Abstract
Significance: The Drosophila larval imaginal discs, which form the adult fly during metamorphosis, are an established model system for the study of epithelial tissue damage. The disc proper is a simple columnar epithelium, but it contains complex patterning and cell-fate specification, and is genetically tractable. These features enable unbiased genetic screens to identify genes involved in all aspects of the wound response, from sensing damage to wound closure, initiation of regeneration, and re-establishment of proper cell fates. Identification of the genes that facilitate epithelial wound closure and regeneration will enable development of more sophisticated wound treatments for clinical use.
Recent Advances: Imaginal disc epithelia can be damaged in many different ways, including fragmentation, induction of cell death, and irradiation. Recent work has demonstrated that the tissue's response to damage varies depending on how the wound was induced. Here, we summarize the different responses activated in these epithelial tissues after the different types of damage.
Critical Issues: These studies highlight that not all wounds elicit the same response from the surrounding tissue. A complete understanding of the various wound-healing mechanisms in Drosophila will be a first step in understanding how to manage damaged human tissues and optimize healing in different clinical contexts.
Future Directions: Further work is necessary to understand the similarities and differences among an epithelial tissue's responses to different insults. Ongoing studies will identify the genes and pathways employed by injured imaginal discs. Thus, work in this genetically tractable system complements work in more conventional wound-healing models.

Rachel Smith-Bolton, PhD
Scope and Significance
The Drosophila imaginal disc is an attractive model tissue for epithelial damage repair because of its high regenerative capacity, well-characterized patterning, conserved genes and signaling pathways, and genetic tractability. This article will compare the different methods of inducing damage, including physical fragmentation of the disc, ablation of large portions of tissue, induction of cell death in small clusters of cells, and irradiation. We will describe what is known to date about the changes in and around the wound, and how they differ based on the method of wound induction.
Translational Relevance
Not all tissues heal well or regenerate after damage, with many human tissues preferentially forming a scar that alters organ function. Thus, the study of a genetically tractable epithelial tissue with rapid healing and remarkable regenerative capacity will provide insights into which cellular responses are involved. This basic research has identified multiple signaling pathways and changes in gene expression that are important for wound closure and induction of regeneration.
Clinical Relevance
After tissue damage is induced in humans by trauma or pathology, the surrounding cells choose which wound-response pathways to activate. In many cases, rapid wound closure is followed by scar formation rather than regenerative growth. The field of regenerative medicine seeks to engineer tissues and scaffolds that will enhance scar-less healing and re-growth of lost structures. A better understanding of the factors that promote these desired responses in epithelia that naturally regenerate will improve these engineered products.
Background
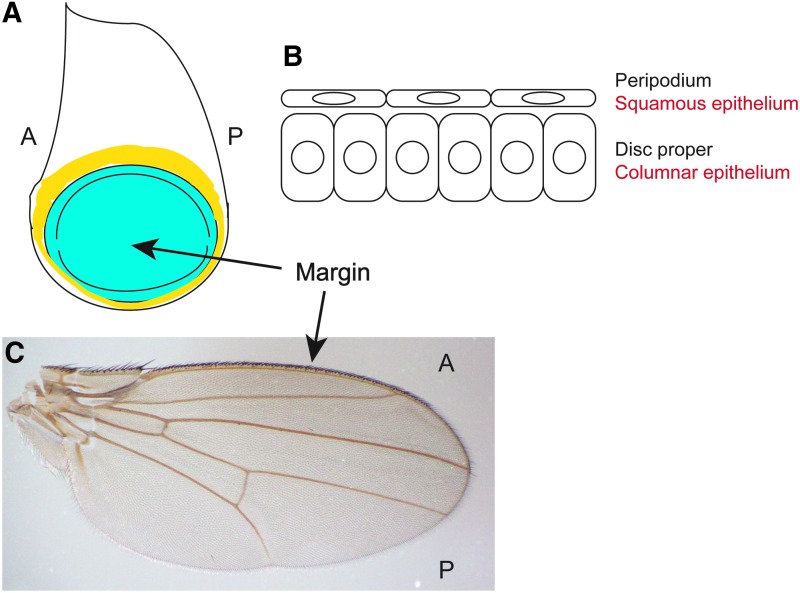
Drosophila larval imaginal discs consist of a simple columnar epithelium adjacent to a squamous epithelium (Fig. 1A, B).1 The primordia of these tissues are specified during embryonic development, and achieve their proper size and shape during larval growth. Extensive work has elucidated the genes that regulate the complex patterning and cell-fate specifications that are established during this juvenile phase of life. On metamorphosis, these tissues transform into the structures of the adult fly such as the wings, legs, eyes, and antennae (Fig. 1C).
Figure 1.
The wing imaginal disc. (A) Drawing of a wing imaginal disc. The blue area will give rise to the wing blade. The yellow area will give rise to the wing hinge. The uncolored areas contribute to the notum and pleura (dorsal and lateral portions of the thorax). A, anterior; and P, posterior. (B) When viewed in cross-section, imaginal discs are made of two epithelial layers: the disc proper, a simple columnar epithelium, and the peripodium, a squamous epithelium. (C) During metamorphosis, the disc proper of the wing imaginal disc becomes the wing blade, shown here. A, anterior; and P, posterior. The disc folds such that the center midline becomes the outer edge of the wing, or margin. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
Seminal work by Ernst Hadorn, Gerold Schubiger, Peter Bryant, and others in the 1960s through 1980s demonstrated that these imaginal epithelia have a remarkable capacity to replace lost tissue and patterning when fragmented. This extensive body of work has been recently comprehensively reviewed.2,3 These early experiments involved removal of discs from the larva, fragmentation of the tissue, and in vivo culture of the pieces in host adult animals during regeneration. Subsequent development of genetic tools enabled systems that induce damage and allow wound healing and regeneration to occur in situ.4,5 In addition to regenerating large portions of lost tissue, Drosophila imaginal discs are capable of replacing dying cells that are scattered throughout the tissue through a process called compensatory proliferation (CP).2,6
While recent exciting work has explored systemic interactions between damaged imaginal discs and the whole animal,7–10 this article will focus on the local signals and changes in gene expression that occur in the epithelium near the tissue damage. These cells close the wound, proliferate to replace lost area, and re-pattern to replace lost cell types. Remarkably, the tissue appears to respond differently depending on the method of induction of damage and the extent of the damage. Here, we describe the similarities and differences among the different methods, and review what is known about the responses observed in the wounded epithelium. We will discuss spatial organization of regenerative proliferation, changes in patterning gene expression, activation of signal transduction pathways, and the role of programmed cell death.
Discussion of Findings and Relevant Literature
Regeneration after fragmentation or tissue ablation
Background
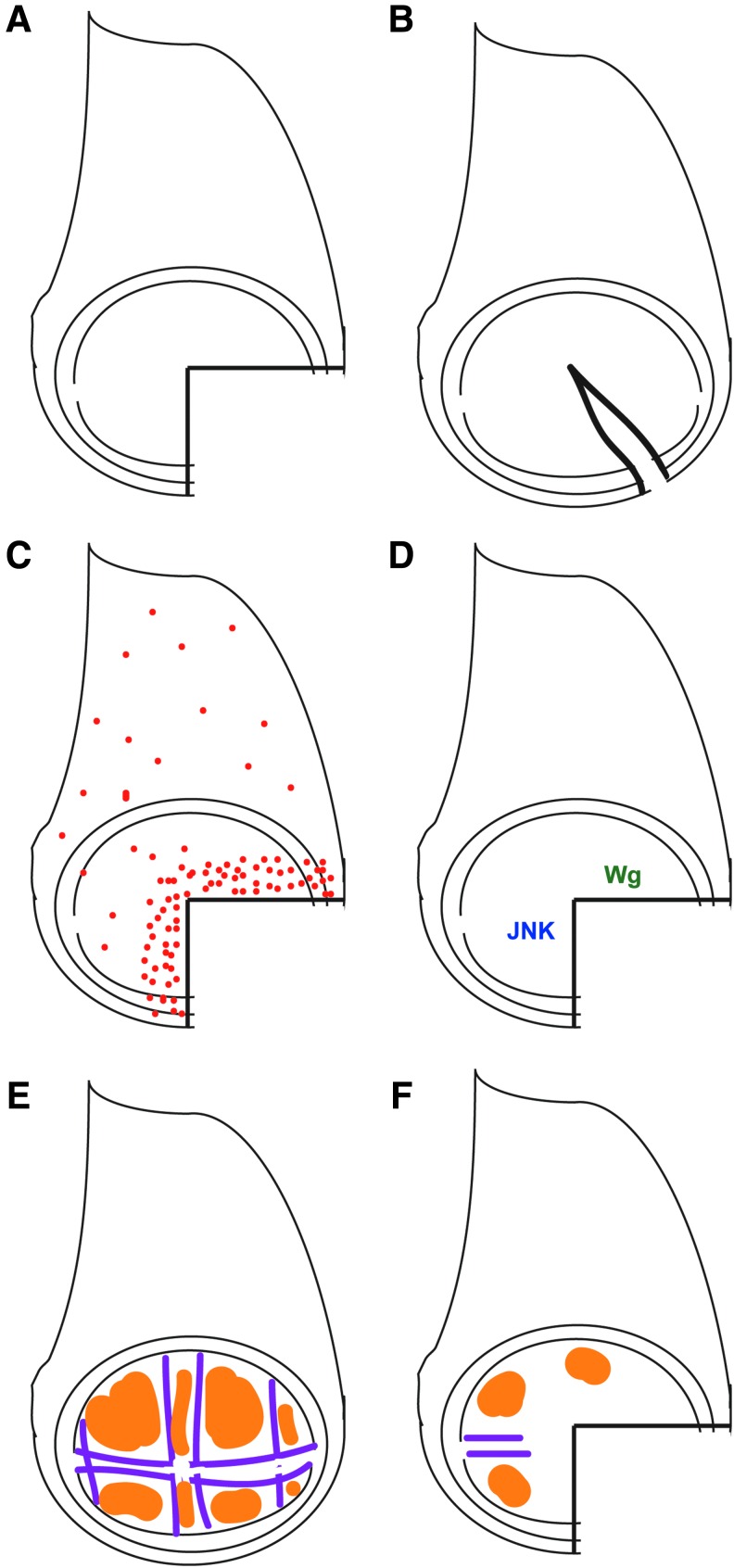
Early wounding and regeneration experiments involved fragmentation of the imaginal disc and culture in host animals (Fig. 2A).2 While this method most closely approximates a traumatic wound or amputation, removing the tissue from the animal and subsequently culturing it likely induces additional stress responses. More recently, wounds in the tissue have been induced by pinching through the intact larval cuticle to make a cut4,11,12 or remove a fragment13 while leaving the wounded disc in place to heal and regenerate (Fig. 2B).
Figure 2.
Regeneration after fragmentation. (A) Drawing showing a typical three-fourth fragment of a wing imaginal disc. (B) Drawing showing a cut in a wing imaginal disc. (C) Red dots represent proliferating cells. Proliferation is concentrated near the wound edges before and after the wound closes.26–28 (D) JNK signaling and Wg expression occur near the wound.4,22,23,30,39,40 Other putative regeneration genes include myc, matrix metalloproteinase 1 (mmp1), regeneration (rgn), augmenter of liver regeneration (alr), cabut, and ash2.4,40,45 (E) Cells have been specified to be vein (purple) or intervein (orange). (F) After fragmentation, vein and intervein markers are no longer expressed near the wound.13 JNK, Jun N-terminal kinase; Wg, wingless. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
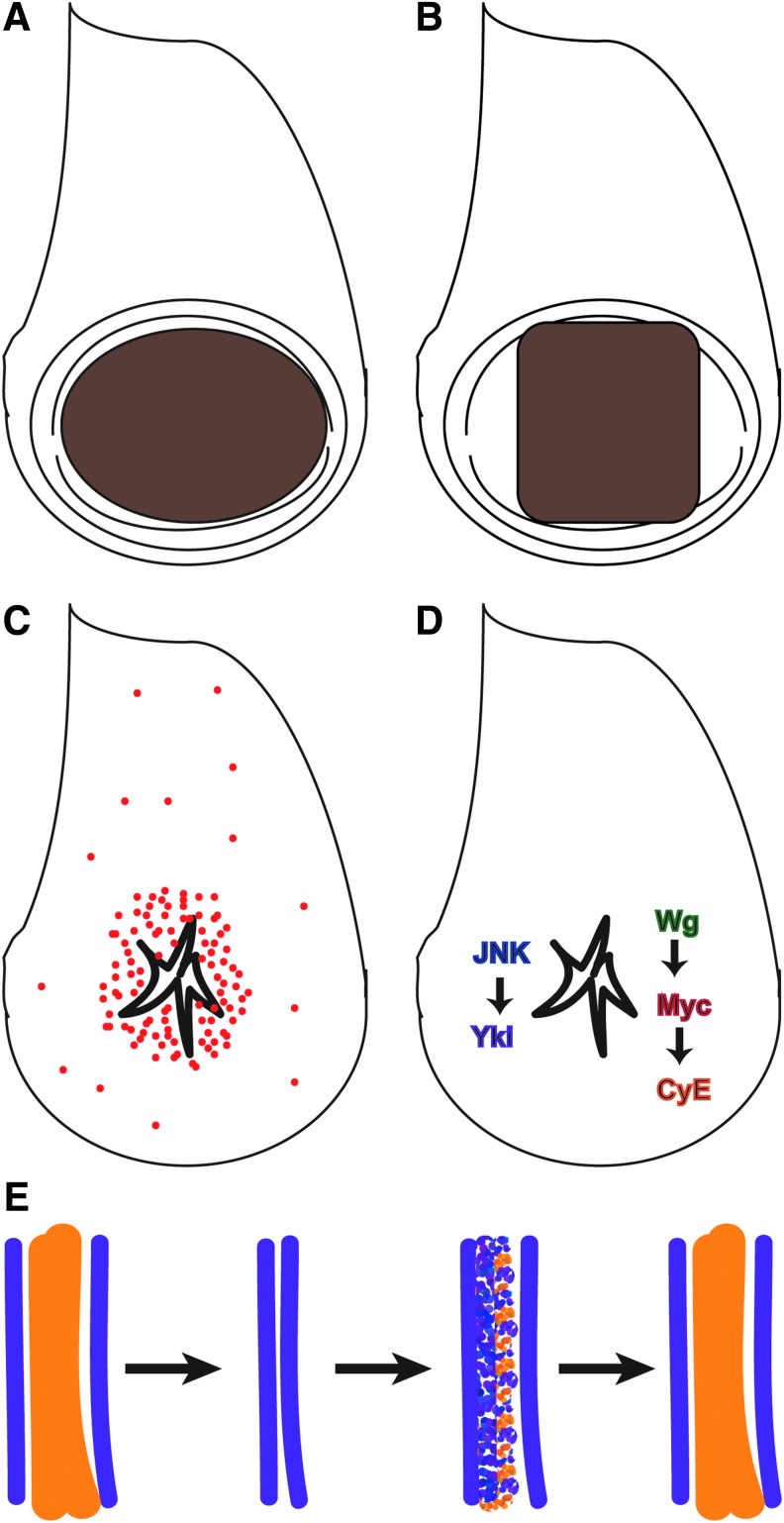
Because physical fragmentation of imaginal discs is impractical for genetic screens, several groups have developed methods of inducing rapid ablation of tissue using genetic tools (Fig. 3A, B).4,5 These techniques use the TARGET GAL4/GAL80TS transcriptional regulatory system14 to control ablation spatially and temporally in a defined portion of the wing imaginal disc. Ablation is carried out by expression of the pro-apoptotic genes reaper4,5 or eiger.4 Ablation is rapid, occurring within 10–24 h when reaper expression is used, and nearly complete, as >95% of cells are eliminated.4,5 Cellular debris moves basally, allowing wound healing to occur.5 Thus, these systems approximate conditions after physical fragmentation, with the advantages of being able to damage hundreds of discs simultaneously simply by shifting temperature, while allowing the tissue to regenerate in situ.
Figure 3.
Regeneration after tissue ablation. (A) Drawing showing area targeted for ablation in brown, which is almost the entire wing primordium, as defined by the expression domain of the gene rotund.4 (B) Drawing showing area targeted for ablation in brown, as defined by the expression domain of the gene spalt major.5 (C) Red dots represent proliferating cells. Proliferation is concentrated near the ablation site.4,5 (D) JNK signaling and Yki activation, as well as Wg, Myc, and CycE are important for regenerative growth after tissue ablation.4,5,31 Other putative regeneration genes include ajuba, capicua, fat, and crumbs.4,32,48 (E) After ablation of an entire intervein (orange) between two veins (purple), the vein cells proliferate and change fate to replace the lost intervein cells.48 Yki, Yorkie. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
Apoptosis
One emerging theme in regenerating tissues is the role of apoptosis. Programmed cell death is important for regeneration in Xenopus tails,15 Hydra,16 Planaria,17 zebrafish,18 and mouse skin and liver.19 As discussed later, apoptosis can also induce CP in Drosophila imaginal discs.6 However, the role of apoptotic cells in recovery from imaginal disc fragmentation is less clear. Scanning electron microscopy of wound edges showed cells that appeared to be apoptotic by morphology.20 Furthermore, cells at the edge of a cut induced by pinching through the larval cuticle contained cleaved caspase 3 and fragmented DNA as assessed by terminal dUTP nick-end labeling (TUNEL)*. However, quantification of TUNEL-labeled cells in a fragmented disc after culture in a host animal showed no increase in cell death at the wound edge.21 A functional analysis, in which initiation of cell death is inhibited in cut discs, has yet to be published. However, signals emitted from the relatively small number of dying cells in these tissues likely do not account for all the regenerative growth that takes place. Likewise, when tissue is removed by ablation, the regenerative growth continues for several days after removal of the cellular debris.4,5 Thus, it is unlikely that the dying cells provide the sole source of regenerative signaling.
Wound closure
Tissue that has been damaged by fragmentation or cutting closes the wound and re-establishes epithelial continuity. An actin cable forms at the edge of the wound a few minutes after damage is induced.22,23 Actin-rich cellular processes extend into the wound.20,22–24 Columnar epithelial cells first make contact with the adjacent peripodial (squamous) epithelium before establishing continuity with the columnar cells of the opposite edge.24 The wound is then closed in a zippering motion.22
Wound closure after tissue ablation occurs in a similar manner. Actin-rich processes are extended into the wound by cells at the wound edge.5 Connections are first made at the apical edges of the healing cells, before the healing extends basally.5 Interestingly, for wounds that extend the length of the disc along the anterior-posterior axis, the wound is first closed in the middle, at the dorsal-ventral boundary. Wound closure then continues in a zippering motion extending toward the edges of the disc.5,22
Proliferation
Regenerative proliferation after disc fragmentation occurs in a zone of cells at the wound edge called a blastema, and begins before the completion of wound closure (Fig. 2C).21,23,25–28 However, the proliferation at the wound edge may require contact between the columnar and peripodial epithelia.26 Lineage tracing using the TIE-DYE multicolor system demonstrated that cells in the blastema produce more progeny and generate larger marked clones than cells outside the blastema or cells in undamaged discs.29 Remarkably, cells outside the blastema proliferate less, with most of the cells at a distance from the wound arrested in G1 or G2.26,28 Marked clones of cells near the wound had an elongated shape, suggesting orientation of cell division toward the wound edge, as well as separation of clonally related cells, suggesting that cells are displaced by intercalating neighbors.30
Regenerative proliferation after ablation is similarly localized to a blastema near the wound (Fig. 3C).4,5 When the ablation was restricted to one compartment, the blastema was also restricted to that compartment.5 When the region ablated was less than a full region of the wing disc, the blastema spread throughout the remainder of that region, in a compartment-specific manner.5 Interestingly, proliferation at a distance from the wound is reduced, in a manner similar to the cell-cycle arrest observed after disc fragmentation.4,28
Jun N-terminal kinase signaling
The Jun N-terminal kinase (JNK) signal transduction pathway is activated at the wound edge in a fragmented disc and is essential for closing the wound (Fig. 2D).22,23 Experiments that permanently labeled all cells that experienced JNK signaling demonstrated that most of the blastema and subsequent regenerated tissue were descended from these JNK-positive cells.21 JNK signaling is also activated in dying and surviving cells at the wound edge after tissue ablation, and is required for wound closure and regenerative growth (Fig. 3D).5 Blocking JNK activity in the dying cells did not impair wound closure or regeneration, indicating that the requirement for JNK signaling is in the blastema cells.5 JNK signaling promotes regenerative growth after tissue ablation, in part, through activating the Ajuba LIM domain protein, which targets components of the Hippo signaling pathway that negatively regulate the transcription factor Yorkie (Fig. 3D) (Yki, Yes-associated protein [YAP] in vertebrates).31,32 Yki activity promotes growth in many contexts, and is required for full wing disc regeneration.33,34 Importantly, JNK signaling is commonly activated in damaged or stressed tissue, confirming that the wound response in Drosophila bears similarities to the wound response in vertebrates.29,35 Furthermore, recent studies have confirmed that is important for vertebrate limb bud regeneration and skin wound healing,36,37 demonstrating that regeneration and wound-healing genes identified in model systems such as Drosophila will yield new factors that regulate vertebrate wound healing.
Mitogen signaling
Signaling downstream of hedgehog (Hh), the wingless/Int family of proteins (WNT)-class molecule Wingless (Wg), and the BMP-class molecule Decapentaplegic (Dpp) control the growth and patterning of imaginal discs.1 Thus, it is possible that these signals are redeployed to direct and control regenerative growth. While the expression of Dpp does not expand toward a cut wound edge,38 ablation of almost the entire primordial wing (Fig. 3A) led to expanded Dpp expression and signaling.4 However, the significance of this expansion remains unclear. Ectopic Wg expression occurs near cut edges in leg, wing, and eye imaginal discs (Fig. 2D).4,30,39,40 Furthermore, the genomic enhancer element that regulates wg expression in response to fragmentation has been identified.41 In addition, Wg is upregulated in the wing cells that form the blastema after tissue ablation (Fig. 3D).4 Expression of Myc and Cyclin E, which promote growth and proliferation, is upregulated in response to Wg signaling after ablation (Fig. 3D), and Myc is also strongly expressed at cut edges in wing imaginal discs.4 Wg signaling and Myc activity are important for blastema proliferation, and overexpression of Myc enhances regeneration.4 Interestingly, WNT signaling is a key factor involved in the regeneration of multiple vertebrate tissues.42
Cell fate and patterning
On fragmentation, large portions of tissue with specified cell fates are removed. How these cell fates are restored after regeneration is an open question. Often, regenerating tissue is assumed to repeat normal development.42,43 However, after removal of the most distal portions of a leg imaginal disc, proper patterning and specification of leg segments were restored in an order that was different from normal development.44 Thus, the mechanisms underlying pattern and cell-fate replacement after tissue fragmentation may be complex and are not yet understood.
Identification of regeneration genes
The strength of Drosophila as a model system lies in its genetic tractability. However, the methods used to damage imaginal discs physically are impractical for high-throughput genetic screening. Instead, these methods have been used in secondary screens for wound healing and regeneration genes after primary screening was carried out using an easier system. For example, genes identified in a screen as modifiers of altered tissue identity and outgrowth caused by aberrant signaling (transdetermination), including matrix metalloproteinase 1 (mmp1), regeneration (rgn), and augmenter of liver regeneration (alr), were subsequently shown to be important for blastema formation after disc fragmentation.40
With the advent of microarrays and high-throughput sequencing, obtaining transcriptional profiles of tissues of interest has become routine and affordable. To identify novel regeneration genes, fragmented discs have been used in conjunction with microarrays to generate a transcriptional profile of regenerating tissue.45 Key genes identified include cabut (cbt) and the chromatin modifier ash2. Since Drosophila is genetically tractable, genes identified in this way can be functionally validated using fragmentation experiments in mutants.
The development of genetic ablation systems has enabled genetic screens for regeneration genes. Pilot screens identified chromosomal deficiencies and growth-regulating mutants that enhanced or suppressed regeneration, demonstrating the feasibility of this approach.4 In particular, mutations in the growth regulator capicua led to enhanced regeneration.4,46 Thus, genetic screens using these systems have the potential to identify undiscovered regeneration genes.
Recent findings
Two recent papers have used different methods of inducing damage to characterize cell fate in the regeneration blastema. To avoid the complications inherent in culturing regenerating tissue, Díaz-García and Baonza have developed a method to cut fragments out of imaginal discs by applying pressure through the larva cuticle.13 This method leaves the animal intact and enables regeneration to occur in situ. They have observed dramatic changes in cell fate close to the wound edge, as well as at a distance from the wound if the fragment removed is large. For example, genes and reporters expressed specifically in pro-vein cells and cells that will constitute the intervein regions of the adult wing were no longer expressed by 20 h after wounding (Fig. 2E, F).13 These results are consistent with findings using tissue ablation and irradiation to induce damage.4,47 This de-specification did not require Wg signaling or regenerative proliferation.13
Consistent with a loss of cell fate and pattern, Díaz-García and Baonza also observed a loss of normal Wg expression. However, they did not detect an upregulation of Wg at the cut site, which has been previously observed in cut leg, eye, and wing discs both in culture and in situ.4,30,39,40 It is possible that different methods of inducing the wound and maintaining the regenerating tissue elicit different responses in gene expression. It is also possible that discs in mid-late third instar, when this in situ fragmentation was carried out, are unable to activate the full regeneration program. Indeed, in many of these damaged wing discs, the lost tissue was not fully restored. Such a loss of regenerative capacity and robust Wg expression at the wound site has been noted in mid-late third instar discs damaged by tissue ablation.4 Thus, more work is needed to define the regenerative capacity of maturing imaginal discs, as well as the role of Wg signaling in the regenerative response.
Using tissue ablation to induce large wounds, Repiso et al.48 closely characterized what happens to proliferating cells at a wound edge. They found that cell division orients toward the wound, similar to cell division near cut edges.30,48 This orientation requires the cell polarity genes fat and crumbs, as well as the growth-promoting transcription factor Yki.48 To answer how lost cell fates are replaced, they specifically ablated a single intervein region that was flanked by pro-veins. Through observation of vein and intervein markers, as well as lineage-tracing experiments, they determined that during regenerative growth, vein cells become re-specified as intervein cells to replace the lost region (Fig. 3E).48 Thus, ablated tissue is replaced through a combination of proliferation that is oriented toward the position of the closed wound as well as re-specification of cell identity.
Compensatory proliferation
Background
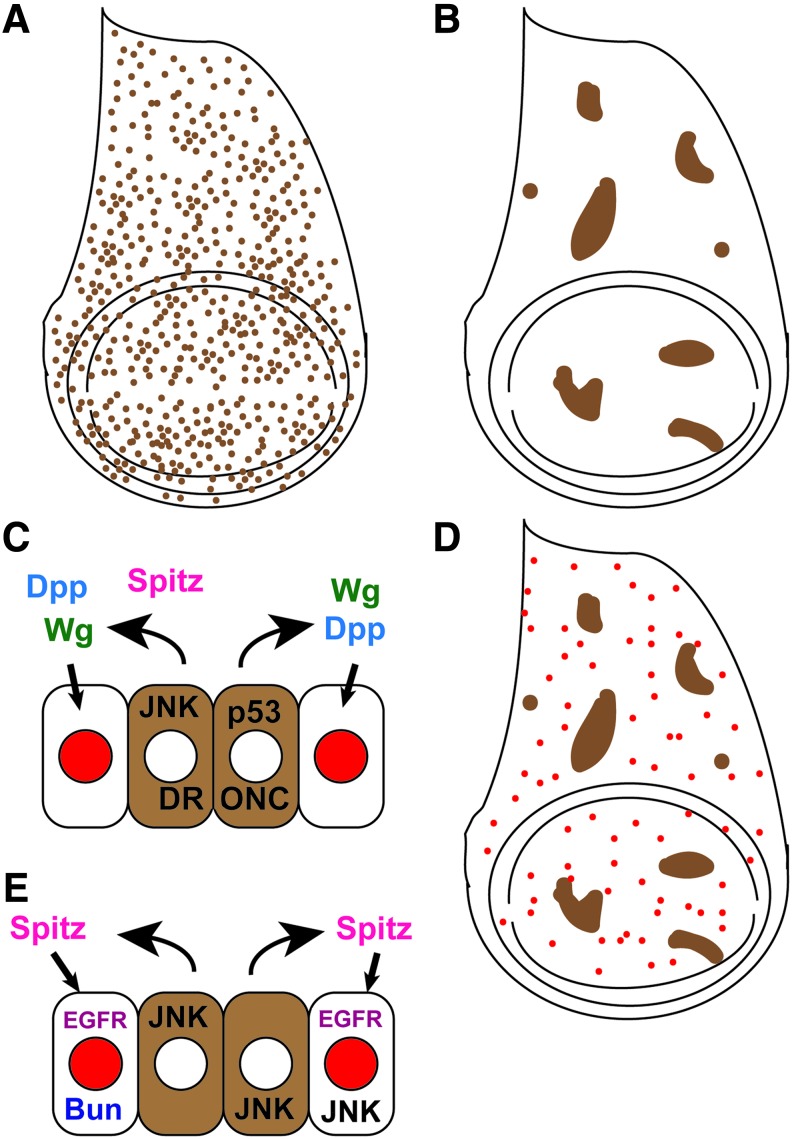
The term CP refers to proliferation that returns the tissue to proper size and shape after sporadic or scattered damage in an imaginal disc. Such damage can be caused by irradiation, induction of apoptosis in clones of cells, or induction of apoptosis that removes some but not all cells in a compartment (Fig. 4A, B). These experimental methods are thought to model cell loss due to irradiation damage, pathology, or activation of tumor-suppressor mechanisms in cancerous cells. Some definitions of CP include mass ablation of tissue as described earlier.49 However, the physical and molecular responses to the loss of large contiguous areas of tissue compared with loss of individual cells or small groups of cells are sufficiently different in that it is helpful to discuss them separately.
Figure 4.
Compensatory proliferation. (A) Irradiation causes death of cells (brown dots) scattered throughout the wing imaginal disc. (B) Death can also be induced in clones of cells (brown) in the disc. (C) Working model of AiP caused by “undead” cells: JNK signaling and p53 and DRONC activity in the undifferentiated “undead” cells of the wing disc (brown) induce production of Wg and Dpp and Spitz, which signal nearby cells to proliferate (red).51–54,66 (D) Proliferation during CP occurs throughout the affected tissue, and it is not localized to a blastema.60 (E) Working model of CP after “genuine” cell death combining findings from recent work, in which JNK can act upstream of Spitz, which signals through the EGFR.66 Bunched and JNK are also required in the cells carrying out the CP.58,60 Additional CP genes include drice, dcp-1, hh, kekken, mtRNApol, top3α, and cmet.55,60,66 AiP, apoptosis-induced proliferation; CP, compensatory proliferation; Dpp, decapentaplegic; DRONC, Drosophila Nedd2-like caspase; EGFR, epidermal growth factor receptor. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
In many models of CP, proliferation requires activation of the apoptotic pathway in the dying cells. This mechanism has been termed apoptosis-induced proliferation (AiP).49 Because all current experimental models of CP involve the death of cells, it is possible that all of them act through AiP, though this is not yet demonstrated. When such dying cells are kept alive by blocking completion of apoptosis, generating “undead” cells, the result is overgrowth of the surrounding tissue, rather than re-establishment of proper tissue size and shape.50 Here, we will discuss CP and mechanisms of AiP stimulated by both “undead” cells and “genuine” cell loss, pointing out differences and similarities where they are known.
Apoptosis
The ability of dying cells to stimulate growth in surrounding tissue was shown by several groups using various methods to induce apoptosis in imaginal discs, with several studies also simultaneously blocking completion of cell death, generating “undead” cells.51–56 These studies showed that the apoptotic pathway components DRONC,51,54,56 DrICE, and Dcp-1,55 and the damage response factor p5354 can play a role in stimulating AiP, depending on the cell type (Fig. 4C). Interestingly, “genuine” dying cells confer protection on nearby surviving cells, which become harder to kill through a mechanism that involves the micro-RNA bantam.57
Wound closure
Because the damage that stimulates CP is scattered throughout the tissue, there are no large discontinuities of the epithelium that require closing. This lack of need for wound closure is true even when cell loss is significant throughout an entire compartment.58 The absence of gaps in the epithelium is one difference between CP and regeneration after fragmentation or rapid tissue ablation.
Proliferation
During CP, cell divisions near clones of dying cells are oriented toward the clone, similar to cell division near a wound edge.59 However, loss of clones of cells does not induce a significant increase in localized proliferation or blastema at the clone edge, but does lead to elevated proliferation throughout the affected compartment (Fig. 4D).60
JNK signaling
JNK signaling is activated by “undead” cells when cell death is induced by expression of the DIAP-1 inhibitors Hid or Reaper.52 However, unlike JNK activity in the regeneration blastema, JNK in AiP induced by “undead” cells is thought to act in the dying cells to induce expression of mitogens that signal to nearby surviving cells (Fig. 4D).52 JNK in “genuine” CP will be discussed further in “recent findings” later on in this article.
Mitogen signaling
AiP involves secretion of signals from “genuine” or “undead” apoptotic cells to stimulate growth in surrounding tissue. Both Dpp and Wg are expressed in arrested apoptotic cells that are generated through a variety of techniques in undifferentiated, proliferating wing and eye imaginal tissue (Fig. 4D).51,53,55 By contrast, inducing apoptosis among differentiating photoreceptors in the eye imaginal disc led to Hh secretion that caused proliferation in surrounding undifferentiated cells.55 To investigate the role of these mitogenic signals further, irradiation was used to induce cell death in wing discs that lacked Wg or Dpp.61 CP occurred normally in the absence of these signals. Thus, while Wg and Dpp are crucial for overgrowth induced by “undead” cells, some other mechanism must induces “genuine” CP that respects normal organ-size controls.
Cell fate and patterning
Wells and Johnston47 asked what happens to cell-fate specification and patterning when large numbers of cells in the wing disc undergo apoptosis after irradiation damage, resulting in extensive CP. While existing patterning was not disrupted, specification of cell fate such as veins, interveins, and sensory organ precursors were significantly delayed until CP was largely complete.47 This delay in patterning required activity of the damage sensor p53. However, no reports show that CP can cause loss of established vein and intervein gene expression as is observed after fragmentation or ablation of contiguous tissue.
Identification of regeneration genes
Because the mechanisms that induce CP in response to sporadic clusters of dying cells are unknown, forward genetic screens have been employed to identify the genes involved. The first such screen was an “enhancer-trap” screen, which identified loci with increased expression after induction of death in clones of cells.62 A subset of these loci also showed increased expression after irradiation as well as fragmentation.62 Thus, these loci contain factors expressed during CP and regeneration.
A second genetic screen used a “mosaic” technique, in which mitotic recombination in the developing eye generated two types of genetically distinct cells: cells that died due to a temperature-sensitive mutation as in the enhancer-trap screen above, and cells that were homozygous mutant for genes to be screened.57 This screen sought mutants in which the living cells failed to carry out enough CP to replace the dying cells. Many mutations isolated affect genes that are also required for normal developmental growth, but CP was particularly sensitive to their loss. One gene identified in this screen was bunched, a growth regulator63 that was also identified in the enhancer trap screen described earlier64 as well as in the transcriptional profile of regenerating discs,45 suggesting at least some similarities between CP and regeneration (Fig. 4E).60
Recent findings
Two recent studies have taken a close look at what occurs in imaginal discs undergoing CP. The first, by Herrera et al.,58 induced cell death by expressing the pro-apoptotic gene hid throughout the primordial wing under spatiotemporal control. After 24 h, only half of the cells had died, and no “wound” or discontinuity in the epithelium was observed, distinguishing this system from tissue ablation.4,5,58 Lineage tracing demonstrated that over 48 h, 75% of the cells could be replaced while maintaining epithelial integrity and proper patterning. The initial proliferative response was not localized in a blastema, similar to other examples of CP,60 though by 48 h after induction of cell death higher levels of proliferation occurred in the region of the dying cells.58 The replacement cells were descended from surviving cells within the targeted region as well as from cells within the primordial wing hinge, which is adjacent to the primordial wing.58 This re-specification of hinge to wing is reminiscent of the vein-to-intervein transition observed after complete tissue ablation of an intervein region.48 In a subsequent study, conversion of cells from anterior to posterior and vice versa was also detected, indicating temporary loss of a lineage restriction that is not violated during normal development.65 Importantly, unlike in AiP caused by “undead” cells, JNK signaling was clearly required in the cells carrying out CP (Fig. 4E).58 However, in contrast to regeneration in response to tissue ablation or fragmentation, Wg was neither expressed nor required in the cells carrying out CP.58
A second recent study, published by Fan et al.,66 used a two-stage screening process to identify genes required for CP. The first stage used a model for “undead” AiP in the eye imaginal disc to screen for mutations that can modify the proliferative response. The screen identified Spitz, a ligand for the Epidermal Growth Factor Receptor (EGFR), as important for AiP. Spitz acts downstream of JNK signaling and activates EGFR signaling in surviving cells.66 The second stage used a model of “genuine” AiP, in which the targeted cells were allowed to complete death, to confirm the results obtained through the “undead” studies. This model is very similar to that used in Herrera et al.,58 with the pro-apoptotic gene hid expressed in the dorsal half of the eye imaginal disc in a spatiotemporally controlled manner, killing a portion of the dorsal eye disc cells.66 Experiments using this model confirmed a requirement for JNK signaling and Spitz, as well as activation of EGFR signaling in “genuine” apoptosis-dependent CP, and confirmed that the restorative proliferation was evenly distributed throughout the dorsal half of the eye disc, similar to the even distribution of proliferation when cell death is induced via hid in the wing disc (Fig. 4E).58,66
Interestingly, this genetic screen covered the same region of the genome covered in the mosaic screen for CP genes described earlier,60 but isolated distinct loci. This discrepancy could be because (1) the CP in the mosaic screen may not be activated by the apoptosis pathway in the dying cells but through another mechanism; (2) the different systems caused cell loss in different spatial distributions, which may activate different CP mechanisms; or most likely (3) the technical differences in screening methods likely identified different classes of CP regulators.
Summary
Drosophila imaginal discs are a genetically tractable model used to study epithelial healing, regeneration, and CP. Tissue damage can be caused by cutting, ablating, irradiating, or inducing apoptosis. In regeneration, a wound closes and localized proliferation occurs. In CP, there is no wound and proliferation is uniform in the repopulating tissue. JNK signaling is important for both regeneration and CP, though it may play different roles depending on the type of tissue damage. Regenerating tissue also requires Wg signaling and Yki activation to promote growth. The growth-promoting signals in CP models that do not use “undead” cells are likely different, and may include Spitz. In both cases, damage induces changes in cell fate: temporary loss of cell fate markers, delay in specification of cell types, or trans-specification of cell types to replace lost structures. Ongoing work using the power of Drosophila genetics will clarify how epithelial cells carry out regeneration and CP to produce fully functional adult body parts of the proper size and form after sustaining significant damage during the juvenile phase of life.
Take-Home Messages.
• Drosophila imaginal discs are a genetically tractable model for epithelial wound closure, regeneration, and CP.
• Tissue damage can be induced by fragmentation, tissue ablation, and scattered cell death.
• Proliferation is localized in regeneration and evenly distributed in CP.
• All methods of damage induce JNK signaling.
• Regeneration requires Wg, Yki, and Myc.
• CP after “genuine” cell death requires Bunched and Spitz, while tissue overgrowth induced by “undead” cells requires Wg, Dpp, Spitz, and EGFR.
• Genetic screens for genes that regulate wound closure, regeneration, and CP will identify novel genes required for these processes.
Abbreviations and Acronyms
- AiP
apoptosis-induced proliferation
- BMP
bone morphogenetic protein
- CP
compensatory proliferation
- Dcp-1
death caspase-1
- DIAP-1
Drosophila Inhibitor of apoptosis 1
- Dpp
decapentaplegic
- DrICE
Drosophila Ice, caspase 3
- DRONC
Drosophila Nedd2-like caspase
- EGFR
epidermal growth factor receptor
- Hh
hedgehog
- JNK
Jun N-terminal kinase
- TARGET
temporal and regional gene expression targeting
- TIE-DYE
three independent excisions-dye
- TUNEL
terminal dUTP nick-end labeling
- Wg
wingless
- WNT
wingless/Int family of proteins
- YAP
yes-associated protein
- Yki
Yorkie
Acknowledgments and Funding Sources
The author would like to thank Andrea Skinner, Keaton Schuster, and Amanda Brock for a critical reading of this article. R.S.-B. is funded by a Young Investigator Award from the Roy J. Carver Charitable Trust (#12-4041).
Author Disclosure and Ghostwriting
Rachel Smith-Bolton has no commercial associations and did not use a ghostwriter.
About the Author
Rachel Smith-Bolton, PhD, obtained her BA from Harvard University in Biochemical Sciences and her PhD from Stanford University under the direction of Michael Simon, PhD. She carried out her Postdoctoral research with Iswar Hariharan, MBBS, PhD, at both the Massachusetts General Hospital Cancer Center and the University of California, Berkeley. She is currently an Assistant Professor at the University of Illinois Urbana-Champaign in the Department of Cell and Developmental Biology, School of Molecular and Cell Biology, as well as an Affiliate of the Institute for Genomic Biology within the Regenerative Biology & Tissue Engineering Theme and the Gene Networks in Neural & Developmental Plasticity Theme.
unpublished experiment at University of California, Berkeley, 2009.
References
- 1.Held LI., Jr Imaginal Discs. Cambridge, MA: Cambridge University Press, 2005 [Google Scholar]
- 2.Worley MI, Setiawan L, Hariharan IK. Regeneration and transdetermination in Drosophila imaginal discs. Annu Rev Genet 2012;46:289–310 [DOI] [PubMed] [Google Scholar]
- 3.Bergantiños C, Vilana X, Corominas M, Serras F. Imaginal discs: renaissance of a model for regenerative biology. Bioessays 2010;32:207–217 [DOI] [PubMed] [Google Scholar]
- 4.Smith-Bolton RK, Worley MI, Kanda H, Hariharan IK. Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev Cell 2009;16:797–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergantiños C, Corominas M, Serras F. Cell death-induced regeneration in wing imaginal discs requires JNK signalling. Development 2010;137:1169–1179 [DOI] [PubMed] [Google Scholar]
- 6.Morata G, Shlevkov E, Pérez-Garijo A. Mitogenic signaling from apoptotic cells in Drosophila. Dev Growth Differ 2011;53:168–176 [DOI] [PubMed] [Google Scholar]
- 7.Halme A, Cheng M, Hariharan IK. Retinoids regulate a developmental checkpoint for tissue regeneration in Drosophila. Curr Biol 2010;20:458–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colombani J, Andersen DS, Léopold P. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science 2012;336:582–585 [DOI] [PubMed] [Google Scholar]
- 9.Garelli A, Gontijo AM, Miguela V, Caparros E, Dominguez M. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science 2012;336:579–582 [DOI] [PubMed] [Google Scholar]
- 10.Katsuyama T, Paro R. Innate immune cells are dispensable for regenerative growth of imaginal discs. Mech Dev 2013;130:112–121 [DOI] [PubMed] [Google Scholar]
- 11.Bryant PJ. Regeneration and duplication following operations in situ on the imaginal discs of Drosophila melanogaster. Dev Biol 1971;26:637–651 [DOI] [PubMed] [Google Scholar]
- 12.Pastor-Pareja JC, Wu M, Xu T. An innate immune response of blood cells to tumors and tissue damage in Drosophila. Dis Model Mech 2008;1:144–154; discussion 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Díaz-García S, Baonza A. Pattern reorganization occurs independently of cell division during Drosophila wing disc regeneration in situ. Proc Natl Acad Sci U S A 2013;110:13032–13037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE 2004;2004:pl6. [DOI] [PubMed] [Google Scholar]
- 15.Tseng A-S, Adams DS, Qiu D, Koustubhan P, Levin M. Apoptosis is required during early stages of tail regeneration in Xenopus laevis. Dev Biol 2007;301:62–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chera S, Ghila L, Dobretz K, et al. Apoptotic cells provide an unexpected source of Wnt3 signaling to drive hydra head regeneration. Dev Cell 2009;17:279–289 [DOI] [PubMed] [Google Scholar]
- 17.Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sánchez Alvarado A. Cell death and tissue remodeling in planarian regeneration. Dev Biol 2010;338:76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gauron C, Rampon C, Bouzaffour M, et al. Sustained production of ROS triggers compensatory proliferation and is required for regeneration to proceed. Sci Rep 2013;3:2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F, Huang Q, Chen J, et al. Apoptotic cells activate the “phoenix rising” pathway to promote wound healing and tissue regeneration. Sci Signal 2010;3:ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reinhardt CA, Hodgkin NM, Bryant PJ. Wound healing in the imaginal discs of Drosophila. I. Scanning electron microscopy of normal and healing wing discs. Dev Biol 1977;60:238–257 [DOI] [PubMed] [Google Scholar]
- 21.Bosch M, Baguñà J, Serras F. Origin and proliferation of blastema cells during regeneration of Drosophila wing imaginal discs. Int J Dev Biol 2008;52:1043–1050 [DOI] [PubMed] [Google Scholar]
- 22.Bosch M, Serras F, Martín-Blanco E, Baguñà J. JNK signaling pathway required for wound healing in regenerating Drosophila wing imaginal discs. Dev Biol 2005;280:73–86 [DOI] [PubMed] [Google Scholar]
- 23.Mattila J, Omelyanchuk L, Kyttälä S, Turunen H, Nokkala S. Role of Jun N-terminal kinase (JNK) signaling in the wound healing and regeneration of a Drosophila melanogaster wing imaginal disc. Int J Dev Biol 2005;49:391–399 [DOI] [PubMed] [Google Scholar]
- 24.Reinhardt CA, Bryant PJ. Wound healing in the imaginal discs of Drosophila. II. Transmission electron microscopy of normal and healing wing discs. J Exp Zool 1981;216:45–61 [DOI] [PubMed] [Google Scholar]
- 25.Kiehle CP, Schubiger G. Cell proliferation changes during pattern regulation in imaginal leg discs of Drosophila melanogaster. Dev Biol 1985;109:336–346 [DOI] [PubMed] [Google Scholar]
- 26.O'Brochta DA, Bryant PJ. Distribution of S-phase cells during the regeneration of Drosophila imaginal wing discs. Dev Biol 1987;119:137–142 [DOI] [PubMed] [Google Scholar]
- 27.Bryant PJ, Fraser SE. Wound healing, cell communication, and DNA synthesis during imaginal disc regeneration in Drosophila. Dev Biol 1988;127:197–208 [DOI] [PubMed] [Google Scholar]
- 28.Sustar A, Schubiger G. A transient cell cycle shift in Drosophila imaginal disc cells precedes multipotency. Cell 2005;120:383–393 [DOI] [PubMed] [Google Scholar]
- 29.Worley MI, Setiawan L, Hariharan IK. TIE-DYE: a combinatorial marking system to visualize and genetically manipulate clones during development in Drosophila melanogaster. Development 2013;140:3275–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sustar A, Bonvin M, Schubiger M, Schubiger G. Drosophila twin spot clones reveal cell division dynamics in regenerating imaginal discs. Dev Biol 2011;356:576–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun G, Irvine KD. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev Biol 2011;350:139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun G, Irvine KD. Ajuba family proteins link JNK to Hippo signaling. Sci Signal 2013;6:ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staley BK, Irvine KD. Hippo signaling in Drosophila: recent advances and insights. Dev Dyn 2012;241:3–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grusche FA, Degoutin JL, Richardson HE, Harvey KF. The Salvador/Warts/Hippo pathway controls regenerative tissue growth in Drosophila melanogaster. Dev Biol 2011;350:255–266 [DOI] [PubMed] [Google Scholar]
- 35.Sabapathy K. Role of the JNK pathway in human diseases. Prog Mol Biol Transl Sci 2012;106:145–169 [DOI] [PubMed] [Google Scholar]
- 36.Hayashi S, Tamura K, Yokoyama H. Yap1, transcription regulator in the Hippo signaling pathway, is required for Xenopus limb bud regeneration. Dev Biol 2014;388:57–67 [DOI] [PubMed] [Google Scholar]
- 37.Lee M-J, Ran Byun M, Furutani-Seiki M, Hong J-H, Jung H-S. YAP and TAZ regulate skin wound healing. J Invest Dermatol 2014;134:518–525 [DOI] [PubMed] [Google Scholar]
- 38.Mattila J, Omelyanchuk L, Nokkala S. Dynamics of decapentaplegic expression during regeneration of the Drosophila melanogaster wing imaginal disc. Int J Dev Biol 2004;48:343–347 [DOI] [PubMed] [Google Scholar]
- 39.Gibson MC, Schubiger G. Hedgehog is required for activation of engrailed during regeneration of fragmented Drosophila imaginal discs. Development 1999;126:1591–1599 [DOI] [PubMed] [Google Scholar]
- 40.McClure KD, Sustar A, Schubiger G. Three genes control the timing, the site and the size of blastema formation in Drosophila. Dev Biol 2008;319:68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schubiger M, Sustar A, Schubiger G. Regeneration and transdetermination: the role of wingless and its regulation. Dev Biol 2010;347:315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoick-Cooper CL, Moon RT, Weidinger G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes Dev 2007;21:1292–1315 [DOI] [PubMed] [Google Scholar]
- 43.Nacu E, Tanaka EM. Limb regeneration: a new development? Annu Rev Cell Dev Biol 2011;27:409–440 [DOI] [PubMed] [Google Scholar]
- 44.Bosch M, Bishop S-A, Baguñà J, Couso J-P. Leg regeneration in Drosophila abridges the normal developmental program. Int J Dev Biol 2010;54:1241–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanco E, Ruiz-Romero M, Beltran S, et al. Gene expression following induction of regeneration in Drosophila wing imaginal discs. Expression profile of regenerating wing discs. BMC Dev Biol 2010;10:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tseng A-SK, Tapon N, Kanda H, et al. Capicua regulates cell proliferation downstream of the receptor tyrosine kinase/ras signaling pathway. Curr Biol 2007;17:728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wells BS, Johnston LA. Maintenance of imaginal disc plasticity and regenerative potential in Drosophila by p53. Dev Biol 2012;361:263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Repiso A, Bergantiños C, Serras F. Cell fate respecification and cell division orientation drive intercalary regeneration in Drosophila wing discs. Development 2013;140:3541–3551 [DOI] [PubMed] [Google Scholar]
- 49.Mollereau B, Perez-Garijo A, Bergmann A, et al. Compensatory proliferation and apoptosis-induced proliferation: a need for clarification. Cell Death Differ 2013;20:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan Y, Bergmann A. Apoptosis-induced compensatory proliferation. The Cell is dead. Long live the Cell! Trends Cell Biol 2008;18:467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huh JR, Guo M, Hay BA. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr Biol 2004;14:1262–1266 [DOI] [PubMed] [Google Scholar]
- 52.Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell 2004;7:491–501 [DOI] [PubMed] [Google Scholar]
- 53.Pérez-Garijo A, Martín FA, Morata G. Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development 2004;131:5591–5598 [DOI] [PubMed] [Google Scholar]
- 54.Wells BS, Yoshida E, Johnston LA. Compensatory proliferation in Drosophila imaginal discs requires Dronc-dependent p53 activity. Curr Biol 2006;16:1606–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan Y, Bergmann A. Distinct mechanisms of apoptosis-induced compensatory proliferation in proliferating and differentiating tissues in the Drosophila eye. Dev Cell 2008;14:399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kondo S, Senoo-Matsuda N, Hiromi Y, Miura M. DRONC coordinates cell death and compensatory proliferation. Mol Cell Biol 2006;26:7258–7268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bilak A, Uyetake L, Su TT. Dying cells protect survivors from radiation-induced cell death in Drosophila. PLoS Genet 2014;10:e1004220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herrera SC, Martín R, Morata G. Tissue homeostasis in the wing disc of Drosophila melanogaster: immediate response to massive damage during development. PLoS Genet 2013;9:e1003446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W, Kale A, Baker NE. Oriented cell division as a response to cell death and cell competition. Curr Biol 2009;19:1821–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gerhold AR, Richter DJ, Yu AS, Hariharan IK. Identification and characterization of genes required for compensatory growth in Drosophila. Genetics 2011;189:1309–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pérez-Garijo A, Shlevkov E, Morata G. The role of Dpp and Wg in compensatory proliferation and in the formation of hyperplastic overgrowths caused by apoptotic cells in the Drosophila wing disc. Development 2009;136:1169–1177 [DOI] [PubMed] [Google Scholar]
- 62.Brook WJ, Ostafichuk LM, Piorecky J, Wilkinson MD, Hodgetts DJ, Russell MA. Gene expression during imaginal disc regeneration detected using enhancer-sensitive P-elements. Development 1993;117:1287–1297 [DOI] [PubMed] [Google Scholar]
- 63.Gluderer S, Oldham S, Rintelen F, et al. Bunched, the Drosophila homolog of the mammalian tumor suppressor TSC-22, promotes cellular growth. BMC Dev Biol 2008;8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Addison WR, Brook WJ, Querengesser LD, Tiong SY, Russell MA. Analysis of an enhancer trap expressed in regenerating Drosophila imaginal discs. Genome 1995;38:724–736 [DOI] [PubMed] [Google Scholar]
- 65.Herrera SC, Morata G. Transgressions of compartment boundaries and cell reprogramming during regeneration in Drosophila. eLIFE 2014;3:e01831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fan Y, Wang S, Hernandez J, et al. Genetic models of apoptosis-induced proliferation decipher activation of JNK and identify a requirement of EGFR signaling for tissue regenerative responses in Drosophila. PLoS Genet 2014;10:e1004131. [DOI] [PMC free article] [PubMed] [Google Scholar]