Abstract
Purpose
Although CD49d is an unfavorable prognostic marker in chronic lymphocytic leukemia (CLL), definitive validation evidence is lacking. A worldwide multicenter analysis was performed using published and unpublished CLL series to evaluate the impact of CD49d as an overall (OS) and treatment-free survival (TFS) predictor.
Patients and Methods
A training/validation strategy was chosen to find the optimal CD49d cutoff. The hazard ratio (HR) for death and treatment imposed by CD49d was estimated by pooled analysis of 2,972 CLLs; Cox analysis stratified by center and stage was used to adjust for confounding variables. The importance of CD49d over other flow cytometry–based prognosticators (eg, CD38, ZAP-70) was ranked by recursive partitioning.
Results
Patients with ≥ 30% of neoplastic cells expressing CD49d were considered CD49d+. Decrease in OS at 5 and 10 years among CD49d+ patients was 7% and 23% (decrease in TFS, 26% and 25%, respectively). Pooled HR of CD49d for OS was 2.5 (2.3 for TFS) in univariate analysis. This HR remained significant and of similar magnitude (HR, 2.0) in a Cox model adjusted for clinical and biologic prognosticators. Hierarchic trees including all patients or restricted to those with early-stage disease or those age ≤ 65 years always selected CD49d as the most important flow cytometry–based biomarker, with negligible additional prognostic information added by CD38 or ZAP-70. Consistently, by bivariate analysis, CD49d reliably identified patient subsets with poorer outcome independent of CD38 and ZAP-70.
Conclusion
In this analysis of approximately 3,000 patients, CD49d emerged as the strongest flow cytometry–based predictor of OS and TFS in CLL.
INTRODUCTION
Chronic lymphocytic leukemia (CLL) shows a highly variable clinical course,1 which can be predicted by a number of biologic markers, including the mutational status of immunoglobulin heavy variable (IGHV) gene,2–4 genomic abnormalities,5 and expression of the surface marker CD382,6 and of the intracytoplasmic protein ZAP-70.7–9
CD49d, the α4 subunit of the integrin heterodimer α4β1, is a surface molecule, the expression of which promotes microenvironment-mediated proliferation of CLL leukemic cells10–13 and identifies a subgroup of patients characterized by progressive course and short survival.14–20 To translate CD49d expression assessment at the clinical level, a number of open issues remain to be clarified, including: one, definition and validation of the optimal CD49d cutoff for predicting CLL survival and progression; two, robustness of CD49d expression as a prognostic biomarker when challenged against potential confounding variables; and three, estimation of the relative prognostic power of CD49d in comparison with CD38 and ZAP-70, two flow cytometry–based biomarkers widely used for CLL outcome stratification.
To address these issues, we carried out a worldwide multicenter initiative to evaluate the prognostic utility of CD49d in CLL, which combined individual patient data (IPD) from all previously reported studies with a large unpublished validation series.
PATIENTS AND METHODS
Identification of Previous Studies and Collection of Data
A Medline search (http://www.ncbi.nlm.nih.gov/pubmed/) was performed on April 30, 2011, using the terms “CD49d” AND “chronic lymphocytic leukemia.” References of all the retrieved articles were also scanned for additional relevant citations. Abstracts of American Society of Hematology annual meetings (years 2006 to 2010) were also screened for “CD49d” OR “integrin” keywords. A total of 44 studies and 70 abstracts were retrieved through Medline search and manual screening, respectively. Studies were included in the pooled analysis if they met the following criteria: one, included at least 50 CLLs; two, evaluated CD49d expression of CLL cells by flow cytometry; three, reported the association between CD49d expression and survival (overall survival [OS]) and/or disease progression (treatment-free [TFS] and/or progression-free survival [PFS]). To avoid inclusion of duplicated data, patient series published multiple times were considered only once.
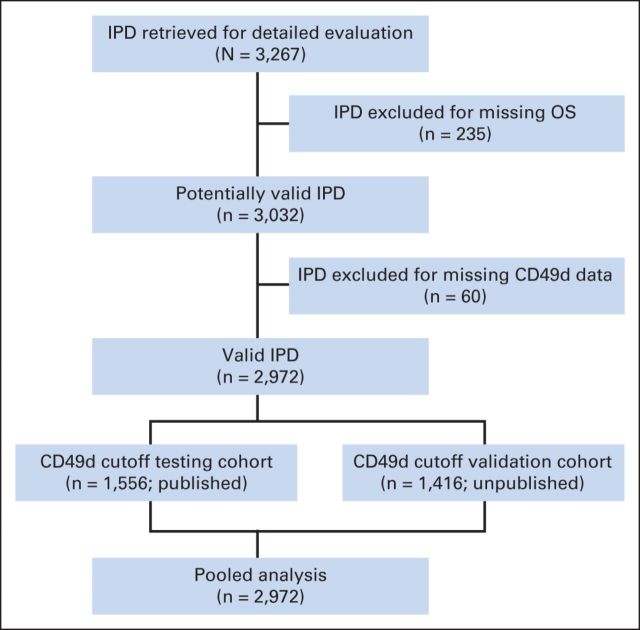
Overall, eight published studies (seven full-length reports and one abstract) were identified for inclusion in pooled analysis, all fulfilling the eligibility criteria and reporting independent results. Median quality score among the published full-length studies was judged high (median value, 5; range, 5 to 7). Of these studies, two were prospective,15,19 two recruited consecutive patients,16,20 and one recruited patients from multiple centers.20 Corresponding authors were contacted and invited to participate. In May 2011, the proposed analysis plan was shared with all authors, and IPD were requested. All authors agreed to send IPD in spreadsheet format, which were merged after variable renaming and/or recoding as needed. Variables are listed in Table 1. Contributors from seven clinical centers located in either the United States (two centers) or the European Union (three centers in Italy, one in Germany, and one in United Kingdom) provided IPD from the published cohorts. Additionally, four centers added updated follow-up and unpublished data on additional patients. The final data set included IPD from 3,267 patients. IPD of 295 patients were excluded for missing data (235 missing end point, 60 missing CD49d). IPD of the remaining 2,972 patients with CLL were included in the final analysis (1,556 published; 1,416 unpublished; CONSORT diagram shown in Fig 1).
Table 1.
Patients Demographic and Clinical Characteristics
| Characteristic | Published Cohort (n = 1,556) |
Unpublished Cohort (n = 1,416) |
||
|---|---|---|---|---|
| No. | % | No. | % | |
| Age, years | ||||
| Median | 64 | 63 | ||
| IQR | 56-71 | 55-71 | ||
| Male sex | 961 | 62 | 898 | 63 |
| Year of diagnosis | ||||
| Median | 2003 | 2007 | ||
| IQR | 1998-2005 | 2002-2009 | ||
| Follow-up, years* | ||||
| Median | 6.9† | 4.0† | ||
| Range | 0-36 | 0-43 | ||
| Deaths | 324† | 21 | 171† | 12 |
| Stage‡ | ||||
| Early | 843 | 55 | 823 | 58 |
| Intermediate | 568 | 37 | 513 | 36 |
| Advanced | 134† | 9 | 75† | 5 |
| ALC, × 109/L | ||||
| Median | 15 | 13 | ||
| IQR | 9-34 | 8-27 | ||
| B-ALC, × 109/L | ||||
| Median | 10 | 9 | ||
| IQR | 5-19 | 5-17 | ||
| β2M > ULN | 544 | 52 | 439 | 57 |
| del11q | 120 | 9 | 103 | 10 |
| del17p | 75 | 6 | 75 | 7 |
| Unmutated IGHV | 487 | 37 | 478 | 42 |
| ZAP-70 positive | 449 | 40 | 555 | 40 |
| CD38+ | 464 | 31 | 406 | 29 |
| CD49d+ | 587 | 38 | 511 | 36 |
| OS | ||||
| 5 year | 90 | 92 | ||
| 10 year | 73 | 80 | ||
| TFS | ||||
| 5 year | 57 | 59 | ||
| 10 year | 39 | 42 | ||
NOTE. Differences between published and unpublished cohorts were evaluated using χ2 or Wilcoxon test.
Abbreviations: ALC, absolute lymphocyte count; B-ALC, absolute B-cell count; β2M, β2-microglobulin; IQR, interquartile range; OS, overall survival; TFS, treatment-free survival; ULN, upper limit of normal.
Estimated by inverted censoring.
P < .01.
Early stage, Binet A or Rai 0; intermediate stage, Binet B or Rai I to II; advanced stage, Binet C or Rai III to IV.
Fig 1.
Study flow diagram. IPD, individual patient data; OS, overall survival.
Stability of CD49d was evaluated in paired samples collected at different time points (interval between sampling, 0.7 to 88.8 months; mean, 23.7 months) in 104 patient cases from the published cohort. In these patient cases, the first sample was collected at diagnosis, and the second sample was collected before therapy (n = 99) or at relapse (n = 5).
The study received approval from each local institutional review board and was conducted in accordance with the Declaration of Helsinki. The meta-analysis plan is available on request. This report has been checked with PRISMA checklist (http://www.prisma-statement.org; Data Supplement).
Statistical Analysis
We adopted a training/validation strategy to determine the optimal cutoff value of CD49d to predict OS (Data Supplement). The 1,556 patients from the published cohorts served as training series, and the 1,416 patients from the unpublished cohorts served as validation series. After having established and validated the optimal cutoff value, the training and validation series were merged for pooled analysis. Study heterogeneity and quality as well as publication bias were evaluated as detailed in the Data Supplement. Log hazard ratio (HR) of CD49d expression and corresponding SE were estimated in each study using univariate Cox proportional regression and combined in a pooled analysis with the fixed-effect model.21–23 To test the independence of the prognostic value of CD49d expression, we performed a comprehensive multivariable Cox model with backward stepwise elimination of nonsignificant covariates. All Cox models were stratified for study site and clinical stage of disease. Models with and without CD49d expression and other covariates were compared using log-likelihood/log-likelihood ratio, concordance index (C-index), and net reclassification improvement (NRI).24–26
Recursive partitioning27,28 was used to study the hierarchic importance of CD38, ZAP-70, and CD49d expression in predicting CLL survival. Survival-tree stability was assessed using the random survival model method.29 Additional details are reported in the Data Supplement. Statistical analyses were carried using R software (http://www.r-project.org).
RESULTS
General Characteristics of Patients
Characteristics of patients with CLL in the training and validation series were comparable. Although the validation series had shorter follow-up resulting from a more recent year of diagnosis, 5- and 10-year OS and TFS were similar (Table 1).
In both the training and validation series, distribution of prognostic biomarkers (eg, presence of del11q, del17p, unmutated IGHV, positivity for ZAP-70 and CD38) was similar and consistent with that expected in newly diagnosed patients with CLL.4,9 Prevalence of CD49d expression, as computed according to the original studies in the published series or using the 30% cutoff in the unpublished series, was also comparable.
Determination of the Optimal Cutoff of CD49d Expression to Predict Survival in CLL
High CD49d expression was always associated with shorter OS and/or disease progression (TFS) in CLL.14–20,30 However, heterogeneous cutoffs (ie, 30% and 45%) and procedures for cutoff determination were employed (Data Supplement).
To determine the optimal cutoff value of CD49d expression to predict OS in CLL, we adopted a training/validation strategy. In the training series, percentage of CD49d-expressing CLL cells followed a U-shaped distribution, with the vast majority (92%) of cases clustering between 0% and 30% or 50% and 100% (Fig 2A); moreover, CD49d values seemed stable over time in consecutive samples (Data Supplement). Both features have been considered evidence of the robustness of CD49d as a clinically useful prognosticator in CLL.14,31 Multiple statistical approaches were applied in the training series to identify the optimal cutoff for CD49d expression. When examining the functional form of the relationship between CD49d expression and OS in martingale residual plot,32 the local regression line failed to indicate a clear step suggestive of a cutoff point, although a slight inflection could be recognized between 20% and 40% (Fig 2B). Similarly, both a data-driven (ie, median) and three outcome-driven methods (ie, Maxstat, Rpart, C-index) failed to agree on a single common cutoff for CD49d, although a value close to 30% was selected by two methods (Data Supplement). Therefore, we decided to validate both the 30% and 45% cutoff values for CD49d expression even if the shift from 30% to 45% changed the classification of CD49d expression status in a minority of patients (3%; Data Supplement). According to a Cox analysis for OS stratified by center and disease stage in the validation series, the 30% cutoff had a slightly higher HR (2.50 v 2.49; Data Supplement) and discrimination of patient outcome (C-index, 0.61 v 0.59; P < .001) compared with the 45% cutoff. These data validate 30% as the best cutoff to code CD49d expression status to predict OS in patients with CLL.
Fig 2.
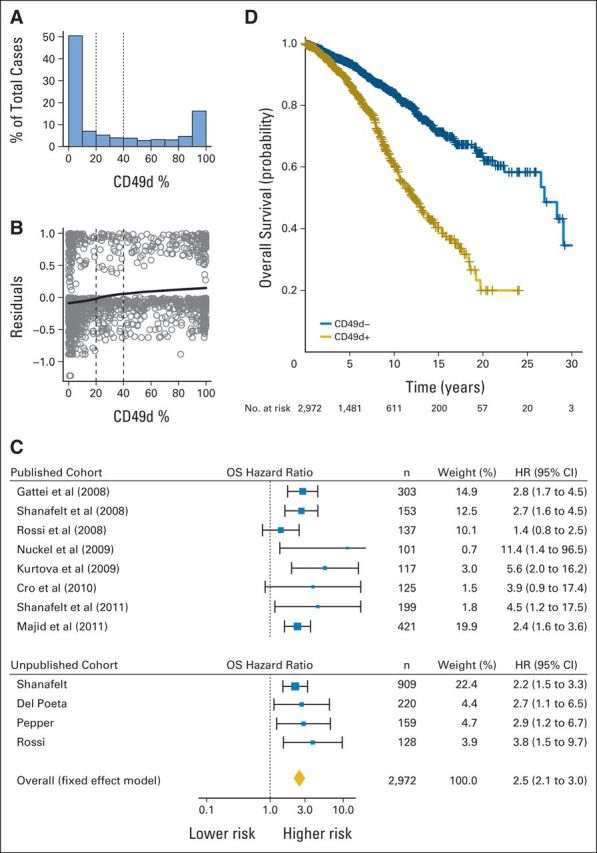
(A) Histogram of CD49d expression for published cohort. (B) Martingale residual plot: CD49d (x-axis) versus martingale residuals (y-axis) for published cohort. (C) Forest plot of overall survival (OS) hazard ratio (HR) in CD49d+ versus CD49d− patients (individual patient data [IPD] for 2,972 patients; CD49d recoded at cutoff ≥ 30%). Solid boxes indicate HR in each study with dimensions proportional to weights (inverse of variance); horizontal lines indicate 95% CIs; diamond indicates pooled HR. (D) Kaplan-Meier curves of OS of IPD for 2,972 patients; CD49d recoded at cutoff ≥ 30% (log-rank P < .001).
Prognostic Impact of CD49d in Pooled Analysis
Training and validation cohorts were merged for pooled analysis of OS and TFS using the 30% cutoff for recoding CD49d expression status. All single-center series contributing to the training and validation cohorts were confined within the tolerance limit bands in funnel plots, thus ruling out publication biases (Data Supplement). No significant heterogeneity was documented by either fixed (Q test, P = .39) or random (I2, 5%) effect models, suggesting little variation in study outcomes among series.
In the pooled analysis, CD49d+ patients had a significantly increased hazard of death (HR, 2.5; 95% CI, 2.1 to 3.0; Fig 2C), translating into a significantly lower OS probability at both 5 (87% v 94%) and 10 years (62% v 84%) compared with CD49d− patients (Fig 2D). Also, CD49d+ patients had a significantly increased hazard of being treated (HR, 2.3; 95% CI, 2.0 to 2.6), translating into a significantly lower probability of remaining treatment free at both 5 (42% v 68%) and 10 years (24% v 50%) compared with CD49d− patients (Fig 3). Estimates of 5- and 10-year OS and TFS were similar when analysis was restricted to the validation cohort (Data Supplement). These data confirm that CD49d expression in ≥ 30% of CLL cells has a detrimental impact on both OS and TFS that is reproducible in patient cohorts from different institutions.
Fig 3.
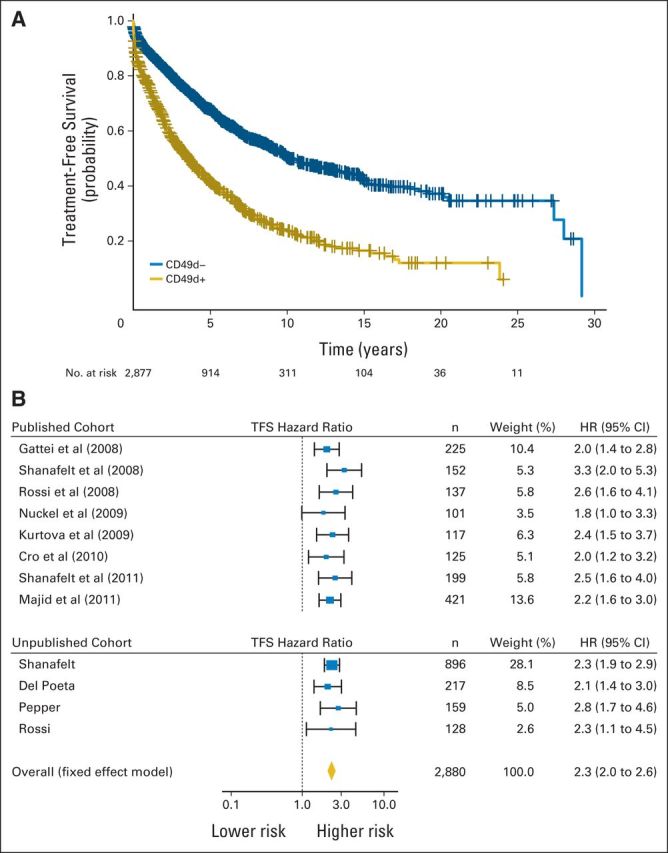
(A) Kaplan-Meier curves of treatment-free survival (TFS) in individual patient data (IPD) for 2,880 patients; CD49d recoded at cutoff ≥ 30% (log-rank P < .001). (B) Forest plot of TFS hazard ratio (HR) in CD49d+ versus CD49d− patients (IPD for 2,880 patients; CD49d recoded at cutoff ≥ 30%). Solid boxes indicate HR in each study with dimensions proportional to weights (inverse of variance); horizontal lines indicate 95% CIs; diamond indicates pooled HR.
Finally, in the pooled series, biologic covariates used for comparison with CD49d (eg, CD38, ZAP-70, IGHV, del11q, and del17p) had the expected association with OS and TFS (Data Supplement).4–9
Prognostic Impact of CD49d in Multivariable Analyses
To study the potentially confounding effect of other variables on the prognostic power of CD49d expression, we performed various adjustments to a Cox proportional hazard model stratified for study site in a cohort of 1,117 patient cases with all prognostic information available. No major differences were observed when comparing this 1,117–patient case cohort with remaining patient cases with respect to: one, trend of OS curves; two, distribution of the main clinicobiologic features; or three, 5- and 10-year OS estimates (Data Supplement).
By Cox analysis for OS, CD49d+ CLL had a two-fold increased risk of death (HR, 2.0; 95% CI, 1.4 to 3.0) after adjusting for disease stage, sex, age, and other clinical and biologic prognostic factors (eg, absolute lymphocyte count [ALC] and β2-microglobulin [β2M] values, presence of del11q and del17p, IGHV mutation status, CD38 and ZAP-70 expression). In the final multivariable model, CD49d turned out to be the sole flow cytometry based–marker with independent prognostic relevance for OS. The only other independent predictors of OS were age, sex, IGHV mutation status, del17p, and ALC (Table 2).
Table 2.
Multivariable Cox Regression Analysis of OS
| Factor | Final Reduced Model |
Initial Full Model |
Univariate Model |
||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | |
| Age > 65 years | 3.25 | 2.18 to 4.84 | < .001 | 3.03 | 2.00 to 4.57 | < .001 | 2.75 | 1.87 to 4.02 | < .001 |
| Unmutated IGHV | 2.48 | 1.68 to 3.66 | < .001 | 1.99 | 1.29 to 3.07 | < .001 | 2.64 | 1.81 to 3.85 | < .001 |
| CD49d ≥ 30% | 2.26 | 1.57 to 3.25 | < .001 | 2.01 | 1.36 to 2.97 | < .001 | 2.31 | 1.62 to 3.29 | < .001 |
| del17p | 2.13 | 1.37 to 3.32 | < .001 | 2.22 | 1.41 to 3.50 | < .001 | 2.90 | 1.87 to 4.49 | < .001 |
| Male sex | 1.83 | 1.24 to 2.69 | .0022 | 1.73 | 1.18 to 2.56 | .0055 | 1.53 | 1.06 to 2.22 | .0241 |
| ALC > 15 × 109/L | 1.66 | 1.11 to 2.48 | .0133 | 1.62 | 1.08 to 2.43 | .0202 | 1.55 | 1.05 to 2.30 | .0290 |
| ZAP-70 | — | — | — | 1.45 | 0.95 to 2.21 | .0864 | 2.27 | 1.57 to 3.27 | < .001 |
| β2M > ULN | — | — | — | 1.26 | 0.80 to 1.97 | .3217 | 2.23 | 1.47 to 3.40 | < .001 |
| CD38 | — | — | — | 1.21 | 0.82 to 1.78 | .3447 | 1.98 | 1.38 to 2.82 | < .001 |
| del11q | — | — | — | 1.19 | 0.70 to 2.04 | .5244 | 1.35 | 0.82 to 2.23 | .24 |
NOTE. Final model was multivariable analysis with backward stepwise elimination of nonsignificant predictors. All models were stratified by study site and clinical stage. Total patient cases included, 1,117; events, 137.
Abbreviations: ALC, absolute lymphocyte count; β2M, β2-microglobulin; HR, hazard ratio; OS, overall survival; ULN, upper limit of normal.
In a model excluding CD49d, both ZAP-70 (HR, 1.60; 95% CI, 1.06 to 2.41) and CD38 (HR, 1.56; 95% CI, 1.08 to 2.26) regained independent prognostic value (Data Supplement). Moreover, ad hoc meta-analyses for ZAP-70 (2,504 patient cases) and CD38 (2,876 patient cases) failed to find heterogeneity in the investigated cohorts, and the pooled effect, estimated in a fixed-effect model, was as expected (Data Supplement).6,9,33–35 Finally, correlation among CD49d, CD38, and ZAP-70 expression levels was mild, and no evidence of collinearity was found among these flow cytometry–based markers or between CD49d and IGHV gene status (Data Supplement). Of note, according to our multivariable analysis, IGHV mutation status seemed to be a better prognostic marker than ZAP-70, in keeping with some reports35–37 but in disagreement with others.8,9
Comparison of the models with and without CD49d by several prediction performance measures (log-likelihood/log-likelihood ratio, C-index, NRI) indicated that omission of CD49d significantly reduced the prognostic power of the model. In this regard, the impact of CD49d exclusion from the model was similar to that of the exclusion of age or IGHV gene status and stronger than the exclusion of del17p, sex, and ALC (Data Supplement).
Similar results were obtained in a model in which β2M and ALC values were excluded to increase the cohort of patient cases with all variables available (n = 1,655; Data Supplement). Multivariable analyses also selected CD49d expression as an independent predictor of TFS in CLL (Data Supplement).
Prognostic Impact of CD49d Among Flow Cytometry–Based Markers
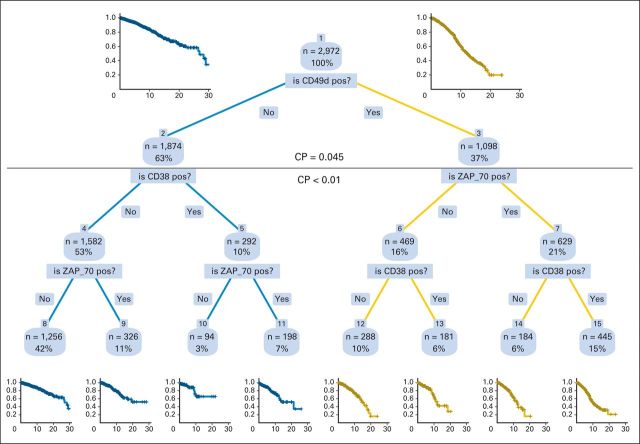
To test the relative importance of the three main flow cytometry–based prognostic variables CD49d, CD38, and ZAP-70 as predictors of OS in CLL, we built a hierarchic full-grown tree model by recursive partitioning analysis (Appendix Fig A1, online only). CD49d was selected at the first split as the most important variable. The optimal tree-pruning level was at the first branching (complexity parameter, 0.045), confirming the results of multivariable Cox model and showing that in a hierarchic model, no additional prognostic power was given by CD38 or ZAP-70 after CD49d was considered. CD49d was also at the first split in tree models developed in 1,666 patients with early-stage disease and in 1,664 patients age ≤ 65 years as well as when using patient cases from the validation cohort only (Data Supplement). These results were internally validated by tree analysis in 1,000 bootstrap samples in which CD49d was always selected over CD38 and ZAP-70 as the most important variable associated with the greatest reduction in predictive error (data not shown).
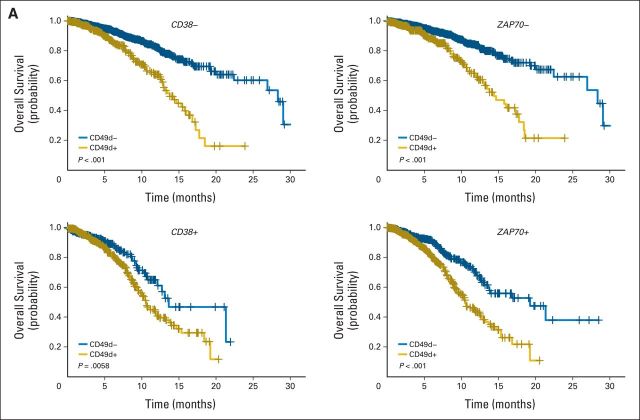
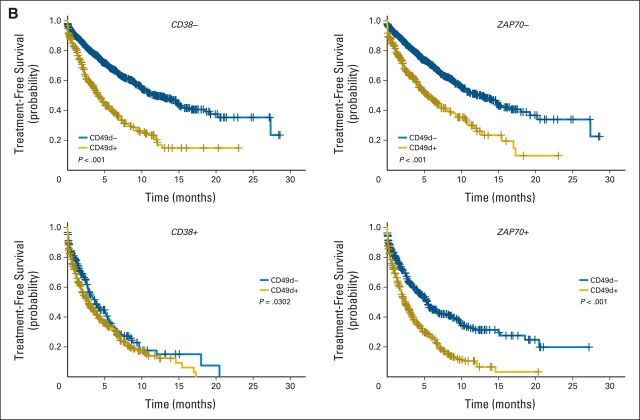
Consistent with recursive partitioning analysis, bivariate survival curves showed that CD49d, when expressed over the established cutoff, also reliably identified subsets with worse prognosis in term of OS and TFS regardless of CD38 or ZAP-70 status (Appendix Fig A2, online only).
Overall, these data show that among CLL phenotypic markers, CD49d has a greater prognostic importance than CD38 or ZAP-70 and demonstrate the additional value of CD49d testing when CD38 and ZAP-70 information is available.
DISCUSSION
In this pooled analysis of 2,972 patients with CLL, CD49d was an independent predictor of OS and TFS and retained its prognostic value after adjusting for other well-established biologic prognosticators (β2M, ALC, CD38, ZAP-70, IGHV gene status, cytogenetic aberrations). Indeed, CD49d was among the most important prognostic variables, along with age and IGHV gene status. CD49d emerged as the strongest flow cytometry–based prognostic marker, with greater prognostic value than CD38 and ZAP-70.
Using a training/validation strategy, we established the 30% cut point as the most useful threshold for classification of CD49d+ CLL. Notably, in this large series as well as in previous studies,14,15,17 CD49d expression showed a bimodal distribution, with most patients showing either very high or very low levels of expression. This fact minimized the number of patient cases with borderline CD49d expression clustered around the cutoff, making CD49d a pragmatic choice of biomarker for reliable prognostication of CLL. In contrast, other markers (eg, ZAP-70) often have expression values distributed around the cutoff,7,8,38 making confident prediction of clinical outcome more difficult.
Methodologic factors of the flow cytometric procedure for CD49d detection investigated here, including mAb clone, mAb fluorochrome conjugation, or immunofluorescence strategy (two v three colors), did not have any effect on the overall percentage of CD49d+ patient cases, again underscoring the robustness of CD49d as a prognosticator. Nevertheless, for clinical routine analyses, we suggest at least a three-color strategy that includes CD49d in a single combination along with CD19 and CD5. This will exclude from analysis the majority of contaminating normal CD5− B cells, known to express high levels of CD49d.39 Altogether, CD49d emerges as a surface marker, the expression of which can be easily investigated in routine clinical practice, is highly reproducible, and is not subject to variation resulting from the application of different methodologies.
As shown by multivariable analyses of the pooled data, CD49d was selected as an independent predictor of OS, along with age, IGHV mutation status, del 17p, sex, and ALC in models including all investigated variables and in models excluding β2M and ALC to increase the number of patient cases. ZAP-70 and CD38 were discarded from the comprehensive final multivariable model but were both retained when CD49d was excluded from analysis, in agreement with literature data.2,4,6–8 In accordance with these results, a hierarchic model restricted to flow cytometric prognostic markers (ZAP-70, CD38, and CD49d) located CD49d at the top of the branching among the entire cohort of patients as well as among young patients and those with early-stage disease. Therefore, CD49d turned out to be the best flow cytometry–based marker to stratify prognosis of patients with CLL. Moreover, as demonstrated by bivariate analyses, CD49d expression reliably identified subsets of patients with poorer outcome independent of CD38 or ZAP-70 status. On this basis, we believe that testing CD49d expression in routine clinical practice should be considered in the baseline prognostic assessment of patients with newly diagnosed CLL as well as to refine prognostic evaluation in patients already stratified by CD38 and ZAP-70. Stability of CD49d expression level over time, demonstrated both here and previously,14,31 should allow reliable evaluation of CD49d at any point during the watch-and-wait interval.
The microenvironmental dependency of CLL cells for prosurvival signals, both integrin and B cell–receptor (BCR) mediated, has been recently highlighted by the emerging role of Bruton's tyrosine kinase (BTK) inhibitors as promising new therapeutic agents in CLL and other B-cell malignancies.40 In particular, both clinical studies and in vitro observations have concordantly indicated that BTK inhibition results in a reduced microenvironmental retention of CLL cells in tissue sites because of impaired integrin- and chemokine receptor–mediated tissue homing.41–43 In this context, it is perhaps not surprising that CD49d (α4 subunit of integrin heterodimer α4β1; one of the most potent mediators of cell-matrix and cell-cell interactions10) and IGHV gene mutational status of BCR both emerge as the strongest predictors of short OS and TFS in CLL. Given the retrospective nature of this study, a prospective validation of the prognostic impact of CD49d is still warranted to definitively introduce this marker into the routine clinical assessment of patients with CLL.
Supplementary Material
Glossary Terms
- Concordance index (C-index):
The concordance index is the probability that given two randomly selected patients, the patient with the worst outcome is, in fact, predicted to have the worst outcome. The measure is similar to an area under the receiver operating characteristics curve and ranges from 0.5 (chance or coin flip) to 1.0 (perfect ability to rank patients).
- Integrin:
Integrins are cell-adhesion receptors that are involved in two important pathways: cell-extracellular matrix and cell-cell interactions, thus allowing for cell-cell and cell-extracellular matrix communication. Functionally, integrins are composed of two subunits (heterodimers), one belonging to the α family and the other belonging to the β family.
- Recursive partitioning:
Multivariable analysis that generates a clinically intuitive decision tree model in which the population is divided into prognostic subgroups. This is achieved through multiple dichotomous divisions on the basis of a set of independent variables.
- ZAP-70:
Involved in T-cell activation and differentiation signals. Early events in the activation of the T-cell receptor (CD3/TCR) include the activation of non–receptor tyrosine kinases (eg, Src family) leading to the phosphorylation of immunoreceptor, tyrosine-based activation motifs (ITAMs) in the cytoplasmic domains of the CD3/TCR ξ chains. The phosphorylation of ITAMs is responsible for the recruitment and activation of ξ-associated protein (ZAP)-70 via its SH2 domain, with subsequent recruitment and activation of other proteins.
Appendix
Fig A1.
Decision tree resulting from recursive partitioning analysis of flow cytometry–measured prognostic variables applied to individual patient data for 2,972 patients. CD49d recoded at cutoff ≥ 30%; CD38 and ZAP-70 coded as originally performed in each center. CD49d was the most significant covariate for entire study population and was selected by algorithm to split patient population into two terminal nodes. Horizontal line shows optimal split point and corresponding values (above and below) of the complexity parameter (CP). Additional splits below generated daughter nodes without significantly different survival probability (shown for clarity and iconographic purposes). Right branch of each split represents positivity of the variable; left branch, negativity. Kaplan-Meier curves at top left and top right estimate overall survival (OS) of patients belonging to the CD49d− and CD49d+ nodes, respectively. Kaplan-Meier curves at bottom estimate OS of patients belonging to terminal nodes of the full-grown, unpruned tree. pos, positive.
Fig A2.
Bivariate analysis by Kaplan-Meier curves of CD49d expression in the context of chronic lymphocytic leukemia (CLL) subset expressing or not CD38 (left panels) or ZAP70 (right panels). (A) Overall survival. Blue lines indicate CD49d− CLL; gold lines, CD49d+ CLL. P values determined using log-rank test. (B) Treatment-free survival. Blue lines indicate CD49d− CLL; gold lines, CD49d+ CLL. P values determined using log-rank test.
Footnotes
See accompanying editorial on page 869
Supported in part by the Ministero Della Salute, Rome, Italy (Ricerca Finalizzata Istituti di Ricovero e Cura a Carattere Scientifico [IRCCS], Progetto Giovani Ricercatori No. GR-2010-2317594 and No. GR-2008-1138053); Fondazione Internazionale di Ricerca in Medicina Sperimentale (FIRMS), Turin; Associazione Italiana Contro le Leucemie, Linfomi e Mielomi (AIL), Venezia Section, Pramaggiore Group, Venice; Ricerca Scientifica Applicata, Regione Friuli Venezia Giulia (“Linfonet” project), Trieste; Associazione Italiana Ricerca Cancro (AIRC), Special Program Molecular Clinical Oncology, 5 × 1000, No. 10007, Milan; Investigator Grant No. IG-13227; 5 × 1000 Intramural Program, Centro di Riferimento Oncologico, Aviano; Leukaemia Lymphoma Research Grant No. 10003; and Bando Progetti di Ricerca di Ateneo, Compagnia di San Paolo, Turin, Italy.
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Pietro Bulian, Tait D. Shanafelt, Jan A. Burger, Gianluca Gaidano, Giovanni Del Poeta, Chris Pepper, Davide Rossi, Valter Gattei
Provision of study materials or patients: Tait D. Shanafelt, Chris Fegan, Holger Nückel, Luca Baldini, Jan A. Burger, Gianluca Gaidano, Giovanni Del Poeta, Chris Pepper, Davide Rossi
Collection and assembly of data: Pietro Bulian, Tait D. Shanafelt, Chris Fegan, Lilla Cro, Holger Nückel, Luca Baldini, Antonina V. Kurtova, Alessandra Ferrajoli, Jan A. Burger, Giovanni Del Poeta, Chris Pepper, Davide Rossi, Valter Gattei
Data analysis and interpretation: Pietro Bulian, Tait D. Shanafelt, Antonella Zucchetto, Holger Nückel, Luca Baldini, Antonina V. Kurtova, Alessandra Ferrajoli, Jan A. Burger, Chris Pepper, Davide Rossi, Valter Gattei
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 2.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- 3.Hamblin TJ, Davis Z, Gardiner A, et al. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- 4.Kröber A, Seiler T, Benner A, et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100:1410–1416. [PubMed] [Google Scholar]
- 5.Döhner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 6.Hamblin TJ, Orchard JA, Ibbotson RE, et al. CD38 expression and immunoglobulin variable region mutations are independent prognostic variables in chronic lymphocytic leukemia, but CD38 expression may vary during the course of the disease. Blood. 2002;99:1023–1029. doi: 10.1182/blood.v99.3.1023. [DOI] [PubMed] [Google Scholar]
- 7.Crespo M, Bosch F, Villamor N, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med. 2003;348:1764–1775. doi: 10.1056/NEJMoa023143. [DOI] [PubMed] [Google Scholar]
- 8.Rassenti LZ, Huynh L, Toy TL, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351:893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 9.Rassenti LZ, Jain S, Keating MJ, et al. Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood. 2008;112:1923–1930. doi: 10.1182/blood-2007-05-092882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burger JA. Targeting the microenvironment in chronic lymphocytic leukemia is changing the therapeutic landscape. Curr Opin Oncol. 2012;24:643–649. doi: 10.1097/CCO.0b013e3283589950. [DOI] [PubMed] [Google Scholar]
- 11.Zucchetto A, Bomben R, Dal Bo M, et al. CD49d in B-cell chronic lymphocytic leukemia: Correlated expression with CD38 and prognostic relevance. Leukemia. 2006;20:523–525. doi: 10.1038/sj.leu.2404087. author reply 528-529. [DOI] [PubMed] [Google Scholar]
- 12.Zucchetto A, Vaisitti T, Benedetti D, et al. The CD49d/CD29 complex is physically and functionally associated with CD38 in B-cell chronic lymphocytic leukemia cells. Leukemia. 2012;26:1301–1312. doi: 10.1038/leu.2011.369. [DOI] [PubMed] [Google Scholar]
- 13.Woyach JA, Johnson AJ, Byrd JC. The B-cell receptor signaling pathway as a therapeutic target in CLL. Blood. 2012;120:1175–1184. doi: 10.1182/blood-2012-02-362624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gattei V, Bulian P, Del Principe MI, et al. Relevance of CD49d protein expression as overall survival and progressive disease prognosticator in chronic lymphocytic leukemia. Blood. 2008;111:865–873. doi: 10.1182/blood-2007-05-092486. [DOI] [PubMed] [Google Scholar]
- 15.Shanafelt TD, Geyer SM, Bone ND, et al. CD49d expression is an independent predictor of overall survival in patients with chronic lymphocytic leukaemia: A prognostic parameter with therapeutic potential. Br J Haematol. 2008;140:537–546. doi: 10.1111/j.1365-2141.2007.06965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossi D, Zucchetto A, Rossi FM, et al. CD49d expression is an independent risk factor of progressive disease in early stage chronic lymphocytic leukemia. Haematologica. 2008;93:1575–1579. doi: 10.3324/haematol.13103. [DOI] [PubMed] [Google Scholar]
- 17.Nückel H, Switala M, Collins CH, et al. High CD49d protein and mRNA expression predicts poor outcome in chronic lymphocytic leukemia. Clin Immunol. 2009;131:472–480. doi: 10.1016/j.clim.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Cro L, Ferrario A, Lionetti M, et al. The clinical and biological features of a series of immunophenotypic variant of B-CLL. Eur J Haematol. 2010;85:120–129. doi: 10.1111/j.1600-0609.2010.01454.x. [DOI] [PubMed] [Google Scholar]
- 19.Shanafelt TD, Drake MT, Maurer MJ, et al. Vitamin D insufficiency and prognosis in chronic lymphocytic leukemia. Blood. 2011;117:1492–1498. doi: 10.1182/blood-2010-07-295683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majid A, Lin TT, Best G, et al. CD49d is an independent prognostic marker that is associated with CXCR4 expression in CLL. Leuk Res. 2011;35:750–756. doi: 10.1016/j.leukres.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 21.Blettner M, Sauerbrei W, Schlehofer B, et al. Traditional reviews, meta-analyses and pooled analyses in epidemiology. Int J Epidemiol. 1999;28:1–9. doi: 10.1093/ije/28.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Barza M, Trikalinos TA, Lau J. Statistical considerations in meta-analysis. Infect Dis Clin North Am. 2009;23:195–210. doi: 10.1016/j.idc.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Katsahian S, Latouche A, Mary JY, et al. Practical methodology of meta-analysis of individual patient data using a survival outcome. Contemp Clin Trials. 2008;29:220–230. doi: 10.1016/j.cct.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: Issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 25.Harrell FE. Regression Modeling Strategies. New York, NY: Springer; 2001. [Google Scholar]
- 26.Pencina MJ, D'Agostino RB, Sr, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30:11–21. doi: 10.1002/sim.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Therneau T, Atkinson E. Rochester, MN: Department of Health Science Research, Mayo Clinic; 1997. Technical Report Series No. 61: An Introduction to Recursive Partitioning Using the RPART Routines. [Google Scholar]
- 28.Venables W, Ripley B. Modern Applied Statistics With S. New York, NY: Springer-Verlag; 2002. [Google Scholar]
- 29.Ishwaran H, Koglaur UB, Blackstone EH, et al. Random survival forests. Ann Appl Stat. 2008;2:841–860. [Google Scholar]
- 30.Kurtova A, Sivina M, Quiroga M, et al. The immunophenotype signature CD49d+CD38+ identifies chronic lymphocytic leukemia cases with a higher potential for migration beneath marrow stromal cell. Blood. 2009:114. (abstr 356) [Google Scholar]
- 31.Shanafelt TD, Hanson C, Dewald GW, et al. Karyotype evolution on fluorescent in situ hybridization analysis is associated with short survival in patients with chronic lymphocytic leukemia and is related to CD49d expression. J Clin Oncol. 2008;26:e5–e6. doi: 10.1200/JCO.2008.16.7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams B, Mandrekar J, Mandrekar S, et al. Rochester, MN: Department of Health Science Research, Mayo Clinic; 2006. Technical Report Series No. 79: Finding Optimal Cutpoints for Continuous Covariates With Binary and Time-to-Event Outcomes. [Google Scholar]
- 33.Oscier DG, Gardiner AC, Mould SJ, et al. Multivariate analysis of prognostic factors in CLL: Clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood. 2002;100:1177–1184. [PubMed] [Google Scholar]
- 34.Del Poeta G, Maurillo L, Venditti A, et al. Clinical significance of CD38 expression in chronic lymphocytic leukemia. Blood. 2001;98:2633–2639. doi: 10.1182/blood.v98.9.2633. [DOI] [PubMed] [Google Scholar]
- 35.Kröber A, Bloehdorn J, Hafner S, et al. Additional genetic high-risk features such as 11q deletion, 17p deletion, and V3-21 usage characterize discordance of ZAP-70 and VH mutation status in chronic lymphocytic leukemia. J Clin Oncol. 2006;24:969–975. doi: 10.1200/JCO.2005.03.7184. [DOI] [PubMed] [Google Scholar]
- 36.Josefsson P, Geisler CH, Leffers H, et al. CLLU1 expression analysis adds prognostic information to risk prediction in chronic lymphocytic leukemia. Blood. 2007;109:4973–4979. doi: 10.1182/blood-2006-11-054916. [DOI] [PubMed] [Google Scholar]
- 37.Pepper C, Majid A, Lin TT, et al. Defining the prognosis of early stage chronic lymphocytic leukaemia patients. Br J Haematol. 2012;156:499–507. doi: 10.1111/j.1365-2141.2011.08974.x. [DOI] [PubMed] [Google Scholar]
- 38.Orchard JA, Ibbotson RE, Davis Z, et al. ZAP-70 expression and prognosis in chronic lymphocytic leukaemia. Lancet. 2004;363:105–111. doi: 10.1016/S0140-6736(03)15260-9. [DOI] [PubMed] [Google Scholar]
- 39.Lucio PJ, Faria MT, Pinto AM, et al. Expression of adhesion molecules in chronic B-cell lymphoproliferative disorders. Haematologica. 1998;83:104–111. [PubMed] [Google Scholar]
- 40.Advani RH, Buggy JJ, Sharman JP, et al. Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765) has significant activity in patients with relapsed/refractory B-cell malignancies. J Clin Oncol. 2013;31:88–94. doi: 10.1200/JCO.2012.42.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spaargaren M, Beuling EA, Rurup ML, et al. The B cell antigen receptor controls integrin activity through btk and PLCgamma2. J Exp Med. 2003;198:1539–1550. doi: 10.1084/jem.20011866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Rooij MF, Kuil A, Geest CR, et al. The clinically active BTK inhibitor PCI-32765 targets B-cell receptor- and chemokine-controlled adhesion and migration in chronic lymphocytic leukemia. Blood. 2012;119:2590–2594. doi: 10.1182/blood-2011-11-390989. [DOI] [PubMed] [Google Scholar]
- 43.Ponader S, Chen SS, Buggy JJ, et al. The bruton tyrosine kinase inhibitor PCI-32765 thwarts chronic lymphocytic leukemia cell survival and tissue homing in vitro and in vivo. Blood. 2012;119:1182–1189. doi: 10.1182/blood-2011-10-386417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.