Abstract
Survivors of pediatric acute lymphoblastic leukemia (ALL) are at increased risk for cognitive impairments that disrupt everyday functioning and decrease quality of life. The specific biological mechanisms underlying cognitive impairment following ALL remain largely unclear, but previous studies consistently demonstrate significant white matter pathology. We aimed to extend this literature by examining the organization of the white matter connectome in young patients with a history of ALL treated with chemotherapy only. We applied graph theoretical analysis to diffusion tensor imaging obtained from 31 survivors of ALL age 5–19 years and 39 matched healthy controls. Results indicated significantly lower small-worldness (p = 0.007) and network clustering coefficient (p = 0.019), as well as greater cognitive impairment (p = 0.027) in the ALL group. Regional analysis indicated that clustered connectivity in parietal, frontal, hippocampal, amygdalar, thalamic, and occipital regions was altered in the ALL group. Random forest analysis revealed a model of connectome and demographic variables that could automatically classify survivors of ALL as having cognitive impairment or not (accuracy = 0.89, p < 0.0001). These findings provide further evidence of brain injury in young survivors of ALL, even those without a history of central nervous system (CNS) disease or cranial radiation. Efficiency of local information processing, reorganization of hub connectivity, and cognitive reserve may contribute to cognitive outcome in these children. Certain connectome properties showed U-shaped relationships with cognitive impairment suggesting an optimal range of regional connectivity.
Key words: : cancer, cognition, connectome, diffusion tensor imaging (DTI)
Introduction
Cognitive impairment is a common neurotoxicity associated with cancer and/or its therapies. This neurotoxicity is often especially debilitating for pediatric cancer survivors due to disruption of vulnerable, developing neural circuitry. Candidate mechanisms for cancer-related cognitive impairment include suppression of neural progenitor proliferation, dysregulation of proinflammatory cytokine cascades, oxidative stress, microvascular damage, and genetic vulnerabilities (Krull et al., 2013a; Monje and Dietrich, 2012; Seigers et al., 2010). Neuroimaging studies indicate that alterations of brain structure and function represent the final common biological pathway resulting in cognitive deficit. Few neuroimaging studies have been conducted to date, but white matter pathology is the most consistent finding (Bhojwani et al., 2014; Edelmann et al., 2014; ElAlfy et al., 2014; Kesler et al., 2010; Morioka et al., 2013).
White matter properties are commonly measured using diffusion tensor imaging (DTI), a noninvasive, magnetic resonance imaging technique. DTI measures the diffusion of water molecules along white matter tracts, which is high along tracts and low perpendicular to them. A variable to quantify this contrast is the fractional anisotropy (FA), with a value between 0 for fully isotropic and 1 for fully anisotropic diffusivity. FA is used as a marker for the degree of myelination and axonal integrity (Mukherjee et al., 2008). DTI is also capable of detecting the diffusion direction and can thereby be used to map the trajectory of virtual white matter fibers, or streamlines, in three-dimensional (3D) space. This technique is referred to as tractography, or fiber tracking.
We applied graph theoretical analysis, also known as graph theory, to DTI data to construct connectomes, which are models of the brain network comprised of regions and their connections. Cognitive functions are believed to be supported by distributed, parallel neural networks that must balance the competing demands of segregation (specialization) and integration (Catani et al., 2012). Connectomes tend to display a “small-world” organization, wherein specialized groups or clusters of neurons are highly connected to each other while being economically connected to other clusters (Bassett and Bullmore, 2006). Thus, connectome properties provide unique insights regarding the efficiency of information processing.
In addition to better representing the distributed, interdependent nature of neural interactions that support cognition, connectome methods have other distinct advantages over traditional voxel-wise neuroimaging approaches. Connectome analyses involve multivariate rather than mass univariate models and tend to combine multiple sources of imaging information. For example, DTI connectomes incorporate FA, tractography, and regional volume. Connectomes also provide both global and local metrics of brain structure and function. Thus, connectomes are multimodal, comprehensive models that improve characterization of the brain's significant complexity.
Small-world topology is established early in brain development (Fan et al., 2011) and disrupted connectome organization has been associated with several other conditions in children that have similar cognitive profiles to pediatric acute lymphoblastic leukemia (ALL) survivors. These include attention deficit hyperactivity disorder (Wang et al., 2009), mild traumatic brain injury (Yuan et al., 2015), and learning disability (Hosseini et al., 2013). We previously showed that the gray matter connectome of young ALL survivors had lower small-world organization compared to healthy controls (Hosseini et al., 2012). However, the aim of this previous study was to test our connectome analysis toolbox and therefore had limitations with respect to its clinical application. In this study, we expand upon our previous research in a larger sample, and by using individual level, DTI-based connectomes. We hypothesized that connectome organization would be lower in ALL survivors and that these properties would be associated with cognitive deficits.
Materials and Methods
Participants
This study included 32 children with a history of ALL who were off therapy for at least 6 months at the time of enrollment. This group was compared with 40 healthy children. We recruited ALL participants through physician referrals and a recruitment liaison in the local oncology clinic. Specifically, children that met our eligibility criteria, based on medical records, were approached by the recruitment liaison or given the study flyer by their physician during a routine, oncology follow-up appointment. Eight refused to participate and an additional three were approached who were found to be ineligible. Control participants were recruited through community postings. Demographic and medical data are shown in Table 1. Exclusion criteria for participants with ALL were a history of cranial radiation, central nervous system involvement, or gross neuropathology, whereas major sensory impairments, magnetic resonance imaging (MRI) contraindications, or any significant medical or psychiatric condition known to affect cognitive function (diagnosed before or unrelated to cancer) were exclusion criteria for both groups. Participants with ALL received intrathecal chemotherapy through Children's Oncology Group (COG) AALL0331 (N = 10), COG AALL0232 (N = 3), Pediatric Oncology Group (POG) 9904 (N = 6), and POG 9905 (N = 13) protocols. Of these, 23 participants received standard dose and nine received high-dose treatment. Informed consent was obtained from the parent/legal guardian and assent was obtained from all participants. Stanford University's Institutional Review Board approved this study, which was carried out in accordance with the Declaration of Helsinki.
Table 1.
Demographic and Medical Data Shown as Mean (SD) Unless Otherwise Noted
| ALL (N = 31) | Controls (N = 39) | t/χ2 | p | |
|---|---|---|---|---|
| Age at evaluation | 11 (3.4) | 12 (2.9) | 1.08 | 0.284 |
| Maternal education (years) | 15 (3.9) | 16 (2.4) | 0.221 | 0.826 |
| Gender (female) | 55% | 39% | 1.87 | 0.172 |
| Minority statusa | 52% | 44% | 0.446 | 0.504 |
| Age at diagnosis (years) | 5.4 (3.7); range: 2–14 | |||
| Treatment intensity (high) | 29% | |||
| Time since treatment (months) | 35 (31); range: 6–111 |
Minority groups included Black, Asian, Hispanic, and Mixed Race.
ALL, acute lymphoblastic leukemia; SD, standard deviation.
Cognitive status
We administered the following standardized measures to all participants: Coding (processing speed), Vocabulary (expressive language/verbal comprehension), Letter–Number Sequencing (working memory), and Matrix Reasoning (perceptual reasoning) subtests of the Wechsler Intelligence Scale for Children 4th Edition (Wechsler, 2003) and Picture Memory (visual learning) and Verbal Learning subtests of the Wide Range Assessment of Memory and Learning, 2nd Edition (Sheslow and Adams, 2005).
Neuroimaging acquisitions and preprocessing
MRI was performed on the same day as the cognitive testing using a GE Discovery MR750 3.0 Tesla whole-body scanner (GE Medical Systems). High-resolution, T1-weighted images were acquired using one of two 3D spoiled gradient echo pulse sequences: TR = 5.9 or 8.5 msec, TE = 1.452 or 3.396, TI = 300 or 500 msec, flip angle = 11 or 15, FOV = 200 or 240 mm, number of excitation = 1, and acquisition matrix = 256 × 192 or 256 × 256 mm. Totally, 124 or 176 contiguous coronal slices were acquired with slice thickness of 1.5 mm and in-plane resolution of 0.859 × 0.859 mm or 0.781 × 0.781. Diffusion-weighted images were acquired from 32 or 40 contiguous axial slices (thickness = 3 or 6.6 mm) using a single-shot echo planar imaging (EPI) sequence (TE = 62.0 or 76.6, TR = 5000 or 7000 msec, field of view = 240 mm, matrix size = 128 × 128 mm). The diffusion tensor acquired for each slice included either four or six images without diffusion weighting (b = 0 sec/mm2) and diffusion-weighted images along 46 or 48 orthogonal directions (b = 850 sec/mm2). The EPI sequence was repeated twice. The number of participants who received the older pulse sequences (ALL = 17, controls = 18) was equally distributed between the two groups (Chi-squared = 0.52, p = 0.47).
DTI data were visually inspected for quality and the data of one participant in the ALL group and one in the control group were excluded from all further analyses due to excessive motion and/or distortion artifact. DTI volumes were corrected for eddy current distortion and tensor reconstruction was performed using linear least-squares fitting in FMRI Software Library v5.0 (Smith et al., 2004). Deterministic tractography was performed in TrackVis v0.5.2.2 (Wang et al., 2007) using the fiber assignment by continuous tracking algorithm (Mori and van Zijl, 2002). We employed a curvature threshold of 60° and streamlines were smoothed using a spline filter (Kesler et al., 2015).
Connectome construction and measurement
We obtained 90 cortical and subcortical regions of interest (ROIs) in Montreal Neurological Institute space from the Automated Anatomical Labeling Atlas (Tzourio-Mazoyer et al., 2002). ROIs were warped into DTI native space as described previously (Kesler et al., 2015). We determined the number of DTI streamlines connecting each pair of ROIs as well as streamline average FA. Regions were considered connected if one streamline end-point terminated within one region and the other end-point terminated within the other region. This resulted in a 90 × 90 weighted, undirected connectivity matrix for each participant, which was corrected for DTI pulse sequence (−0.50 = older sequence, 0.50 = newer sequence) using linear regression. A threshold of three streamlines was applied to minimize false positive connections (Kesler et al., 2015). We weighted each valid edge using the product of the streamline number and FA divided by average ROI volume (extracted from T1 images) to correct for individual differences in brain size. Brain graphs were constructed for each participant with N = 90 nodes, network degree of E = number of edges, and a network density of D = E/[(N × (N − 1))/2] representing the fraction of present connections to all possible connections.
Several metrics were calculated to evaluate the small-world organization of these brain graphs using Brain Connectivity Toolbox (Rubinov and Sporns, 2010). The clustering coefficient of a node is defined as the proportion of actual connections to possible connections between a node's neighbors. The clustering coefficient of a network is the average of clustering coefficients across nodes and is a measure of network segregation. We refer to the clustering coefficient of the network as “network clustering coefficient” to distinguish it from nodal clustering coefficient. Path length is the number of edges that must be traversed between two nodes. Because efficient information processing is assumed to follow the shortest path between nodes, the characteristic path length of a network is the average shortest path length between all pairs of nodes in the network and is a measure of network integration.
Small-worldness is associated with high network clustering and low characteristic path length (Bassett and Bullmore, 2006). Thus, small-worldness index is calculated as CN/LN, where CN is the normalized network clustering coefficient and LN is the normalized characteristic path length of the network. These values were normalized by the mean network clustering coefficient and characteristic path length of 20 random networks with the same number of nodes, total edges, and degree distribution as the networks of interest (Hosseini and Kesler, 2013; Hosseini et al., 2012). Connectome properties were computed at minimum network connection density (0.12) as well as across a range of densities (0.12–0.27) using the area under the curve (AUC).
We defined a node as a hub if its degree was at least one standard deviation (SD) higher than the mean network degree (Sporns et al., 2007). We also evaluated connectome modularity to provide insight regarding subnetwork organization. This is accomplished by decomposing the network into nonoverlapping groups of regions (modules) that have maximal within-group connections and minimal between-group connections (Sporns and Betzel, 2016). Within-module degree Z-score and participation coefficient were calculated for hub regions that showed altered nodal connectivity (Guimera and Amaral, 2005).
Statistical analyses
Connectome properties
Global and regional small-world connectome properties, including small-worldness index, network clustering coefficient, characteristic path length, nodal clustering, and modularity, were compared between groups using nonparametric permutation analysis with 2000 repetitions (Bruno et al., 2012). This results in a permutation distribution of difference under the null hypothesis. The actual between-group difference in network measure of interest was then placed in the corresponding permutation distribution and a two-tailed p-value was calculated based on its percentile position.
Cognitive status
Group differences in individual cognitive test scores were evaluated using analysis of variance. Z-scores were calculated for each test score based on the control group's mean and SD. A participant was rated as having impaired cognitive function if two or more tests had a Z-score at or below −1.5 and/or one test had a Z-score at or below −2.0 (Wefel et al., 2011). The number of impaired participants was compared between groups using Chi-squared analysis.
Predictors of cognitive impairment
We used random forest (Breiman, 2001) classification to determine an algorithm for automatically predicting cognitive impairment (1 = impaired, 0 = unimpaired) in the ALL group. This approach involves an ensemble of decision trees that are decorrelated through bootstrap sampling of training sets. Specifically, the tree learning algorithm uses random subsets of features to grow multiple trees (a forest) that predict the class of the subject. The overall result of the random forest is the class with the highest number of votes across all trees. Random forest models are versatile, high-performance machine learning methods that are able to handle both continuous and categorical predictors, able to learn complex interactions between predictors and less prone to overfitting than other methods (Kingsford and Salzberg, 2008).
Only significant connectome properties were included as predictors (features). A correlation matrix was calculated to identify redundant features (those with 0.75 or higher absolute correlation). Because of their high correlation (r = 0.94), small-worldness index was included, but network clustering was not. Demographic variables were age at enrollment (years), female gender (1 = female, 0 = male), minority status (1 = minority, 0 = nonminority), and maternal education level (years) as a proxy of cognitive reserve (Kesler et al., 2010). Medical/treatment features were age at diagnosis (years), high-dose treatment (1 = high dose, 0 = standard dose), and time since treatment completion (months).
Recursive feature elimination was used to remove minimally contributing features and optimize the classification. The validity of the classifier was tested using bootstrapping (Efron and Tibshirani, 1997). Modeling steps, including recursive feature elimination, training, tuning, testing, and evaluation of feature importance, were carried out with the R statistical package (R Foundation) using the Classification and Regression Training (caret) (Kuhn, 2008) and randomForest (Liaw and Wiener, 2002) packages.
Results
Global connectome properties
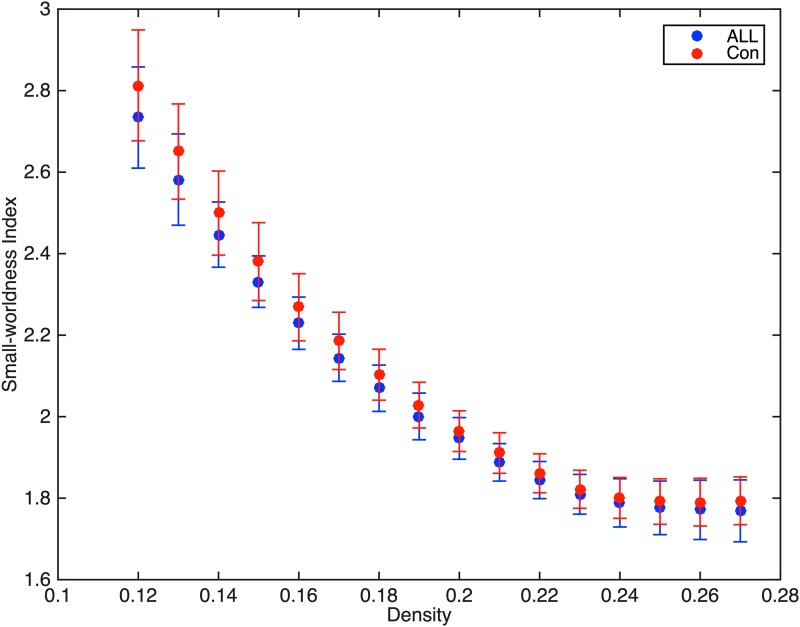
As shown in Figure 1, both groups demonstrated expected small-world connectome organization as indicated by small-worldness index greater than one across network densities (Humphries and Gurney, 2008). There were no individual participants in either group who demonstrated a small-worldness index less than 1. At minimum network connection density, the ALL group showed significantly lower network clustering coefficient (p = 0.019) and small-worldness index (p = 0.007) (Table 2). Characteristic path length was not significantly different between groups (p = 0.78). These results were confirmed by AUC analysis across network densities (Table 2).
FIG. 1.
Both groups demonstrated expected small-world connectome organization with small-worldness index being greater than 1 across network densities. The ALL group demonstrated significantly lower small-worldness index compared to controls (Con). Error bars show standard deviation. ALL, acute lymphoblastic leukemia.
Table 2.
Global Connectome Properties Shown as Mean (SD)
| ALL (N = 31) | Controls (N = 39) | pa | |
|---|---|---|---|
| At minimum connection density (0.12) | |||
| Normalized network clustering coefficient | 3.23 (0.16) | 3.31 (0.17) | 0.019 |
| Normalized characteristic path length | 1.18 (0.02) | 1.18 (0.02) | 0.78 |
| Small-worldness index | 2.73 (0.12) | 2.81 (0.14) | 0.007 |
| Modularity | 0.441 (0.01) | 0.445 (0.01) | 0.499 |
| AUC across densities (0.12–0.27) | |||
| Normalized network clustering coefficient | 0.351 (0.01) | 0.356 (0.01) | 0.048 |
| Normalized characteristic path length | 0.170 (0.002) | 0.170 (0.002) | 0.251 |
| Small-worldness index | 0.308 (0.01) | 0.314 (0.01) | 0.013 |
| Modularity | 0.057 (0.002) | 0.057 (0.002) | 0.150 |
Two-tailed, based on percentile position in permutation distribution.
AUC, area under the curve.
Regional connectome properties
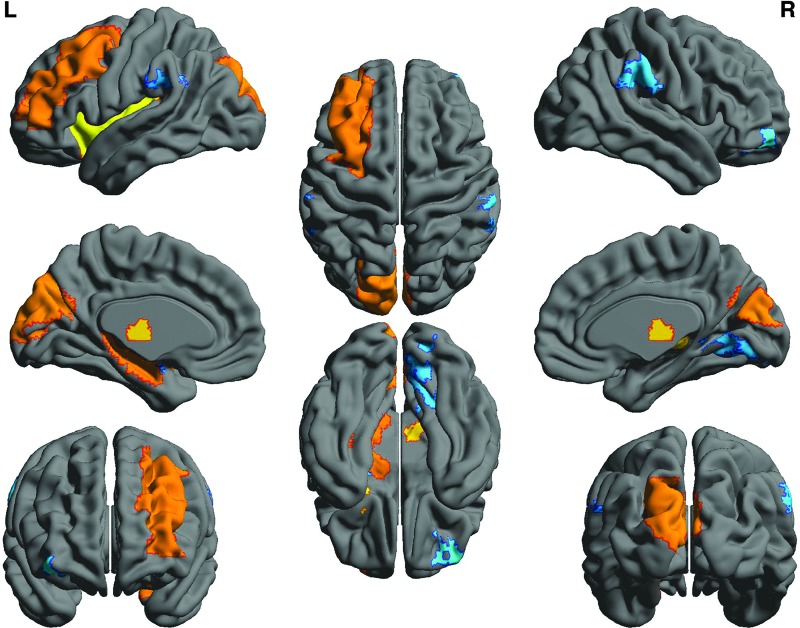
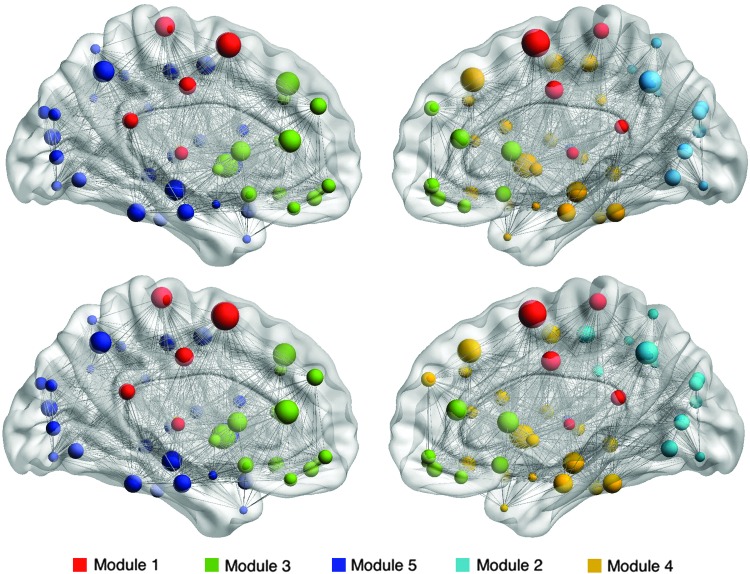
The two groups demonstrated a similar profile of hub regions, including supplementary motor area, putamen, anterior cingulate, superior frontal gyrus, insula, precuneus, cuneus, and hippocampus. However, the ALL group demonstrated significantly altered nodal clustering coefficient in several of these hub regions. Specifically, nodal clustering was lower in left amygdala, right middle frontal orbital gyrus, right lingual gyrus, and bilateral supramarginal gyri. The ALL group also demonstrated significantly higher clustering in left cuneus, left middle frontal gyrus, left hippocampus, left insula, left superior occipital gyrus, and right thalamus (Fig. 2). Modularity was not different between the two groups at minimum density (p = 0.499) or across densities (p = 0.150). Five nearly identical modules were detected for each group (Fig. 3). Module 1 consists of bilateral dorsal mesial regions surrounding somatosensory cortices. Module 2 consists of right hemisphere posterior parietal–occipital regions. Module 3 includes bilateral frontal–striatal regions. Modules 4 and 5 were lateralized to right and left hemisphere, respectively, and consisted of several frontal–parietal and mesial temporal regions. Hubs with altered nodal clustering (left cuneus, left hippocampus, and left insula) demonstrated significantly lower participation coefficients (p < 0.0001) and altered within-module degree (p < 0.0001, Table 3).
FIG. 2.
Compared to controls, the ALL group demonstrated significantly altered nodal clustering coefficient in several regions. Warm colors indicate areas of higher clustering, including left cuneus, left middle frontal gyrus, left hippocampus, left insula, left superior occipital gyrus, and right thalamus. Cool colors indicate regions of lower clustering, including left amygdala, right middle frontal orbital gyrus, right lingual gyrus, and bilateral supramarginal gyri.
FIG. 3.
Brain graphs for the control group (top row) and ALL group (bottom row) with edges represented by lines and nodes represented by spheres. Sphere size reflects nodal clustering. Edges are shown with equal weight for illustration purposes. Node color corresponds to module membership. Both groups demonstrated five nearly identical modules, although certain nodes within each module, particularly module 5, showed altered clustering in the ALL group compared to controls.
Table 3.
Modular Properties of Hub Regions Showing Significantly Altered Clustered Connectivity Shown as Mean (SD)
| ALL (N = 31) | Controls (N = 39) | pa | |
|---|---|---|---|
| Participation coefficient | |||
| Left cuneus | 0.50 (0.09) | 0.70 (0.06) | <0.0001 |
| Left hippocampus | 0.50 (0.06) | 0.62(0.07) | <0.0001 |
| Left insula | 0.49 (0.07) | 0.63 (0.03) | <0.0001 |
| Within-module degree Z-score | |||
| Left cuneus | 0.35 (0.75) | −0.64 (0.51) | <0.0001 |
| Left hippocampus | −0.70 (0.30) | 0.28 (0.44) | <0.0001 |
| Left insula | −0.69 (0.41) | −0.003 (0.37) | <0.0001 |
Two-tailed, based on percentile position in permutation distribution.
Cognitive performance
As shown in Table 4, the ALL group demonstrated significantly greater cognitive impairment (p = 0.027) with lower scores compared with controls on measures of verbal memory, processing speed, and vocabulary (p < 0.034).
Table 4.
Cognitive Data Shown as Mean (SD)
| ALL (N = 31) | Controls (N = 39) | F/χ2 | p | |
|---|---|---|---|---|
| Coding | 50 (9.2) | 54 (8.6) | 4.71 | 0.034 |
| Digits | 51 (8.4) | 54 (9.9) | 3.04 | 0.086 |
| List learning | 48 (11) | 56 (7.3) | 16.4 | <0.0001 |
| Picture memory | 43 (8.6) | 47 (10) | 3.91 | 0.052 |
| Vocabulary | 54 (13) | 60 (12) | 5.35 | 0.024 |
| Matrix reasoning | 52 (11) | 55 (8.6) | 2.17 | 0.146 |
| Impaired | 39% | 15% | 4.92 | 0.027 |
Predictors of cognitive performance
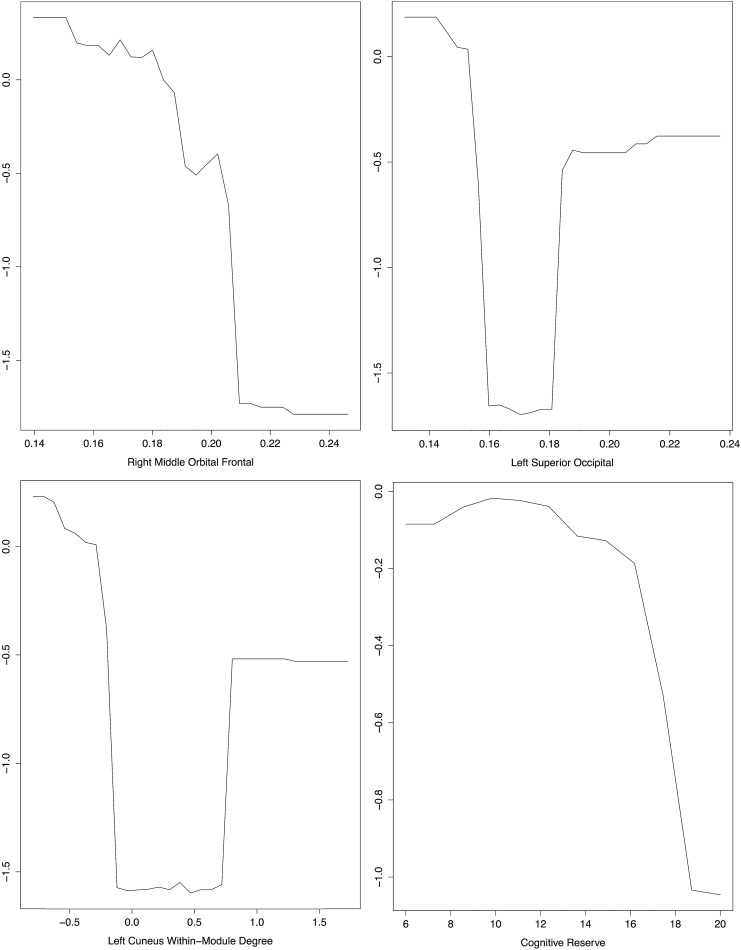
Random forest classification indicated a significantly accurate model for predicting cognitive impairment in the ALL group. Classification accuracy was 89.39% (p < 0.0001). Sensitivity was 95.83% and specificity was 85.71%. As detailed in Figure 4, variables in this model included nodal clustering (right middle orbital frontal gyrus, left superior occipital gyrus), left cuneus within-model degree Z-score and maternal education.
FIG. 4.
Partial dependence plots from random forest classification for features (predictors) in the final model. These plots illustrate the marginal effect of the feature on the class probability (“impaired”) after partialling out the influence of all other variables in the model. Cognitive impairment in the ALL group was associated with lower right middle orbital frontal clustering coefficient and lower cognitive reserve (maternal education). Nonlinear relationships were noted for left superior occipital clustering coefficient and left cuneus within-module degree Z-score, wherein extreme values tended to be associated with greater probability of impairment.
Discussion
This is the first study, to our knowledge, to examine the white matter connectome in young survivors of pediatric ALL. Our findings indicate that connectome small-worldness is lower in these children compared to same-age peers. This reflected lower network clustering coefficient. A small-world network is organized in such a manner that most regions are connected to their neighbors (high clustering) and can be reached by every other region through a small number of steps (low path length). High connectivity of neighboring regions supports efficient local processing (Simon, 1962). Therefore, our findings suggest decreased connectivity within neighboring brain regions such that efficiency of local information processing is reduced in young survivors of ALL. Accordingly, the ALL group also showed greater cognitive impairment than controls.
Our analysis identified five modules in each group, consistent with those observed in a previous study of typically developing children (Chen et al., 2013). Although the capacity of the brain network to be decomposed into modules did not differ between groups, nodal clustering in the ALL group was significantly altered in regions within all five modules. Module 5 included the highest percentage of atypically clustered regions (23% vs. 5–10%). This module was a left lateralized subnetwork of frontal–parietal and mesial temporal regions, including amygdala and hippocampus. These regions are known to support the executive and memory functions that were lower than typical in the ALL group. Module 5 also included regions shown to be associated with vocabulary skills in children such as inferior frontal, inferior temporal, inferior parietal, and precentral gyri (Lee et al., 2014).
A previous study by Edelmann et al. (2014) in slightly older ALL survivors demonstrated a primarily left lateralized profile of alterations in FA in comparison between chemotherapy-treated patients and healthy controls. Given that others have shown bilateral alteration of FA (Morioka et al., 2013) as well as no group differences in FA (Genschaft et al., 2013), the significance, if any, of left lateralized findings remains unclear. However, increased left hemisphere susceptibility has been consistently noted in several other neurological conditions. Potential reasons for this preferential vulnerability include asymmetry in hemodynamics and developmental trajectory (Njiokiktjien, 2006). The language-dominant hemisphere tends to be more highly connected (Parker et al., 2005) which is associated with greater energy demands (Lord et al., 2013). Left hemisphere may therefore be less resilience to disruption of physiological resources due to injury and disease. Although nonsignificant, there were more left-handed controls, so we repeated analyses post hoc with handedness as a covariate. This did not change our results and handedness was not a significant contributor to the models (p > 0.33). Waber et al. (1992) previously noted that children who were younger than 36 months of age, when treated for ALL, tended to demonstrate right hemisphere-based cognitive difficulties, whereas children treated at older ages, demonstrated left hemisphere-related difficulties. We did not detect a significant effect of age at diagnosis on cognitive impairment, but our sample consisted of children who were diagnosed at age 5 years, on average. Therefore, our results seem to support the findings of Waber and colleagues.
Although the two groups showed a similar profile of hub regions, clustered connectivity of several of these hubs (i.e., left hippocampus, left insula, left cuneus) was atypically high in the ALL group. Hubs play an important role in network resilience and regulation of information flow (Vertes and Bullmore, 2015). Hubs with higher intramodular connectivity are known as provincial hubs (high within-module degree, low participation coefficient), whereas those that are more connected to other modules (low within-module degree, high participation coefficient) are connectors. The left hippocampal and left insular hubs in the ALL group demonstrated lower participation in the entire network as well as lower intramodular connectivity compared to controls, but these alterations were not significantly predictive of cognitive impairment. In the ALL group, the left cuneus showed lower participation coefficient and higher within-module degree compared to controls and the latter was associated with intact cognitive function. These results suggest that problematic regional hyperconnectivity of the left cuneus might be offset to some extent by increased intramodular connectivity of this region.
A previous study from our group involving adult onset cancer also suggested that increased connectivity of certain regions may confer a cognitive performance benefit in some cases (Hosseini and Kesler, 2014). In the present study, differences in nodal connectivity involved a profile of both higher and lower nodal clustering compared to controls, which is consistent with our previous study of pediatric ALL survivors showing both hyper- and hypoconnectivity of regions in intrinsic functional networks (Kesler et al., 2014). The study by Edelmann et al. (2014) showed that increased FA in certain regions was associated with poorer cognitive functioning. They suggested that this unexpected relationship may reflect glial scarring and/or white matter compaction.
Our findings contribute novel information by highlighting the complexity of interpreting atypically high versus low white matter connectivity. As shown in Figure 4, the relationship of regional connectivity with cognitive impairment tended to be nonlinear, approximating a U-shaped function such that extreme values were associated with impairment. This suggests that cognition is associated with an optimal range of regional connectivity. Additionally, these findings may help explain the general tendency for correlations between neuroimaging metrics and neuropsychological test scores to be lacking, middling, and/or inconsistent across various populations. As suggested by a reviewer, we examined, post hoc, traditional multiple regression between cognitive test scores and connectome properties. No significant relationships were identified (p > 0.47). Such linear approaches would have missed our important findings that suggest cognitive preservation or impairment likely depends on the balance between various competing neural systems and processes. This information could potentially inform treatment strategies by suggesting that blindly increasing or decreasing certain neural features may result in unwanted side effects. Further research is required to determine the profile of compensatory versus pathological connectome changes following pediatric ALL.
Our results also indicated that maternal education, a proxy for cognitive reserve, helped predict cognitive impairment among young survivors of ALL. Consistent with the cognitive reserve hypothesis, those with higher maternal education levels (i.e., higher reserve) demonstrated an advantage relative to those with lower reserve. Previous research from our group has demonstrated the role of cognitive reserve as a moderator of individual cognitive outcomes in ALL, such that those with higher cognitive reserve prove robust to brain damage up until a critical level for impairment (Kesler et al., 2010).
Frontal–parietal and temporal regions are noted as robustness-related brain regions with respect to cognitive reserve (Santarnecchi et al., 2015). Interestingly, we identified many of these same regions as having altered clustering in the ALL group. This may suggest that altered connectivity in these areas is associated with cognitive impairment that becomes manifest after a certain threshold of damage. Furthermore, the present findings are remarkable in that the expected advantage of reserve on cognition was not conferred unless maternal education was ∼17 years or more (Fig. 4). It is possible that reserve factors must occur at high levels to offer sufficient resilience against cognitive impairments from ALL and its treatments. However, alternative proxies could yield different results. Further investigation regarding this association between cognitive reserve and resilience to ALL is needed, particularly using longitudinal designs.
The specific molecular and cellular mechanisms underlying white matter damage following pediatric ALL are currently unclear. Leukoencephalopathies immediately following intrathecal methotrexate treatments are a well-known risk (Reddick et al., 2005). There is some indication that susceptibility may relate to particular gene variants, many of which are involved in neurogenesis (Bhojwani et al., 2014). Methotrexate has been shown to disrupt oligodendroglial progenitor cells and phospholipids important for myelination and white matter development (Krull et al., 2013b; Monje and Dietrich, 2012). Progenitor cells in white matter appear to be more sensitive to methotrexate than those in gray matter (Wood et al., 2014), which may help explain why white matter injury tends to be the more common finding in survivors of pediatric ALL. Methotrexate may also increase microglial activation (Seigers et al., 2010; Wood et al., 2014), which is associated with several neurotoxic effects, including oxidative and nitrosative stress (Lull and Block, 2010). These mechanisms would be expected to have diffuse rather than focal effects, which is consistent with our findings.
This study has several limitations, including small sample size and cross-sectional design. Without longitudinal data, the differential effects of cancer pathology, chemotherapy, and/or other disease and treatment-related factors cannot be determined. We did not observe significant effects of medical/treatment variables on connectome organization or cognitive function. These have historically been inconsistent predictors perhaps due, in part, to the heterogeneity and small sample sizes involved in many studies. We also did not have available data regarding specific methotrexate dose or exposure to other relevant medication such as glucocorticoids, for example, and, therefore, further research is required. We had a large number of potential predictors of cognitive outcome in a small sample and, therefore, our study may have lacked power to detect certain effects. Our study involved two different DTI pulse sequences including an older, less optimal sequence. However, we employed careful quality assurance methods; pulse sequence was equally distributed between the groups and was not associated with connectome properties. Our cognitive battery was very limited in the number of tests and breadth of cognitive domains covered. We chose a very brief battery to reduce burden and increase compliance in our young cohort, but additional tests may have provided further insights regarding the clinical significance of connectome alterations. Finally, alternative methods such as the use of probabilistic rather than deterministic tractography and/or different regional parcellation schemes may yield different results.
Conclusion
Despite these limitations, we provide further evidence of brain injury in children with ALL who were without CNS disease and treated with chemotherapy alone. We contribute novel findings to this field of research by presenting the first evidence of altered connectome properties and their association with cognitive dysfunction. We also illustrate an interesting preliminary model for predicting cognitive impairment from a combination of connectome properties and cognitive reserve. Continued research regarding the mechanisms by which ALL and chemotherapy disrupt connectome topology and cognitive function could help identify interventions that will protect against these neurotoxicities without reducing the anticancer efficacy of treatment regimens. Small-world connectomes are highly associated with gene expression networks and also appear to be preserved across species (Calabrese et al., 2015; Fakhry and Ji, 2015). Thus, connectome studies potentially provide unique translational opportunities to identify intervention targets for ALL-related cognitive impairment.
Acknowledgment
This research was supported by a grant from the National Institutes of Health (1K07CA134639 and 1R03CA191559 to S.R.K.).
Author Disclosure Statement
No competing financial interests exist.
References
- Bassett DS, Bullmore E. 2006. Small-world brain networks. Neuroscientist 12:512–523 [DOI] [PubMed] [Google Scholar]
- Bhojwani D, Sabin ND, Pei D, Yang JJ, Khan RB, Panetta JC, Krull KR, Inaba H, Rubnitz JE, Metzger ML, Howard SC, Ribeiro RC, Cheng C, Reddick WE, Jeha S, Sandlund JT, Evans WE, Pui CH, Relling MV. 2014. Methotrexate-induced neurotoxicity and leukoencephalopathy in childhood acute lymphoblastic leukemia. J Clin Oncol 32:949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. 2001. Random forests. Mach Learn 45:5–32 [Google Scholar]
- Bruno J, Hosseini SM, Kesler S. 2012. Altered resting state functional brain network topology in chemotherapy-treated breast cancer survivors. Neurobiol Dis 48:329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese E, Badea A, Cofer G, Qi Y, Johnson GA. 2015. A Diffusion MRI tractography connectome of the mouse brain and comparison with neuronal tracer data. Cereb Cortex 25:4628–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catani M, Dell'acqua F, Bizzi A, Forkel SJ, Williams SC, Simmons A, Murphy DG, Thiebaut de Schotten M. 2012. Beyond cortical localization in clinico-anatomical correlation. Cortex 48:1262–1287 [DOI] [PubMed] [Google Scholar]
- Chen Z, Liu M, Gross DW, Beaulieu C. 2013. Graph theoretical analysis of developmental patterns of the white matter network. Front Hum Neurosci 7:716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann MN, Krull KR, Liu W, Glass JO, Ji Q, Ogg RJ, Sabin ND, Srivastava DK, Robison LL, Hudson MM, Reddick WE. 2014. Diffusion tensor imaging and neurocognition in survivors of childhood acute lymphoblastic leukaemia. Brain 137:2973–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, Tibshirani R. 1997. Improvements on cross-validation: The 632+ bootstrap method. J Am Stat Assoc 92:548–560 [Google Scholar]
- ElAlfy M, Ragab I, Azab I, Amin S, Abdel-Maguid M. 2014. Neurocognitive outcome and white matter anisotropy in childhood acute lymphoblastic leukemia survivors treated with different protocols. Pediatr Hematol Oncol 31:194–204 [DOI] [PubMed] [Google Scholar]
- Fakhry A, Ji S. 2015. High-resolution prediction of mouse brain connectivity using gene expression patterns. Methods 73:71–78 [DOI] [PubMed] [Google Scholar]
- Fan Y, Shi F, Smith JK, Lin W, Gilmore JH, Shen D. 2011. Brain anatomical networks in early human brain development. Neuroimage 54:1862–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschaft M, Huebner T, Plessow F, Ikonomidou VN, Abolmaali N, Krone F, Hoffmann A, Holfeld E, Vorwerk P, Kramm C, Gruhn B, Koustenis E, Hernaiz-Driever P, Mandal R, Suttorp M, Hummel T, Ikonomidou C, Kirschbaum C, Smolka MN, 2013. Impact of chemotherapy for childhood leukemia on brain morphology and function. PLoS One 8:e78599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimera R, Amaral LA. 2005. Cartography of complex networks: modules and universal roles. J Stat Mech 2005:P02001-1–P02001-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini SM, Black JM, Soriano T, Bugescu N, Martinez R, Raman MM, Kesler SR, Hoeft F. 2013. Topological properties of large-scale structural brain networks in children with familial risk for reading difficulties. Neuroimage 71:260–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini SM, Hoeft F, Kesler SR. 2012. GAT: a graph-theoretical analysis toolbox for analyzing between-group differences in large-scale structural and functional brain networks. PLoS One 7:e40709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini SM, Kesler SR. 2013. Influence of choice of null network on small-world parameters of structural correlation networks. PLoS One 8:e67354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini SM, Kesler SR. 2014. Multivariate pattern analysis of FMRI in breast cancer survivors and healthy women. J Int Neuropsychol Soc 20:391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries MD, Gurney K. 2008. Network “small-world-ness”: a quantitative method for determining canonical network equivalence. PLoS One 3:e0002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Gugel M, Pritchard-Berman M, Lee C, Kutner E, Hosseini SM, Dahl G, Lacayo N. 2014. Altered resting state functional connectivity in young survivors of acute lymphoblastic leukemia. Pediatr Blood Cancer 61:1295–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Tanaka H, Koovakkattu D. 2010. Cognitive reserve and brain volumes in pediatric acute lymphoblastic leukemia. Brain Imaging Behav 4:256–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler SR, Watson CL, Blayney DW. 2015. Brain network alterations and vulnerability to simulated neurodegeneration in breast cancer. Neurobiol Aging 36:2429–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsford C, Salzberg SL. 2008. What are decision trees? Nat Biotech 26:1011–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull KR, Bhojwani D, Conklin HM, Pei D, Cheng C, Reddick WE, Sandlund JT, Pui CH. 2013a. Genetic mediators of neurocognitive outcomes in survivors of childhood acute lymphoblastic leukemia. J Clin Oncol 31:2182–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull KR, Hockenberry MJ, Miketova P, Carey M, Moore IM. 2013b. Chemotherapy-related changes in central nervous system phospholipids and neurocognitive function in childhood acute lymphoblastic leukemia. Leuk Lymphoma 54:535–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M. 2008. Building predictive models in R using the caret package. J Stat Soft 28:1–26 [Google Scholar]
- Lee NR, Raznahan A, Wallace GL, Alexander-Bloch A, Clasen LS, Lerch JP, Giedd JN. 2014. Anatomical coupling among distributed cortical regions in youth varies as a function of individual differences in vocabulary abilities. Hum Brain Mapp 35:1885–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A, Wiener M. 2002. Classification and regression by random forest. R News 2:18–22 [Google Scholar]
- Lord LD, Expert P, Huckins JF, Turkheimer FE. 2013. Cerebral energy metabolism and the brain's functional network architecture: an integrative review. J Cereb Blood Flow Metab 33:1347–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lull ME, Block ML. 2010. Microglial activation and chronic neurodegeneration. Neurotherapeutics 7:354–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje M, Dietrich J. 2012. Cognitive side effects of cancer therapy demonstrate a functional role for adult neurogenesis. Behav Brain Res 227:376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, van Zijl PC. 2002. Fiber tracking: principles and strategies—a technical review. NMR Biomed 15:468–480 [DOI] [PubMed] [Google Scholar]
- Morioka S, Morimoto M, Yamada K, Hasegawa T, Morita T, Moroto M, Isoda K, Chiyonobu T, Imamura T, Nishimura A, Morimoto A, Hosoi H. 2013. Effects of chemotherapy on the brain in childhood: diffusion tensor imaging of subtle white matter damage. Neuroradiology 55:1251–1257 [DOI] [PubMed] [Google Scholar]
- Mukherjee P, Berman JI, Chung SW, Hess CP, Henry RG. 2008. Diffusion tensor MR imaging and fiber tractography: theoretic underpinnings. AJNR Am J Neuroradiol 29:632–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njiokiktjien C. 2006. Differences in vulnerability between the hemispheres in early childhood and adulthood. Hum Physiol 32:37–42 [PubMed] [Google Scholar]
- Parker GJ, Luzzi S, Alexander DC, Wheeler-Kingshott CA, Ciccarelli O, Lambon Ralph MA. 2005. Lateralization of ventral and dorsal auditory-language pathways in the human brain. Neuroimage 24:656–666 [DOI] [PubMed] [Google Scholar]
- Reddick WE, Glass JO, Helton KJ, Langston JW, Li CS, Pui CH. 2005. A quantitative MR imaging assessment of leukoencephalopathy in children treated for acute lymphoblastic leukemia without irradiation. AJNR Am J Neuroradiol 26:2371–2377 [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. 2010. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52:1059–1069 [DOI] [PubMed] [Google Scholar]
- Santarnecchi E, Rossi S, Rossi A. 2015. The smarter, the stronger: intelligence level correlates with brain resilience to systematic insults. Cortex 64:293–309 [DOI] [PubMed] [Google Scholar]
- Seigers R, Timmermans J, van der Horn HJ, de Vries EF, Dierckx RA, Visser L, Schagen SB, van Dam FS, Koolhaas JM, Buwalda B. 2010. Methotrexate reduces hippocampal blood vessel density and activates microglia in rats but does not elevate central cytokine release. Behav Brain Res 207:265–272 [DOI] [PubMed] [Google Scholar]
- Sheslow D, Adams W. 2005. Wide Range Assessment of Memory and Learning, Second Edition (WRAML2). Lutz, FL: Psychological Assessment Resources [Google Scholar]
- Simon H. 1962. The architecture of complexity. Proc Am Philos Soc 106:467–482 [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. 2004. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23 Suppl 1:S208–S219 [DOI] [PubMed] [Google Scholar]
- Sporns O, Betzel RF. 2016. Modular brain networks. Annu Rev Psychol 67:613–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Honey CJ, Kotter R. 2007. Identification and classification of hubs in brain networks. PLoS One 2:e1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. 2002. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15:273–289 [DOI] [PubMed] [Google Scholar]
- Vertes PE, Bullmore ET. 2015. Annual research review: growth connectomics—the organization and reorganization of brain networks during normal and abnormal development. J Child Psychol Psychiatry 56:299–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waber DP, Bernstein JH, Kammerer BL, Tarbell NJ, Sallan SE. 1992. Neuropsychological diagnostic profiles of children who received CNS treatment for acute lymphoblastic-leukemia—the systemic approach to assessment. Dev Neuropsychol 8:1–28 [Google Scholar]
- Wang L, Zhu C, He Y, Zang Y, Cao Q, Zhang H, Zhong Q, Wang Y. 2009. Altered small-world brain functional networks in children with attention-deficit/hyperactivity disorder. Hum Brain Mapp 30:638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Benner T, Sorensen AG, Wedden VJ. 2007. Diffusion toolkit: a software package for diffusion imaging data processing and tractography. Proc Int Soc Mag Reson Med 15:3720 [Google Scholar]
- Wechsler D. 2003. Wechsler Intelligence Scale for Children, 4th ed. San Antonio: Psychological Corporation [Google Scholar]
- Wefel JS, Vardy J, Ahles T, Schagen SB. 2011. International Cognition and Cancer Task Force recommendations to harmonise studies of cognitive function in patients with cancer. Lancet Oncol 12:703–708 [DOI] [PubMed] [Google Scholar]
- Wood L, Miller S, Vogel H, Monje M. 2014. Differential sensitivity of oligodendrocyte precursor cells and microglia to methotrexate in grey versus white matter (P7.011). Neurology 82:P7.011 [Google Scholar]
- Yuan W, Wade SL, Babcock L. 2015. Structural connectivity abnormality in children with acute mild traumatic brain injury using graph theoretical analysis. Hum Brain Mapp 36:779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]