Abstract
Euglena gracilis, a microalgal species of unicellular flagellate protists, has attracted much attention in both the industrial and academic sectors due to recent advances in the mass cultivation of E. gracilis that have enabled the cost-effective production of nutritional food and cosmetic commodities. In addition, it is known to produce paramylon (β-1,3-glucan in a crystalline form) as reserve polysaccharide and convert it to wax ester in hypoxic and anaerobic conditions–a promising feedstock for biodiesel and aviation biofuel. However, there remain a number of technical challenges to be solved before it can be deployed in the competitive fuel market. Here we present a method for efficient selective breeding of live oil-rich E. gracilis with fluorescence-activated cell sorting (FACS). Specifically, the selective breeding method is a repetitive procedure for one-week heterotrophic cultivation, staining intracellular lipids with BODIPY505/515, and FACS-based isolation of top 0.5% lipid-rich E. gracilis cells with high viability, after inducing mutation with Fe-ion irradiation to the wild type (WT). Consequently, we acquire a live, stable, lipid-rich E. gracilis mutant strain, named B1ZFeL, with 40% more lipid content on average than the WT. Our method paves the way for rapid, cost-effective, energy-efficient production of biofuel.
Microalgae-based bioproducts have gained much attention in both the industrial and academic sectors. Microalgae photosynthesize with a higher efficiency than higher plants and can be cultivated in agriculturally unused soils. These features of microalgae have helped us advance the cultivation of microalgae such as Arthrospira and Chlorella which are nutrient-rich as a source of food for us and other animals1. The biomass from mass-cultured microalgae is also exploited as a source of specific chemicals, such as β-carotene, astaxanthin, and polyunsaturated fatty acids, to be used for nutritional supplements, pharmaceutical drugs, and cosmetic products1,2. Furthermore, many microalgal species are known to produce and store oil in response to nutritional and environmental conditions such as nitrogen deficiency3,4,5. Although there remain a number of technical difficulties to be solved, the economically sustainable production of biofuel from mass-cultured microalgae is highly anticipated as a next-generation energy source6,7.
Euglena gracilis, a species of single-celled flagellate protists found in freshwater, is one of the aforementioned industrially exploited microalgae. E. gracilis is neutrient-rich and known to accumulate paramylon (β-1,3-glucan in a crystalline form) as reserve polysaccharide in response to nitrogen deficiency or heterotrophic carbon sources8,9. Recent reports suggest the functionality of paramylon on hepatoprotection10, atopy treatment11, and colon cancer suppression12. Based on these features, mass-cultured E. gracilis has been commercially supplied as an ingredient of functional food and a source of paramylon extraction. E. gracilis is also known to consume the intracellular paramylon in a hypoxic condition to obtain energy without oxygen13. This process is accompanied by the production of wax ester–an ester mainly composed of C14:0 saturated fatty acid, myristic acid, and myristyl alcohol14,15. The composition of a relatively short chain of fatty acids and alcohols is especially suited for its conversion to biodiesel and aviation biofuel.
To improve E. gracilis’ productivity of the wax ester and further promote the utility of its biomass to biofuel, effective breeding methods need to be developed before it can be deployed in the competitive fuel market. Although a number of E. gracilis mutants have previously been developed, most of them have been derived from inefficient mutations in its chloroplast genomes16. Furthermore, its killing curve by UV irradiation indicates that E. gracilis is polyploid, making it difficult to alter its nuclear genome17,18. In contrast, heavy-ion irradiation has been identified as an effective approach to producing E. gracilis mutants different from those generated by exposing them to classical mutagenesis such as chemical mutagens and UV19. Previously, heavy-ion irradiation has been employed for higher plants20 and tested on the mutant production of various microalgal species21,22 and is expected to be of use for the development of E. gracilis mutants as well.
Fluorescence-activated cell sorting (FACS) is a powerful tool for counting and characterizing a large heterogeneous population of cells, including microalgal mutants, with a high throughput of more than 10,000 cells/sec by detecting fluorescent light from each laser-excited cell during flow23. While FACS is commonly employed in basic medical research and clinical practice, it has also been used to separate a single microalgal species from a mixed culture24 and to produce an axenic microalgal culture25. Staining with a fluorescent reagent, Nile Red, has been used in conjunction with FACS for detection and sorting of algal cells with high oil content26,27,28,29. For evaluation and characterization of E. gracilis, while FACS has been used to analyze its cell cycle by staining with propidium iodide or Hoechst 33258 after fixation30,31, it has been difficult to apply FACS to live E. gracilis cells due to the toxicity of the staining reagents and its low cell viability after FACS.
In this Article, we present a method that overcomes the aforementioned technical difficulties and hence enables efficient selective breeding of live oil-rich E. gracilis with FACS. Specifically, the breeding method is a repetitive procedure for one-week heterotrophic cultivation, staining intracellular lipids with BODIPY505/515, and FACS-based isolation of top 0.5% lipid-rich E. gracilis cells with high viability, after inducing mutation with Fe-ion irradiation to the wild type (WT). The entire breeding process only takes several weeks. As a result of using the breeding method, we acquire a live, stable, lipid-rich E. gracilis mutant strain, named B1ZFeL, with 40% more lipid content on average than the WT. We anticipate that a further understanding of lipid metabolism combined with a further optimization of the breeding method will enable rapid, cost-effective, energy-efficient production of biofuel.
Results
Autofluorescence of live E. gracilis cells
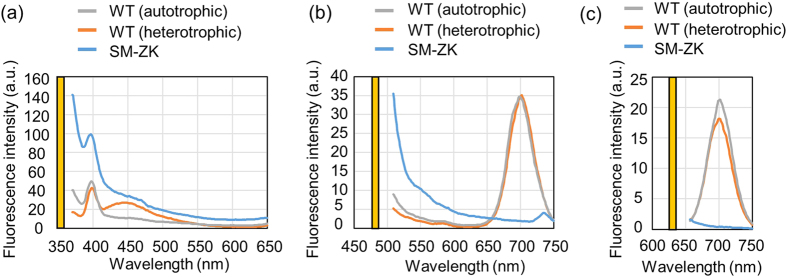
Fluorescence by staining target cells with fluorescent dyes and specific probes is commonly used for the identification of cells of interest and the key requirement for FACS. Since in the case of microalgae, their intracellular photosynthetic pigments often emit strong autofluorescence that becomes noise in FACS-based recognition of target cells, it is an essential requirement to carefully select appropriate fluorescent dyes and detection techniques28. To determine the requirement, we investigated the autofluorescence of E. gracilis cells, in particular the WT (Z strain) cultured in an autotropic and heterotrophic condition and a chloroplast-less mutant strain, SM-ZK, cultured in a heterotrophic condition when exciting them with laser light at 350 nm, 488 nm, and 635 nm (commonly used wavelengths in FACS). As shown in Fig. 1a, the excitation at 350 nm induced each strain to emit strong fluorescence at 400 nm and weak fluorescence around 450 nm. The cause of the strong fluorescence is unclear while that of the weak fluorescence is presumably NAD(P)H. Also, as shown in Fig. 1b, the excitation at 488 nm induced the WT to emit fluorescence at 700 nm. This fluorescence is caused by chlorophyll as the chloroplast-less SM-ZK strain did not emit fluorescence at this excitation wavelength. Finally, as shown in Fig. 1c, the excitation at 635 nm induced the WT to emit fluorescence at 700 nm.
Figure 1. Characterization of the autofluorescence of E. gracilis cells.
(a–c) Autofluorescence spectrum of the WT (Z strain) in the autotrophic and heterotrophic cultures and the chloroplast-less SM-ZK strain in the heterotrophic culture. The wavelength of the excitation laser is 350 nm (a), 488 nm (b), and 635 nm (c). The yellow bars indicate the wavelength of the excitation laser.
Staining of live E. gracilis cells with BODIPY505/515
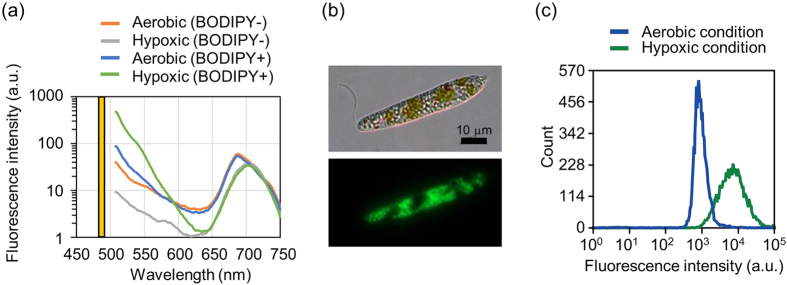
Following the spectral profile of the autofluorescence from E. gracilis cells, we examined the capability of the lipophilic green fluorescent dye, BODIPY505/515, which stains neutral lipids stored in the cells just like Nile Red which is toxic and causes interference with natural cellular functions such as lipid production yield. BODIPY505/515 has been used to stain intracellular lipids in microalgae32,33,34. When excited by laser light at 488 nm, BODIPY505/515-stained E. gracilis cells emit fluorescence at 515 nm, which is compatible to the autofluorescence profile of E. gracilis. To test if the intracellular lipids in E. gracilis can be stained by BODIPY505/515, we incubated a heterotrophic culture of E. gracilis cells in a hypoxic condition to induce the conversion of paramylon to wax ester. After a sequence of condition studies, we reached an optimum staining procedure for exposing the cells to 5 μM of BODIPY505/515 for 5 min in water. Figure 2a shows the fluorescence spectrum of the stained cells. Here the fluorescence signal from BODIPY505/515-stained cells was enhanced by increasing the duration of hypoxic conditioning (Supplementary Figure 1A). The fluorescence signal was found to be well correlated with the actual lipid content (Supplementary Figures 1B,C). Also, as microscope images of the stained cells show in Fig. 2b, the cells have a granular spatial pattern of fluorescence. In contrast to staining with Nile Red, staining with BODIPY505/515 clearly showed the lipid granules of live cells (Supplementary Figure 2). Furthermore, to quantify the fluorescence, we performed population analysis of the stained cells with FACS. As Fig. 2c shows, the cells in the hypoxic condition show stronger fluorescence than the cells cultured in the aerobic condition. Here the fluorescence is the light that passed through the 529/28 filter.
Figure 2. Characterization of the fluorescence of BODIPY505/515-stained E. gracilis cells.
(a) Fluorescence spectrum of BODIPY505/515-stained E. gracilis cells. The yellow bar indicates the wavelength of the excitation laser. (b) DIC (upper) and fluorescent (lower) images of a BODIPY505/515-stained E. gracilis cell which was in the hypoxic condition for 2 days. (c) Histogram of BODIPY505/515-stained E. gracilis cells with and without hypoxic incubation obtained by flow cytometry analysis. The fluorescence intensity is the strength of the light that passed through the 529/28 bandpass filter.
High-throughput sorting of live E. gracilis cells with high viability
Following the results about the autofluorescence and fluorescence of E. gracilis, we performed FACS of live E. gracilis cells, but with an improved nozzle. Previously, it has been difficult to conduct high-throughput sorting of Euglena cells with conventional cell sorters since they can easily be damaged by their nozzle with a typical diameter of 70 μm. This is presumably because E. gracilis does not have a cell wall, is relatively large in size, and is vulnerable to shear force compared to other microalgae. For this reason, the throughput was required to be reduced for sorting of E. gracilis with high viability. In our FACS analysis, more than 40% of E. gracilis cells were killed during the analysis and sorting process with the 70-μm nozzle (Supplementary Figure 3A). In order to minimize the damage to the cells, we investigated and determined the optimum conditions for sorting live E. gracilis cells using multi-well plates filled with KH medium with nozzles of 70 μm, 100 μm, and 120 μm in diameter. After two weeks of static cultivation for proliferation, we enumerated the wells in which the cells survived and determined the survival rate of the cells. Only 57% of the wells (110 wells/192 wells) showed proliferation with the 70-μm nozzle while 97% (187 wells/192 wells) and 99% (190 wells/192 wells) were recovered with the 100-μm and 120-μm nozzles, respectively (Supplementary Figure 3B).
Production of a lipid-rich E. gracilis mutant strain
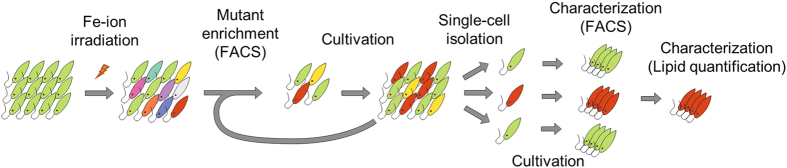
The results from lipid staining with BODIPY505/515 and high-throughput sorting suggest that FACS is an effective high-throughput tool for selectively breeding E. gracilis based on its lipid content. To demonstrate this, we designed a screening procedure for enriching mutants which produce wax ester even in the aerobic culture condition (Fig. 3). Specifically, E. gracilis cells were irradiated by a 50 Gy Fe-ion beam to induce mutation and were then separated to four independent groups, each of which consists of 105 cells. Each group was let recover for a week in a heterotrophic condition to accumulate paramylon (which was converted to wax ester in the mutants) and then stained with BODIPY505/515 and subjected to cell sorting. The top 0.5% of the total population of the stained cells with the BODIPY505/515 fluorescence signal was collected and cultured in the same condition. After repeating this procedure for enriching the mutants 4 times with a one-week interval between the consecutive sorting steps, 15 mutant candidates were randomly isolated from each mutant pool. The phenotype of the progenies of each candidate was evaluated by FACS with BODIPY505/515 staining. Finally, we selected the strain with the largest fluorescence strength, which we named B1ZFeL after the nomenclature16 with the addition of the phenotypic designation “B” for BODIPY505/515 staining and the mutagen designation “Fe” for Fe-ion irradiation, and applied it to further phenotypic characterization.
Figure 3. Method for selective breeding of a lipid-rich E. gracilis mutant strain.
The procedure of the method is as follows. (i) A pool of WT (Z strain) E. gracilis cells are irradiated with a Fe-ion beam for mutation induction. (ii) Mutants are enriched by isolation with FACS and BODIPY505/515 staining. (iii) The selected mutants are cultivated for a week. (iv) The process of the mutant enrichment and cultivation is repeated four times. (v) After four rounds of the mutant enrichment and cultivation, 15 individual mutants are randomly isolated and cultivated independently. (vi) Each established mutant strain is analyzed by FACS, such that the best stained strain is selected for further characterization. (vii) The lipid content of the selected strain is quantified. The entire breeding process only takes several weeks.
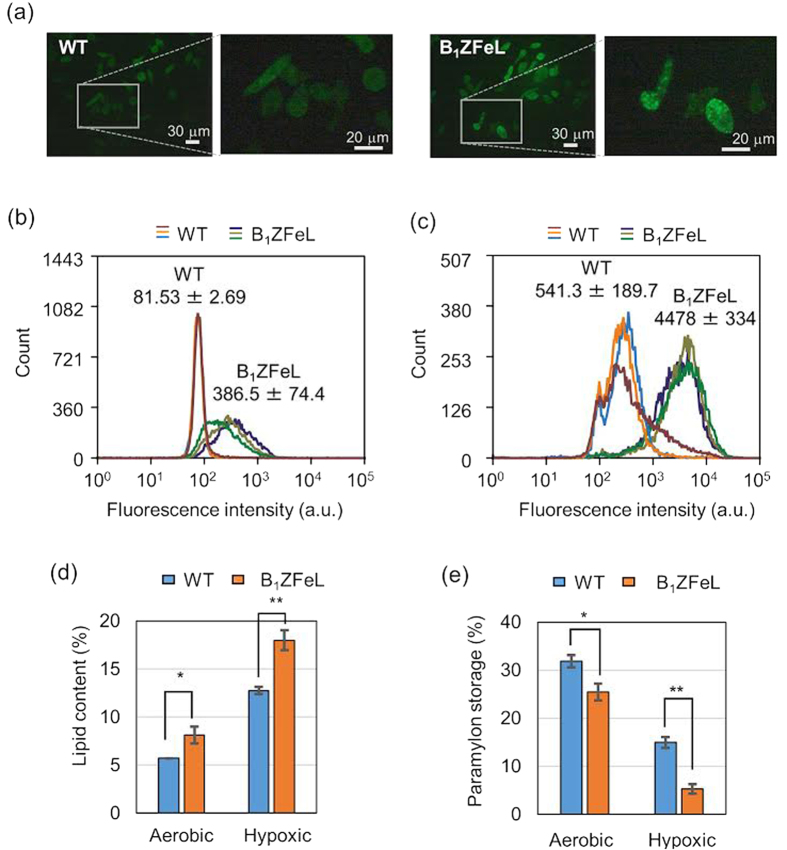
As the fluorescence microscope images in Fig. 4a show, the B1ZFeL strain showed granule-like stained properties without hypoxic conditioning while the growth rate of the B1ZFeL strain in a conical flask was found to be slightly lower than that of the WT (Z strain) (Supplementary Figure 4). As shown in Fig. 4b, the B1ZFeL strain shows a factor of 4.7 stronger fluorescence in our flow cytometry analysis (FACS measurements without cell sorting) after BODIPY505/515 staining than the WT. Here the values in the figure indicate the mean and the standard error of the mean of each strain. As shown in Fig. 4c, after hypoxic incubation, the fluorescence intensity of both the WT and B1ZFeL distributions increased while the difference between the two strains increased to a factor of 8.3.
Figure 4. Characterization of the lipid-rich strain B1ZFeL.
(a) Fluorescence images of BODIPY505/515-stained WT (Z strain) and B1ZFeL cells in the aerobically heterotrophic culture. (b,c) Histogram of the BODIPY505/515-stained WT and B1ZFeL cells. The cells without (b) and with (c) hypoxic incubation were stained with BODIPY505/515 and analyzed by FACS analysis. The fluorescence intensity is the strength of the light that passed through the 529/28 bandpass filter. The three histograms represent the results of independent cultures for each strain. The values in the plots are the average fluorescence value of each strain with the standard error of the mean N = 3. (d,e) Lipid (d) and paramylon (e) contents of the WT and B1ZFeL in the heterotrophic culture with and without hypoxic incubation. N = 3. *p < 0.05, **p < 0.005, t-test with Bonferroni’s correction.
In order to directly quantify the actual neutral lipid content of the harvested cells, we measured the weight proportion of the neutral lipids extracted by n-hexane. As shown in Fig. 4d, the results are consistent with those from the flow cytometry analysis with BODIPY505/515-stained E. gracilis cells. It is shown that the B1ZFeL mutant has 1.4 times higher lipid content than the WT both in the aerobic condition and after the hypoxic incubation. Finally, to study the underlying mechanism of the high lipid content, we also examined the weight proportion of paramylon in the cells. As shown in Fig. 4e, the intracellular content of paramylon was found to be lower in the B1ZFeL mutant than the WT both in the aerobic condition and after the hypoxic incubation. The increase in the B1ZFeL mutant’s lipid content accompanied by the decrease in its paramylon storage suggests that the higher lipid content is derived from a constantly enhanced wax fermentation pathway in the cell. Also, the proportion of fatty acids and alcohols which account for the extracted lipids were found to be nearly identical in the WT and B1ZFeL mutant (Supplementary Figures 5A,B, Supplementary Tables 1A,B). These results indicate that the B1ZFeL mutant’s phenotype was not derived from a malfunction of the specific lipid metabolic pathway, but from a general upregulation of the wax fermentation pathway.
Discussion
In our proof-of-principle demonstration of the breeding method, we obtained the industrially useful mutant strain, B1ZFeL, with 40% more lipid content than the WT. The produced strain was found to store wax ester at the expense of lower paramylon storage, which implies the constant upregulation of the wax fermentation process in the mutant strain. The BODIPY505/515-based fluorescence was 4.7 and 8.3 times higher in the B1ZFeL strain than the WT in the aerobic condition and after the hypoxic incubation, respectively, whereas the neutral lipid content was measured to be only 1.4 times higher in the B1ZFeL than the WT both in the aerobic condition and after the hypoxic incubation. These discrepancies are presumably due to the reason that either the BODIPY505/515-stained granules include only a fraction of the entire lipid content that was extracted by n-hexane or the B1ZFeL mutant has an additional phenotype that results in a higher permeability for the staining reagents.
It is worthwhile to stress that while B1ZFeL mutants showed the higher lipid content than the WT in the aerobic condition, the lipid content further increased after the hypoxic incubation. Such a phenotypic alteration may be accomplished by modifying its environment recognition mechanism. B1ZFeL may have an incompletely altered recognition of the environment, meaning that the cells recognize the aerobic condition as a hypoxic condition, which possibly manifests a weak allele of the predetermined target gene. While the obtained mutant also requires a wax fermentation process for an efficient conversion of paramylon into lipid, the efficiency of the process can be enhanced significantly by using the strain. In our demonstration, we performed screening of mutants from four groups of 105 independent genomes (cells) modified by the Fe-ion irradiation, but in order to obtain better mutant strains, it is desirable to expand the size of screening with the same process.
The wax ester fermentation process is an energy-generating reaction in which ATP is produced in the process of decomposing glucose, which is formed by the hydrolysis of paramylon, into pyruvic acid. The following E. gracilis-specific fatty acid synthesis pathway in mitochondria to form acyl CoA from pyruvic acid is break-even in total13,35. Because the paramylon storage was decreased in B1ZFeL (Fig. 4e), its primary carbon source is considered to be the paramylon or surplus glucose. However, some of the glucose which was to be supplied to the TCA cycle may also be used for the wax production because the B1ZFeL mutant showed a slower growth than the WT (Supplementary Figure 4). In fact, on the 6th day of heterotrophic cultivation, the number of B1ZFeL cells was about 85% of that of the WT while the lipid content after the hypoxic incubation was 1.4 times higher in B1ZFeL mutants (Fig. 4d). These numbers indicate that even after the same duration of cultivation, the B1ZFeL mutant strain yields an about 20% higher lipid amount with an increased productivity.
Finally, the utility of our FACS method is not limited to the live cell sorting of E. gracilis, but is also applicable to other Euglena species. Other than E. gracilis, there exist more than 800 Euglena species and related species with common traits of accumulating paramylon and wax36. While E. gracilis is unique in that they proliferate at a high rate, many of the other species also have characteristics useful for industrial applications. For example, E. anabaena var. minor precipitates easily37, E. mutabilis grows in acid mine drainages38, and E. sanguinea produces carotenoid astaxantin39. While not included in this Article, we have attempted and verified that FACS analysis can also be applied to E. anabaena var. minor and E. mutabilis. While E. gracilis is currently the only Euglena species available for industrial use and other species still have a number of technical and economical difficulties to overcome before industrial use, selective breeding of such species is expected to broaden the utility of Euglena.
Materials and Methods
Strains and culture media
The E. gracilis strains used in the study are a WT Z strain and chloroplast-less SM-ZK strain40 originally provided by IAM (JAPAN) and maintained at euglena Co., Ltd. for 10 years. The E. gracilis Z strain were maintained in CM medium (pH 3.5)41 while the E. gracilis SM-ZK strain was maintained in KH medium (pH 3.5)42. For an autotrophic culture and heterotrophic culture of E. gracilis, CM medium and KH medium were used, respectively.
Autofluorescence measurements
The autofluorescence measurements were performed on the E. gracilis WT Z strain cultured in CM medium or KH medium and the chloroplast-less SM-ZK strain cultured in KH medium. Each culture was centrifuged (2,000 g, 30 sec) and washed by water three times. A sample of 105 E. gracilis stain cells was diluted to 1 mL in water. The fluorescence from the strains was measured by a fluorophotometer (F-2500, Hitachi) with excitation light at 350 nm, 488 nm, or 635 nm. For the autofluorescence measurements of BODIPY505/515-stained cells, the cells with and without hypoxic incubation were used. The hypoxic incubation was performed by filling a 1.5 mL tube with the cells at a concentration of 107 cells/mL in the KH medium and incubating the tube in the dark for 2 days at 23 °C.
BODIPY505/515 staining for fluorescence measurements
The stock solution of BODIPY505/515 was provided by dissolving it to DMSO at 1 mM. The stock solution was diluted in water to 10 μM just before use. The cells for staining were cultured as noted. The centrifuged cells at 2,000 g for 30 sec were suspended in water. 200 μL of the prepared 10 μM BODIPY505/515 solution was added to the 200 μL cell suspension which includes 2 × 105 cells in water. The mixture was gently mixed and incubated in the dark for 5 min. Then, the cells were washed with water three times by centrifuging the sample at 2,000 g for 30 sec and re-suspended in water. The stained cells were protected from light until the time of use.
FACS
A fluorescence-activated cell sorter, MoFlo XDP (Beckman Coulter), was used for FACS analysis and cell sorting. This FACS machine is equipped with two semiconductor lasers at wavelengths of 488 nm (Sapphire, Coherent) and 642 nm (CUBE, Coherent) which was not used. The fluorescence signal from each cell was dissected to each wavelength range by optical filters and detected by a series of photomultiplier tubes (H957-27, HAMAMATSU). The fluorescence from BODIPY505/515-stained cells was evaluated by the FACS machine’s FL1-log-height value, which represents the fluorescence intensity that passed through a 529/28 bandpass filter. As described above, the 100-μm nozzle tip was used for high-throughput sorting of E. gracilis. To obtain unbiased results from FACS tests, the sample needed to be tested as soon as it was mixed and set to the suction tube of the FACS machine because wax-ester-rich cells are smaller in specific gravity than paramylon-rich cells which tend to precipitate. Otherwise, the proportion of wax-ester-rich cells to the overall sample volume would vary over the measurement duration. For this reason, every sample was well mixed before it was set to the FACS machine and subjected to FACS tests. The acquisition of FACS data was completed within a minute. Cell sorting was performed with suspending the precipitated cells every few minutes.
Fe-ion irradiation
Fe-ion irradiation was conducted in the same way as in a previous report19 with minor modifications. E. gracilis cells were mutagenized by Fe-ion irradiation. 1 mL of E. gracilis cells (4 × 105 cells/mL) in CM medium were encapsulated in 5 × 7 cm sized hybridization bags and irradiated by Fe ions (LET: 650 keV/μm) at 50 Gy in the RIKEN RI-beam factory (Wako, Saitama, Japan; http://www.rarf.riken.go.jp/Eng/facilities/RIBF.html). The cultures were let recover for a week in 1 mL of KH medium on 24 well plates before being subjected to the mutant screen. The recovery cultures were put under 100 μmol photons/m2/sec of constant illumination at 29 °C.
Growth test
The growth test on E. gracilis cells in the heterotrophic condition was conducted by culturing them in conical flasks on a rotary shaker with about 100 μmol photons/m2/sec of constant illumination at 29 °C. The strength of light illuminated on each flask was averaged by changing the positions of the conical flasks every day. The cells precultured in KH medium were washed. The culture was started with 106 cells in 100 ml of KH medium (OD680 ≈ 0.09). The cell density was measured by a particle analyzer (CDA-1000, Sysmex) each day. On the 6th day, half of the culture was harvested by centrifugation (2,000 g, 2 min) for FACS analysis and quantification of the intracellular paramylon and lipid content. The other half was subjected to hypoxic incubation by sealing the top of the conical flask and protected from ambient light. After 4 days of hypoxic incubation, the remaining half was harvested and analyzed in the same way.
Quantification of paramylon and lipids
The intracellular paramylon and lipids were quantified to verify the changes in their content. The harvested cells were dried in a freeze dryer (FDV-1200, EYELA). Approximately 10 mg and 100 mg of samples of dried cells were used for quantifying their intracellular paramylon and lipid content, respectively. The paramylon content was evaluated in the same way as in previous reports13,37. The dried cells were briefly suspended in 10 mL of acetone and homogenized twice for 90 sec by using a sonicator (UD-201, TOMY). The extract was collected by centrifugation (800 g, 5 min), boiled for 30 min in 10 mL of 1% sodium dodecyl sulfate aqueous solution, and washed twice with 10 mL of 0.1% sodium dodecyl sulfate aqueous solution and then with water. The extracted insoluble carbohydrate, which consists almost entirely of paramylon in E. gracilis13, was quantified using the phenol-sulfuric acid method43. Likewise, The neutral lipid was extracted from the dried cells by using traditionally used n-hexane as a solvent44. 10 mL of n-hexane was added to the dried cells. The suspending solution was homogenized for 90 sec by using a sonicator (UD-201, TOMY). The liquid phase was collected by filtering it with a piece of glass fiber filter paper. Another round of hexane extraction was conducted on the residue. The collected lipid dissolving hexane was evaporated using a rotary evaporating system (N-1100 and NVC-2100, EYELA). After drying, the residue’s weight left in the flask was quantified as the extracted total neutral lipid.
Additional Information
How to cite this article: Yamada, K. et al. Efficient selective breeding of live oil-rich Euglena gracilis with fluorescence-activated cell sorting. Sci. Rep. 6, 26327; doi: 10.1038/srep26327 (2016).
Supplementary Material
Acknowledgments
We thank Dr. Yamagishi (RIKEN) for helping us with the work, Dr. Adachi (Kochi University) for helping us with BODIPY staining, and IAM culture collection for providing E. gracilis strains. The ion beam irradiation was performed at the RIBF operated by the RIKEN Nishina Center and Center for Nuclear Study, University of Tokyo. This work was funded by the ImPACT Program and SIP Program (Cabinet Office, Government of Japan).
Footnotes
Author Contributions K.Y., H.S., T.T., Y.K. and S.M. performed the experiments. K.Y., H.S., K.S., K.G., T.A. and O.I. designed the experiments. K.Y. and H.S. analyzed the data. All participated in writing the manuscript.
References
- Spolaore P., Joannis-Cassan C., Duran E. & Isambert A. Commercial applications of microalgae. J. Biosci. Bioeng. 101, 87–96 (2006). [DOI] [PubMed] [Google Scholar]
- Pulz O. & Gross W. Valuable products from biotechnology of microalgae. Appl. Microbiol. Biotechnol. 65, 635–648 (2004). [DOI] [PubMed] [Google Scholar]
- Tornabene T. G., Holzer G., Lien S. & Burris N. Lipid composition of the nitrogen starved green alga Neochloris oleoabundans. Enzyme Microb. Technol. 5, 435–440 (1983). [Google Scholar]
- Weers P. M. M. & Gulati R. D. Growth and reproduction of Daphnia galeata in response to changes in fatty acids, phosphorus, and nitrogen in Chlamydomonas reinhardtii. Limnol. Ocean. 42, 1584–1589 (1997). [Google Scholar]
- Hirooka S., Higuchi S., Uzuka A., Nozaki H. & Miyagishima S. Acidophilic green alga Pseudochlorella sp. YKT1 accumulates high amount of lipid droplets under a nitrogen-depleted condition at a low-pH. PLoS One 9, e107702 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraan S. Mass-cultivation of carbohydrate rich macroalgae, a possible solution for sustainable biofuel production. Mitig. Adapt. Strateg. Glob. Chang. 18, 27–46 (2013). [Google Scholar]
- Chisti Y. Biodiesel from microalgae. Biotechnol. Adv. 25, 294–306 (2007). [DOI] [PubMed] [Google Scholar]
- Sumida S., Ehara T., Osafune T. & Hase E. Ammonia- and light-induced degradation of paramylum in Euglena gracilis. Plant Cell Physiol 28, 1587–1592 (1987). [Google Scholar]
- Briand J. & Calvayrac R. Paramylon synthesis in heterotrophic and photoheterotrophic Euglena (euglenophyceae). J. Phycol. 16, 234–239 (1980). [Google Scholar]
- Sugiyama A. et al. Hepatoprotective effects of paramylon, a beta-1,3-D-glucan isolated from Euglena gracilis Z, on acute liver injury induced by carbon tetrachloride in rats. J. Vet. Med. Sci. Japanese Soc. Vet. Sci. 71, 885–890 (2009). [DOI] [PubMed] [Google Scholar]
- Sugiyama A. et al. Oral administration of paramylon, a beta-1,3-D-glucan isolated from Euglena gracilis Z inhibits development of atopic dermatitis-like skin lesions in NC/Nga mice. J. Vet. Med. Sci. 72, 755–763 (2010). [DOI] [PubMed] [Google Scholar]
- Watanabe T. et al. Antitumor activity of the β-glucan paramylon from Euglena against preneoplastic colonic aberrant crypt foci in mice. Food Funct. 4, 1685–90 (2013). [DOI] [PubMed] [Google Scholar]
- Inui H., Miyatake K., Nakano Y. & Kitaoka S. Wax ester fermentation in Euglena gracilis. FEBS Lett. 150, 89–93 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi T. et al. Wax ester and lipophilic compound profiling of Euglena gracilis by gas chromatography-mass spectrometry: toward understanding of wax ester fermentation under hypoxia. Metabolomics 11, 175–183 (2014). [Google Scholar]
- Hulanicka D., Erwin J. & Bloch K. Lipid Metabolism of Euglena gracilis. J. Biol. Chem. 239, 2778–2787 (1964). [PubMed] [Google Scholar]
- Schiff J. A., Lyman H. & Russell G. K. [2] Isolation of mutants of Euglena gracilis: An addenum. Methods Enzymol. 69, 23–29 (1980). [Google Scholar]
- Schiff J. A. & Epstein H. T. In Reprod. Mol. subcellular, Cell. (Locke M.) 131–189 (Academic Press, 1965). [Google Scholar]
- Hill H. Z., Schiff J. A. & Epstein H. T. Studies of chloroplast development in Euglena XIII. Variation of ultraviolet sensitivity with extent of chloroplast development. Biophys. J. 6, 125–133 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K. et al. Production of a thermal stress resistant mutant Euglena gracilis strain using Fe-ion beam irradiation. Biosci. Biotechnol. Biochem. 8451, 1–7 (2016). [DOI] [PubMed] [Google Scholar]
- Abe T., Ryuto H. & Fukunishi N. In Plant Mutat. Breed. Biotechnol. (Shu Q. Y., Forster B. P. & Nakagawa H.) 99–106 (CABI 2012). [Google Scholar]
- Hu G. et al. Enhanced lipid productivity and photosynthesis efficiency in a Desmodesmus sp. mutant induced by heavy carbon ions. PLoS One 8, e60700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota S. et al. Phenotypic spectrum of Parachlorella kessleri (Chlorophyta) mutants produced by heavy-ion irradiation. Bioresour. Technol. 149, 432–8 (2013). [DOI] [PubMed] [Google Scholar]
- Hyka P., Lickova S., Přibyl P., Melzoch K. & Kovar K. Flow cytometry for the development of biotechnological processes with microalgae. Biotechnol. Adv. 31, 2–16 (2013). [DOI] [PubMed] [Google Scholar]
- Sinigalliano C. D. et al. Viable cell sorting of dinoflagellates by multiparametric flow cytometry. Phycologia 48, 249–257 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sensen C. W., Heimann K. & Melkonian M. The production of clonal and axenic cultures of microalgae using fluorescence-activated cell sorting. Eur. J. Phycol. 28, 93–97 (1993). [Google Scholar]
- Montero M. F., Aristizábal M. & Reina G. G. Isolation of high-lipid content strains of the marine microalgae Tetraselmis for biodiesel production by flow cytometry and single-cell sorting. J. appl. phycol. 23, 1053–1057 (2011) [Google Scholar]
- Mendoza H. et al. Characterization of Dunaliella salina strains by flow cytometry: a new approach to select carotenoid hyperproducing strains. Electron. J. Biotechnol. 11, 5–6 (2008). [Google Scholar]
- Terashima M., Freeman E. S., Jinkerson R. E. & Jonikas M. C. A fluorescence-activated cell sorting-based strategy for rapid isolation of high-lipid Chlamydomonas mutants. Plant J. 81, 147–159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen Doan T. T. & Obbard J. P. Enhanced lipid production in Nannochloropsis sp. using fluorescence-activated cell sorting. GCB Bioenergy 3, 264–270 (2011). [Google Scholar]
- Lefort-Tran M., Bre M. H., Pouphile M. & Manigault P. DNA flow cytometry of control Euglena and cell cycle blockade of vitamin B12-starved cells. Cytometry 8, 46–54 (1987). [DOI] [PubMed] [Google Scholar]
- Hagiwara S., Takahashi M., Yamagishi A., Zhang Y. & Goto K. Novel findings regarding photoinduced commitments of G1-, S- and G2-phase cells to cell-cycle transitions in darkness and dark-induced G1-, S- and G2-phase arrests in Euglena. Photochem. Photobiol. 74, 726–733 (2001). [DOI] [PubMed] [Google Scholar]
- Govender T., Ramanna L., Rawat I. & Bux F. BODIPY staining, an alternative to the Nile Red fluorescence method for the evaluation of intracellular lipids in microalgae. Bioresour. Technol. 114, 507–11 (2012). [DOI] [PubMed] [Google Scholar]
- Brennan L., Blanco Fernández A., Mostaert A. S. & Owende P. Enhancement of BODIPY505/515 lipid fluorescence method for applications in biofuel-directed microalgae production. J. Microbiol. Methods 90, 137–43 (2012). [DOI] [PubMed] [Google Scholar]
- Cooper M. S., Hardin W. R., Petersen T. W. & Cattolico R. A. Visualizing‘ green oil’ in live algal cells. J. Biosci. Bioeng. 109, 198–201 (2010). [DOI] [PubMed] [Google Scholar]
- Inui H., Miyatake K., Nakano Y. & Kitaoka S. Fatty acid synthesis in mitochondria of Euglena gracilis. Eur. J. Biochem. 142, 121–126 (1984). [DOI] [PubMed] [Google Scholar]
- Johnson L. P. In Biol. Euglena, Vol. I (Buetow D. E.) 1–25 (Academic Press Inc., New York, 1968). [Google Scholar]
- Suzuki K. et al. Selection and characterization of Euglena anabaena var. minor as a new candidate Euglena species for industrial application. Biosci. Biotechnol. Biochem. 79, 1730–6 (2015). [DOI] [PubMed] [Google Scholar]
- Halter D. et al. Surface properties and intracellular speciation revealed an original adaptive mechanism to arsenic in the acid mine drainage bio-indicator Euglena mutabilis. Appl. Microbiol. Biotechnol. 93, 1735–1744 (2012). [DOI] [PubMed] [Google Scholar]
- Grung M. & Liaaen-Jensen S. Algal carotenoids 52; secondary carotenoids of algae 3; carotenoids in a natural bloom of Euglena sanguinea. Biochem. Syst. Ecol. 21, 757–763 (1993). [Google Scholar]
- Oda Y., Nakano Y. & Kitaoka S. Utilization and Toxicity of Exogenous Amino Acids in Euglena gracilis. J. Gen. Microbiol. 128, 853–858 (1982). [Google Scholar]
- Cramer M. & Myers J. Growth and Photosynthetic Characteristics of Euglena gracilis. 17, 384–402 (1952). [Google Scholar]
- Koren L. E. & Hutner S. H. High-yield media for photosynthesizing Euglena gracilis Z. J. Protozool. 14, (Suppl.), 17 (1967). [Google Scholar]
- Montgomery R. Determination of glycogen. Arch. Biochem. Biophys. 67, 378–386 (1957). [DOI] [PubMed] [Google Scholar]
- Ayers A. L. & Dooley J. J. Laboratory extraction of cottonseed with various petroleum hydrocarbons. J. Am. Oil Chem. Soc. 25, 372–379 (1948). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.