Abstract
Heart failure is a life-threatening and costly ailment characterized by structural and functional impairment of the heart. Despite major advances in understanding protein-mediated transcriptional control and signaling pathways that underlie the cellular and interstitial alterations of heart failure, significant therapeutical breakthroughs for innovative treatments of this disease are still missing. The recent extensive profiling of the mammalian transcriptome has revealed a large number of long non-coding RNAs (lncRNAs) that play a diversity of important regulatory roles in gene expression. In here, we focus on a recent work by Ounzain and colleagues comprising genome-wide profiling of the cardiac transcriptome after myocardial infarction with an emphasis on the identification of novel heart-specific lncRNAs.
Keywords: Long non-coding RNA (lncRNA), heart failure, gene networks
Heart failure is a life-threatening and costly condition categorized by structural and functional impairment of ventricular filling or ejection of blood by the heart. Considering the overall increase in lifespan of the general population and high incidence of risk factors and improved survival from acute cardiovascular events, it is expected that the prevalence of heart failure will increase along with the associated emotional and socio-economic burdens. Upon injury, the heart responds to a diversity of biochemical and hemodynamic stressors through a process that entails molecular, cellular and interstitial alterations that are reflected further in size, shape and function of the heart (1). Although the initial response is adaptive and mainly serves to balance the decay in the pumping capacity of the heart, prolonged stress stimulation progressively contributes to deteriorate ventricular function which ultimately can lead to heart failure. Despite major advances in understanding the mechanisms that underlie cardiac remodeling and the worsened cardiac performance, this did not translate into significant therapeutical breakthroughs for innovative treatments of heart failure in patients.
Once considered transcriptional noise, several transcriptome studies have shown prevalent transcription of a large number of non-coding RNAs (ncRNAs) (2). These ncRNAs are classified into two groups according to their length, whereby small ncRNAs are arbitrarily set at less than 200 nucleotides in size and include several ncRNAs species including small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs) and microRNAs (miRNAs) (3-5). In addition, transcripts of ncRNAs that are larger than 200 nucleotides are called long non-coding RNAs (lncRNAs). Of all currently studied noncoding genes, lncRNAs are a mystery species of growing scientific interest. Just like the extensively studied miRNAs, lncRNAs have been reported to have divers biological functions such as regulation of chromatin remodeling, maintenance of the nuclear structure integrity, and transcriptional and post-transcriptional processing (6,7). Despite their length, lncRNAs are distinct from protein coding genes as they are expressed at lower levels, are less evolutionarily conserved and mainly consist of a distinctive gene structure of usually 1–2 exons (8,9). In contrast to other ncRNAs, some lncRNAs have been shown to give rise to small peptides (10), showcasing the diversity of lncRNAs. The importance is underlined by the fact that specific lncRNA knockout strategies in mice showed severe effects that caused pre- and postnatal lethal phenotypes (11,12). Whereas deletion of lncRNAs Peril and Mdgt resulted in viability defects, mice lacking the lncRNA Fendrr died at the embryonic stage due to defects in several organs including the heart (11). These developmental defects were shown to be independent of adjacent genetic regions, e.g., cis or enhancer-like regulatory domains, as deletion of these lncRNAs did not phenocopy deletion of adjacent genes, which suggests a distinct function for these lncRNAs (12). One of the first reports on ncRNA expression showed that infants with non-syndromic tetralogy of Fallot had higher levels of an ALC-1 antisense lncRNA that disturbed ALC-1 translation in hypertrophied ventricles compared to their healthy counterparts (13,14). Next to the fact that some lncRNAs have been found to be altered in the developing or diseased heart, several single nucleotide polymorphism (SNP) in lncRNAs have shown to be strongly correlated with cardiovascular disease (15,16). For example, SNPs in myocardial infarction associated transcript (MIAT) and antisense non-coding RNA in the INK4 locus (ANRIL) forecast increased risk of cardiovascular disease (15,16). Moreover, lncRNA H19 was significantly upregulated in failing murine hearts, suggesting a role for hypoxia regulated lncRNA expression in heart failure (17). This is further highlighted by a recent study employing a comprehensive myocardial transcript profiling of coding and non-coding RNA species in failing human hearts and hearts that were unloaded with a left ventricular assist device (LVAD) (18). LVAD support has been known to diminish cardiac stress by reducing tissue hypoxia, auxiliary to the concept of restoration of cardiac normoxia by mechanical unloading (19,20). Fascinatingly, a significantly higher proportion of differentially expressed lncRNAs was normalized in hearts that were unloaded compared to mRNA and miRNA levels (18). In fact, lncRNA levels not only responded more sensitively to LVAD support but their expression profile allowed to distinguish left ventricular samples from patients with ischemic and non-ischemic heart failure before and after LVAD support (18). It is no longer a question whether lncRNAs have a role in the cardiovascular system under normal physiological condition and in disease states. However, understanding the comprehensive role of lncRNA played herein is still in its infancy.
In an attempt to elucidate this, Ounzain and colleagues report an exciting genome-wide profiling of the cardiac transcriptome after myocardial infarction with an emphasis on the identification of heart-specific lncRNAs (21). In the article by Ounzain et al., the authors performed RNA sequencing, ab initio transcript reconstruction and integrated genome-wide approaches to supplement gene regulatory networks with lncRNA profile in hearts after myocardial infarction. Of the multi-exonic transcripts, 14.3% were allocated as lncRNAs of which a smaller part (5.6%) corresponded to the UCSC annotation and a larger part (8.7%) was still not annotated. These unannotated transcripts showed several typical lncRNA physiognomies, such as genomic orientations relative to nearby coding genes and the low protein coding potential, and were therefore designated as novel lncRNAs. To verify whether these novel lncRNAs are cardiac specific and are associated with enhancer sequences active in the heart, qRT-PCR verification and association with flanking cis-regulatory proximal coding genes showed that differentially expressed novel lncRNAs were significantly more enriched in the myocardium than other transcript classes. Intriguingly, although gene ontology classification of the proximal genes revealed enrichment in processes associated with cardiac function and transcriptional regulation, novel lncRNAs were significantly associated with transcriptional control. Moreover, these lncRNAs were also located proximal to cardiac developmental genes, implicating a role for these novel lncRNAs in the stress responses and transcriptional reprogramming of the remodeling heart. Indeed, it was suggested that lncRNAs expression profiles in general are more profound to different heart failure etiologies when compared to other RNA transcripts (18). Mechanistically, Ounzain et al. demonstrated in the current study that the lncRNAs act as competitive endogenous RNAs specifically for microRNAs. Correlation network analysis of miRNA-lncRNA-mRNA indicated that lncRNAs correlated positively with miRNAs that have been known to be modulated in the post-infarcted myocardium, whereas mRNA levels correlated negatively. The likelihood that the predominant mechanism of action of cardiac lncRNAs is cis-, rather than trans-, gene regulation implicates a role as decoys for miRNAs. This is further exemplified by a study showing that linc-MD1, a muscle-specific lncRNA, absorbs miR-133 to regulate the expression of transcription factors that activate muscle-specific gene expression such as MAML1 and MEF2C (22). Moreover, linc-MD1 correlated strongly with aberrant muscle loss, as is seen in patients with Duchenne muscular dystrophy (23). In a cardiac setting, Wang et al. reported that cardiac apoptosis-related lncRNA (CARL) indirectly regulated mitochondrial fission and apoptosis, via an endogenous miR-539 sponge and thereby regulates PHB2 expression (24).
To assess whether the cardiac transcriptome, in particular the novel lncRNAs, correlated with the contractile and remodeling parameters of the heart in vivo, the cardiac transcriptome was associated with physiological traits in human patients. Though the novel reported lncRNAs correlated better with all physiological traits assessed, clustering for both coding transcripts and novel lncRNA indicated a group of related lncRNAs that correlated positively with cardiac function and negatively with remodeling parameters. The complexity to delineate how these particular physiological traits contribute to cardiac function and the function of lncRNAs herein is underscored by a single cluster showing diverse correlation to these interlinked functional parameters. Moreover, determining critical regulatory elements of transcriptional activity such as distinct chromatin states and methylation and acetylation of histone lysine residues is a major advance in understanding transcriptional regulation of lncRNAs. In line, although the novel lncRNAs were associated with more active chromatin states, their expressions showed to be more dependent on a canonical promoter signature rather than an exclusively active enhancer signature.
Although the lncRNAs complied with the UCSC annotation and have typical lncRNA physiognomies, validation of these transcripts is necessary to filter out false reads. The authors used qRT-PCR to validate a select group that was associated with functional parameters and to determine timely and regional cardiac expression. In contrast to mRNAs, lncRNA themselves must physically localize to their particular site of action, making knowledge of lncRNA subcellular localization patterns vital for insights into their biology and function. Nuclear and cytoplasmic fractionation could indicate subcellular enrichment of an lncRNA and give insights into the functionality of the lncRNA. Indeed, an exclusive nuclear localization would argue against putative short peptide sequences encoded by the lncRNA, since translation occurs in the cytoplasm. However, to fully delineate the exact role of an lncRNA, further analysis on localization to particular subcellular areas within the cytosol or nucleus may lead to better insights on the role of an lncRNA. For example, lncRNAs such as NEAT1 and MALAT1 have been shown to localize to nuclear bodies within the nucleus and the lncRNA GAS5 was shown to shuttle between the nucleus and cytoplasm (25,26). More elegantly, Clemson and colleagues showed that the lncRNA XIST specifically accumulates on the inactive X-chromosome (27). Hence, RNA fluorescence in situ hybridization (RNA FISH) could better address these questions and reveal potential mechanisms for the described novel lncRNAs. Moreover, this technique would directly discriminate the cell type presence in situ of these novel lncRNA in the injured myocardium instead of the laborious and indirect detection in isolated different cell types
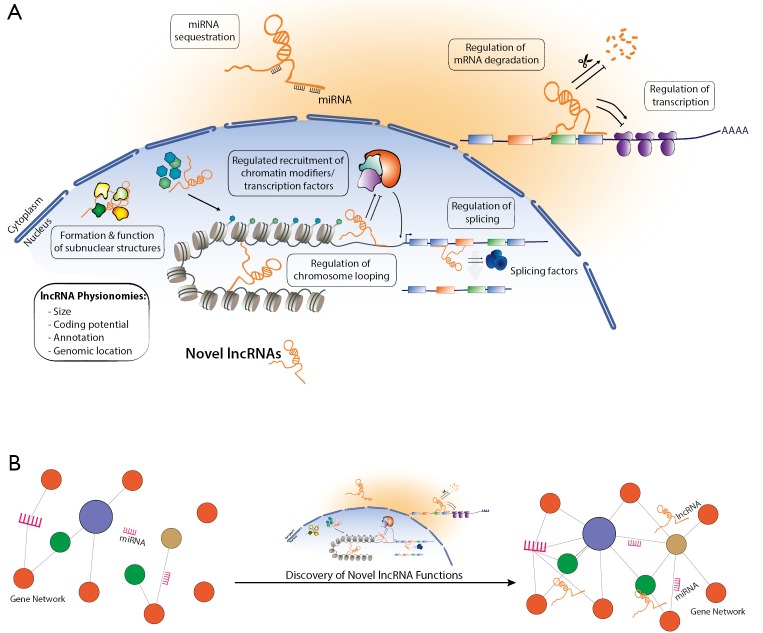
For a long time heart function was considered to be orchestrated through regulatory networks that are composed solely of a protein-mediated transcriptional control and signaling pathways, and which were perturbed during cardiac disease. In line, efforts for therapeutical interventions were systematized around these networks with little translational outcomes so far. Clearly the path of lncRNAs from background noise and serendipitous transcription to key regulators has led to significant insights into gene regulatory networks (Figure 1). This previously hidden layer of gene regulation underpins the sophisticated regulatory networks to not only coordinate physiological responses and environmental adaptation, but also pathophysiological responses in multifaceted heart diseases. In order to take the next step towards modulation of lncRNA levels as an approach for tackling cardiac disease, reviewing their function and role in cardiac biology is of utmost importance. Therefore, efforts to unveil the multiple lncRNAs and their role in cardiac disease, exemplified by the study of Ounzain and colleagues, provides an excellent toolkit to assist in exploring the possibilities for ranging from biomarkers of disease to targets for therapeutical intervention.
Figure 1.
Long non-coding RNAs (lncRNAs) and their functions. (A) Possible functional mechanisms for the novel lncRNAs; (B) lncRNAs as an additional layer of controllers of gene regulatory networks.
Acknowledgements
Funding: This work was supported by the PLN foundation and a HUSTCARE grant (CVON2011-12) from the Netherlands CardioVascular Research Initiative (CVON): the Dutch Heart Foundation, Dutch Federation of University Medical Centers, the Netherlands Organization for Health Research and Development, and the Royal Netherlands Academy of Sciences.
Footnotes
Provenance: This is a Guest Perspective commissioned by Section Editor Zhijun Han, MD (Department of Laboratory Medicine, Wuxi Second Hospital, Nanjing Medical University, Wuxi, China).
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Kehat I, Molkentin JD. Molecular pathways underlying cardiac remodeling during pathophysiological stimulation. Circulation 2010;122:2727-35. 10.1161/CIRCULATIONAHA.110.942268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheuermann JC, Boyer LA. Getting to the heart of the matter: long non-coding RNAs in cardiac development and disease. EMBO J 2013;32:1805-16. 10.1038/emboj.2013.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 2013;14:100-12. 10.1038/nrg3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell 2009;136:656-68. 10.1016/j.cell.2009.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sluijter JP. MicroRNAs in Cardiovascular Regenerative Medicine: Directing Tissue Repair and Cellular Differentiation. Available online: http://dx.doi.org/ 10.1155/2013/593517 [DOI]
- 6.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014;15:7-21. 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- 7.Zhu YG, Feng XM, Abbott J, et al. Human mesenchymal stem cell microvesicles for treatment of Escherichia coli endotoxin-induced acute lung injury in mice. Stem Cells 2014;32:116-25. 10.1002/stem.1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinger ME, Pang KC, Mercer TR, et al. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol 2008;4:e1000176. 10.1371/journal.pcbi.1000176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet 2013;9:e1003569. 10.1371/journal.pgen.1003569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper C, Vincett D, Yan Y, et al. Steroid Receptor RNA Activator bi-faceted genetic system: Heads or Tails? Biochimie 2011;93:1973-80. 10.1016/j.biochi.2011.07.002 [DOI] [PubMed] [Google Scholar]
- 11.Sauvageau M, Goff LA, Lodato S, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2013;2:e01749. 10.7554/eLife.01749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trajkovski M, Hausser J, Soutschek J, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011;474:649-53. 10.1038/nature10112 [DOI] [PubMed] [Google Scholar]
- 13.O'Brien JE, Jr, Kibiryeva N, Zhou XG, et al. Noncoding RNA expression in myocardium from infants with tetralogy of Fallot. Circ Cardiovasc Genet 2012;5:279-86. 10.1161/CIRCGENETICS.111.961474 [DOI] [PubMed] [Google Scholar]
- 14.Ritter O, Haase H, Schulte HD, et al. Remodeling of the hypertrophied human myocardium by cardiac bHLH transcription factors. J Cell Biochem 1999;74:551-61. [DOI] [PubMed] [Google Scholar]
- 15.Carlock LR, Skarecky D, Dana SL, et al. Deletion mapping of human chromosome 5 using chromosome-specific DNA probes. Am J Hum Genet 1985;37:839-52. [PMC free article] [PubMed] [Google Scholar]
- 16.Ishii N, Ozaki K, Sato H, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 2006;51:1087-99. 10.1007/s10038-006-0070-9 [DOI] [PubMed] [Google Scholar]
- 17.Lee JH, Gao C, Peng G, et al. Analysis of transcriptome complexity through RNA sequencing in normal and failing murine hearts. Circ Res 2011;109:1332-41. 10.1161/CIRCRESAHA.111.249433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang KC, Yamada KA, Patel AY, et al. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 2014;129:1009-21. 10.1161/CIRCULATIONAHA.113.003863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambardekar AV, Buttrick PM. Reverse remodeling with left ventricular assist devices: a review of clinical, cellular, and molecular effects. Circ Heart Fail 2011;4:224-33. 10.1161/CIRCHEARTFAILURE.110.959684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alcalay J, Ullrich SE, Kripke ML. Local suppression of contact hypersensitivity in mice by a monofunctional psoralen plus UVA radiation. Photochem Photobiol 1989;50:217-20. 10.1111/j.1751-1097.1989.tb04151.x [DOI] [PubMed] [Google Scholar]
- 21.Ounzain S, Micheletti R, Beckmann T, et al. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur Heart J 2015;36:353-68a. 10.1093/eurheartj/ehu180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011;147:358-69. 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legnini I, Morlando M, Mangiavacchi A, et al. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol Cell 2014;53:506-14. 10.1016/j.molcel.2013.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K, Long B, Zhou LY, et al. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun 2014;5:3596. [DOI] [PubMed] [Google Scholar]
- 25.Ip JY, Nakagawa S. Long non-coding RNAs in nuclear bodies. Dev Growth Differ 2012;54:44-54. 10.1111/j.1440-169X.2011.01303.x [DOI] [PubMed] [Google Scholar]
- 26.Puig JG, Michán AD, Jiménez ML, et al. Female gout. Clinical spectrum and uric acid metabolism. Arch Intern Med 1991;151:726-32. 10.1001/archinte.1991.00400040074016 [DOI] [PubMed] [Google Scholar]
- 27.Clemson CM, McNeil JA, Willard HF, et al. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol 1996;132:259-75. 10.1083/jcb.132.3.259 [DOI] [PMC free article] [PubMed] [Google Scholar]