Supplemental Digital Content is Available in the Text.
Key Words: single genome amplification, pre-exposure prophylaxis, Truvada
Abstract:
We describe HIV-1 evolutionary dynamics in the 4 participants from the TDF2-PrEP trial who became HIV-1 infected while prescribed emtricitabine and tenofovir disoproxil fumarate (FTC/TDF). At seroconversion, virus diversity in the 2 participants with detectable drug was only 0.05% (95% confidence intervals: 0.04 to 0.06) and 0.07% (0.06 to 0.08) compared with 2.25% (1.95 to 2.6) and 0.42% (0.36 to 0.49) in those with no detectable drug and 0.07%–0.69% in 5 placebo recipients (P > 0.5). At 10 months, diversity in adherent participants was only 0.37% (0.31 to 0.41) and 0.86% (0.82 to 0.90) compared with 0.5%–1.7% among participants who did not take FTC/TDF (P > 0.5). Although limited by the small number of infections that reduced the power to detect differences, we found that sequences from seroconverters with detectable drug were more homogeneous than those from placebo or nonadherent seroconverters.
INTRODUCTION
Daily PrEP with FTC/TDF is an effective HIV prevention strategy for persons at high risk of infection.1 In clinical trials, the level of HIV protection by PrEP was strongly correlated with antiretroviral (ARV) drug adherence.2 Across trials, infections in active arms were generally seen in participants with undetectable drug, although some seroconverters had measurable drug at the time of the first laboratory evidence of infection. Suboptimal PrEP adherence and HIV infection create an opportunity for continued drug activity during undiagnosed infection and raise questions on the impact of transient ARV drug pressure on early establishment of infection and acute virus dynamics. In some cohort studies of acute HIV infection, the degree of virus diversity during early infection was found to be predictive of the subsequent disease course3,4. The effect of PrEP on virus evolutionary dynamics and disease course will likely depend on the extent of ARV drug activity at the time of infection and before HIV diagnoses.
The TDF2 was a phase III, randomized, double-blinded, placebo-controlled clinical trial of daily oral FTC/TDF among sexually active heterosexual men and women in Botswana.5 In this trial, the overall protective efficacy in the modified intention-to-treat analysis was 62.2%.5 In the as-treated analysis, in which follow-up data for participants were censored 30 days after their last reported study drug dose, the protective efficacy was 77.9%, with 4 HIV-1 infections in the FTC/TDF group and 19 in the placebo group.
In this study, we used single genome amplification (SGA) and phylogenetic and sequence analysis to create a virological snapshot of HIV-1 in the 4 cases in the FTC/TDF arm of the TDF2 trial. We defined the number of transmitted/founder (T/F) viruses in these 4 infections and compared virus evolution dynamics during the first year of infection among the FTC/TDF arm cases and 5 seroconverters who were not taking study drug.
METHODS
Study Samples
We characterized HIV-1 sequences from the 4 participants from TDF2 that seroconverted in the FTC/TDF arm (1069H, 4108P, 3104V, and 1273P) and from 5 seroconverters that were not taking study drug (1249F, 3184X, 1177E, 3205B, and 4123Y). Table S1 (see Supplemental Digital Content, http://links.lww.com/QAI/A779) shows the characteristics of these individuals including gender, plasma drug levels, and virus load at the seroconversion visit. Two of the 4 FTC/TDF-arm cases (1069H and 4108P) had detectable tenofovir (TFV) and FTC in plasma at a visit before and closest to their estimated seroconversion date, and 2 (3104V and 1273P) did not have measurable drug in plasma.5 None of these seroconverters had detectable drug resistance mutations at the first RNA-positive sample or in any of their samples from subsequent study visits6. The 5 seroconverters that were not taking study drug were selected based on sample availability, plasma HIV-1 viral load, and ability to amplify sequences and perform SGA. For each participant, HIV RNA was extracted from plasma samples collected at the seroconversion visit and a median of 10 months (range: 5–12) later, with the exception of 1273P who was lost to follow up. None of the 9 seroconverters initiated HAART during this period of observation.
SGA and Sequence Analysis
SGA was performed after a standard protocol obtained from the Center for HIV/AIDS Vaccine Immunology using primers specific for HIV subtype C. Serial dilutions of the reverse transcriptase-generated cDNA were made to identify dilutions of cDNA resulting in <30% of positive wells by polymerase chain reaction. Based on a Poisson distribution, amplifications at this dilution represent sequences derived from a single cDNA copy.7 Env sequences (∼1 kb) representing the V1-V5 and flanking regions of HIV env were generated using an ABI3130xl-automated sequencer. For each participant, we generated an average of 21 sequences from the seroconversion visit specimens (range: 8–28) and 23 sequences (range: 17–27) from the specimens collected at a follow-up study visit. All sequences clustered with an HIV subtype C reference sequence (see Figure S1, Supplemental Digital Content, http://links.lww.com/QAI/A779).
Phylogenetic Analysis
Sequences were aligned and edited using Geneious software (version 6.1.6). Phylogenetic analysis was performed using the MEGA (version 5.0) software package. Nucleotide alignments were prepared using ClustalW; all indels were removed from the alignments before phylogenetic analysis. Phylogenetic trees were inferred using the neighbor joining (NJ) method and Tamura-Nei substitution model and rooted with reference HIV sequences from multiple subtypes (A to K). The number of T/F variants that established infection was determined by exploring the pattern of env diversity at the seroconversion visit using the highlighter tool (http://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter.html). For this analysis, we excluded hypermutated sequences and gaps, and we assumed that variants with 2 or less unique nucleotide changes arise from the same T/F variant.7 The Poisson-Fitter tool (http://www.hiv.lanl.gov/content/sequence/POISSON_FITTER/poisson_fitter.html) was used to confirm infection with single variants and calculate the time since infection.8 Estimates of evolutionary divergence were performed by pairwise analysis of T/F virus and later time point sequences using the maximum composite likelihood method in MEGA5. Results were expressed as number of nucleotide substitutions per site per year.9,10 A repeated-measures marginal model with compound symmetry covariance structure was used to compare differences in virus diversity or diversification among participants who seroconverted in the FTC/TDF arm with participants who were not taking study drug.11 Differences in within-subject variation were studied using the Wilcoxon test.
RESULTS
Virus Diversity and Estimated Time Since Infection in the 4 Participants Assigned to FTC/TDF
Both 1069H and 4108P had detectable TFV and FTC in plasma at a visit before and closest to their estimated seroconversion date.5 Figure S2 (see Supplemental Digital Content, http://links.lww.com/QAI/A779) shows the highlighter plots for these 2 participants who demonstrates low diversity at the seroconversion visit consistent with infection with a single variant. Poisson-Fitter analysis of seroconversion visit specimens also supported homogeneous infections with 1 T/F variant and provided an estimated time since infection of 24 days [95% confidence intervals (CI): 11 to 37] for 1069H and 27 days (95% CI: 11 to 43) for 4108P (Table 1).
TABLE 1.
Nucleotide Diversity and Diversification in Seroconverters From the TDF2 Trial
HIV infection in the 2 seroconverters without detectable FTC or TFV (3104V and 1273P), was initiated with multiple variants; 3 in 3104V and 5 in 1273P (Table 1; see Figure S2, Supplemental Digital Content, http://links.lww.com/QAI/A779). Poisson-Fitter analysis of sequences from 1273P further supported heterogeneous infection, which did not allow an estimation of the time since infection. We could not apply the Poisson-Fitter tool to 3104V because the number of bases at which any 2 sequences differed was too high (mean Hamming distance = 17.23).
Nucleotide Diversity and Estimated Time Since Infection in Seroconverters Assigned to Placebo
Infections in seroconverters assigned to placebo were initiated with 1 (1249F, 3205B, and 4123Y) or 2 T/F variants (3184X and 1177E) (see Figure S3, Supplemental Digital Content, http://links.lww.com/QAI/A779). Estimated time since infection was 43 (95% CI: 24 to 62) days for 1249F, 23 (12–33) days for 3205B, and 18 (5 to 31) days for 4123Y (Table 1). We could not calculate the estimated time since infection in 3184X and 1177E due to heterogeneous infections.
Virus Diversity and Diversification in TDF2 Infections
Nucleotide sequences derived from the seroconversion visit specimens from 1069H and 4108P (detectable plasma drug levels) were very homogenous with mean nucleotide diversity values of only 0.05% (95% CI: 0.04 to 0.06) and 0.07% (0.06 to 0.08), respectively (Table 1). These values were higher for the 2 participants assigned to FTC/TDF but who had no detectable plasma drug levels [2.25% (1.95 to 2.60) in 3104V and 0.42% (0.36 to 0.49) in 1273P] and in the seroconverters that were not taking study drug (median = 0.13%; range = 0.07–0.69).
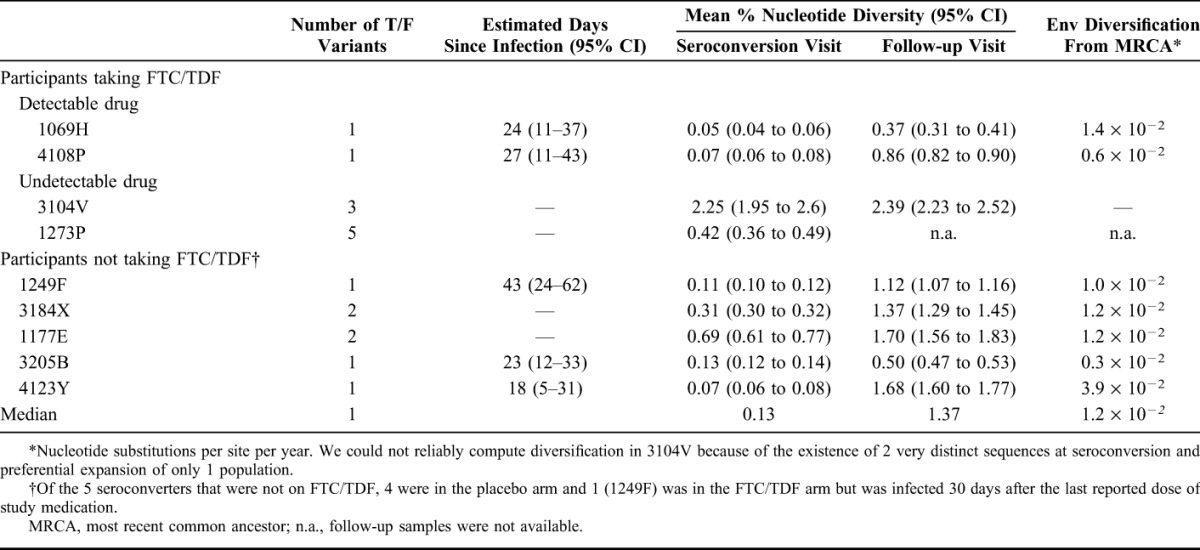
At study follow-up, nucleotide diversity in 1069H and 4108P was 0.37% (95% CI: 0.31 to 0.41) and 0.86% (0.82 to 0.90) compared with a median of 1.37% (min, max = 0.50 to 1.70) in seroconverters that were not on study drug (Table 1). A comparison of virus diversity among seroconverters with detectable plasma drug levels with those that were not on study drug showed no significant differences either at baseline or at follow-up (P > 0.5 for each comparison), although this analysis was limited by the small number of individuals in each group. Figure 1 shows the cumulative frequencies of nucleotide substitutions per site and illustrates how the 2 FTC/TDF participants with detectable plasma drug levels at the seroconversion visit (1069H and 4108P) had low virus diversity both at baseline and at follow-up.
FIGURE 1.
Cumulative frequencies of nucleotide substitutions per site in seroconverters from the TDF2 trial. A, Cumulative frequencies of nucleotide substitutions per site at seroconversion. B, Cumulative frequencies of nucleotide substitutions per site after a median of 10 (range = 5–12) months.
We also investigated nucleotide divergence from the most recent common ancestor after a median of 10 months of infection. Mean nucleotide divergences in 1069H and 4108P fell within the range observed in seroconverters not taking study drug (P > 0.5) (Table 1).
DISCUSSION
We investigated acute HIV dynamics in participants who become infected with HIV during the TDF2-PrEP trial. We defined virus diversity at seroconversion and after 1 year of infection using SGA and phylogenetic and sequence analysis. We found that infections in the 2 participants with measurable FTC and TFV at their seroconversion visit were more homogeneous and evolved more slowly than those from participants who became infected while not taking study drug. These findings are consistent with early observations in macaques infected with SHIV during intermittent oral PrEP with FTC/TDF.12,13 In those studies, continued drug exposure with 1–2 weekly drug doses during acute SHIV infection was associated with limited virus diversity and evolution and a better preservation of immune responses.12–15 This scenario in macaques could easily mimic persons who acquire HIV because of inconsistent PrEP adherence and who continue on the PrEP regimen until diagnosis. We hypothesize that transient PrEP exposure may have also limited HIV diversity in the 2 infections from TDF2 that had detectable FTC and TDF in plasma. Our estimate of 24 and 27 days from HIV infection to seroconversion is consistent with the frequency of HIV testing (monthly) and provides an opportunity for continuous ARV drug exposure between infection and diagnoses. Perhaps not surprisingly, viruses for these 2 participants diversified from the most recent common ancestor at rates statistically indistinguishable from the rates observed for the infections that occurred in the absence of plasma FTC/TDF, likely reflecting FTC/TDF discontinuation after confirmed HIV diagnoses.
A major limitation of our study was the small number of seroconversions in the FTC/TDF arm that reduced statistical power to <30% and limited our ability to compare virus diversity and diversification between treatment groups. Sample size calculations found that for such analysis to have sufficient power, we would require 12 subjects per group for a subject-specific comparison of diversity or 6 subjects per group for a repeated-measures analysis. A recent study also found statistically insignificant lower viremias in FTC/TDF seroconverters from TDF2 compared with those assigned to placebo, although sample size limitations affected the ability to detect small but potentially significant differences in virus load set points.6 Thus, although not definitive, all these findings point to potential virological differences between PrEP breakthrough infections and HIV infections acquired in the absence of ARV drugs and stress the need to expand this type of analysis to other PrEP trials. These studies should also evaluate if PrEP use at the time of infection may favor single variant transmission or monospecific expansions of viruses with fitness advantages in the presence of ARVs.
Our definition of adherence was based on having detectable FTC and TFV in the plasma of the seroconversion-visit blood specimen. All clinical trials of PrEP have demonstrated that efficacy is strongly associated with adherence, especially when assessed by plasma drug concentrations.2 A limitation of this approach is that drug concentrations are measured at a single time associated with a scheduled clinical encounter, which may not reflect consistent adherence and is also subject to a form of social desirability bias whereby less adherent subjects may be prone to take the study product immediately before their study visit so as not to disclose their nonadherence.
In summary, we found that among persons who became HIV-1 infected while taking FTC/TDF PrEP and who at that time had detectable plasma concentrations of drug, env sequences were more homogeneous and evolved more slowly than env sequences isolated from HIV-1 seroconverters who had no detectable FTC or TFV, or were not taking PrEP. If confirmed in other PrEP trials, these findings suggest that transient PrEP exposure during acute infection may have a measurable effect on virus evolution. Our findings underscore the need to better understand the potential impact of PrEP on control of HIV-1 infection.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Rachael Aubert and Janet McNicholl for their help with sample inventory, specimen selection, and study coordination, Jim Smith and Bin Li for sharing the HIV env subtype C primers, and Bill Switzer and Anupama Shankar for their help with the phylogenetic analysis.
Footnotes
Supported by CDC intramural funds.
Work was presented partially at the 8th IAS Conference on HIV Pathogenesis, Treatment, and Prevention (IAS 2015), July 19–22, 2015, Vancouver, Canada.
W.H. and J.G.G.-L. are named on a US Government patent on Inhibition of HIV infection through chemoprophylaxis (US9044509 B2 granted on June 2, 2015). The other authors have no funding or conflicts of interest to disclose.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.jaids.com).
REFERENCES
- 1.Krakower DS, Jain S, Mayer KH. Antiretrovirals for primary HIV prevention: the current status of pre- and post-exposure prophylaxis. Curr HIV/AIDS Rep. 2015;12:127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amico KR, Stirratt MJ. Adherence to preexposure prophylaxis: current, emerging, and anticipated bases of evidence. Clin Infect Dis. 2014;59(suppl 1:S55–S60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rachinger A, Kootstra NA, Gijsbers EF, et al. HIV-1 envelope diversity 1 year after seroconversion predicts subsequent disease progression. AIDS. 2012;26:1517–1522. [DOI] [PubMed] [Google Scholar]
- 4.Markham RB, Wang WC, Weisstein AE, et al. Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci U S A. 1998;95:12568–12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thigpen MC, Kebaabetswe PM, Paxton LA, et al. ; the TDF2 Study Group. Antiretroviral preexposure prophylaxis for heterosexual HIV transmission in Botswana. N Engl J Med. 2012;367:423–434. [DOI] [PubMed] [Google Scholar]
- 6.Chirwa LI, Johnson JA, Niska RW, et al. CD4(+) cell count, viral load, and drug resistance patterns among heterosexual breakthrough HIV infections in a study of oral preexposure prophylaxis. AIDS. 2014;28:223–226. [DOI] [PubMed] [Google Scholar]
- 7.Salazar-Gonzalez JF, Bailes E, Pham KT, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giorgi EE, Funkhouser B, Athreya G, et al. Estimating time since infection in early homogeneous HIV samples using a poisson model. BMC Bioinformatics. 2010;11:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura K, Dudley J, Nei M, et al. MEGA4: Molecular evolutionary Genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. [DOI] [PubMed] [Google Scholar]
- 11.Verbeke G, Molenberghs G. Linear Mixed Models in Practice: A SAS-oriented Approach. New-York: Springer-Verlag; 1997. [Google Scholar]
- 12.Zheng Q, Ruone S, Switzer WM, et al. Limited SHIV env diversification in macaques failing oral antiretroviral pre-exposure prophylaxis. Retrovirology. 2012;9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Lerma JG, Cong M, Mitchell J, et al. Intermittent prophylaxis with oral truvada protects macaques form rectal SHIV infection. Sci Transl Med. 2010;2:14ra14. [DOI] [PubMed] [Google Scholar]
- 14.Curtis KA, Kennedy MS, Luckay A, et al. Delayed maturation of antibody avidity but not seroconversion in Rhesus macaques infected with SHIV during oral pre-exposure prophylaxis. J Acquir Immune Defic Syndr. 2011;57:355–362. [DOI] [PubMed] [Google Scholar]
- 15.Kersh EN, Luo W, Zheng Q, et al. Reduced inflammation and CD4 loss in acute SHIV infection during oral pre-exposure prophylaxis. J Infect Dis. 2012;206:770–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.