Abstract
Specialized chromatin domains contribute to nuclear organization and regulation of gene expression. Gene-poor regions are di- and trimethylated at lysine 9 of histone H3 (H3K9me2/3) by the histone methyltransferase, Suv39h1. This enzyme harnesses a positive feedback loop to spread H3K9me2/3 over extended heterochromatic regions. However, little is known about how feedback loops operate on complex biopolymers such as chromatin, in part because of the difficulty in obtaining suitable substrates. Here we describe the synthesis of multi-domain ‘designer chromatin’ templates and their application to dissecting the regulation of human Suv39h1. We uncovered a two-step activation switch where H3K9me3 recognition and subsequent anchoring of the enzyme to chromatin allosterically promotes methylation activity, and confirmed that this mechanism contributes to chromatin recognition in cells. We propose that this mechanism serves as a paradigm in chromatin biochemistry since it enables highly dynamic sampling of chromatin state combined with targeted modification of desired genomic regions.
Hierarchical organization within the nucleus enables cell-type specific interpretation of eukaryotic genomes. A fundamental feature of this architecture is the packing of DNA through cationic protein scaffolds, histone octamers, to yield nucleosome core particles1. Post-translational modifications (PTMs) of histones can affect both the structure and function of chromatin regions through biophysical and biochemical means2. For instance, the formation of tightly compacted chromatin domains across gene-poor regions (heterochromatin) is catalyzed by the histone methyltransferase Suv39h1 (KMT1A) and the closely related enzyme Suv39h2 (KMT1B), which install the classic heterochromatin modifications, H3K9me2/3 (Fig. 1a)3,4. These histone marks recruit heterochromatin protein 1 (HP1) to mediate downstream silencing of affected chromatin domains5–7. Importantly, for proper nuclear organization, heterochromatin must be propagated by Suv39h1 and HP1 over large, defined regions of the genome — a process referred to as spreading8. Excessive or insufficient spreading results in genome instability9 and can cause diseases due to gene mis-regulation10.
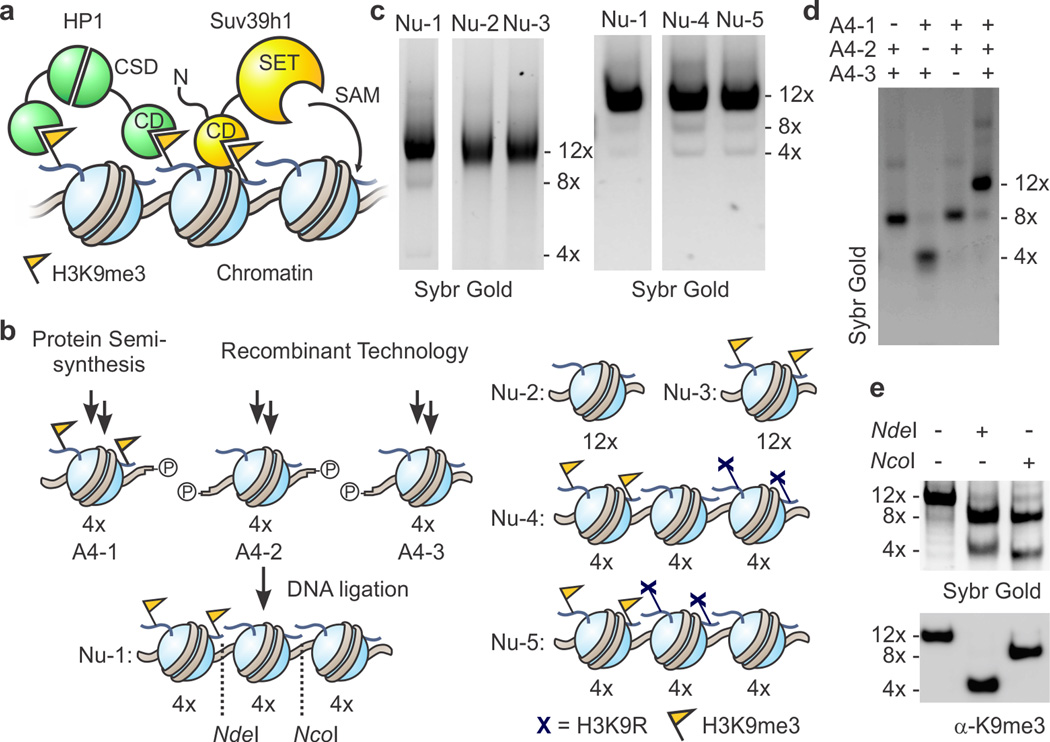
Figure 1. Heterotypic designer chromatin substrates.
(a) Simplified model for heterochromatin spreading. H3K9me3-modified (yellow FLAGs) nucleosomes serve to recruit the scaffolding protein HP1 and the histone methyltransferase (HMT) Suv39h1 through their chromodomains (CDs). The latter catalyzes the S-adenosylmethionine (SAM)-dependent methylation of H3K9 via its SET domain, thus providing a positive feedback loop. HP1 forms a dimer through interactions of its chromo-shadow domain (CSD). (b) Schematic for the synthesis of heterotypic designer nucleosome arrays. NdeI and NcoI sites for restriction analysis are indicated by dotted lines. See supporting information for more detail. Cartoon representations of control arrays are shown on the right. (c) Purity of designer chromatin assessed by native gel electrophoresis. For additional data, see Supplementary Fig. 3. (d) Ligations of purified 4-mer arrays proceed selectively as indicated by native gel electrophoresis of crude products from ligation reactions of designated array fragments. (e) PTM-containing ‘priming’ domain remains intact through designer chromatin synthesis. Ligated arrays (Nu-1) were digested and the resulting fragments analyzed by native gel electrophoresis and western blotting for H3K9me3. See supplementary Figure 3c for more detail.
Suv39h1 contains a chromodomain (CD) capable of binding the product of its own reaction, H3K9me2/3 (Fig. 1a)11. The CD is important for chromatin binding and methyltransferase activity of Suv39h1 in cells11,12, suggesting that a positive feedback loop controls heterochromatin formation. Indeed, semi-synthetic dinucleosome substrates have been employed to demonstrate that Clr4, the yeast homolog of Suv39h1, is stimulated by H3K9me3 marks in vitro13. Despite these insights, the mechanism of how the CD and the enzymatic core cooperate to enact propagation of H3K9me3 along the chromatin fiber has remained enigmatic, especially for the mammalian spreading system where the regulation of the methyltransferase has yet to be explored in a defined biochemical system. Allosteric regulation of enzymes is an important aspect of many processes in chromatin biochemistry14,15, and particularly interesting from an enzymology standpoint given that, in this case, the substrate and allosteric modulator are tethered in a heteropolymeric template. This feature raises questions relating to the geometry of the feedback system, which have largely remained unexplored.
To address these issues, we engineered nucleosome arrays with distinct subdomains, resembling the complex architecture of chromatin in cells16. We have applied this type of ‘designer chromatin’17 to reconstitute Suv39h1-dependent H3K9me3 spreading in vitro, providing insight into the geometric and enzymatic mechanism of this process. We found that Suv39h1 preferentially takes small spreading steps within a single chromatin template, but long-range heterochromatin propagation can occur upon chromatin folding. Furthermore, our experiments unveiled that Suv39h1 is controlled by an unanticipated two-step activation switch involving a latent chromatin binding feature.
RESULTS
Synthesis of multi-domain designer chromatin
To enable biochemical reconstitution of Suv39h1-dependent heterochromatin spreading, we devised the synthesis of 12-mer nucleosome arrays that contain a stretch of four initiating nucleosomes, followed by a block of eight substrate nucleosomes (Nu-1, Fig. 1b). Notably, homogeneous arrays of this length have served for numerous biochemical and biophysical studies as they encompass the length of a small gene17,18. Moreover, tetranucleosomes appear to be a defined subunit of higher order chromatin structure observed in models derived from electron microscopy19 and crosslinking in cells20. Inspired by methods for the synthesis of heterotypic di- and tetranucleosomes21,22, we chose to use DNA ligation to assemble chemically defined arrays from homogeneous 4-mer building blocks (Fig. 1b). The 4-mers were prepared from histone octamers (consisting of unmodified, recombinant histones and semi-synthetic H3K9me3, Supplementary Results, Supplementary Fig. 1) and four repeats of a strong nucleosome positioning sequence23, separated by 30 bp linker DNA (Supplementary Fig. 2). To enable directed ligation, each building block was furnished with specific, non-palindromic 3- or 4-bp overhangs. A one-pot ligation of all three fragments, followed by purification by MgCl2-precipitation and sucrose gradient centrifugation provided 12-mer array Nu-1 in good purity (Fig. 1c and Supplementary Fig. 3). Ligations were specific as the omission of fragments leads to the expected truncated products (Fig. 1d), and digestion of purified arrays revealed that the H3K9me3 marks were retained at their designated location (Fig. 1e).
An in vitro heterochromatin spreading system
With designer chromatin substrates in hand, we turned our attention to the issue of reconstituting H3K9me2/3 spreading in vitro. Recombinant human Suv39h1 was isolated from an insect cell expression system (Supplementary Fig. 4) and used in a scintillation-based histone methyltransferase (HMT) assay employing 3H-SAM and a designer chromatin substrate. Consistent with a functional positive feedback loop, the presence of the H3K9me3 priming domain within array Nu-1 led to robust stimulation of HMT activity in comparison to control substrate Nu-2 which lacks this domain (Fig. 2a and Supplementary Fig. 5a) — this corresponds to a 25-fold activation when considering available methylation sites between substrates. Importantly, a nucleosome array carrying H3K9me3 throughout (Nu-3) was not further methylated under the same conditions, in line with the expected specificity of Suv39h1 (Fig. 2a).
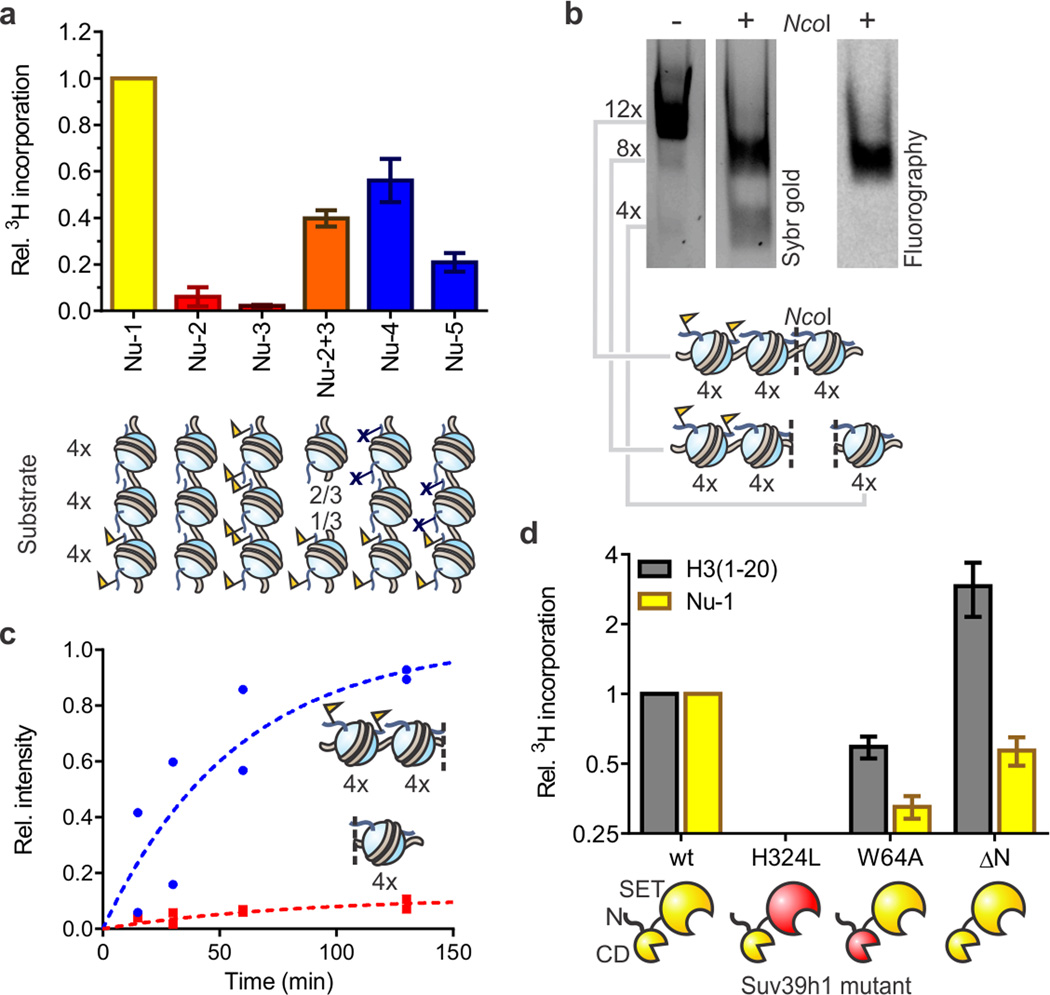
Figure 2. Reconstitution of Suv39h1-dependent heterochromatin spreading in vitro.
(a) Substrate preference of Suv39h1-catalyzed spreading. Arrays Nu-2 and Nu-3 represent homotypic templates (red) carrying either H3K9me0 or H3K9me3, respectively. Inter-fiber spreading is evaluated with a 2:1 mixture of arrays Nu-2 and Nu-3 (orange) to match the stoichiometry of histones in array Nu-1 (yellow). Arrays Nu-4 and Nu-5 (blue) contain H3K9R mutations to block spreading. HMT activity was measured using 3H-SAM as the co-factor in the presence of 5 mM MgCl2. Scintillation counts are normalized to the values determined with heterotypic array Nu-1, without additional normalization for number of substrate sites. Error bars, s.e.m. (n = 3). (b) Suv39h1 preferentially methylates nucleosomes adjacent to preinstalled H3K9me3 marks. Arrays were methylated with 3H-SAM in the presence of 0.5 mM MgCl2 and subsequently digested with NcoI, separated by native gel electrophoresis and analyzed by Sybr gold staining (left) and fluorography (right). See Supplementary Fig. 8 for more details. (c) Time course of Suv39h1-dependent spreading of the H3K9me3 mark. Individual values, normalized to the sum of the values for the 4mer and 8mer obtained at 130 min, from two independent measurements are shown. (d) HMT activity of Suv39h1 variants with an H3 peptide encompassing residues 1–20 (gray) or array Nu-1 (yellow). Measurements are normalized to the values obtained with wild-type Suv39h1 and the respective substrate, and plotted on a log2 scale. Error bars, s.e.m. (n = 3).
To explore whether this spreading phenomenon can operate in an inter-array fashion, we performed an HMT assay using a 1:2 mixture of homogeneous arrays with (Nu-3) and without (Nu-2) the H3K9me3 mark. The absolute concentrations of priming vs. substrate sites match those of array Nu-1. Under the conditions of the assay, arrays are predominantly self-assembled (Supplementary Fig. 6), thus resembling densely packed chromatin from cells24. We observed elevated levels of Suv39h1 activity on this substrate mixture compared to the individual components (Fig. 2a), indicating that HMT activation can extend across chromatin fibers that are self-assembled in vitro, consistent with physical models of heterochromatin spreading25. As expected, trans spreading is strongly reduced when the assay is performed under conditions where chromatin assembly is unfavorable (0.5 mM MgCl2, Supplementary Fig. 6 and 7). Even when chromatin is self-assembled (5 mM MgCl2), inter-array activation cannot account for all the HMT activity seen with heterotypic array Nu-1 (at most ∼40%) implying that the cis spreading mechanism still dominates in this case.
We next turned our attention to the geometry of Suv39h1-catalyzed cis-spreading. This investigation was enabled by designer chromatin arrays Nu-4 and Nu-5 that contain three distinct subdomains; an H3K9me3 priming region, an unmodified substrate region and a region containing H3K9R nucleosomes that serves as a barrier for Suv39h1 (Figs. 1b and c). Higher levels of HMT activity were observed on array Nu-4, which contains the substrate region proximal to the priming region, compared to array Nu-5, which has the reverse configuration (Fig. 2a). This result demonstrates that Suv39h1 prefers small spreading steps. To test whether residual activity on distal nucleosomes (as seen in array Nu-5) is due to trans-spreading or the possibility of Suv39h1 reaching over 4 intermittent nucleosomes, we analyzed the methylation distribution in a spreading reaction on array Nu-1 in the presence of 0.5 mM MgCl2. A unique NcoI restriction site in the array allowed us to interrogate the substrate regions proximal or distal to the priming site by digestion, followed by fluorography (Figs. 2b,c and Supplementary Fig. 8). Consistent with directional spreading, Suv39h1 activity is almost exclusively localized on nucleosomes proximal to the pre-installed H3K9me3 mark, with only a small amount observed on the distal subdomain. Collectively, these experiments establish the utility of our reconstituted system for probing PTM spreading and reveal that the presence of pre-installed H3K9me3 priming marks is sufficient to stimulate Suv39h1 activity on proximal nucleosomes.
The CD of Suv39h1 promotes spreading in vitro
To gain further insight into the spreading mechanism, we compared the HMT activity of Suv39h1 variants on both designer chromatin array Nu-1 as well as histone peptides (residues 1–20 of H3). As expected, a Suv39h1 variant with the active site mutation H324L was inactive on both substrates (Fig. 2d)3. Disabling the positive feedback loop through mutation of the chromodomain of Suv39h1 (W64A, Supplementary Fig. 9) has only a small effect on peptide substrates, in agreement with previous findings26, but leads to a more profound reduction of activity on array Nu-1 (Fig. 2d). We also investigated whether the N-terminus of the methyltransferase, known to interact with HP127, affects HMT activity. Interestingly, the Suv39h1 variant lacking the N-terminal 39 residues (ΔN) displays enhanced activity on peptide substrates, but reduced activity on chromatin (Fig. 2d) suggesting that the N-terminus contributes to chromatin recognition and/or HMT activity. We note that, despite the diminished activity, the ΔN variant is still capable of directional spreading (Supplementary Fig. 8). Inclusion of recombinant HP1, which engages H3K9me3 through its own CD (Supplementary Fig. 10a), in our spreading system did not lead to further stimulation of Suv39h1 activity on array 1 (Supplementary Fig. 10b,c).
Allosteric activation of Suv39h1 on chromatin substrates
Conceivably, H3K9me3 could stimulate Suv39h1 activity through recruitment to chromatin, allosteric activation of the enzymatic activity, or a combination of both. Indeed, an intra-fiber allosteric model has previously been proposed to regulate Clr4, the yeast homolog of Suv39h1.13 We explored the mechanism of Suv39h1-stimulation by performing the HMT assay on unmodified peptide or unmodified nucleosome array (Nu-2) substrates in the presence of an additional H3K9me3-containing peptide. We observed no trans-stimulation of HMT activity in the context of the H3 peptide substrate (Fig. 3a). In sharp contrast, addition of H3K9me3 peptide strongly enhanced HMT activity on unmodified chromatin (Fig. 3a), with an approximately 10-fold increase in vmax, and a 4-fold reduction in K1/2 (Supplementary Fig. 5b,c). This stimulation was only observed in a chromatin context as a mixture of DNA and H3K9me3 peptides did not stimulate HMT activity on H3K9me0 peptide substrates (Supplementary Fig. 11). This chromatin-specific stimulation required an intact chromodomain within Suv39h1, which presumably functions as a sensor for the methylation mark (Fig. 3b). More surprisingly, the H3K9me3 peptide was also unable to stimulate the ΔN variant of Suv39h1, further implicating this region of the enzyme in the spreading feedback mechanism.
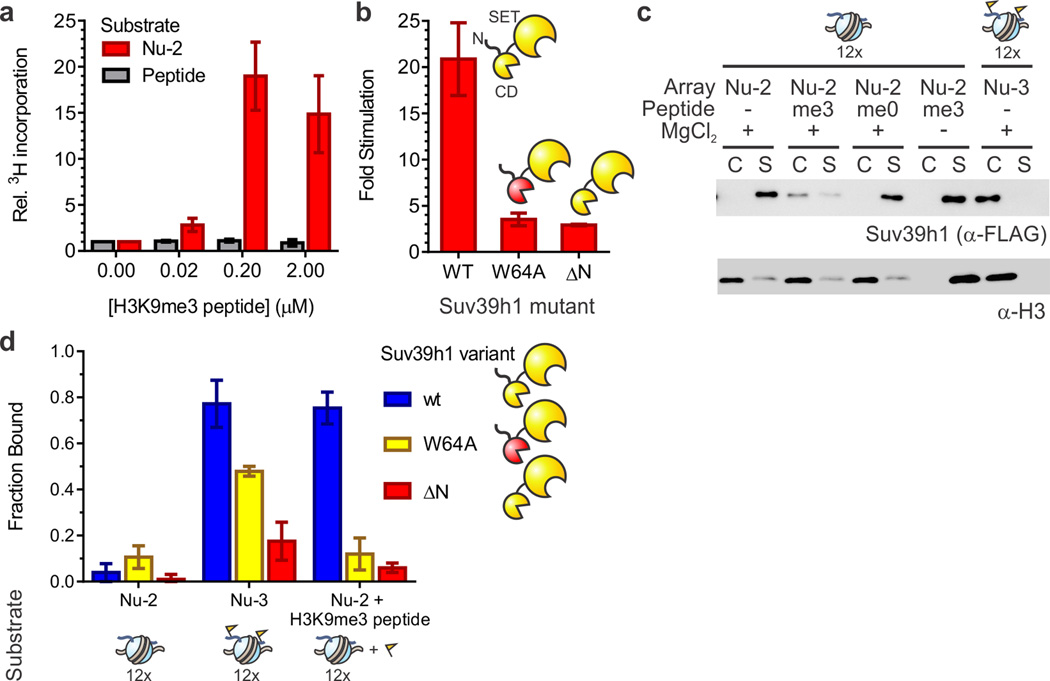
Figure 3. Trans-activation of Suv39h1.
(a) Allosteric activation of Suv39h1 by H3K9me3 peptides occurs only on chromatin substrates. 3H-SAM-based HMT assays were performed on either unmodified peptide (gray) or array Nu-2 (red) substrates in the presence of increasing amounts of H3K9me3 peptide added in trans. In each case, 3H-me incorporation was normalized to the value obtained in the absence of H3K9me3 peptide. Error bars, s.e.m. (n = 4). (b) Suv39h1-activation requires an intact chromodomain and N-terminus. Stimulation of HMT activity is defined as the ratio of scintillation counts obtained on unmodified chromatin Nu-2 in the presence of 0.2 µM H3K9me3 peptide vs. its absence. Error bars, s.e.m. (n = 3). (c) The H3K9me3 mark promotes chromatin binding in cis and in trans. Western blot of a binding assay of wild-type Suv39h1 with array Nu-2 or Nu-3. Where appropriate, H3 peptides are added at a concentration of 0.2 µM. A control reaction in the absence of MgCl2 is included (lanes 7 and 8). See Supplementary Fig. 14 for full image of the membrane. (d) Chromatin binding is promoted by the H3K9me3 mark and requires an intact chromodomain and N-terminus. Bound fractions are determined by densitometry of Suv39h1 western blots (α-FLAG) bands corresponding to the re-dissolved pellet (chromatin associated) and supernatant (unbound) of binding reactions. Error bars, s.e.m. (n = 3).
The ability of H3K9me3 peptides to trans-activate Suv39h1 activity on chromatin reveals an allosteric component to the spreading process. Because this effect does not extend to peptide substrates, we asked whether the H3K9me3 mark induces chromatin binding by the methyltransferase. To this end, we developed a chromatin binding assay wherein nucleosome arrays containing either H3K9me0 or H3K9me3 were incubated with wild-type Suv39h1 or its variants in the presence of 5 mM MgCl2, causing self-assembly and precipitation of chromatin (Supplementary Fig. 6). Resuspension of the pellet in the absence of MgCl2 allowed us to assess the extent to which Suv39h1 associates with chromatin. Suv39h1 did not associate appreciably with unmodified chromatin array Nu-2, but readily bound to homogeneously H3K9-methylated arrays Nu-3 (Figs. 3c & d). Importantly, chromatin association was strongly enhanced by H3K9me3 not only in cis, but also when this mark was added as a peptide in trans. Control experiments confirmed that chromatin binding depends on the H3K9me3 mark, and that in the absence of MgCl2, chromatin and consequently also Suv39h1 remain in the supernatant. As expected, chromatin binding was attenuated when either the CD of Suv39h1 was crippled or the N-terminus removed, confirming their role in the spreading mechanism (Fig. 3d). Partial binding of the W64A variant to array Nu-3 may originate from residual affinity of the mutant CD to H3K9me3 (ref 11) in context of the full-length protein.
The N-terminus contributes to chromatin binding
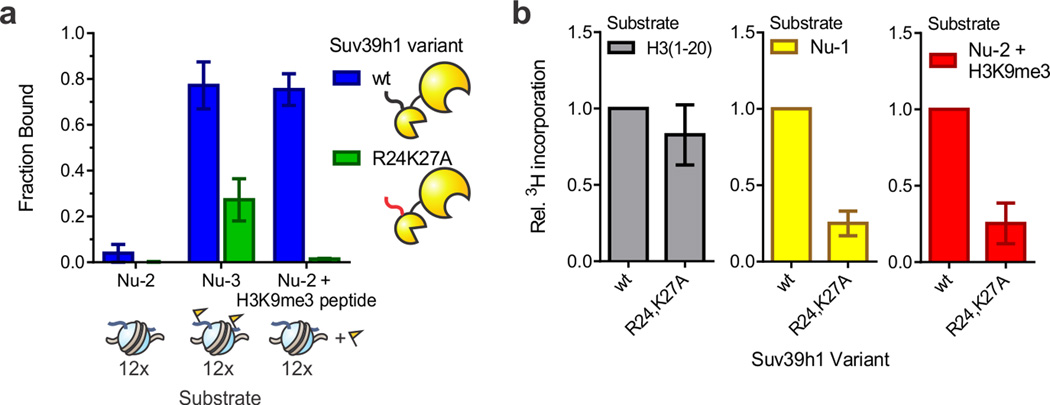
We noticed that the sequence of the Suv39h1 N-terminus displays a remarkable similarity to the N-terminal helix of the Zn-finger from the DNA-binding protein Gal4 (Supplementary Fig. 12a). Consequently, we wondered whether this N-terminal motif in Suv39h1 was important for function. Specifically, we prepared a variant where the postulated DNA binding residues (R24 and K27, Supplementary Fig. 12a) are mutated to Ala. Chromatin binding of the R24,K27A double mutant was strongly diminished in the context of fully methylated array Nu-3, as well as unmodified array Nu-2 in the presence of stimulatory H3K9me3 peptide (Fig. 4a). We note that, in the context of a truncated construct encompassing the N-terminus and the CD, the R24,K27A double mutant retains the ability to bind H3K9me3 peptides, albeit with a modest loss in affinity (Supplementary Fig. 9). While we therefore cannot exclude partial contributions from changes to the CD, the strong loss in chromatin binding observed for the R24,K27A mutant compared to the W64A mutant argues that additional factors control chromatin engagement in the former. Finally, the R24A and K27A mutations severely compromised activity in HMT assays with array Nu-1 and with array Nu-2 in the presence of H3K9me3 peptide (Fig. 4b).
Figure 4. The N-terminus of Suv39h1 contributes to chromatin binding in vitro.
(a) Residues R24 and K27 of Suv39h1 contribute to chromatin binding. Bound fractions are determined by densitometry of Suv39h1 western blots (α-FLAG) bands corresponding to the re-dissolved pellet (chromatin associated) and supernatant (unbound) of co-precipitation reactions. Error bars, s.e.m. (n = 3). (b) The R24,K27A mutant displays reduced activity on chromatin substrates. HMT measurements with H3 peptides (left), array Nu-1 (middle), or array Nu-2 in the presence of 0.2 µM H3K9me3 peptide (right) were normalized to the values obtained for wt Suv39h1. Error bars, s.e.m. (n = 5).
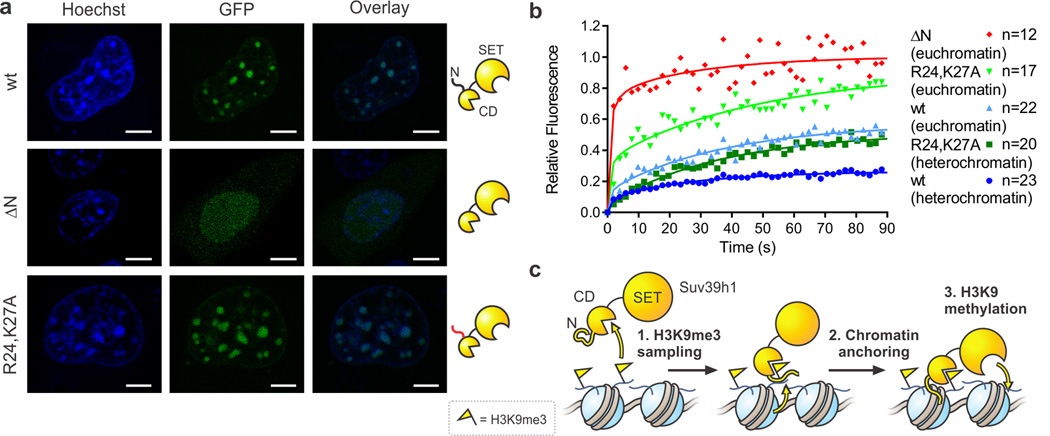
Finally, we tested whether the N-terminus of Suv39h1 contributes to chromatin binding in cells. To this end, Suv39h1-GFP fusions were transfected into NIH-3T3 cells, and the subnuclear localization of the constructs visualized by fluorescence microscopy. Wild-type Suv39h1-GFP and the R24,K27A mutant localize to dense heterochromatin foci, whereas a variant lacking the entire N-terminus (ΔN) is diffusely distributed through the nucleus (Fig. 5a). The difference in localization between the R24,K27A and the ΔN variant presumably reflects the ability of the former, but not the latter, to interact with HP1 (Supplementary Fig. 10d)12,27,28.
Figure 5. The Suv39h1 N-terminus contributes to chromatin binding in vivo.
(a) Fluorescence microscopy images of indicated Suv39h1-GFP fusion proteins in NIH-3T3 cells. Cells were stained with Hoechst 33342 dye to visualize nuclei and heterochromatin foci. Scale bar = 5 µM. (b) FRAP analysis of Suv39h1-GFP fusion proteins in NIH-3T3 cells. FRAP profiles were recorded upon bleaching heterochromatin foci or a circular spot in the low intensity region of nuclei (euchromatin). For visualization purposes, the average of 12–23 individual measurements were fit to the coupled reaction-diffusion model of Sprague et al. (ref 43). (c) Model for a two-state activation of Suv39h1 methyltransferase activity. Freely diffusing Suv39h1 exhibits low HMT activity (left). Chromodomain (CD)-dependent recognition of nucleosomal H3K9me3 and subsequent anchoring to chromatin involving the N-terminus of Suv39h1 (center) enhances HMT activity in the vicinity of the stimulating mark (right).
Fluorescence recovery after photobleaching (FRAP) confirmed that wild-type Suv39h1 exchanges at the timescale of seconds, with a subpopulation remaining immobile on the timescale of minutes, in agreement with previous reports29,30. As expected, exchange occurs more readily in euchromatin than in heterochromatin (Fig. 5b). The ΔN variant interacts with chromatin only transiently, as reflected by the short residence time and the complete recovery (Fig. 5b), consistent with its inability to interact with HP1 (Supplementary Fig. 10d)27 as well as the reduced chromatin binding we observed in vitro. Notably, deletion of the N-terminus of Suv39h1 has also been shown to derepress major satellite repeats in vivo under steady-state conditions28. Critically, the R24,K27A mutant of Suv39h1 exhibits more extensive exchange than the wild-type enzyme in both euchromatin and heterochromatin, presumably reflecting a loss in chromatin binding despite the retention of HP1 binding ability (Fig. 5b, Supplementary Fig. 10d). These results support the idea that specific residues present in the putative Zn-finger-like motif contribute to chromatin binding in vivo.
DISCUSSION
Although the heteropolymeric nature of chromatin has long been recognized16, few model systems have been established to test in a controlled manner how information (for example a histone PTM) is interpreted as a function of the location relative to a reporter site. One approach to this problem has been to artificially tether chromatin-modifying enzymes to specific genomic loci and then analyze the chromatin state and transcriptional output in the vicinity of the targeted enzymes31,32. However, cell-based approaches of this type, while informative, do not establish direct causal relationships. Hence, there is a pressing need for compositionally defined in vitro facsimiles of cellular chromatin that can be used to fully understand molecular mechanisms attendant to processes such as spreading33,34. In this study, we have developed a means to access semi-synthetic designer chromatin that mimics natural chromatin with unprecedented detail in terms of array length and composition. The tripartite nature of arrays Nu-1, Nu-4 and Nu-5 enables incorporation of up to three distinct chromatin states at the level of tetranucleosomes, important subunits of chromatin structure19,20. As such, these arrays promise to aid in elucidating the spatial determinants of how (far) histone modifications control chromatin structure and function, exemplified here by their application to investigate the enzymology of heterochromatin spreading.
Collectively, our biochemical data converge on a two-stage activation mechanism for Suv39h1 (Fig. 5c). In our model, Suv39h1, inactive in its free form, first samples chromatin through its chromodomain. Recognition of H3K9me3 then allosterically activates a latent chromatin binding motif to anchor the enzyme, likely involving a Zn-finger-like segment at the N-terminus. This second step in turn stimulates H3K9 methylation, specifically targeted to spatially close nucleosomes due to the multivalent nature of Suv39h1’s chromatin engagement. This model features a highly mobile state of Suv39h1, characterized by low HMT activity to quickly sample chromatin states, which can convert into an immobile population, engaging chromatin with its chromodomain. The N-terminus plays a role in this transition, through additional chromatin binding activity or by mediating cooperative interactions involving the CD and SET domain. This model complements orthogonal pathways to recruit and activate Suv39h1 for initiation of heterochromatin spreading, such as binding to RNA transcripts35 or heterochromatin proteins27,32,36.
Previously, a ‘guided state’ mechanism has been proposed for spreading by Clr4, the yeast homolog of Suv39h113. In this model, Clr4 binds to the chromatin template independent of the presence of H3K9me3, but is able to use the mark to reposition its active site, thus increasing the methyltransferase activity. Our studies of the reconstituted mammalian spreading system reveal that the H3K9me3 mark can allosterically activate Suv39h1 on chromatin. Here, the N-terminus of Suv39h1 could act as an anchor to strengthen the interaction with the substrate. Indeed, we favor a model where direct binding to DNA accounts for this property, although a mechanism in which the N-terminus orchestrates the interplay between the chromodomain and the active site cannot be excluded. The apparent difference in mechanism between the yeast and mammalian systems most likely arises because of differences in the domain architecture in the respective enzymes (Supplementary Fig. 12b). Clr4 does not contain any N-terminal extension, thus lacking the putative anchor present in Suv39h1. The connection between the CD and the SET domain also varies dramatically between the two homologs: the mammalian enzyme contains a 13-residue linker between the extended C-terminal helix of the CD and the N-terminal helix of the pre-SET domain37,38, whereas the yeast enzyme features a >100 amino acid insertion at that position39.
How are chromatin binding and activation of catalysis coupled in Suv39h1? The significantly increased activity of the ΔN variant on peptide substrates (Fig. 2c) hints at the intriguing possibility that the N-terminus auto-inhibits the SET domain, either allosterically or through direct binding. Upon chromatin binding, auto-inhibition is relieved, thus increasing the HMT activity. This effect could explain the difference in activity between the wild-type enzyme, the ΔN variant and the R24,K27A mutant on chromatin substrates (Supplementary Fig. 13). The latter exhibits low HMT activity because it is auto-inhibited and cannot engage chromatin. By contrast, the ΔN variant suffers from poor chromatin binding, but displays intermediate catalytic proficiency because it is not auto-inhibited. Structural studies on the full-length enzyme might shed light on this issue.
We propose that the 2-state mechanism delineated above represents a common theme in chromatin biochemistry. Firstly, coupling of fast sampling of the chromatin environment with strong local activation of enzymatic activities at designated regions through a positive feedback loop provides an ideal mechanistic foundation for the formation of dynamic PTM domains. Moreover, many histone-modifying enzymes contain subunits that bind their respective products. Prominent examples include the methyltransferases PRC2 (which is also regulated through interactions with nucleic acids), G9a, and GLP14,40, the acetyltransferases p300 and GCN541,42, and the demethylase KDM5A15. Sophisticated designer chromatin substrates of the type generated herein provide a foundation on which to test these ideas further.
ONLINE METHODS
General laboratory methods
Amino acid derivatives and coupling reagents were purchased from Novabiochem (Darmstadt, Germany). Dimethylformamide (DMF), dichloromethane (DCM), triisopropylsilane (TIS) were purchased from Sigma-Aldrich and used without further purification. Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) was purchased from Thermo Scientific. 2-(7-Aza-1H-Benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU) and O-(Benzotriazol-1-yl)-N,N,N’,N’-tetramethyluronium hexafluorophosphate (HBTU) were purchased from Genscript. Trifluoroacetic acid (TFA) was purchased from Halocarbon. N,N-Diisopropylethylamine (DIPEA) was purchased from Acros Organics. Sephacryl S-200 columns were obtained from GE Healthcare. T4 DNA ligase and restriction enzymes were obtained from New England BioLabs. Primer synthesis and DNA sequencing were performed by Integrated DNA Technologies and Genewiz, respectively. Centrifugal filtration units were from Sartorius, and MINI dialysis units and ECL solution were from Pierce. Kodak XAR film, anti-FLAG M2 affinity gel, and anti-FLAG peptide were purchased from Sigma Aldrich. Size exclusion chromatography was performed on an AKTA FPLC system from GE Healthcare equipped with a P-920 pump and UPC-900 monitor. Analytical reversed-phase HPLC (RP-HPLC) was performed on a Agilent 1200 series instrument with a Vydac C18 column (5 micron, 4 x 150 mm), employing 0.1% TFA in water (HPLC solvent A), and 90% acetonitrile, 0.1% TFA in water (HPLC solvent B), as the mobile phases. Analytical gradients were 0–70% HPLC buffer B over 30 min at a flow rate of 1 mL/min, unless stated otherwise. Preparative scale purifications were conducted on a Waters prep LC system comprised of a Waters 2545 Binary Gradient Module and a Waters 2489 UV detector (Waters). A Vydac C18 preparative column (15–20 micron, 20 x 250 mm) or a semi-preparative column (12 micron, 10 mm x 250 mm) was employed at a flow rate of 20 mL/min or 4 mL/min, respectively. ESI-MS analysis was conducted on a MicrOTOF-Q II ESI-Qq-TOF mass spectrometer (Bruker Daltonics). UV spectrometry was performed on an Agilent 8453 UV-Vis spectrophotometer (Agilent, Santa Clara, CA).
Statistical methods
Error bars indicate the standard error of the mean. Typically, values from independent replicates were normalized to measurements obtained using wild-type Suv39h1 and Nu-1 substrates. Errors were subsequently calculated for relative values.
Solid phase peptide synthesis
Peptides were synthesized on Rink Amide-ChemMatrix resin (PCAS Biomatrix), either using manual addition of the reagents (using a stream of dry N2 to agitate the reaction mixture) or on a Liberty Peptide Synthesizer equipped with a Discovery microwave module (CEM).
For manual solid phase synthesis, typical cycles were: (a) Fmoc group deprotection with 3 mL of 20% piperidine in DMF (1 x 1 min, 2 x 8 min) and (b) coupling of 5 eq. of amino acid to the growing peptide chain with 4.9 eq. of either HATU or HOBt/HBTU and 10 eq. of DIPEA for 20 – 45 min. Double couplings were used when necessary to ensure complete acylation. Between each step, the resin was thoroughly washed with DMF. Upon completion of the synthesis, peptides were cleaved from the resin with 95% TFA, 2.5% TIS and 2.5% H2O. The crude peptide was then precipitated with diethyl ether, dissolved in water with 0.1% TFA and analyzed via RP-HPLC. Subsequently, the peptide was purified by preparative RP-HPLC, lyophilized and stored at −20 °C.
H3 peptides
Unmodified and H3K9me3-containing histone peptides (encompassing residues 1–20 with the sequence H-ARTKQ TARK(me0/3)S TGGKA PRKQL GYK(alloc)-NH2) were prepared by automated solid phase peptide synthesis. A C-terminal GYK(alloc) motif was included as a spectroscopic handle and for potential modification. Peptides were purified by reverse phase HPLC on a C18 column (Vydac 218TP1022 10 µM; 22 x 250 mm) at a flow rate of 18 mL/min using a 10–30% B gradient. The purified peptides were analyzed by reverse phase HPLC and ESI-MS: H3(1–20) unmodified (M+H)+ observed: 2,614.9 Da, expected 2,614.5 Da; H3(1–20)K9me3 (M+H)+ observed: 2,656.6 Da, expected 2,656.5 Da). Peptides were lyophilized and stored at −20 °C. Aliquots were dissolved in H2O and concentrations determined by the absorbance of the tyrosine residue (ε280nm = 1,490 M−1cm−1). Biotinylated peptides were a generous gift from Peter W. Lewis and C. David Allis (Rockefeller University, New York, USA).
Synthesis of hH3.1K9me3
Homogeneously modified hH3.1K9me3 (H3K9me3) was prepared by protein semi-synthesis according to previously published procedures42. Briefly, the N-terminal tail encompassing residues 1–28 and the K9me3 mark was synthesized on solid phase with a C-terminal diaminobenzoic acid moiety. This handle enabled facile conversion of the cleaved peptide into an α-thioester for native chemical ligation44. Trimethyllysine at position 9 was incorporated using Fmoc-Lys(me3,Cl)-OH. The C-terminal residue, Fmoc-Ser(tBu)-OH, was coupled to diaminobenzoic acid in solution as described44, and subsequently loaded onto a rink amide ChemMatrix resin. SPPS was carried out using standard Fmoc-protocols (see above). Upon completion of the peptide synthesis, the diaminobenzoyl linker was activated by acylation with p-nitrophenylchloroformate for 2 x 40 min. at room temperature, followed by incubation with 0.5 M DIPEA in DMF to yield a cyclic N-acylurea intermediate. The peptides were then cleaved from the resin and deprotected with TFA/triisopropylsilane (TIS)/H2O (95:2.5:2.5) for 3 h, precipitated with cold ether and collected by centrifugation. Crude peptides were then converted into α-thioesters by incubation in 100 mM phosphate buffer, pH 7.5 containing 150 mM 2-mercaptoethanesulfonate (MESNa) for 1 h at room temperature and purified by RP-HPLC on a preparative scale using a 5–20% B gradient over 60 min. The resulting peptide was characterized by ESI-MS: (M+H)+ observed: 3,165 Da; expected: 3,165 Da.
The C-terminal fragment of H3.1 (encompassing residues 29–135 and containing the mutations A29C, C96A, and C110A) was produced in E. coli as a SUMO fusion as previously described42. The protein was isolated from inclusion bodies and purified by Ni-NTA chromatography under denaturing conditions. Upon refolding of SUMO, the fusion protein was incubated with Ulp1 SUMO protease, and the cleaved histone fragment purified by RP-HPLC (45–60 % HPLC buffer B gradient, 20 mL/min, over 60 min). The final product was characterized by ESI-MS: H3.1 (29–135, A29C,C96A,C110A) (M+H)+ observed: 12,263 Da, calculated: 12,261 Da).
The α-thioester (4.5 mg, 1.4 µmoles) was ligated to the C-terminal fragment (8 mg, 0.65 µmoles) in 650 µL ligation buffer (100 mM sodium phosphate pH 7.5 containing 6 M guanidinium chloride (GdmCl), 30 mM 4-mercaptophenylacetic acid (MPAA) and 60 mM TCEP) for 6 hours at RT. The product was purified by reverse phase HPLC and subjected to radical desulfurization45 with 40 mM glutathione, 200 mM TCEP, 16 mM VA-61 at 37 °C overnight. hH3.1K9me3 was purified by reverse phase HPLC in 20 % isolated yield (2 mg) over 2 steps: (M + H)+ observed: 15,250 Da; expected 15,251 Da (Supplementary Fig. 1).
Expression of recombinant histones
Unmodified, recombinant human histones H2A, H2B, H3.1(C96,C110A), and H4 were expressed in E. coli and purified by ion exchange and reverse chromatography as previously described42. The purified histones were analyzed by reverse phase HPLC and ESI-MS: H2A (M+H)+ observed: 13,964 Da, expected 13,964 Da; H2B (M+H)+ observed: 13,758 Da, expected 13,759 Da; H3.1(C96,110A) (M+H)+ observed: 15,208 Da, expected 15,209 Da; H4 (M+H)+ observed: 11,235 Da, expected 11,236 Da.
The H3K9R mutant was generated by Quik Change Site-Directed Mutagenesis (Agilent) using primers CAAACCGCGCGTAGGTCCACCGGCG and CGCCGGTGGACCTACGCGCGGTTTG according to manufacturer’s instructions. The protein was produced analogously to the wild type. ESI-MS: (M+H)+ observed: 15,236.1 Da, expected 15,236.7 Da.
Octamer formation
Octamers containing the desired histone variants (wild-type H3, H3K9me3, H3K9R) were prepared as previously described46. Briefly, recombinant and semi-synthetic histones were dissolved in histone unfolding buffer (20 mM Tris-HCl, 6 M GdmCl, 0.5 mM DTT, pH 7.5), combined (1.1 eq H2A, 1.1 eq H2B, 1.0 eq H3 variant, 1 eq H4), and the total histone concentration was adjusted to 1 mg/mL. Octamers were assembled by dialysis at 4 °C against 3x 1 L of octamer refolding buffer (10 mM Tris-HCl, 2 M NaCl, 0.5 mM EDTA, 1 mM DTT, pH 7.5) and subsequently purified by size exclusion chromatography on a Superdex S200 10/300 column. Fractions containing octamers were combined, concentrated, diluted with glycerol to a final 50 % v/v and stored at −20 °C.
Preparation of nucleosomal DNA
Dodecameric repeats of the 601 sequence23 separated by 30 bp linkers were produced from pWM530 by EcoRV digestion and PEG-6000 precipitation according to previously published procedures47. Tetrameric repeats (A4-1, A4-2, A4-3) were generated from a plasmid harboring the 601 sequence using a standard PCR- and restriction enzyme-based molecular cloning approaches. Full sequences of the tetrameric repeats, including the location of the DraIII, BsaI and AlwNI non-palindromic sites used in chromatin assembly, are provided in Supplementary Fig. 2. Note, for the central segment, A4-2, peripheral NdeI and NcoI restriction sites were included to enable selective cleavage of the resulting arrays for analytical purposes. The 4-mer repeats were excised from the respective WM530-based vectors with AlwNI and BsaI (A4-1) or BsaI and DraIII (A4-2, A4-3). Subsequently, the backbone was digested with EcoRV to enable selective PEG precipitation of the 4-mer repeats. Typically, two or three iterations of precipitation were conducted. The purified DNA was dissolved in TE buffer and stored at −80 °C in aliquots.
Formation of homotypic nucleosome arrays
Homotypic dodecameric arrays were assembled from purified octamers (containing either recombinant H3 (Array Nu-2) or semi-synthetic H3K9me3 (Array Nu-3)) and recombinant DNA in the presence of buffer DNA by salt gradient dialysis as previously described 48. Homotypic tetramers were prepared in the absence of buffer DNA on a 50–200 pmole scale. The resulting arrays were purified by sucrose gradient centrifugation (5–40% sucrose in 4.5 mL TEK buffer (10 mM Tris, pH 8, 0.1 mM EDTA, 10 mM KCl)) for 5h at 40,000 rpm in a 55Ti Rotor (Beckman Coulter) at 4 °C. Fractions containing saturated tetrameric arrays were pooled, concentrated on a Vivaspin 500 centrifugal concentrator (30 kDa cutoff) and washed three times with TEK containing 0.2 mM PMSF to remove the sucrose. Six different types of homotypic tetramers were produced: A4-1 wt H3; A4-2 wt; A4-3 wt; A4-1 H3K9R; A4-2 H3K9R; A4-3 H3K9me3. Recoveries for purified fragments ranged from 20–60 %.
Formation of heterotypic nucleosome arrays by DNA ligation
Heterotypic arrays were synthesized by DNA ligation from homotypic fragments. Unique non-palindromic overhangs generated by AlwNI, BsaI and DraIII enabled one-pot, site-specific ligation. Typically, 1.25 equivalents of outer fragments (40 nM per 601 site) and 1.0 equivalent of the middle segment (32 nM per 601 site) were joined using T4 DNA ligase (NEB, 5 U/µL) in 70 mM Tris-HCl, pH 7.5 containing 6 mM MgCl2, 1 mM ATP and 10 mM DTT. After 2h at 16 °C, the reactions were centrifuged (17,000 g for 10 min at 4 °C). The pellet containing predominantly 12-mer products was redissolved in TEK buffer and subjected to sucrose gradient purification as described for the 4-mer intermediates. Isolated yields were typically around 20 %.
Array integrity was assessed by ScaI- and MNase digestion. The former was performed with 0.9 pmoles 601 site and 500 U ScaI in 3.5 µL 10 mM Tris-HCl, pH 7.5 containing 0.1 M KCl, 0.5 mM MgCl2, 1 mM DTT for 4h at RT. Digested products were analyzed by native gel electrophoresis on a 5% acrylamide gel. MNase digestion was performed with 0.9 pmoles 601 site and 0.2 U MNase in 20 µL 10 mM Tris-HCl, pH 7.5 containing 1 mM CaCl2 at 4 °C. The reaction was quenched with SDS after 30 s, and the digested DNA was isolated with a Qiagen PCR purification kit and analyzed by acrylamide gel electrophoresis. Representative examples of ScaI and MNase digestions of dodecameric arrays Nu-1, Nu-2 and Nu-3 are shown in Supplementary Figure 3.
The integrity of H3K9me3 domains installed through chromatin ligations were tested by restriction digestion, native gel electrophoresis and western blotting. Specifically, 0.125 pmoles array Nu-1 (1 pmoles H3K9me3 and 2 pmoles of unmodified H3 on the histone level) were incubated for 1 hour at room temperature in 1x digestion buffer (10 mM Tris, pH 8, 50 mM KCl, 1 mM MgCl2) in the presence of 10 U NdeI or NcoI. Subsequently, array fragments were separated by native gel electrophoresis (2 % acrylamide, 1% agarose), and transferred to a PVDF membrane on a semi-dry blotter in the absence of SDS and MeOH. The resulting blots were developed with anti-H3K9me3 antibodies (Abcam, ab8898, 1:200).
Cloning, expression, and purification of Suv39h1
The gene coding for Suv39h1 was obtained from Open Biosystems (MHS1011-61443) and subcloned into a pFastBac vector (Life Technologies) with an N-terminal FLAG Tag. Variants were produced by site directed mutagenesis using the QuikChange Site-Directed Mutagenesis kit (Agilent). Bacmids were generated in DH10Bac cells according to manufacturer’s instructions and transfected into Sf-9 cells. Proteins were produced in Sf-9 cells infected with desired baculovirus constructs in Sf-900 III serum-free media on a 50–500 mL scale in shaker flasks. After 2 days of infection, cells were harvested by centrifugation. The cells were either stored at −80 °C or directly lysed by dounce homogenization in lysis buffer (20 mM Tris-HCl, 500 mM NaCl, 4 mM MgCl2, 0.4 mM EDTA, 20% glycerol, 1 mM PMSF, 2 mM DTT, pH 7.9) and the solution cleared by centrifugation. The soluble extracts were incubated with anti-FLAG M2 affinity gel (100 µL resin per 100 mL cell culture) in wash buffer (20 mM Tris, pH 7.9, 150 mM NaCl, 2 mM MgCl2, 0.2 mM EDTA, 15% (v/v) glycerol, 0.1% NP-40, 1 mM PMSF, 1 mM DTT) for 1 hour at 4 °C. The resin was collected by centrifugation and washed three times with wash buffer. Bound proteins were subsequently eluted in wash buffer containing 0.25 mg/mL FLAG peptide (2x 20 min at 4 °C). Eluted fractions were concentrated using Vivaspin 500 filters (10 kDa MW cutoff), and stored at −80 °C. Wild-type and mutant Suv39h1 were analyzed by SDS-PAGE and silver staining (Supplementary Fig. 4a) and/or western blotting (Supplementary Fig. 4b). Concentrations of wild-type Suv39h1 were estimated from silver-stained gels including a series of BSA standards. Relative concentrations of variants were estimated from α-FLAG western blots.
Histone methyltransferase assays
The methyltransferase activity of Suv39h1 and its variants was measured using a scintillation based assay. Typically, arrays (40 nM 601 site) were incubated with approximately 10 nM Suv39h1 in 10 µL HMT buffer (50 mM Tris-HCl, pH 8.5 containing 5 mM MgCl2, 5 mM DTT, 1 mM PMSF) in the presence of 1.4 µM 3H-S-Adenosylmethionine (3H-SAM Perkin Elmer, Waltham, MA) for 2 h at room temperature. Note that under these conditions, arrays are reversibly self-assembled. Reactions were quenched by spotting on Whatman P81 phosphocellulose filter discs (Sigma). The filters were dried for 1 h at RT, washed 3x with 0.2 M NaHCO3, pH 9, and finally dried on a gel dryer for 30 min at 40 °C. Scintillation counting was performed with 1 mL Ultima Gold scintillation cocktail on a MicroBeta2 scintillation counter (Perkin Elmer). Alternatively, 10 µM H3(1–20) peptide was used as substrates instead of nucleosome arrays. When appropriate, H3(1–20)K9me3 peptide was added at 0.2 µM, unless otherwise stated.
Where indicated, HP1 or its variants were added at 1 µM, approximating the concentration of HP1β in euchromatin.29 We chose the HP1β isoform, also known as Cbx1, because it exhibits robust binding to H3K9me3 marks in vitro.49
To determine the location of HMT activity on array Nu-1, an HMT reaction was performed as above, but with higher concentrations of arrays (75 nM 601 site), Suv39h1 (15 nM) and 2.1 µM 3H-SAM in 5 µL HMT buffer containing 0.5 mM MgCl2. The lower concentration of Mg2+ facilitates downstream analysis, and reduces chromatin self-assembly, ensuring that spreading occurs in an intra-array fashion. After intervals of 15, 30, 60 and 130 min at RT, 10 µL of digestion buffer containing 20 U of NcoI and 200 µM cold SAM were added. Digestions were allowed to proceed for 15 min at RT. Subsequently, the resulting array fragments (an 8-mer containing the pre-installed H3K9me3 marks and the adjacent 4 nucleosomes and a 4-mer containing the four nucleosomes distal to the pre-modified sites) were resolved by native gel electrophoresis (1% agarose and 2 % acrylamide). Gels were stained with Sybr Gold (Invitrogen), followed by fixing (40 % EtOH, 10 % AcOH, 45 min, RT) and incubation in Amplify solution (GE healthcare, 30 min, RT). Next, the gels were dried (2 h, 70 °C) and 3H-methyl incorporation was visualized by fluorography using XAR films. Bands corresponding to the 8-mer and 4-mer fragments were quantified by densitometry and relative values of 3H-incorporation were normalized by the total Sybr Gold signal in each lane. Time courses were conducted in duplicate.
Chromatin self-assembly assay
Self-assembly of chromatin in HMT assay buffer was assessed by incubating dodecameric arrays (40 nM 601 site) in 10 µL HMT buffer with varying concentrations of MgCl2 (0–5 mM) for 10 min at 4 °C. Array aggregates were pelleted by centrifugation (10 min at 17,000 g at 4 °C) and the amount of chromatin in the supernatant was determined by UV spectroscopy or Sybr Gold fluorescence (SpectraMax M3 plate reader, Molecular Devices, Sunnyvale, CA, at 495 nm excitation, 540 nm emission).
Chromatin-Suv39h1 binding assays
Binding of Suv39h1 variants to chromatin was evaluated with a co-precipitation assay. Dodecameric arrays (37.5 nM 601 site) were precipitated in the presence of approximately 10 nM Suv39h1 variant in 10 µL HMT buffer (5 mM MgCl2 unless stated otherwise) supplemented with 0.1 mg/mL BSA. Where appropriate, histone peptides were added at 0.2 µM. After 30 min at RT, the samples were centrifuged for 10 min at 4 °C. The supernatants were collected and the pellets were redissolved in 10 µL TEK for 10 min at RT and centrifuged again for 10 min at 4 °C to remove insoluble species. Individual fractions were denatured with SDS-loading buffer (5 min at 95 °C) and the presence of Suv39h1 variants analyzed by western blot (α-FLAG).
Cloning, expression and purification of HP1
The gene coding for human Cbx1 (HP1β) was amplified from a cDNA library (Invitrogen Superscript, produced according to manufacturer’s instructions) by PCR using primers CAGCcatatgGGGAAAAAACAAAACAAGAAG and AGCCggatccTTAATTCTTGTCATCTTTTTTGTCATCATCC. The PCR product was digested with NdeI and BamHI and ligated into an appropriately digested pET15b vector. The resulting vector was transformed into the E. coli strain BL21 for protein production (1L expression for 3h at 37 °C with 0.6 mM IPTG). Cells were collected by centrifugation (20 min. at 3,500 g at 4°C) and lysed in 15 mL lysis buffer (20mM Tris-HCl, pH 7.6, 0.5 mM EDTA, 1 mM β-ME, 10% glycerol, 100 mM NaCl, 10 mM imidazole, 0.1% NP-40, Roche protease inhibitors) by sonication. The lysate was cleared by centrifugation (20 min at 15,000 g at 4°C) and the supernatant was purified by IMAC (3 mL Ni-NTA agarose, eluted with 20mM Tris-HCl, pH 7.6, 1 mM β-ME, 10% glycerol, 100 mM NaCl, 500 mM imidazole, 0.1% NP-40, 1 mM PMSF). Folded Cbx1 was further purified by size exclusion chromatography on a Superdex S200 column (running buffer: 20mM Tris-HCl, pH 7.6, 1 mM DTT, 10% glycerol, 100 mM NaCl, 1 mM EDTA, 0.1 mM PMSF). Pure fractions were combined, concentrated to approximately 0.1 mM and stored at −20 °C with 30 % glycerol. Cleaving the N-terminal His6-tag with thrombin had no effect on the ability of HP1 to compete with Suv39h1 variants (data not shown). Purified proteins were analyzed by ESI-MS: (M + H)+ observed: 23,450 Da; expected 23,450 Da. ESI-MS without His6-tag: (M + H)+ observed: 21,699 Da; expected 21,699 Da.
A variant of HP1 with a crippled chromodomain (K44A, W45A) was generated by site directed mutagenesis using primers TCCTCATCTGAGAATCCCTTCGCCGCTAGGAGGTACTCCACTTTGCC and GGCAAAGTGGAGTACCTCCTAGCGGCGAAGGGATTCTCAGATGAGGA. The protein was produced and purified as described above. ESI-MS: (M + H)+ observed: 23,278 Da; expected 23,278 Da.
HP1 peptide binding experiments
To verify the binding activity of HP1 variants, pull-down experiments with biotinylated H3 peptides were conducted. 10 µM unmodified or H3K9me3 peptides were incubated with 14 µM HP1 variant in 20 µL PD buffer (20 mM HEPES, pH 7.5, 80 mM KCl, 0.1 % Igepal, 0.2 mM EDTA, 5 % glycerol, 2 mM DTT) at RT. After 1h, reactions were added to 25 µL Streptavidin-Agarose Resin (Pierce Biotech) and incubated in a rotator for another 30 min. The beads were collected by centrifugation and washed 2x with 100 µL PD buffer. Bound proteins were eluted in 100 µL SDS loading buffer (50 mM Tris-HCl pH 6.8, 2 % (w/v) SDS, 10 % (v/v) glycerol, 1 % (v/v) 2-mercaptoethanol, and 12.5 mM EDTA, 0.02 % (w/v) bromophenol blue). 15 µL of the elution (15%) were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. For comparison, 2 µL of the input (10%) were included in the analysis (Supplementary Fig. 8).
Fluorescence microscopy and FRAP
NIH-3T3 cells were cultured as monolayers in Dulbecco’s Minimal Essential Medium (Gibco BRL, Invitrogen Corporation), supplemented with 10% fetal calf serum, antibiotics (penicillin, streptomycin), and L-glutamine at 37°C in an atmosphere of 5% CO2 in air. Cells were transiently transfected with Suv39h1-GFP fusion constructs (in pcDNA-3.1) using lipofectamine 2000 (Life Technologies). After 6 hours of transfection, cells were trypsinized and applied to a MaTek glass bottom dish. Cells were allowed to attach and grow for another 24 hours before staining with Hoechst 33342 (5 µg/µL). Fluorescence microscopy was performed on a Nikon R1 confocal microscope using a 100x/1.40 objective. Samples were kept at 37 °C in the presence of 5 % CO2. DNA was visualized at 405 nm, GFP fusions at 488 nm. 5 frames were imaged before photobleaching was conducted on a 1 µm circular spot with a 488 nm laser operating at 20 % (heterochromatin) or 100% intensity (euchromatin) for 120 ms. Recovery was monitored for 96 s at 2 s intervals. For visualization purposes, the resulting FRAP traces were fit to the coupled binding-diffusion model of Sprague et al. using MATLAB (Mathworks, Natick, MA)43.
Preparation of truncated Suv39h1 variants
The N-terminal domain and the CD (residues 1–101) of Suv39h1 variants were fused to an MBP tag, and the resulting proteins were produced in E. coli strain BL21 DE3 (overnight at 18 °C). Cells were subsequently lysed in a buffer containing 25 mM Tris, pH 7.5, 200 mM NaCl, 5 mM β-ME, 5% glycerol, 10 µM ZnCl2. Fusion proteins were purified by amylose affinity chromatography and subsequently denatured and refolded. Specifically, unfolding was performed in the presence of 6 M GdmCl for 1 hour at room temperature and refolding was achieved by stepwise dialysis in a buffer containing 50 mM Tris 7.8, 300 mM NaCl, 5 mM β-ME, 20% glycerol, and 50 µM ZnCl2 in the presence of 4 M, 2 M and 0 M Urea for 2h at 4 °C each step. Monomeric proteins were isolated by size exclusion chromatography in a buffer containing 25 mM Tris, pH 7.5, 200 mM NaCl, 10% glycerol and 1 mM DTT, concentrated to approximately 10 µM and stored at 4 °C.
Suv39h1 peptide binding assays
100 pmoles of biotinylated H3 peptides (with or without the K9me3 mark) were immobilized on 20 µL MyOne Streptavidin Dynabeads (Invitrogen) for 15 min at room temperature. Beads were washed with 3x500 µL binding buffer (25 mM Tris, pH 7.5, 150 mM NaCl, 0.1 % BSA, 0.1 % NP40, 10 % glycerol, 1 mM DTT). Subsequently, 20 pmoles of MBP-Suv(1–101) variants were added, and binding was allowed to proceed for 30 min at room temperature. Finally, beads were washed 2x with 100 µL binding buffer, and bound material was eluted in 10 µL SDS loading buffer (5 min at 95 °C) and analyzed by SDS PAGE followed by coomassie staining. For comparison, 4 pmoles of each MBP-Suv(1–101) mutant were included on the gel.
Suv39h1-HP1 interaction assays
Binding between Suv39h1 variants and HP1 was assessed using a pulldown approach. A mixture of the binding partners was incubated in 25 µL binding buffer (25 mM Tris, pH 7.5, 150 mM NaCl, 0.1 % BSA, 0.1 % NP40, 10 % glycerol, 1 mM DTT) for 1 h at room temperature. The reactions were subsequently added to Protein G beads (Life Technologies, corresponding to 75 µL slurry), preloaded with anti-Cbx1 antibody (Abcam ab10478, 5 µg). After 3 hours at 4 °C, the supernatant was removed and the beads washed 2x with 100 µL binding buffer. Bound proteins were eluted with SDS loading buffer and analyzed by western blot against the His6-tag (HP1, Millipore HIS.H8, 1:1000) and FLAG-tag (Suv39h1, Sigma f1804, 1:10000). Pulldowns were performed in duplicate.
Supplementary Material
Acknowledgments
We thank Dr. Uyen Nguyen and Dr. Yael David for help with tissue culture; Dr. Gary Laevsky for advice on microscopy; Dr. Peter Lewis for generously providing biotinylated H3 peptides; Dr. C. David Allis, Dr. Galia Debelouchina, Dr. Zachary Brown, Boyuan Wang and Christopher Jenness for helpful discussions; and Krupa Jani for carful proofreading of this manuscript. NIH-3T3 cells were a generous gift from the Schwarzbauer lab (Princeton University). Funding from the Swiss National Science Foundation (postdoctoral fellowships to M.M.M. and B.F.) and NIH (RO1-GM107047) is gratefully acknowledged.
Footnotes
AUTHOR CONTRIBUTIONS
M.M.M., B.F., T.W.M. conceived the project; M.M.M., B.F., L.B., G.L. designed and performed experiments with supervision from T.W.M.; all authors analyzed data; M.M.M. and T.W.M wrote the manuscript with contributions from all authors.
SUPPLEMENTARY MATERIALS
COMPETING INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 2.Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol. 2010;2:1–25. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 4.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 5.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 6.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 7.Noma K-i, Allis DC, Grewal SIS. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 8.Talbert P, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nature Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 9.Peters A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 10.Hahn M, Dambacher S, Schotta G. Heterochromatin dysregulation in human diseases. J Appl Physiol. 2010;109:232–242. doi: 10.1152/japplphysiol.00053.2010. [DOI] [PubMed] [Google Scholar]
- 11.Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 12.Melcher M, Schmid M, Aagaard L, Selenko P, Laible G, Jenuwein T. Structure-Function Analysis of SUV39H1 Reveals a Dominant Role in Heterochromatin Organization, Chromosome Segregation, and Mitotic Progression. Mol Cell Biol. 2000;20:3728–3741. doi: 10.1128/mcb.20.10.3728-3741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Sady B, Madhani HD, Narlikar GJ. Division of labor between the chromodomains of HP1 and Suv39 methylase enables coordination of heterochromatin spread. Mol Cell. 2013;51:80–91. doi: 10.1016/j.molcel.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres IO, Kuchenbecker KM, Nnadi CI, Fletterick RJ, Kelly MJ, Fujimori DG. Histone demethylase KDM5A is regulated by its reader domain through a positive-feedback mechanism. Nat Commun. 2015;6:6204. doi: 10.1038/ncomms7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rando OJ. Global patterns of histone modifications. Curr Opin Genet Dev. 2007;17:94–99. doi: 10.1016/j.gde.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Müller MM, Muir TW. Histones: at the crossroads of peptide and protein chemistry. Chem Rev. 2015;115:2296–2349. doi: 10.1021/cr5003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGinty RK, Tan S. Nucleosome structure and function. Chem Rev. 2015;115:2255–2273. doi: 10.1021/cr500373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song F, Chen P, Sun D, Wang M, Dong L, Liang D, Xu R-M, Zhu P, Li G. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh TH, Weiner A, Lajoie B, Dekker J, Friedman N, Rando OJ. Mapping nucleosome resolution chromosome folding in Yeast by Micro-C. Cell. 2015;162:108–119. doi: 10.1016/j.cell.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng C, Hayes JJ. Intra- and inter-nucleosomal protein-DNA interactions of the core histone tail domains in a model system. J Biol Chem. 2003;278:24217–24224. doi: 10.1074/jbc.M302817200. [DOI] [PubMed] [Google Scholar]
- 22.Blacketer MJ, Feely SJ, Shogren-Knaak MA. Nucleosome interactions and stability in an ordered nucleosome array model system. J Biol Chem. 2010;285:34597–34607. doi: 10.1074/jbc.M110.140061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 24.Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 25.Erdel F, Müller-Ott K, Rippe K. Establishing epigenetic domains via chromatin-bound histone modifiers. Ann N Y Acad Sci. 2013;1305:29–43. doi: 10.1111/nyas.12262. [DOI] [PubMed] [Google Scholar]
- 26.Chin H, Patnaik D, Estève P-O, Jacobsen S, Pradhan S. Catalytic properties and kinetic mechanism of human recombinant Lys-9 histone H3 methyltransferase SUV39H1: participation of the chromodomain in enzymatic catalysis. Biochemistry. 2006;45:3272–3284. doi: 10.1021/bi051997r. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto K, Sonoda M. Self-interaction of heterochromatin protein 1 is required for direct binding to histone methyltransferase, SUV39H1. Biochem Biophys Res Commun. 2003;301:287–292. doi: 10.1016/s0006-291x(02)03021-8. [DOI] [PubMed] [Google Scholar]
- 28.Muramatsu D, Singh P, Kimura H, Tachibana M, Shinkai Y. Pericentric Heterochromatin Generated by HP1 Protein Interaction-defective Histone Methyltransferase Suv39h1. J Biol Chem. 2013;288:25285–25296. doi: 10.1074/jbc.M113.470724. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Müller-Ott K, et al. Specificity, propagation, and memory of pericentric heterochromatin. Mol Sys Biol. 2013;10:746. doi: 10.15252/msb.20145377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krouwels IM, Wiesmeijer K, Abraham TE, Molenaar C, Verwoerd NP, Tanke HJ, Dirks RW. A glue for heterochromatin maintenance: stable SUV39H1 binding to heterochromatin is reinforced by the SET domain. J Cell Biol. 2005;170:537–549. doi: 10.1083/jcb.200502154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stewart M, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol. 2005;25:2525–2538. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown ZZ, Müller MM, Kong H-E, Lewis PW, Muir TW. Targeted Histone Peptides: Insights into the Spatial Regulation of the Methyltransferase PRC2 by using a Surrogate of Heterotypic Chromatin. Angew Chem Int Ed. 2015;54:6457–6461. doi: 10.1002/anie.201500085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson A, Li G, Sikorski TW, Buratowski S, Woodcock CL, Moazed D. Reconstitution of Heterochromatin-Dependent Transcriptional Gene Silencing. Mol Cell. 2009;35:769–781. doi: 10.1016/j.molcel.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porro A, Feuerhahn S, Delafontaine J, Riethman H, Rougemont J, Lingner J. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat Commun. 2013;5:5379. doi: 10.1038/ncomms6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall IM, Shankaranarayana GD, Noma K-I, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 37.Wu H, et al. Structural biology of human H3K9 methyltransferases. PLoS One. 2010;5:e8570. doi: 10.1371/journal.pone.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang T, et al. Crystal structure of the human SUV39H1 chromodomain and its recognition of histone H3K9me2/3. PLoS One. 2012;7:e52977. doi: 10.1371/journal.pone.0052977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min J, Zhang X, Cheng X, Grewal SI, Xu R-MM. Structure of the SET domain histone lysine methyltransferase Clr4. Nat Struct Biol. 2002;9:828–832. doi: 10.1038/nsb860. [DOI] [PubMed] [Google Scholar]
- 40.Liu N, et al. Recognition of H3K9 methylation by GLP is required for efficient establishment of H3K9 methylation, rapid target gene repression, and mouse viability. Genes Dev. 2015;29:379–393. doi: 10.1101/gad.254425.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S, Shogren-Knaak MA. The Gcn5 bromodomain of the SAGA complex facilitates cooperative and cross-tail acetylation of nucleosomes. J Biol Chem. 2009;284:9411–9417. doi: 10.1074/jbc.M809617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen UTT, Bittova L, Müller MM, Fierz B, David Y, Houck-Loomis B, Feng V, Dann GP, Muir TW. Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries. Nat Meth. 2014;11:834–840. doi: 10.1038/nmeth.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sprague BL, Pego RL, Stavreva DA, McNally JG. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys J. 2004;86:3473–3495. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
ONLINE METHODS REFERENCES
- 44.Blanco-Canosa JB, Dawson PE. An Efficient Fmoc-SPPS Approach for the Generation of Thioester Peptide Precursors for Use in Native Chemical Ligation. Angew Chem Int Ed. 2008;47:6851–6855. doi: 10.1002/anie.200705471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wan Q, Danishefsky SJ. Free-radical-based, specific desulfurization of cysteine: a powerful advance in the synthesis of polypeptides and glycopolypeptides. Angew Chem Int Ed. 2007;46:9248–9252. doi: 10.1002/anie.200704195. [DOI] [PubMed] [Google Scholar]
- 46.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 47.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: Requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 48.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hiragami-Hamada K, Shinmyozu K, Hamada D, Tatsu Y, Uegaki K, Fujiwara S, Nakayama J-I. N-terminal phosphorylation of HP1α promotes its chromatin binding. Mol Cell Biol. 2011;31:1186–1200. doi: 10.1128/MCB.01012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.