Abstract
Alterations in the expression of microRNAs (miRNAs) have been proposed to play a role in the pathogenesis of acute lymphoblastic leukemia (ALL) and chronic lymphocytic leukemia (CLL). Dicer is one of the main regulators of miRNA biogenesis, and deregulation of its expression has been indicated as a possible cause of miRNA alterations observed in various cancers. Our aim was to analyze the expression of the Dicer protein and its relationship with ALL and CLL. This cross-sectional study was performed from 2010 to 2012 in Shahid Faghihi Hospital, Shiraz, Iran. In this study, 30 patients with CLL, 21 patients with ALL, 10 child healthy donors, and 19 adult healthy donors were recruited. The patients’ samples were checked via flow cytometry, immunohistochemistry, and immunocytochemistry. The controls’ samples were also examined in the hematology ward. Total RNA was extracted from the bone marrow and peripheral blood samples of the patients and controls. Then, reverse-transcription polymerase chain reaction was used to estimate the level of Dicer miRNA. The outcomes of the expression analysis of Dicer revealed statistically significant differences between the ALL patients/child healthy controls (mean±SD, 0.19±0.28 vs. 0.73±0.12; P<0.001) and the CLL patients/adult healthy controls (mean±SD, 0.24±0.25 vs. 0.41±0.28; P=0.033). This is the first piece of evidence showing that the expression of the Dicer gene greatly decreased in the patients with ALL in comparison to the child controls. The expression of the Dicer gene was also downregulated in the patients with CLL compared to the adult controls. Given the above findings, the expression of Dicer may play an important role in the progression and prognosis of these diseases.
Keywords: DICER1 protein, Gene expression, Leukemia
What’s Known
The downregulation of various molecules and signaling pathways have been implicated in the pathogenesis of ALL and CLL; nevertheless, their exact pathogenesis has not been completely explained.
The discovery of microRNAs (miRNAs) has brought a new insight into the pathogenesis of many diseases, including ALL and CLL.
What’s New
We found that the expression level of the Dicer gene in patients with ALL and CLL was significantly lower than that in child and adult normal controls, indicating that the abnormal expression of the Dicer gene may be involved in the development of ALL and CLL. In ALL, this is a new finding.
Introduction
Leukemia is a heterogeneous group of cancers caused by neoplastic transformation in blood cell progenitors during their differentiation in the bone marrow. Acute lymphoblastic leukemia (ALL) and chronic lymphocytic leukemia (CLL) are major subtypes of leukemia in most cancer registries.1 ALL progresses rapidly without treatment. Although it is the most common type of leukemia diagnosed in children, 49% of the cases are diagnosed in patients aged 20 years and older.2 The American Cancer Society3 estimated 6,020 new cases and 1,440 deaths from ALL in the United States for 2014. Today, ALL is still the sixth most common malignancy in Iran.4 The risk factors associated with the development of this malignancy include environmental, genetic, and infectious factors. Identifying the risk factors for ALL is an important step in the reduction of the overall burden of the disease.5 CLL usually progresses slowly. It is the most common type of leukemia in adults in Western countries, with 95% of the cases occurring in those aged 50 and older. CLL is more common in men6 with a male-to-female ratio of approximately 1.7:1. It was predicted that there would be 15,720 new cases of CLL in 2014, 9,100 cases in men and 6,620 cases in women.7 By 2010, the incidence rate of CLL had reached 23.9% in Iran.8 There are very few known risk factors for CLL. Some studies have shown that exposure to certain chemical materials, family history, gender, and ethnicity can increase the risk of CLL.3
The downregulation of various molecules and signaling pathways have been implicated in the pathogenesis of ALL and CLL; nevertheless, their exact pathogenesis has not been completely explained. The discovery of microRNAs (miRNAs) has brought a new insight into the pathogenesis of many diseases, including ALL and CLL. With the ability to regulate gene expression at the post-transcriptional level and to become involved in important biological processes, miRNAs are small non-coding RNAs. There is increasing evidence that the various abnormal expressions of multiple miRNAs can play a critical role in ALL and CLL.9, 10 However, the reasons cited for the alterations in the expression of miRNAs found in ALL and CLL have been varied. One possible theory suggests the defect of the miRNA biogenesis machinery involving Dicer and other key regulators of miRNA.11 Dicer is a cytoplasmic endonuclease, a fundamental protein component of the miRNA loading complex (miRLC), with a key role in the production of miRNAs and small interfering RNAs which inhibits gene expression by causing the destruction of specific miRNA molecules. This protein is encoded by DICER1 gene, which is located on chromosome 14 (14q32.13).12 It has been reported that a decreased expression of Dicer is associated with greater invasiveness and poorer survival in ovarian cancer.13 Similarly, low levels of Dicer and other regulators of miRLC have been observed in high-risk neuroblastomas with a poor outcome.14,15
The aim of the present study was to investigate altered levels of Dicer miRNA in patients with ALL and CLL.
Patients and Methods
This cross-sectional study was done from 2010 to 2012 in the Molecular Pathology and Cytogenetic Ward of Shahid Faghihi Hospital, Shiraz, Iran. The study included 80 samples: 30 previously untreated patients with CLL, 21 untreated patients with ALL, 10 child controls, and 19 adult controls. All the patients provided informed consent, and the study was approved by the local ethics committee of the hospitals affiliated to Shiraz Medical University. Diagnosis and response were based on the criteria recommended by the revised criteria of the National Cancer Institute (NCI).16 All the patients were referred to our ward by oncologists. The diagnosis was based on peripheral smear and bone marrow examination subsequently confirmed with molecular study. Specimens with a total white blood cell count of 20,000 to 30,000 were chosen, and more than 70% of them were leukemic cells. The patients with CLL had at least 5 × 109/L B lymphocytes with expressions of monoclonal surface immunoglobulin, CD20, CD5, CD19, and CD23. Patients under treatment were excluded from the current study. Definite diagnosis in all the patients was established based on morphology, immunocytochemistry, immunohistochemistry, and flow cytometry.
The normal controls’ samples were checked using cell counter devices (Sysmex XS-800i, Japan), and peripheral smears (all of them normal) were made from them. A description of the study population by gender and age is depicted in table 1.
Table 1.
Information on the patients and controls
| Variables | Cases-ALL N, 21 | Child controls N, 10 | P value | CLL | Cases-CLL N, 30 | Adult controls N, 19 | P value |
|---|---|---|---|---|---|---|---|
| Gender | |||||||
| Female | 7 (33.33%) | 4 (40%) | 0.71 | 12 (40%) | 8 (42.1%) | 0.88 | |
| Male | 14 (66.67%) | 6 (60%) | 0.56 | 18 (60%) | 11 (57.9%) | 0.11 | |
| Age | |||||||
| 1-10 | 13 | 6 | 1-21 | 0 | 0 | ||
| 10-20 | 5 | 4 | 21-42 | 2 | 5 | ||
| 20-30 | 2 | 0 | 42-63 | 14 | 9 | ||
| 30-40 | 1 | 0 | 63-84 | 14 | 5 |
ALL: Acute lymphoblastic leukemia; CLL: Chronic lymphocytic leukemia
Quantitative Reverse Transcription Polymerase Chain Reaction (Qrt-Pcr) Analysis for Dicer Mirna Expresion
The leukemic cells were separated by Ficoll-Histopaque to the bottom of Falcon tubes. Total RNA was extracted from the bone marrow and peripheral blood mononuclear cells using AccuZol (Bioneer, Seoul, South Korea) following the manufacturer’s protocol. The quality and quantity of RNA were assessed with a smart spectrophotometer (Bio-Rad, Berkeley, CA, U.S.A.). Retrotranscription reaction was performed with a High-Capacity cDNA Reverse Transcription Kit (Fermentas, Waltham, Massachusetts, U.S.A.) according to the manufacturer’s recommendations. Comparability between the quantitative polymerase chain reaction (qPCR) assays was ensured by obtaining standard curves through quantitative reverse transcription polymerase chain reaction (qRT-PCR) amplification of the serial dilutions of cDNA templates. Quantitative real-time PCR was performed using a SYBR Kit (Bioneer, Seoul, South Korea) on an AB15700 sequence detection system (Applied Biosystems, Foster City, CA, U.S.A.) using the primers shown in table 2. In a total volume of 25 µL of the reaction mixture, 5 µL of complementary DNA templates was mixed with 12.5 µL of SYBR Green PCR Master Mix (Bioneer, Seoul, South Korea) and each pair of primers at a final concentration of 200 nM. Reactions were run with the following thermal cycling parameters: 95 °C for 15 minutes followed by 50 cycles of 95 °C for 30 seconds (denaturation) and 60 °C for 60 seconds, melting curve program (60 to 95 °C) with a heating rate of 0.1 °C/s. The miRNA expression levels of ABL were used as an internal standard.
Table 2.
Real-time primer sequences and annealing temperature
| Genes | Sequence (5′→3′) | Product size (bp) | Annealing temperature (°C) |
|---|---|---|---|
| Dicer | F: CCCGGCTGAGAGAACTTACG R: CTGTAACTTCGACCAACACCTTTAAA |
103 | 60 |
| ABL | F: TGGAGATAACACTCTAAGCATAACTAA AGGT R: GATGTAGTTGCTTGGGACCCA |
117 | 60 |
Statistical Analysis
The relative expression of Dicer in the four groups was calculated via the 2-∆∆ct method: ∆ct=ct(Dicer)-ct(ABL). The efficiency of both of the Dicer and ABL genes was between 98 and 102%. The Mann–Whitney U-test was used to identify statistically significant differences between the groups. All the statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS), version 19.0 (SPSS, Chicago, IL). The significance level was set at a P≤0.05.
Results
All the 21 patients with ALL, 10 child controls, 30 patients with CLL, and 19 adult controls were analyzed. The diagnoses were confirmed using conventional methods such as flow cytometry. No effort was spared to detect any change in the expression of Dicer in the patients. The age of the patients with ALL was between 1 and 35 years with a median of 9.5 years, the age of the child controls was between 5 and 14 years with a median of 9.5 years, the age of the patients with CLL was between 40 and 84 years with a median of 62.9 years, and the age of the adult controls was between 25 and 64 years with a median of 51.1 years. The expression of the Dicer and ABL genes in the four groups was observed using qPCR. Taking ABL as a control sample, the relative expression levels of Dicer in the ALL patients/child controls and the CLL patients/adult controls were analyzed. There was a significant statistical difference between the ALL patients/child controls and the CLL patients/adult controls. The results of the real-time PCR analysis were confirmed on 2% agarose gel via electrophoresis and visualized with ethidium bromide staining (figure 1). The results of the expression analysis of the Dicer gene in the ALL patients/child controls and the CLL patients/adult controls are shown in table 3 and figure 2.
Figure 1.
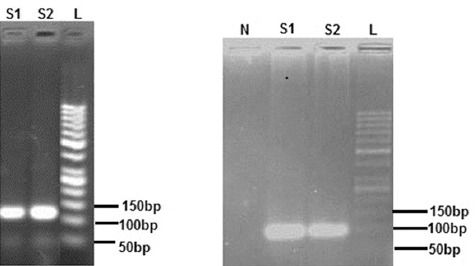
These pictures show the results of the real-time polymerase chain reaction (PCR) analysis on agarose gel. a) ABL gene amplification. S1: Patients with acute lymphoblastic leukemia (ALL); S2: Patients with chronic lymphocytic leukemia (CLL); L: Ladder. b) Dicer gene amplification. N: Control negative; S1: Patients with ALL; S2: Patients with CLL; L: Ladder.
Table 3.
Comparison of relative gene expression for the Dicer gene between the ALL patients/child controls and the CLL patients/adult controls
| Gene | Group | N | Mean±SD | Range | P value |
|---|---|---|---|---|---|
| Dicer | ALL | 21 | 0.19±0.28 | 0.00±0.93 | <0.001 |
| Child controls | 10 | 0.73±0.12 | 0.56±0.94 | ||
| CLL | 30 | 0.24±0.25 | 0.00±0.93 | 0.033 | |
| Adult controls | 19 | 0.41±0.28 | 0.01±0.88 |
*Mann–Whitney test, SD: Standard deviation
Figure 2.
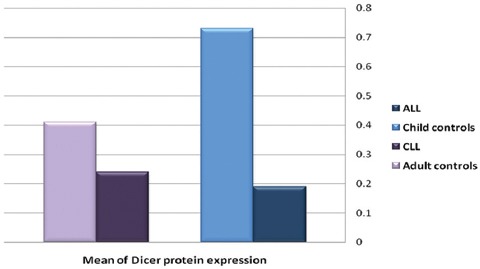
It depicts the frequency of the expression of the Dicer protein in the patients with acute lymphoblastic leukemia (ALL) and chronic lymphocytic leukemia (CLL) and the child/adult controls. The expression mean of the Dicer protein in the patients with ALL was lower than that of the child controls. The expression level of this protein also showed a decrease in the patients with CLL compared with the adult controls.
Discussion
There are several dysregulations of miRNAs in ALL and CLL. The downregulation of miR-15a/16-1 because of a genomic deletion in patients with CLL showed a relationship between disordered miRNA expression and developed hematopoietic cancer. Mice with this deletion expanded CLL or another leukemia type, probably by increasing B-cell proliferation via regulating cell cycle-associated genes.17 Increased B-cell proliferation was also considered in miR-155 transgenic mice, which finally developed ALL and high-grade lymphoma. These studies demonstrate that the deregulated expression of miRNAs may contribute to leukemogenesis through disturbing the strong control of normal hematopoietic processes.18 The miRNA dysregulation may also be caused by different reasons. One reason that was also investigated in the present study is the dysregulation of the Dicer gene. Dicer is one of the key components of miRLC. The synthesis of mature endogenous interfering RNAs involves a cascade of events that are inescapably linked to the functions of Dicer and other components of miRLC. The erosion of Dicer fails the generation of most mature miRNAs. Koralov et al.19 showed that the erosion of Dicer prevented the differentiation of pro-B cells to pre-B cells and meaningfully modified antibody diversity through increasing the diversification of Igк variable regions and changing DH element usage of IgH variable regions. Afterward, Xu et al.20 demonstrated the role of Dicer in regulating B-cell terminal differentiation through a specific deletion of Dicer in murine antigen-activated B cells. Remarkably, after immunization with antigen, these mice were not capable of producing high-affinity class-switched antibodies, memory B cells, and long-lived plasma cells. The results of a cohort study by Zhu et al.11 chime in with our study in so far as the downregulation of Dicer decreased the survival of patients with CLL; nonetheless, the authors extended their work with a karyotype study of their patients and role of cytogenetic change in CLL prognosis. Grelier et al.21 examined 104 patients with breast cancer and showed that the downregulation of Dicer decreased 5-year survival. The authors also demonstrated the downregulation of the Dicer gene in mesenchymal breast cancer. Therefore, Dicer is essential for functional miRNAs, which perform many cellular processes. Based on the above results, the dysregulation of Dicer in CLL can affect miRNAs, which play a crucial role in the regulation of T and B lymphocytes’ terminal differentiation. We also found patients with CLL in the south of Iran, which is in line with other studies showing a decreased expression of Dicer. Nevertheless, the current literature contains no evidence apropos patients with ALL; we seem to be the first to reveal a decreased expression of Dicer also in patients with ALL. The findings of the present study showed that the relative expression of Dicer significantly decreased in the patients with CLL compared to the healthy controls. Our finding concerning the downregulation of Dicer in CLL is consistent with several recent reports showing that the expression of Dicer is significantly reduced in a variety of human cancers, including breast cancer, prostate cancer, ovarian cancer, and neuroblastomas. For example, Wu et al.15 reported that the Dicer miRNA level was significantly lower in hepatocellular carcinoma corresponding to nonneoplastic liver tissues, but the expression level of this gene was not associated with clinical characteristics. Interestingly, we found a low expression of Dicer also in our patients with ALL, which has not been previously mentioned in the literature. In another study, it was demonstrated that the downregulation of Dicer in CLL resulted in a strong association with shorter overall survival and with reduced treatment-free survival.11 Moreover, the findings of a study by Zighelboim et al.22 showed that lower DICER1 transcript levels were associated with disease recurrence and worse disease-free survival in patients with endometrioid endometrial carcinoma. He et al.23 studied 90 patients with cervical cancer and found that the Dicer miRNA levels in 90 cervical cancer tissues decreased by comparison with normal cervix tissues. Additionally, they found that low expressions of the Dicer miRNA and protein correlated with a poor prognosis and relapse (including distance metastasis) of cervical cancer. Compared to our study, they found that a decreased expression of Dicer could be a cause of cancer.
All the above-mentioned studies are concordant with the current study with regard to miRNA levels, but they did not mention the expression of Dicer in ALL. However, based on previous studies, Dicer appears to play a tumor suppressor role. There is evidence for this notion. First, the ablation of Dicer enhanced tumor development in a K-ras-induced mouse model of lung cancer.24 Second, a low expression of Dicer was able to augment neuroblastoma tumor proliferation.14 Moreover, aggressive tumors are also thought to be able to decrease total miRNA levels and contribute to their poor differentiation, suggesting a decreased rather than an increased expression of Dicer in aggressive cancers.24 So far, the mechanism that regulates the expression of Dicer has remained unclear. One probable reason may be linked with the Dicer gene location at the subtelomeric region on chromosome 14 (14q32.13), which was detected to be affected by allelic deletion in several tumors, including endometrial cancer, lung cancer, primary neuroblastomas, chronic myelogenous leukemia, and lymphoma.25-28 The other reason for the altered expression of Dicer is genomic mutations in this gene. The mutations of the Dicer gene have been detected in ovarian cancer and in familial multinodular goiter.13,29 Furthermore, epigenetic mechanisms may constitute another possible reason. Pampalakis et al.30 reported DNA methylation as a probable mechanism for the downregulation of Dicer based on the presence of the strong CpG island spanning the first exon of the gene. In light of the results of the present study and those reported previously, the Dicer gene functions as a cancer suppressor and the downregulation of this gene produces various cancers. There was no limitation to the current study.
Conclusion
Our findings demonstrated that the expression level of the Dicer gene in the patients with ALL and CLL was significantly lower than that in the child and adult normal controls, indicating that the abnormal expression of the Dicer gene may be involved in the development of ALL and CLL. The above results indicated that the downregulation of the Dicer gene is a frequent event in ALL and CLL and that it may be associated with clinicopathologic features. Our study adds new data to the variable expression patterns of Dicer in different tumor types, especially ALL.
Conflict of Interest: None declared.
References
- 1.Cancer Treatment and Survivorship Facts & Figures 2014-2015 [Internet] American Cancer Society. [cited 2014 Dec 8]. Available from: http://www.cancer.org/acs/groups/content/@research/documents/document/acspc-042801.pdf .
- 2.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–43. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Facts & Figures 2014 [Internet] American Cancer Society. [Cited 2014 Dec 8]. Available from: http://www.cancer.org/acs/groups/content/@research/documents/webcontent/acspc-042151.pdf .
- 4.Mehrabani D, Tabei S, Heydari S, Shamsina S, Shokrpour N, Amini M, et al. Cancer occurrence in Fars Province, Southern Iran. Iran Red Crescent Med J. 2008;10:314–22. [Google Scholar]
- 5.Belson M, Kingsley B, Holmes A. Risk factors for acute leukemia in children: a review. Environ Health Perspect. 2007;115:138–45. doi: 10.1289/ehp.9023. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang S, Kipps TJ. The pathogenesis of chronic lymphocytic leukemia. Annu Rev Pathol. 2014;9:103–18. doi: 10.1146/annurev-pathol-020712-163955. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 8.Mozaheb Z. Epidemiology of Lymphoid Malignancy in Asia. Croatia: INTECH Open Access Publisher; 2012. [Google Scholar]
- 9.Xu L, Liang YN, Luo XQ, Liu XD, Guo HX. Association of miRNAs expression profiles with prognosis and relapse in childhood acute lymphoblastic leukemia. Zhonghua Xue Ye Xue Za Zhi. 2011;32:178–81. [PubMed] [Google Scholar]
- 10.Moussay E, Wang K, Cho JH, van Moer K, Pierson S, Paggetti J, et al. MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2011;108:6573–8. doi: 10.1073/pnas.1019557108. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu DX, Fan L, Lu RN, Fang C, Shen WY, Zou ZJ, et al. Downregulated Dicer expression predicts poor prognosis in chronic lymphocytic leukemia. Cancer Sci. 2012;103:875–81. doi: 10.1111/j.1349-7006.2012.02234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji X. The mechanism of RNase III action: how dicer dices. Curr Top Microbiol Immunol. 2008;320:99–116. doi: 10.1007/978-3-540-75157-1_5. [DOI] [PubMed] [Google Scholar]
- 13.Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–50. doi: 10.1056/NEJMoa0803785. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin RJ, Lin YC, Chen J, Kuo HH, Chen YY, Diccianni MB, et al. microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res. 2010;70:7841–50. doi: 10.1158/0008-5472.CAN-10-0970. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu JF, Shen W, Liu NZ, Zeng GL, Yang M, Zuo GQ, et al. Down-regulation of Dicer in hepatocellular carcinoma. Med Oncol. 2011;28:804–9. doi: 10.1007/s12032-010-9520-5. [DOI] [PubMed] [Google Scholar]
- 16.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–56. doi: 10.1182/blood-2007-06-093906. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Costinean S, Sandhu SK, Pedersen IM, Tili E, Trotta R, Perrotti D, et al. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood. 2009;114:1374–82. doi: 10.1182/blood-2009-05-220814. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–74. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 20.Xu S, Guo K, Zeng Q, Huo J, Lam KP. The RNase III enzyme Dicer is essential for germinal center B-cell formation. Blood. 2012;119:767–76. doi: 10.1182/blood-2011-05-355412. [DOI] [PubMed] [Google Scholar]
- 21.Grelier G, Voirin N, Ay AS, Cox DG, Chabaud S, Treilleux I, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. 2009;101:673–83. doi: 10.1038/sj.bjc.6605193. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zighelboim I, Reinhart AJ, Gao F, Schmidt AP, Mutch DG, Thaker PH, et al. DICER1 expression and outcomes in endometrioid endometrial adenocarcinoma. Cancer. 2011;117:1446–53. doi: 10.1002/cncr.25665. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He L, Wang HY, Zhang L, Huang L, Li JD, Xiong Y, et al. Prognostic significance of low DICER expression regulated by miR-130a in cervical cancer. Cell Death Dis. 2014;5:e1205. doi: 10.1038/cddis.2014.127. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 25.Chiosea S, Jelezcova E, Chandran U, Luo J, Mantha G, Sobol RW, et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–50. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 26.Fujino T, Risinger JI, Collins NK, Liu FS, Nishii H, Takahashi H, et al. Allelotype of endometrial carcinoma. Cancer Res. 1994;54:4294–8. [PubMed] [Google Scholar]
- 27.Sercan HO, Sercan ZY, Kizildag S, Undar B, Soydan S, Sakizli M. Consistent loss of heterozygosity at 14Q32 in lymphoid blast crisis of chronic myeloid leukemia. Leuk Lymphoma. 2000;39:385–90. doi: 10.3109/10428190009065838. [DOI] [PubMed] [Google Scholar]
- 28.Agueli C, Cammarata G, Salemi D, Dagnino L, Nicoletti R, La Rosa M, et al. 14q32/miRNA clusters loss of heterozygosity in acute lymphoblastic leukemia is associated with up-regulation of BCL11a. Am J Hematol. 2010;85:575–8. doi: 10.1002/ajh.21758. [DOI] [PubMed] [Google Scholar]
- 29.Rio Frio T, Bahubeshi A, Kanellopoulou C, Hamel N, Niedziela M, Sabbaghian N, et al. DICER1 mutations in familial multinodular goiter with and without ovarian Sertoli-Leydig cell tumors. JAMA. 2011;305:68–77. doi: 10.1001/jama.2010.1910. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pampalakis G, Diamandis EP, Katsaros D, Sotiropoulou G. Down-regulation of dicer expression in ovarian cancer tissues. Clin Biochem. 2010;43:324–7. doi: 10.1016/j.clinbiochem.2009.09.014. [DOI] [PubMed] [Google Scholar]


