Abstract
Glycolysis is a catabolic process of glucose hydrolysis needed for energy and biosynthetic intermediates, whereas gluconeogenesis is a glucose production process important for maintaining blood glucose levels during starvation. Although they share many enzymes, these two processes are not simply the reverse of each other and are instead reciprocally regulated. Two key enzymes that regulate irreversible steps in these two processes are pyruvate kinase (PK) and phosphoenolpyruvate carboxy kinase (PEPCK), which catalyze the last and first step of glycolysis and gluconeogenesis, respectively, and are both regulated by lysine acetylation. Acetylation at Lys305 of the PKM (muscle form of PK) decreases its activity and also targets it for chaperone-mediated autophagy and subsequent lysosome degradation. Acetylation of PEPCK, on the other hand, targets it for ubiquitylation by the HECT E3 ligase, UBR5/EDD1, and subsequent proteasomal degradation. These studies established a model in which acetylation regulates metabolic enzymes via different mechanisms and also revealed cross talk between acetylation and ubiquitination. Given that most metabolic enzymes are acetylated, we propose that acetylation is a major posttranslational modifier that regulates cellular metabolism.
Glucose is the major energy supply for most cells. For example, the brain primarily relies on glucose for energy production. Moreover, glucose is also the major carbon source for anabolic synthesis. Glycolysis is a catabolic process that converts glucose to pyruvate and produces ATP and NADH, which can be oxidized through oxidative phosphorylation (OXPHO) to produce ATP. Under anaerobic conditions (i.e., in the absence of O2), pyruvate, the last glycolytic metabolite, is reduced to lactate, whereas under aerobic conditions (i.e., in the presence of O2), pyruvate is imported into mitochondria where it is metabolized first through the tricarboxylic acid (TCA) cycle and then completely oxidized to produce maximal amounts of ATP as well as CO2 and water in most nondividing cells. In growing cells such as in tumor cells, even in the presence of normal oxygen supply, many glycolytic intermediates are needed for biosynthesis and, as a result, the flux of glycolysis into TCA and OXPHO is reduced, leading to the accumulation of lactate. This phenomenon, known as the Warburg effect (Warburg 1956), is commonly seen in cancer cells. Although the biochemical mechanism and physiological significance underlying the Warburg effect remain incompletely understood, enhanced glucose uptake has provided the basis for the development of FDG-PET technology that has been widely used clinically for detecting tumors by injecting patients with the radiolabeled glucose analog, 2(18F)-fluoro-2-deoxy-D-glucose (FDG), followed by positron emission tomography (PET). The Warburg effect and the discovery of increasingly more widespread alterations in the expression and function of glycolytic enzymes in tumors (Vander Heiden et al. 2009; Koppenol et al. 2011) illustrate the critical importance of understanding the regulation of the enzymes involved in glucose metabolism.
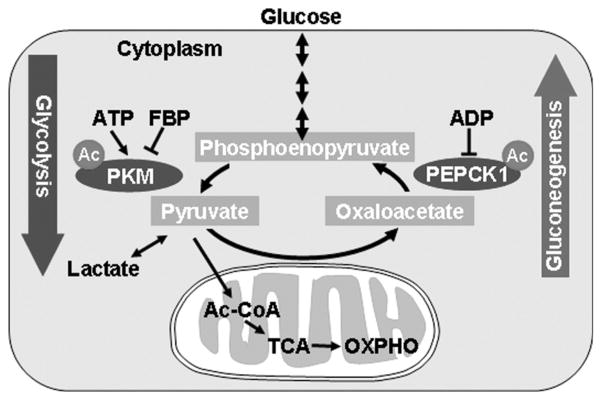
In contrast to glycolysis, many cells, such as hepatocytes, can synthesize glucose from pyruvate, a process known as gluconeogenesis that is important for maintaining blood glucose levels in tissues such as the brain, which almost exclusively relies on glucose. Both glycolysis and gluconeogenesis are tightly and reciprocally regulated in response to the change of energy status and glucose levels in the cell. Although glycolysis and gluconeogenesis share several enzymes that catalyze reversible reactions, the irreversible key steps are catalyzed by separate enzymes that are subjected to different regulations. For example, phosphoenolpyruvate carboxy kinase (PEPCK) is a key regulatory enzyme driving gluconeogenesis, whereas pyruvate kinase (PK) is a key enzyme propelling glycolysis (Fig. 1). The regulation of glycolysis and gluconeogenesis, including in PK and PEPCK, occurs on multiple levels, such as gene expression, allosteric regulation by small metabolites, and posttranslational modification. This chapter discusses one newly discovered regulation, acetylation, on both PEPCK and PK. Much of what we have learned on the acetylation regulation of these two enzymes pertains to the regulation of other metabolic enzymes by acetylation and has direct relevance to tumorigenesis.
Figure 1.
Acetylation regulates two key enzymes, pyruvate kinase (PK) and phosphoenolpyruvate carboxy kinase (PEPCK) and controls glycolysis and gluconeogenesis. PKM, Muscle form of PK.
LYSINE ACETYLATION IS A MAJOR POSTTRANSLATIONAL MODIFICATION IN METABOLIC ENZYMES
Lysine acetylation was first discovered in histone nearly half a century ago (Phillips 1963; Allfrey et al. 1964). For many years, the investigation of acetylation focused primarily on histone and other factors associated with chromatin and transcriptional regulation. Moreover, many transcriptional coactivators and corepressors are associated with or are themselves histone acetyltransferase or deacetylase, respectively. Therefore, acetylation is a major regulator of gene expression and epigenetic control. Recent proteomic studies have identified more than 2000 proteins that are potentially modified by acetylation on lysine residues (Kim et al. 2006; Choudhary et al. 2009; Wang et al. 2010; Zhao et al. 2010; Weinert et al. 2011), which has significantly expanded the scope of cellular regulation by acetylation. Lysine acetylation is found in proteins involved in diverse biological processes, including in particular metabolism, owing in part to the use of liver in two of the acetylation proteomic studies. Notably, virtually all enzymes involved in glycolysis and gluconeogenesis were identified by the proteomic analysis as acetylated (Guan and Xiong 2010). The full scope of this surprising finding is yet to be appreciated, but subsequent investigations on the regulation of individual metabolic enzyme by acetylation, including both PK and PEPCK1 as discussed below, are increasingly pointing to critical and broad roles of acetylation in metabolic regulation.
ACETYLATION INHIBITS PKM ACTIVITY AND PROMOTES CHAPERONE-MEDIATED AUTOPHAGY
Pyruvate kinase catalyzes the transfer of phosphate from phosphoenopyruvate (PEP) to ADP to produce ATP and pyruvate. This is the final and irreversible step in glycolysis and is subjected to multiple regulations. The inhibition of PK by ATP provides a mechanism to link glycolysis to cellular ATP levels. Moreover, PK is activated by fructose bisphosphate (FBP), a glycolytic intermediate that is upstream of the reaction catalyzed by PK (Fig. 1). Thus, the activation of PK by FBP maintains the rate of PK activity to a similar level to the rate of the upstream reaction to prevent the accumulation of glycolytic intermediates when glycolysis is required primarily for energy production. PK is a family of enzymes consisting of four isozymes, L, R, M1, and M2, in human cells (Mazurek et al. 2005). Although the L and R isoforms of PK are specifically expressed in liver and blood cells, respectively, the M1 isoform, PKM1, is commonly expressed in most adult cells. The M2 isoform, PKM2, is encoded by the same gene as PKM1 and differs by one exon as a result of alternative splicing. PKM2 is primarily expressed during embryonic development, turned off in adult cells, and reexpressed in many cancer cells, in part by the oncogene Myc-mediated splicing (David et al. 2010). Our proteomic analyses have identified that PKM (the muscle form of PK) is acetylated at multiple sites, including Lsy305 (K305), which is conserved in all four isoforms of PK. Various assays, including the use of anti-acetyl-PK(K305) antibody, confirmed that PK was acetylated. The mutational analysis and enzyme activity assays showed that acetylation of K305 inhibited PK activity by increasing its Km toward the substrate PEP (Lv et al. 2011). Thus, the K305-acetylated PK is expected to have reduced catalytic activity, which could lead to an accumulation of glycolytic intermediates upstream of PEP, a condition that favors cell growth over maintaining homeostasis of a nondividing state.
PKM2 acetylation is stimulated by high glucose concentrations. Interestingly, glucose also decreased PKM2 protein levels, indicating a possible relationship between PKM2 acetylation and protein levels. The level of PKM2 mRNA remained unaltered in response to the changes in glucose concentrations, and inhibition of proteasome did not have any effect on the high glucose–induced decrease in PKM2 protein level, suggesting a transcription-and proteasome-independent mechanism in the regulation of PKM2 protein levels. Instead, glucose-induced PKM2 degradation was blocked by the inhibition of lysosomal degradation, such as treatment with a protease inhibitor leupeptin. Mutation of Lys305 to an Arg residue stabilized PKM2 and pharmacological inhibition of deacetylases conversely reduced PKM2 protein levels, linking the acetylation at Lys305 to the down-regulation of PKM2 protein by the lysosome (Fig. 2).
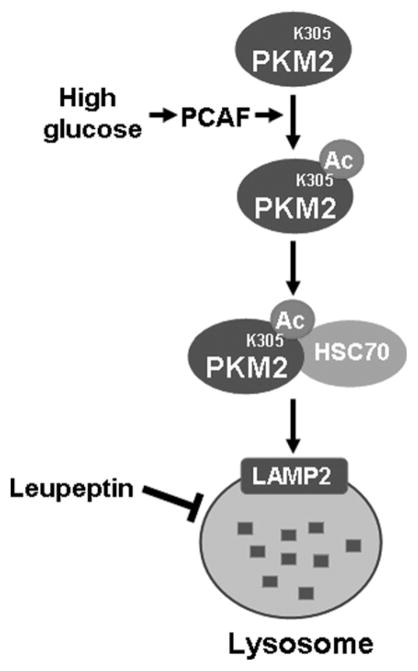
Figure 2.
Acetylation promotes PKM2 degradation by chaperone-mediated autophagy. High glucose stimulates PKM2 acetylation on lysine 305, which decreases PKM2 catalytic activity. Moreover, K403 acetylation increases its ability to interact with HSC70, a chaperone that can be recognized by LAMP2 on the lysosome, thereby bringing K305-acetylated PKM2 to the lysosome for degradation.
Our subsequent studies demonstrated that glucose induced the degradation of PKM2 via a selective lysosome-dependent degradation pathway, chaperone-mediated autophagy (CMA), which targets specific proteins and is different from the macroautophagy that nonselectively sequesters and delivers damaged proteins and organelles to lysosomes for degradation. We found that acetylation of K305 in PKM2 increased the binding between PKM2 and HSC70, a chaperone of the heat shock protein family that transports target proteins to the lysosome by binding to the lysosome surface receptor LAMP2 for degradation (Fig. 2). These results provide the first example of acetylation targeting a protein degradation by the lysosome via CMA.
To investigate the physiological significance of K305-acetylation-targeted PKM2 degradation, endogenous PKM2 was knocked out in cultured H1299 human non-small-cell lung carcinoma cells and the acetylation mimetic K305Q mutant PKM2 was reexpressed in the knockout cells. The glycolytic intermediates present before the PKM2 reaction, such as glucose 6-phosphate and fructose 6-phosphorylate, were accumulated. In contrast, glycolytic metabolites after the PKM2 reaction, such as pyruvate and lactate, were reduced in these cells. Furthermore, H1299 cells expressing the K305Q mutant proliferate more rapidly and form tumors in nude mice that are larger than those in cells expressing the wild-type PKM2. These data provide in vivo evidence supporting an important role of acetylation at Lys305 in the negative regulation of PKM2 function, which leads to the accumulation of glycolytic metabolites and increased cell growth and proliferation.
Altered glycolysis is a metabolic hallmark found in many cancer cells; these cells utilize large amounts of glucose and only metabolize a small fraction of the glucose via the TCA cycle for energy production. The mechanism for the reduction of PKM2 activity in promoting tumor cell growth is not fully understood. One explanation is that the accumulation of glycolytic intermediates increases biosynthesis and thus cell growth. Higher activity of PK is therefore expected to be associated with reduced accumulation of glycolytic intermediates and decreased cell growth. This notion is consistent with the specific expression of PKM2, which has lower basal activity than PKM1, in fast-growing embryonic cells and splicing switch from PKM1 to PKM2 in tumor cells. Our finding that K305 acetylation negatively regulates PK activity provides a novel mechanism for cells to regulate the accumulation of glycolytic metabolites and the rate of cell growth. In theory, such regulation may occur not only in the cells expressing PKM2 but also in cells expressing the other isoform of PK, because Lys305 is conserved in all four isoforms. It will be interesting to determine whether Lys305 acetylation is increased in fast-growing cells, such as during embryogenesis or tumorigenesis.
ACETYLATION PROMOTES UBIQUITIN-MEDIATED PEPCK DEGRADATION
PEPCK catalyzes the key rate-limiting reaction in gluconeogenesis, which is opposite to the reaction catalyzed by PK (Fig. 1). PEPCK has been shown to be the key regulatory enzyme in gluconeogenesis as well as glyceroneogenesis, serine synthesis, and amino acid metabolism (Yang et al. 2009a). Both PEPCK transcription and enzyme activity are tightly regulated (Yang et al. 2009b). Gluconeogenesis should be inhibited when blood glucose levels are high, whereas gluconeogenesis should be high when blood glucose levels are low. Our mass spectrometry analyses identified four acetylation sites in human PEPCK (Zhao et al. 2010). We further showed that PEPCK acetylation is regulated by extracellular nutrients, such as glucose and amino acids. Glucose and amino acids should have different effects on PEPCK function because amino acids can serve as precursors for gluconeogenesis, whereas glucose is the end product of gluconeogenesis. Interestingly, they also display opposing effects on PEPCK acetylation, with glucose and amino acids causing a decrease and increase in PEPCK acetylation, respectively. Although acetylation has no direct effect on PEPCK enzymatic activity, it is strongly correlated with a decrease in PEPCK protein level (Zhao et al. 2010).
Our subsequent studies established that acetylation of PEPCK1 promotes its degradation in a proteasome-dependent manner and induces a significant increase in PEPCK ubiquitination. When the acetylation-defective mutant PEPCK was examined, the glucose-induced PEPCK ubiquitination and destabilization were abolished. To investigate the mechanism behind the PEPCK regulation, we affinity-purified PEPCK immunocomplex and identified numerous PEPCK-interacting proteins by mass spectrometry. Among them are multiple proteasome subunits, which is consistent with the proteasome-dependent degradation of PEPCK and, importantly, UBR5 (ubiquitin protein ligase E3 component n-recognin 5), which is a member of the HECT family of R3 ligases and the only PEPCK-interacting protein with obvious E3 activity. The interaction between PEPCK and UBR5 was stimulated by PEPCK acetylation. Moreover, knocking out UBR5 significantly stabilized PEPCK, especially at high levels of glucose when PEPCK1 is strongly acetylated and destabilized. These results suggest that acetylation promotes PEPCK ubiquitination and degradation by increasing its ability to interact with the UBR5 E3 ligase (Fig. 3) (Jiang et al. 2011).
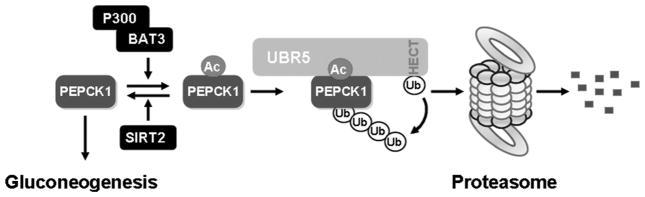
Figure 3.
Acetylation promotes PEPCK ubiquitination and degradation. P300 and SIRT2 function as an acetyltransferase and a deacetylase for PEPCK, respectively. The acetylated PEPCK recruits the UBR5 E3 ubiquitin ligase, which then ubiquitinates PEPCK and leads to its degradation by the proteasome.
One protein identified from mass spectrometric analysis of PEPCK immunocomplex is BAT3 (HLA-B-associated transcripts 3), also known as BAG6 (BCL2-associated athanogene 6 and Scythe), which has been previously reported to target the p300 acetyltransferase to acetylate p53 (Sasaki et al. 2007) and HSP90 (Cerchietti et al. 2010). We found that the knockout of either BAT3 or P300 resulted in a significant decrease in PEPCK acetylation, whereas the overexpression of P300 increased PEPCK acetylation. Consistent with the role of acetylation in promoting PEPCK degradation, the overexpression of P300 and BAT5 decreased PEPCK protein levels, whereas knocking out P300 significantly stabilized PEPCK even in the presence of high glucose. These data indicate that P300 and BAT5 are PEPCK acetyltransferases. We also identified SIRT2 as a PEPCK1 deacetylase based on the observation that PEPCK1 acetylation was increased after treatment of cells with nicotinamide, a selective inhibitor of the SIRT family of deacetylases, but not trichostatin A, an inhibitor of the HDAC family of deacetylases. We found that coexpression of SIRT2, but not the related SIRT1, significantly decreased PEPCK acetylation, strongly reduced PEPCK ubiquitination, and stabilized both ectopically expressed and endogenous PEPCK (Jiang et al. 2011).
The physiological significance of PEPCK acetylation in glucose metabolism was also investigated. Knocking down either P300 or BAT5 in cultured cells increased glucose synthesis and, conversely, knocking out SIRT2 reduced glucose production. Furthermore, when Sirt2 was knocked out in mouse liver, the major gluconeogenesis organ in vivo, the blood glucose level was significantly reduced and liver PEPCK protein levels were reduced. These experiments demonstrate that acetylation of PEPCK played a major role in physiological glucose homeostasis. We propose that, in cells with high levels of glucose, BAT3/BAG6-mediated acetylation of PEPCK by P300 promotes its interaction with UBR5, which then ubiquitinates PEPCK to trigger its degradation by the proteasome. SIRT2 would antagonize the effect of P300, thereby increasing the stability of PEPCK and stimulating gluconeogenesis (Fig. 3).
CONCLUSION
Proteomic studies have revealed that most metabolic enzymes are acetylated in both bacteria and mammalian liver, and subsequent studies have demonstrated the critical role of acetylation in regulation of metabolic enzymes (Kim et al. 2006; Wang et al. 2010; Zhao et al. 2010). The two examples described above illustrate the importance of acetylation in the regulation of glucose metabolism. In both examples, the levels of acetylation in the substrate metabolic enzymes are changed in response to the changes in extracellular nutrient conditions, resulting in changes in enzymatic activity and protein levels. More importantly, the acetylation regulation of these two enzymes has been shown to occur in vivo, to promote tumorigenesis in the case of Lys305 acetylation of PKM2 or to reduce the levels of PEPCK1 and blood glucose after knocking out its deacetylase SIRT2. In both PKM2 and PEPCK1, it appears that acetylation promotes the binding of acetylated substrate with an additional protein: HSC70 in the case of K305-acetylated PKM2 and URB5 E3 ligase for acetylated PEPCK1. The mechanisms by which acetylation bridges these interactions are not clear at present. The bromodomain is well characterized as a protein motif that selectively recognizes acetylated lysines in histones (Mujtaba et al. 2007). Neither UBR5 nor HSC70 contain an apparent bromodomain, leaving it open as to whether acetylation promoting PKM2-HSC70 and PEPCK1-URB5 association involves an additional factor or there exists an additional yet-to-be-characterized protein motif that recognizes acetylated lysine residue.
It should be pointed out that acetylation regulates metabolic enzymes, and probably other proteins, by mechanisms in addition to promoting protein degradation. Examples include inhibition and stimulation of catalytic activity without affecting the stability and levels of many acetylated metabolic enzymes (Yu et al. 2009; Qiu et al. 2010; Tao et al. 2010; Zhao et al. 2010; Chen et al. 2011). Given the prevalence of acetylation, it is possible that acetylation may regulate the function of its substrate proteins by a wide range of mechanisms, as is the case for the regulation of protein function by phosphorylation.
Although the acetylation of PKM2 and PEPCK is affected by glucose, the precise mechanisms for how glucose controls the acetylation levels of these proteins remain to be determined. Does glucose affect acetyltransferase or deacetylase to influence the acetylation of PEPCK and PKM2? Moreover, because only limited numbers of acetyltransferases and deacetylases have been identified, whereas the numbers of acetylated proteins are in thousands, how is the specificity of acetylation maintained? Future studies to determine the specificity of acetylation will be important to understand the mechanism and physiological significance of acetylation in the regulation of not only metabolism, but also many other cellular processes.
References
- Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchietti LC, Hatzi K, Caldas-Lopes E, Yang SN, Figueroa ME, Morin RD, Hirst M, Mendez L, Shaknovich R, Cole PA, et al. BCL6 repression of EP300 in human diffuse large B cell lymphoma cells provides a basis for rational combinatorial therapy. J Clin Invest. 2010;120:4569–4582. doi: 10.1172/JCI42869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang J, Lin Y, Lei Q, Guan KL, Zhao S, Xiong Y. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep. 2011;12:534–541. doi: 10.1038/embor.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther T, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan KL, Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends in Biochem Sci. 2010;36:108–116. doi: 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, Xiong Y, Guan KL, Zhao S. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H, Zha Z, Liu Y, Li Z, Xu Y, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- Phillips DM. The presence of acetyl groups of histones. Biochem J. 1963;87:258–263. doi: 10.1042/bj0870258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Gan EC, Wakeham A, Kornbluth S, Mak TW, Okada H. HLA-B-associated transcript 3 (Bat3)/Scythe is essential for p300-mediated acetylation of p53. Genes Dev. 2007;21:848–861. doi: 10.1101/gad.1534107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- Weinert BT, Wagner SA, Horn H, Henriksen P, Liu WR, Olsen JV, Jensen LJ, Choudhary C. Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Sci Signal. 2011;4:ra48. doi: 10.1126/scisignal.2001902. [DOI] [PubMed] [Google Scholar]
- Yang J, Kalhan SC, Hanson RW. What is the metabolic role of phosphoenolpyruvate carboxykinase? J Biol Chem. 2009a;284:27025–27029. doi: 10.1074/jbc.R109.040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Reshef L, Cassuto H, Aleman G, Hanson RW. Aspects of the control of phosphoenolpyruvate carboxykinase gene transcription. J Biol Chem. 2009b;284:27031–27035. doi: 10.1074/jbc.R109.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Lin Y, Yao J, Huang W, Lei Q, Xiong Y, Zhao S, Guan KL. Lysine 88 acetylation negatively regulates ornithine carbamoyltransferase activity in response to nutrient signals. J Biol Chem. 2009;284:13669–13675. doi: 10.1074/jbc.M901921200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]