such as the defense-responsive elements WRKY71OS and TC-rich repeats, MBSI and the GA-response element GAREAT TC-rich repeats were found in this region. Some identified cis-elements are listed in Table 1.
Table 1.
Putative cis-acting regulatory elements in the GhMPK11 promoter.
| cis-Element | Position | Sequence (5′-3′) | |
|---|---|---|---|
| Stress response elements | HSE | -1299(-) | AAAAAATTTC |
| LTR | -792(-) | CCGAAA | |
| MBSI | -196(-) | aaaAaaC(G/C)GTTA | |
| MBS | -190(-), -995(+) | C/TAACTG | |
| TC-rich repeats | -1021(-) | ATTCTCTAAC | |
| Hormones | GAREAT | -376(+), -1184(-) | TAACAAR |
| TATCCAOSAMY | -95(+) | TATCCA | |
| WRKY71OS | -401(+), -1093(+), -1212(+), 1206(+) | TGAC/GTCA | |
| Sugar | WBOXHVISO1 | -401(+), -1093(+), -1212(+) | TGACT |
| SREATMSD | -94(+) | TTATCC | |
| Light regulation elements | 3-AF1 binding site | -829(+) | AAGAGATATTT |
| ACE | -196(-), -674(+) | AAAACGTTTA | |
| AE-box | -801(-) | AGAAACTT | |
| Box4 | -911(+), -1153(+) | ATTAAT | |
| Development-related elements | EIRE | -950(+) | TTCGACC |
| CAAT-motif | -975(+), -896(+), -599(+), -890(+) | CAAT | |
| Skn-1-motif | -871(-), -1032(+) | GTCAT | |
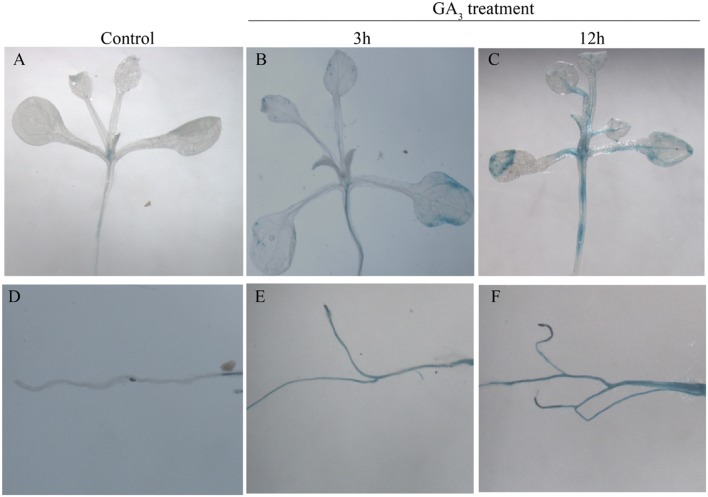
To analyze the promoter activity of GhMPK11, ProGhMPK 11::GUS transgenic A. thaliana plants were obtained. GUS staining results showed that with no treatment, the GUS signal was barely detectable in the leaf and root (Figures 4A and D). However, after the plants were treated with GA3 for 3 and 12 h, GUS activity increased in the leaves and roots over time (Figures 4B–E and F). These data suggested that GhMPK11 is expressed in response to GA and may be involved in the GA signaling pathway.
FIGURE 4.
β-glucuronidase activity analysis in ProGhMPK11::GUS plants in response to GA3. (A,D) No treatment. (B,E) Treated with GA3 for 3 h. (C,F) Treated with GA3 for 12 h.
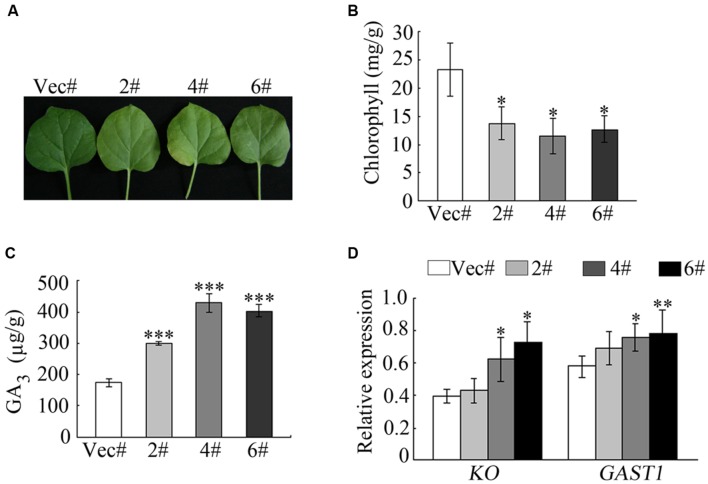
GA3 Influences the Chlorophyll Content of GhMPK11-Overexpressing Plants
To further analyze the function of GhMPK11, transgenic GhMPK11-OE N. benthamiana plants and transgenic vector control N. benthamiana plants were produced. During this process, we observed an interesting phenomenon. When compared with control plants, the leaves of GhMPK11-overexpressing plants (OE plants) were less green (Figure 5A). In accordance with this phenomenon, the chlorophyll content of control plants was higher than that of OE plants (Figure 5B). Considering the sharp increase in GhMPK11 transcription under GA3 treatment and the negative effect of GA on chlorophyll synthesis (Nir et al., 2014), we analyzed the GA3 content in leaves from control and OE plants. Interestingly, OE plant leaves showed a higher level of GA3 than control plant leaves (Figure 5C). To determine whether GhMPK11 could regulate GA-related genes, the expression levels of GAST1 and KO genes were detected. The results revealed that the control plants exhibited lower expression levels of GAST1 and KO compared to OE plants (Figure 5D), which suggested that GhMPK11 regulates GA-related genes.
FIGURE 5.
Overexpression of GhMPK11 influences the chlorophyll content. (A) Detached leaves from 8-week-old OE and control plants. (B) The chlorophyll contents of leaves detached from OE and control plants. (C) The GA3 contents in OE and control plant leaves. (D) GA-related genes expression was detected by qRT-PCR. The data are the means ± SD from three independent experiments. The asterisks indicate statistically significant differences between the transgenic and control plants (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001, Student’s t-test).
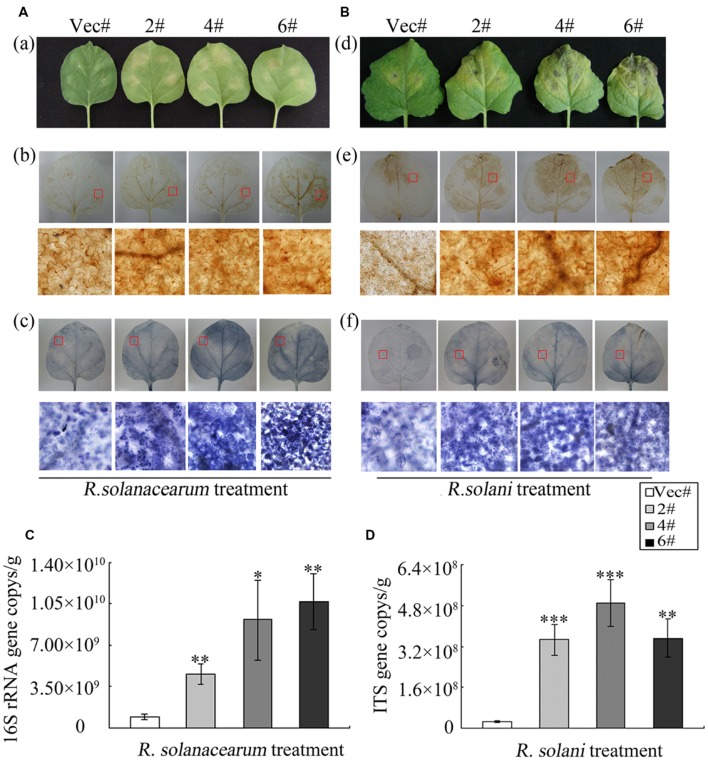
GhMPK11-Overexpressing Plants Show Increased Susceptibility to R. solanacearum and R. solani
To explore the role of GhMPK11 in pathogen resistance in the transgenic tobacco plants, detached leaves from OE and control plants were infiltrated with R. solanacearum (OD600 = 0.6–0.8) and R. solani (105 spores/mL) resuspensions (Figures 6A,B). After 6 days of pathogen inoculation, OE plants showed obvious signs of chlorotic and necrotic lesions, whereas control plants did not. A burst of ROS is a common feature of defense responses (Apel and Hirt, 2004; Jones and Dangl, 2006); H2O2 and superoxide anion (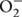 ) are two types of ROS that can be detected by 3, 3-diaminobenzidine (DAB) and NBT staining, respectively (Thordal-Christensen et al., 1997; Fryer et al., 2002, 2003; Ren et al., 2002). Thus, staining and microscopy analyses were also performed to explain this phenomenon. The DAB and NBT staining results revealed greater staining of the OE plant leaves than control plant leaves after pathogen infiltration (Figures 6A,B). Pathogen growth assays performed by qRT-PCR revealed the accumulations of R. solanacearum and R. solani particles in OE plants were higher than that in control plants after 6 days pathogen inoculation (Figures 6C,D). These results indicated that GhMPK11 overexpression enhanced the susceptibility of transgenic plants to pathogens and ROS.
) are two types of ROS that can be detected by 3, 3-diaminobenzidine (DAB) and NBT staining, respectively (Thordal-Christensen et al., 1997; Fryer et al., 2002, 2003; Ren et al., 2002). Thus, staining and microscopy analyses were also performed to explain this phenomenon. The DAB and NBT staining results revealed greater staining of the OE plant leaves than control plant leaves after pathogen infiltration (Figures 6A,B). Pathogen growth assays performed by qRT-PCR revealed the accumulations of R. solanacearum and R. solani particles in OE plants were higher than that in control plants after 6 days pathogen inoculation (Figures 6C,D). These results indicated that GhMPK11 overexpression enhanced the susceptibility of transgenic plants to pathogens and ROS.
FIGURE 6.
GhMPK11 Overexpression influences the diseases responses of plants.
(A) Leaves infiltrated with R. solanacearum and R. solani
(B). The levels of H2O2 and 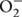 in leaves were determined by DAB (a) and NBT (b) staining, respectively. (C,D) Pathogen accumulation after 6 days cultivation determined by qRT-PCR. Data are the mean ± SD from three independent experiments. The asterisks indicate statistically significant differences between the transgenic and control plants (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001, Student’s t-test).
in leaves were determined by DAB (a) and NBT (b) staining, respectively. (C,D) Pathogen accumulation after 6 days cultivation determined by qRT-PCR. Data are the mean ± SD from three independent experiments. The asterisks indicate statistically significant differences between the transgenic and control plants (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001, Student’s t-test).
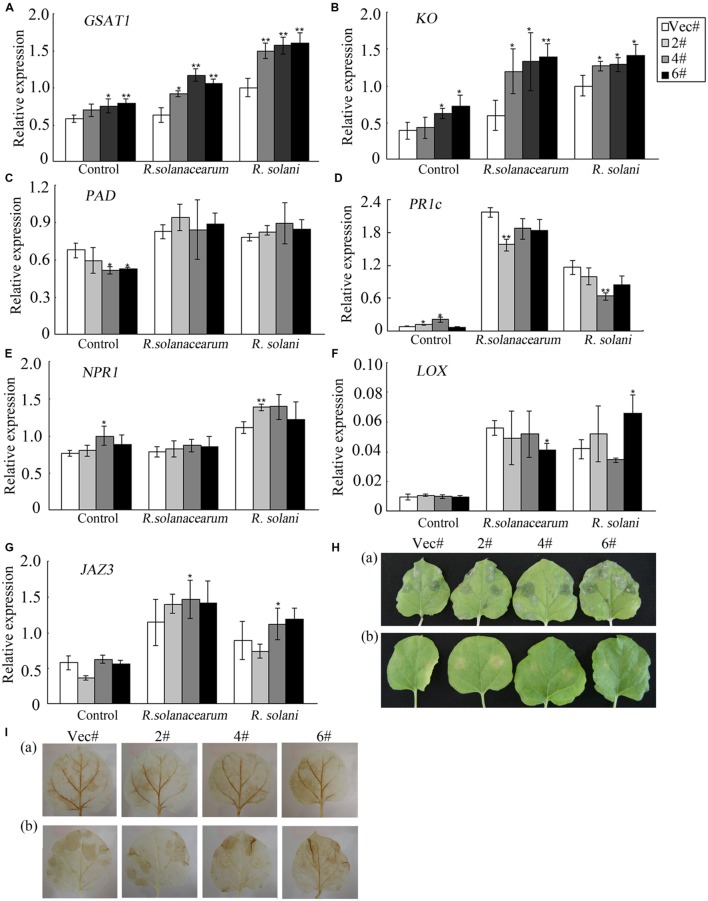
The Increased Susceptibility of Plants to Pathogen Treatment is Related to GA3 Signaling
To evaluate which signaling pathway the GhMPK11 plants depend on, genes involved in GA, SA, and JA signaling pathways were analyzed by qRT-PCR. Total RNA was extracted from leaves before and after pathogen infiltration. After pathogen infiltration, the expression levels of GAST1 and KO, which are GA-related genes (Figures 7A,B), significantly differed, while the expression levels of the SA-related genes PAD, PR1c, and NPR1 and of the JA-related genes LOX and JAZ3 only slightly differed (Figures 7C–G). These findings suggested that GhMPK11 might be associated with the GA signaling pathway.
FIGURE 7.
Expression analysis of hormone-related genes and diseases responses of plants under exogenous GA3 treatment. (A–G) The expression levels of the GA-related genes GAST1 and KO; the SA-related genes PAD, PR1c, and NPR1; and the JA-related genes LOX and JAZ3 were detected by qRT-PCR. (H) (a) Leaves treated with R. solani and 50 mM GA3 and (b) leaves treated with R. solani. (I) DAB staining of leaves shown in (H). The data are the means ± SD from three independent experiments. The asterisks indicate statistically significant differences between the transgenic and control plants (∗P < 0.05; ∗∗P < 0.01, Student’s t test).
Exogenous GA3 Treatment Increase the Susceptibility of Plants to R. solani Treatment
To address the above hypothesis, leaves detached from control and OE plants were infiltrated with R. solani resuspension solution and divided into two groups. Group (b) served as a control [Figure 7H (b)], and group (a) was also treated with exogenous GA3 by placing one piece of cotton soaked in 50 mM exogenous GA3 on the petioles [Figure 7H (a)]. Four days after infiltration, the leaves in group (a) showed enhanced susceptibility to R. solani [Figure 7H (a)] compared to the leaves in group (b) [Figure 7H (b)]. Figure 7I demonstrates increased ROS accumulation in group (a) compared to group (b). Furthermore, control and OE plants showed similar responses. Taken together, these results indicated that exogenous GA3 treatment decreased the resistance of plants to R. solani treatment and enhanced ROS accumulation.
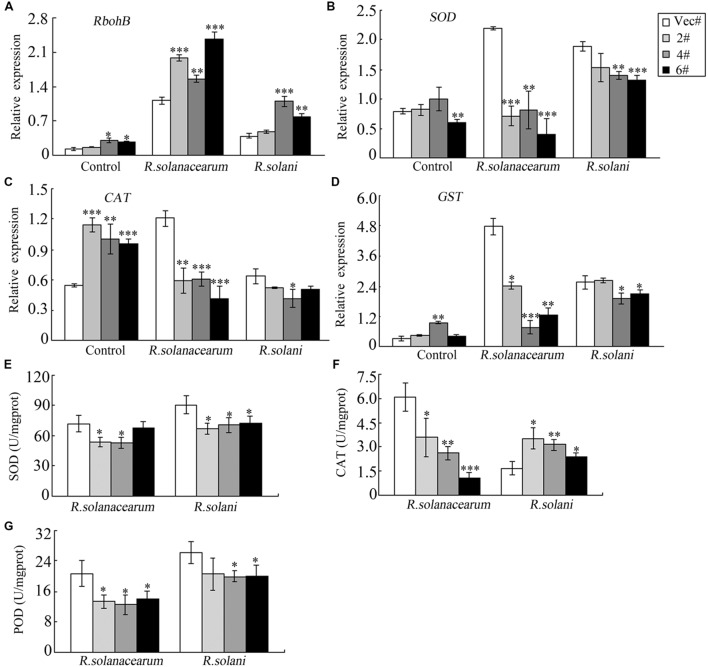
GhMPK11 Overexpression Influences the Transcription of Defense-Related Genes and the Activity of Antioxidant Enzymes
To further explore the underlying mechanism of pathogen sensitivity, the transcription of the ROS-related gene RbohB was analyzed. RbohB showed a higher expression level in OE plants after pathogen infiltration (Figure 8A). To maintain cellular ROS homeostasis, cells have evolved various enzymes to clear excess ROS, such as SODs, PODs, CAT and GST (Willekens et al., 1997; Apel and Hirt, 2004; Cosio and Dunand, 2009). Here, the expression levels of NtSOD, NtCAT and NtGST were measured by qRT-PCR, and all of these enzymes had higher expression levels in control plants than in OE plants (Figures 8B–D) after either R. solanacearum or R. solani treatment. Similarly, after R. solanacearum or R. solani treatment, control plants showed higher activity levels of the antioxidant enzymes SOD and POD (Figures 8E,G). CAT activity was lower in OE plants compared to control plants when treated with R. solanacearum, while CAT activity was higher in OE plants after R. solani treatment (Figure 8F). All these data indicated that GhMPK11 overexpression might enhance ROS accumulation by regulating ROS-related genes and antioxidant enzymes under pathogen infiltration.
FIGURE 8.
Relative expression levels and enzymes activities of defense-related genes following diseases infections. The expression levels of RbohB (A), SOD (B), CAT (C) and GST (D) and the total activities of the antioxidant enzymes SOD (E), CAT (F) and POD (G) in tobacco plants at 4 days after infiltration with R. solanacearum or R. solani. The data are the means ± SD from three independent experiments. The asterisks indicate statistically significant differences between the transgenic and control plants (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001, Student’s t-test).
GhMPK11 Overexpression Reduces the Resistance of Transgenic Plants to Oxidative Stress
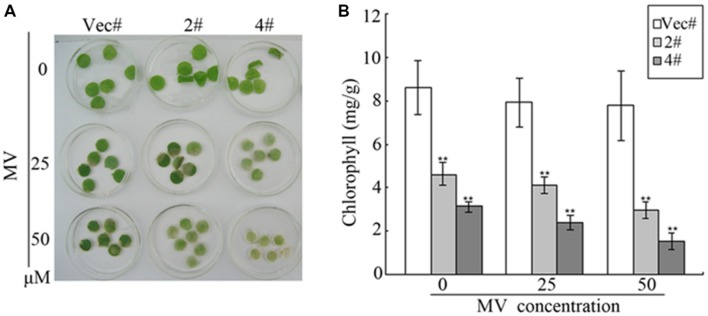
Methyl viologen is an herbicide that can cause chlorophyll degradation and cell membrane leakage through ROS production (Kurepa et al., 1998). In our study, MV was used to treat transgenic plants in order to detect the responses of GhMPK11 to oxidative stress. At the vegetable stage (8-week-old seedlings), after detached leaves were soaked in MV solution for 72 h, leaves from transgenic plants displayed more serious bleaching or chlorosis than those from control plants (Figure 9A). Furthermore, Figure 9B shows the decreased chlorophyll content of transgenic leaves, further validating the difference in oxidative damage between the transgenic and control plants. These results suggested that GhMPK11 has a negative influence on oxidative stress responses.
FIGURE 9.
Oxidative analysis of GhMPK11. (A) Leaf disks from control and OE plants soaked in MV solution for 72 h. (B) Relative chlorophyll content after MV treatment. The data are the means ± SD from three independent experiments. The asterisks indicate statistically significant differences between the transgenic and control plants (∗∗P < 0.01, Student’s t-test).
Discussion
Although many studies have revealed the biological function of specific MAPK proteins in plant defense responses, these studies have mainly focused on MPK3, MPK6 and MPK4, and studies of other MAPK members, especially MAPKs in cotton, are limited. In this study, a group B MAPK gene, GhMPK11, was isolated and characterized in cotton. GhMPK11 transgenic plants showed decreased resistance to the pathogens R. solanacearum and R. solani; this decreased resistance was verified by the larger necrotic lesions and enhanced pathogen growth observed in infiltrated transgenic plants compared to infiltrate control plants.
Multiple alignments and phylogenetic analyses based on MAPK proteins revealed that GhMPK11 and other MAPK members have a similar protein structure; these analyses confirmed that GhMPK11 encodes a MAPK. Glory and Murphy (2007) demonstrated that subcellular localization might indicate how a gene interacts with transcription factors to function, offer valuable clues to uncovering its functions, and help understand the complicated pathways that regulate biological processes at the cellular level. GhMPK11 localized to the nucleus, indicating that it might function with transcription factors. In addition, the observed expression pattern of GhMPK11 in response to GA3, as well as the results of promoter analysis and GUS activity analysis, indicated that GhMPK11 might be involved in the GA signaling pathway (Figures 1–4).
To deeply investigate the biological function of GhMPK11, 35s::GhMPK11 transgenic plants and 35 s vector control plants were obtained. During the process of transgenic plants growth, an interesting leaf color phenotype was observed. GA is known to have a negative influence on chlorophyll levels in tomato plants (Nir et al., 2014), and compared with wild-type (WT) plants, GA-deficient mutants have darker green leaves (Koornneef and van der Veen, 1980; Koornneef et al., 1985; Peng and Harberd, 1997). Furthermore, GA-deficient plants have higher levels of bioactive GAs than do WT plants (Talon et al., 1990; Peng et al., 1999). Considering these previous studies and our experimental results, we hypothesized that light green leaves were probably caused by the altered GA content in these plants. The higher GA3 content and GA-related gene (KO, GAST1) expression levels observed in OE plants supported this hypothesis (Figure 5). The transcriptional suppression of KO has been shown to be responsible for decreased GA content (Fukazawa et al., 2000). GAST1 is a GA-induced gene, and a higher transcript level of this gene correlates with higher GA3 content (Shi and Olszewski, 1998). These findings suggested that GhMPK11 could alter GA3 content by regulating GA-related genes.
Except enhanced expression of GhMPK11 in response to GA3, strong induction of GhMPK11 expression by pathogens was also detected. This expression pattern is consistent with the expression pattern of AtMPK11 in response to PAMPs (Eschen-Lippold et al., 2012), which suggests that GhMPK11 might function in defense responses. Figure 6 showed that GhMPK11 overexpression could reduce pathogen resistance and enhance pathogen growth. The enhanced susceptibility of GhMPK11 transgenic plants to R. solanacearum was consistent with a previous study of MPK4 that demonstrated that an mpk4 mutant exhibited enhanced resistance to Pseudomonas syringae pv. tomato DC3000, a Gram-negative plant pathogenic bacterium (Petersen et al., 2000). Although SA and JA are well-established phytohormones that are involved in disease responses (Yang D.L. et al., 2008), emerging evidence has indicated a relationship between GAs and pathogen infection (Qin et al., 2012). Researchers have found that GAs are actively involved in plant immunity and development (Bari and Jones, 2009; Grant and Jones, 2009); however, the underlying mechanism has been only partially elucidated. To determine which pathway was associated with the enhanced susceptibility of transgenic plants, the expression levels of genes related to SA, JA and GA signaling were detected. Significant changes in the expression levels of GA-related genes and slight changes in the expression levels of SA- or JA-related genes indicate that the enhanced susceptibility of transgenic plants to diseases might correlate with the GA signaling pathway. To confirm this result, plants were treated with exogenous GA3 while being simultaneously infiltrated with R. solani. OE and control leaves displayed enhanced susceptibility and the same phenotype upon R. solani infiltration (Figure 7). Taken together, these results suggest that exogenous GA3 can reduce the influence of endogenous GA3 in transgenic plants on disease responses and that GhMPK11 overexpression can enhance plant susceptibility to pathogen infiltration through the GA signaling pathway. These findings are consistent with the results of a study that found that GA-overproducing eui rice appears more susceptible to bacterial and fungal pathogens in the field (Yang D.L. et al., 2008).
A burst of ROS is a common feature of defense responses (Apel and Hirt, 2004; Jones and Dangl, 2006), and plants have developed intricate pathways to fight various environmental stresses by producing more ROS (Coupe et al., 2006). Furthermore, ROS accumulation has been shown to have a negative effect on resistance to necrotrophic pathogens (Yoshioka et al., 2009). In this study, R. solanacearum and R. solani, which are both necrotrophic pathogens, were used to treat plants (Geraats et al., 2003; Van Loon et al., 2006; Molla et al., 2013; Byth-Illing and Bornman, 2014). Therefore, after pathogen treatment, the accumulation of H2O2 and 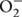 increased greatly (Figure 6). To further determine the mechanism of ROS accumulation, ROS-related genes were detected. A previous study showed that down-regulation of PvRbohB can decrease
increased greatly (Figure 6). To further determine the mechanism of ROS accumulation, ROS-related genes were detected. A previous study showed that down-regulation of PvRbohB can decrease 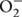 and H2O2 production in P. vulgaris roots (Montiel et al., 2012); conversely, increased expression levels of RbohB will lead to more ROS. In addition, the decreased expression level and activity of ROS detoxification enzymes also contributed to ROS accumulation (Figure 8). The levels of ROS detoxification enzymes can be regulated by the SCFSLY 1/GID2 complex through the GA signaling pathway (Grant and Jones, 2009). Thus, the decreased tolerance of transgenic plants to MV treatment may be associated with elevated ROS levels (Figure 9).
and H2O2 production in P. vulgaris roots (Montiel et al., 2012); conversely, increased expression levels of RbohB will lead to more ROS. In addition, the decreased expression level and activity of ROS detoxification enzymes also contributed to ROS accumulation (Figure 8). The levels of ROS detoxification enzymes can be regulated by the SCFSLY 1/GID2 complex through the GA signaling pathway (Grant and Jones, 2009). Thus, the decreased tolerance of transgenic plants to MV treatment may be associated with elevated ROS levels (Figure 9).
Considering these findings, we conclude that the enhanced susceptibility of transgenic plants to pathogen infiltration is a result of enhanced ROS accumulation, which is regulated by GhMPK11 through the GA signaling pathway. However, although the influence of GhMPK11 on pathogen resistance was studied here, the comprehensive regulatory mechanism of the defense responses to pathogen infiltration in cotton requires further investigation.
Author Contributions
XG designed the experiments. FW performed the experiments and analyzed the results with contributions from CW, YY, and HJ. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China [Grant Number 31171837; 31471424].
Abbreviations
- CaMV
cauliflower mosaic virus
- CAT
catalase
- DAB
3,30-diaminobenzidine
- ET
ethylene
- GA3
gibberellin 3
- GAST1
GA-stimulated transcript 1
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- GUS
β-glucuronidase
- JA
jasmonic acid
- KO
ent-kaurene oxidase
- MS
Murashige and Skoog
- MV
methyl viologen
- OE
overexpressing
- ORF
open reading frame
- qPCR
quantitative real-time PCR
- NBT
nitroblue tetrazolium
- POD
peroxidase
- RbohB
respiratory burst oxidase homolog
- ROS
reactive oxygen species
- SA
salicylic acid
- SOD
superoxide dismutase
- WT
wild-type
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00689
FIGURE S1 | The subcellular localization of GhMPK11 protein in onion epidermal cells. Transiently expression of the 35S-GhMPK11-GFP and 35S-GFP constructs in onion epidermal cells. Green fluorescence was observed using a confocal microscope. Onion cell nuclei were marked by DAPI staining. Bar = 200 mm.
TABLE S1 | Primers used in this study.
References
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., et al. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311 91–94. 10.1126/science.1118642 [DOI] [PubMed] [Google Scholar]
- Adie B. A., Perez-Perez J., Perez-Perez M. M., Godoy M., Sanchez-Serrano J. J., Schmelz E. A., et al. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19 1665–1681. 10.1105/tpc.106.048041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species, metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Bari R., Jones J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69 473–488. 10.1007/s11103-008-9435-0 [DOI] [PubMed] [Google Scholar]
- Brader G., Djamei A., Teige M., Palva E. T., Hirt H. (2007). The MAP kinase kinase MKK2 affects disease resistance in Arabidopsis. Mol. Plant Microbe Interact. 20 589–596. 10.1094/MPMI-20-5-0589 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Petersen M., Nielsen H. B., Zhu S., Newman M. A., Shokat K. M., et al. (2006). Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 47 532–546. 10.1111/j.1365-313X.2006.02806.x [DOI] [PubMed] [Google Scholar]
- Byth-Illing H. A., Bornman L. (2014). Heat shock, with recovery, promotes protection of Nicotiana tabacum during subsequent exposure to Ralstonia solanacearum. Cell Stress Chaperones 19 193–203. 10.1007/s12192-013-0445-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J., Scheffler B. E., Dennis E., Triplett B. A., Zhang T., Guo W., et al. (2007). Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 145 1303–1310. 10.1104/pp.107.107672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebrook E. H., Thomas S. G., Phillips A. L., Hedden P. (2014). The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 217 67–75. 10.1242/jeb.089938 [DOI] [PubMed] [Google Scholar]
- Cosio C., Dunand C. (2009). Specific functions of individual class III peroxidase genes. J. Exp. Biol. 60 391–408. 10.1093/jxb/ern318 [DOI] [PubMed] [Google Scholar]
- Coupe S. A., Palmer B. G., Lake J. A., Overy S. A., Oxborough K., Woodward F. I., et al. (2006). Systemic signaling of environmental cues in Arabidopsis leaves. J. Exp. Biol. 57 329–341. 10.1093/jxb/erj033 [DOI] [PubMed] [Google Scholar]
- Dóczi R., Brader G., Pettkó-Szandtner A., Rajh I., Djamei A., Pitzschke A., et al. (2007). The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell 19 3266–3279. 10.1105/tpc.106.050039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschen-Lippold L., Bethke G., Palm-Forster M. A., Pecher P., Bauer N., Glazebrook J., et al. (2012). MPK11-a fourth elicitor-responsive mitogen-activated protein kinase in Arabidopsis thaliana. Plant Signal. Behav. 7 1203–1205. 10.4161/psb.21323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer M. J., Ball L., Oxborough K., Karpinski S., Mullineaux P. M., Baker N. R. (2003). Control of ascorbate peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J. 33 691–705. 10.1046/j.1365-313X.2003.01656.x [DOI] [PubMed] [Google Scholar]
- Fryer M. J., Oxborough K., Mullineaux P. M., Baker N. R. (2002). Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 53 1249–1254. 10.1093/jexbot/53.372.1249 [DOI] [PubMed] [Google Scholar]
- Fukazawa J., Sakai T., Ishida S., Yamaguchi I., Kamiya Y., Takahashi Y. (2000). Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12 901–915. 10.1105/tpc.12.6.901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E. G., Grau-Enguix F., Abbas M., Locascio A., Thomas S. G., et al. (2012). Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109 13446–13451. 10.1073/pnas.1119992109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., et al. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18 1190–1198. 10.1038/cr.2008.300 [DOI] [PubMed] [Google Scholar]
- Geraats B. P., Bakker P. A., Lawrence C. B., Achuo E. A., Höfte M., van Loon L. C. (2003). Ethylene-insensitive tobacco shows differentially altered susceptibility to different pathogens. Phytopathology 93 813–821. 10.1094/PHYTO.2003.93.7.813 [DOI] [PubMed] [Google Scholar]
- Glory E., Murphy R. F. (2007). Automated subcellular location determination and high-throughput microscopy. Dev. Cell 12 7–16. 10.1016/j.devcel.2006.12.007 [DOI] [PubMed] [Google Scholar]
- Golldack D., Li C., Mohan H., Probst N. (2013). Gibberellins and abscisic acid signal crosstalk, living and developing under unfavorable conditions. Plant Cell Rep. 32 1007–1016. 10.1007/s00299-013-1409-2 [DOI] [PubMed] [Google Scholar]
- Grant M. R., Jones J. D. G. (2009). Hormone (dis) harmony moulds plant health and disease. Science 324 750–752. 10.1126/science.1173771 [DOI] [PubMed] [Google Scholar]
- Group M. A. P. K. (2002). Mitogen-activated protein kinase cascades in plants, a new nomenclature. Trends Plant Sci. 7 301–308. 10.1016/S1360-1385(02)02302-6 [DOI] [PubMed] [Google Scholar]
- Grover C. E., Kim H., Wing R. A., Paterson A. H., Wendel J. F. (2004). Incongruent patterns of local and global genome size evolution in cotton. Genome Res. 14 1474–1482. 10.1101/gr.2673204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G. H., Yu S. X. (2007). Extraction of high-quality total RNA in cotton leaf with improved CTAB method. Cotton Sci. 19 69–70. [Google Scholar]
- Ichimura T., Wakamiya-Tsuruta A., Itagaki C., Taoka M., Hayano T., Natsume T., et al. (2002). Phosphorylation-dependent interaction of kinesin light chain 2 and the 14-3-3 protein. Biochemistry 41 5566–5572. 10.1021/bi015946f [DOI] [PubMed] [Google Scholar]
- Jammes F., Song C., Shin D., Munemasa S., Takeda K., Gu D., et al. (2009). MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. U.S.A. 106 20520–20525. 10.1073/pnas.0907205106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R. A. (1987). Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5 387–405. 10.1007/BF02667740 [DOI] [Google Scholar]
- Jonak C., Okrész L., Bögre L., Hirt H. (2002). Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 5 415–424. 10.1016/S1369-5266(02)00285-6 [DOI] [PubMed] [Google Scholar]
- Jones J. D. G., Dangl J. L. (2006). The plant immune system. Nature 444 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Elgersma A., Hanhart C. J., van Loenen-Martinet E. P., van Rijn L., Zeevaart J. A. D. (1985). A gibberellin-insensitive mutant of Arabidopsis thaliana. Physiol. Plant 65 33–39. 10.1111/j.1399-3054.1985.tb02355.x [DOI] [Google Scholar]
- Koornneef M., van der Veen J. H. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 58 257–263. 10.1007/BF00265176 [DOI] [PubMed] [Google Scholar]
- Kurepa J., Smalle J., Van M. M., Inze D. (1998). Oxidative stress tolerance and longevity in Arabidopsis: the late-flowering mutant gigantea is tolerant to paraquat. Plant J. 14 759–764. 10.1046/j.1365-313x.1998.00168.x [DOI] [PubMed] [Google Scholar]
- Li J., Sima W., Ouyang B., Wang T., Ziaf K., Luo Z., et al. (2012). Tomato SlDREB gene restricts leaf expansion and internode elongation by downregulating key genes for gibberellin biosynthesis. J. Exp. Bot. 63 6407–6420. 10.1093/jxb/ers295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang L., Wang X., Zhang W., Hao L., Chu X., et al. (2013). Cotton GhMPK6a negatively regulates osmotic tolerance and bacterial infection in transgenic Nicotiana benthamiana, and plays a pivotal role in development. FEBS J. 280 5128–5144. 10.1111/febs.12488 [DOI] [PubMed] [Google Scholar]
- Liu Y. G., Chen Y. (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43 649–650. 10.2144/000112601 [DOI] [PubMed] [Google Scholar]
- Lu W., Chu X., Li Y., Wang C., Guo X. (2013). Cotton GhMKK1 induces the tolerance of salt and drought stress, and mediates defence responses to pathogen infection in transgenic Nicotiana benthamiana. PLoS ONE 8:e68503 10.1371/journal.pone.0068503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Zhang S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51 245–266. 10.1146/annurev-phyto-082712-102314 [DOI] [PubMed] [Google Scholar]
- Molla K. A., Karmakar S., Chanda P. K., Ghosh S., Sarkar S. N., Datta S. K., et al. (2013). Rice oxalate oxidase gene driven by green tissue-specific promoter increases tolerance to sheath blight pathogen (Rhizoctonia solani) in transgenic rice. Mol. Plant Pathol. 2013 910–922. 10.1111/mpp.12055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel J., Nava N., Cárdenas L., Sánchez-López R., Arthikala M. K., Santana O., et al. (2012). A Phaseolus vulgaris NADPH oxidase gene is required for root infection by Rhizobia. Plant Cell Physiol. 53 1751–1767. 10.1093/pcp/pcs120 [DOI] [PubMed] [Google Scholar]
- Nakashita H., Yasuda M., Nitta T., Asami T., Fujioka S., Arai Y., et al. (2003). Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 33 887–898. 10.1046/j.1365-313X.2003.01675.x [DOI] [PubMed] [Google Scholar]
- Nir I., Moshelion M., Weiss D. (2014). The Arabidopsis gibberellin methyl transferase 1 suppresses gibberellin activity, reduces whole-plant transpiration and promotes drought tolerance in transgenic tomato. Plant Cell Environ. 37 113–123. 10.1111/pce.12135 [DOI] [PubMed] [Google Scholar]
- Ortiz-Masia D., Perez-Amador M. A., Carbonell P., Aniento F., Carbonell J., Marcote M. J. (2008). Characterization of PsMPK2, the first C1 subgroup MAP kinase from pea (Pisum sativum L.). Planta 227 1333–1342. 10.1007/s00425-008-0705-5 [DOI] [PubMed] [Google Scholar]
- Peng J., Harberd N. P. (1997). Transposonassociated somatic gai-loss sectors in Arabidopsis. Plant Sci 130 181–188. 10.1016/S0168-9452(97)00216-1 [DOI] [Google Scholar]
- Peng J., Richards D. E., Moritz T., Ca no- Delgado A., Harberd N. P. (1999). Extragenic suppressors of the Arabidopsis gai mutation alter the dose-response relationship of diverse gibberellin responses. Plant Physiol. 119 1–10. 10.1104/pp.119.4.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M., Brodersen P., Naested H., Andreasson E., Lindhart U., Johansen B., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103 1111–1120. 10.1016/S0092-8674(00)00213-0 [DOI] [PubMed] [Google Scholar]
- Pitzschke A., Schikora A., Hirt H. (2009). MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12 421–426. 10.1016/j.pbi.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Qin X., Liu J. H., Zhao W., Chen X., Guo Z., Peng Y. (2012). Gibberellin 20-oxidase gene, OsGA20ox3 regulates plant stature and disease development in rice. Mol. Plant Microbe Interact. 26 227–239. 10.1094/MPMI-05-12-0138-R [DOI] [PubMed] [Google Scholar]
- Rasche F., Knapp D., Kaiser C., Koranda M., Kitzler B., Zechmeister-Boltenstern S., et al. (2011). Seasonality and resource availability control bacterial and archaeal communities in soils of a temperate beech forest. ISME J. 5 389–402. 10.1038/ismej.2010.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M. W., Roux M., Petersen M., Mundy J. (2012). MAP Kinase cascades in Arabidopsis innate immunity. Front. Plant Sci. 3:169 10.3389/fpls.2012.00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Yang H., Zhang S. (2002). Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 277 559–565. 10.1074/jbc.M109495200 [DOI] [PubMed] [Google Scholar]
- Rodriguez M. C. S., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61 621–649. 10.1146/annurev-arplant-042809-112252 [DOI] [PubMed] [Google Scholar]
- Šamajová O., Komis G., Šamaj J. (2013). Emerging topics in the cell biology of mitogen-activated protein kinases. Trends Plant Sci. 18 140–148. 10.1016/j.tplants.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Shan D. P., Huang J. G., Yang Y. T., Guo Y. H., Wu C. A., Yang G. D., et al. (2007). Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. New Phytol. 176 70–81. 10.1111/j.1469-8137.2007.02160.x [DOI] [PubMed] [Google Scholar]
- Shi J., An H., Zhang L., Gao Z., Guo X. (2010). GhMPK7, a novel multiple stress-responsive cotton group C MAPK gene, has a role in broad spectrum disease resistance and plant development. Plant Mol. Biol. 74 1–17. 10.1007/s11103-010-9661-0 [DOI] [PubMed] [Google Scholar]
- Shi L., Olszewski N. E. (1998). Gibberellin and abscisic acid regulate GAST1 expression at the level of transcription. Plant Mol. Biol. 38 1053–1060. 10.1023/A:1006007315718 [DOI] [PubMed] [Google Scholar]
- Siemens J., Keller I., Sarx J., Kunz S., Schuller A., Nagel W., et al. (2006). Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development. Mol. Plant Microbe Interact. 19 480–494. 10.1094/MPMI-19-0480 [DOI] [PubMed] [Google Scholar]
- Sinha A. K., Jaggi M., Raghuram B., Tuteja N. (2011). Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 6 196–203. 10.4161/psb.6.2.14701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunilkumar G., Campbell L. M., Puckhaber L., Stipanovic R. D., Rathore K. S. (2006). Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc. Natl. Acad. Sci. U.S.A. 103 18054–18059. 10.1073/pnas.0605389103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon M., Koornneef M., Zeevaart J. A. D. (1990). Accumulation of C19-gibberellins in the gibberellin-insensitive dwarf mutant gai of Arabidopsis thaliana (L) Heynh. Planta 182 501–505. 10.1007/BF02341024 [DOI] [PubMed] [Google Scholar]
- Tanoue T., Adachi M., Moriguchi T., Nishida E. (2000). A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2 110–116. 10.1038/35000065 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H., Zhang Z., Wei Y., Collinge D. B. (1997). Subcellular localization of H2O2 in plants, H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11 1187–1194. 10.1046/j.1365-313X.1997.11061187.x [DOI] [Google Scholar]
- Van Loon L. C., Geraats B. P., Linthorst H. J. (2006). Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 11 184–191. 10.1016/j.tplants.2006.02.005 [DOI] [PubMed] [Google Scholar]
- Wang F., Zhang Y., Yao P., Guo X., Li H., Xu B. (2014). Molecular identification and stress response of the apoptosis-inducing factor gene 3 (AccAIF3) from Apis cerana cerana. Apidologie 45 685–700. 10.1007/s13592-014-0285-2 [DOI] [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, Chap. 38 eds Innis M., Gelfand D., Sninsky J., White T. (Orlando, FL: Academic Press; ), 315–322. [Google Scholar]
- Widmann C., Gibson S., Jarpe M. B., Johnson G. L. (1999). Mitogen-activated protein kinase, conservation of a three-kinase module from yeast to human. Physiol. Rev. 79 143–180. [DOI] [PubMed] [Google Scholar]
- Willekens H., Chamnongpol S., Davey M., Schraudner M., Langebartels C., Van Montagu M., et al. (1997). Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J. 16 4806–4816. 10.1093/emboj/16.16.4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige B. C., Isono E., Richter R., Zourelidou M., Schwechheimer C. (2011). Gibberellin regulates PIN-FORMED abundance and is required for auxin transport-dependent growth and development in Arabidopsis thaliana. Plant Cell 23 2184–2195. 10.1105/tpc.111.086355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Chen J., Wang Q., Yang Y. (2014). Direct phosphorylation and activation of a mitogen-activated protein kinase by a calcium-dependent protein kinase in rice. Plant Cell 26 3077–3089. 10.1105/tpc.114.126441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Yang K. Y., Yoo S. J., Liu Y., Ren D., Zhang S. (2014). Reactive oxygen species in signaling the transcriptional activation of WIPK expression in tobacco. Plant Cell Environ. 37 1614–1625. 10.1111/pce.12271 [DOI] [PubMed] [Google Scholar]
- Yang D. L., Li Q., Deng Y. W., Lou Y. G., Wang M. Y., Zhou G. X., et al. (2008). Altered disease development in the eui mutants and eui overexpressors indicates that gibberellins negatively regulate rice basal disease resistance. Mol. Plant 1 528–537. 10.1093/mp/ssn021 [DOI] [PubMed] [Google Scholar]
- Yang L., Tang R., Zhu J., Liu H., Mueller-Roeber B., Xia H., et al. (2008). Enhancement of stress tolerance in transgenic tobacco plants constitutively expressing AtIpk2β, an inositol polyphosphate 6-/3-kinase from Arabidopsis thaliana. Plant Mol. Biol 66 329–343. 10.1007/s11103-007-9267-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H., Asai S., Yoshioka M., Kobayashi M. (2009). Molecular mechanisms of generation for nitric oxide and reactive oxygen species, and role of the radical burst in plant immunity. Mol. Cells 28 321–329. 10.1007/s10059-009-0156-2 [DOI] [PubMed] [Google Scholar]
- Yu F., Huaxia Y., Lu W., Wu C., Cao X., Guo X. (2012). GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development. BMC Plant Biol. 12:144 10.1186/1471-2229-12-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Guo W., Zhang T. (2002). Molecular linkage map of allotetraploid cotton (Gossypium hirsutum L. × Gossypium barbadense L.) with a haploid population. Theor. Appl. Genet. 105 1166–1174. 10.1007/s00122-002-1100-4 [DOI] [PubMed] [Google Scholar]
- Zhang J., Zou D., Li Y., Sun X., Wang N. N., Gong S. Y., et al. (2014). GhMPK17 a cotton mitogen-activated protein kinase, is involved in plant response to high salinity and osmotic stresses and ABA signaling. PLoS ONE 9:e95642 10.1371/journal.pone.0095642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Li Q., Li Z., Staswick P. E., Wang M., Zhu Y., et al. (2007). Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis–Pseudomonas syringae interaction. Plant Physiol. 145 450–464. 10.1104/pp.107.106021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 | The subcellular localization of GhMPK11 protein in onion epidermal cells. Transiently expression of the 35S-GhMPK11-GFP and 35S-GFP constructs in onion epidermal cells. Green fluorescence was observed using a confocal microscope. Onion cell nuclei were marked by DAPI staining. Bar = 200 mm.
TABLE S1 | Primers used in this study.