Abstract
Breast cancers possess fundamentally altered metabolism that fuels their pathogenicity. While many metabolic drivers of breast cancers have been identified, the metabolic pathways that mediate breast cancer malignancy and poor prognosis are less well understood. Here, we used a reactivity-based chemoproteomic platform to profile metabolic enzymes that are enriched in breast cancer cell-types linked to poor prognosis, including triple-negative breast cancer (TNBC) cells and breast cancer cells that have undergone an epithelial-mesenchymal transition-like state of heightened malignancy. We identified glutathione-S-transferase Pi 1 (GSTP1) as a novel TNBC target that controls cancer pathogenicity by regulating glycolytic and lipid metabolism, energetics, and oncogenic signaling pathways through a protein interaction that activates glyceraldehyde-3-phosphate dehydrogenase activity. We show that genetic or pharmacological inactivation of GSTP1 impairs cell survival and tumorigenesis in TNBC cells. We put forth GSTP1 inhibitors as a novel therapeutic strategy for combatting TNBCs through impairing key cancer metabolism and signaling pathways.
Graphical abstract
eTOC blurb
Using a reactivity-based chemoproteomic platform, Louie et al. have identified GSTP1 as a triple-negative breast cancer target that, when inhibited, impairs breast cancer pathogenicity through inhibiting GAPDH activity and downstream metabolism and signaling pathways.
Introduction
Breast cancers possess fundamentally altered metabolism that drives their pathogenic features. Since Otto Warburg’s seminal discovery in the 1920s that cancer cells have heightened glucose uptake and aerobic glycolysis, recent studies have identified many other biochemical alterations in cancer cells, including heightened glutamine-dependent anaplerosis and de novo lipid biosynthesis that serve as metabolic platforms for breast cancer cells to generate biomass for cell division and metabolites that modulate cancer cell signaling, epigenetics, and pathogenicity (Benjamin et al., 2012; Cantor and Sabatini, 2012; Pavlova and Thompson, 2016). While targeting dysregulated metabolism is a promising strategy for breast cancer treatment, the metabolic pathways that drive pathogenicity in breast cancer subtypes that are correlated with heightened malignancy and poor prognosis remain poorly understood.
Mortality from breast cancer is almost always attributed to metastatic spread of the disease to other organs, thus precluding resection as a treatment method. Unfortunately, conventional chemotherapy fails to eradicate most human cancers, including aggressive breast cancers. Studies over the past decade have uncovered certain breast cancer types and cell-types that are associated with poor prognosis, such as estrogen/progesterone/HER2 receptor-negative (triple-negative) breast cancers (TNBCs) or cancer stem/precursor cells (CSCs) that possess self-renewing and tumor initiating capabilities, epithelial-to-mesenchymal transition (EMT), poor prognosis, and chemotherapy-resistance within breast tumors (Dawson et al., 2009; Dietze et al., 2015; Polyak and Weinberg, 2009). While eliminating these breast cancer types is critical in combatting breast cancer, there are currently few to no therapies that target this malignant population of breast cancer cells.
In this study, we used a reactivity-based chemoproteomic platform to identify metabolic enzymes that are heightened in TNBC cells or upon induction of an EMT-like programming of heightened malignancy in breast cancer cells. Through this profiling effort, we identified glutathione-S-transferase Pi 1 (GSTP1) as a critical metabolic driver that is heightened specifically in TNBCs to control multiple critical nodes in cancer metabolism and signaling pathways to drive breast cancer pathogenicity.
Results
Profiling Dysregulated Metabolic Enzymes in TNBC Cells and CDH1 Knockdown Breast Cancer Cells
To identify metabolic drivers of breast cancer pathogenicity in aggressive breast cancer cell-types associated with malignancy and poor prognosis, we used a reactivity-based chemical proteomic strategy to map cysteine and lysine reactivity in TNBC cells and breast cancer cells with EMT-like features (Fig. 1; Table S1). Both TNBC cells and breast cancer cells that have undergone EMT have been linked to heightened aggressiveness and poor prognosis. Specifically, we wanted: 1) to identify TNBC-specific metabolic enzyme targets by comparing a panel of 4 non-TNBC and 5 TNBC cell lines; and 2) to identify upregulated enzyme targets in MCF7 breast cancer cells upon knockdown of CDH1, a critical mediator of EMT and cell-cell adhesion. We knocked down CDH1 in MCF7 cells with short-hairpin oligonucleotides (shCDH1 cells) to induce an EMT-like state. These cells show upregulation of the mesenchymal marker vimentin and concordant increases in serum-free cell survival, proliferation, and migration (Fig. S1), consistent with EMT-like characteristics.
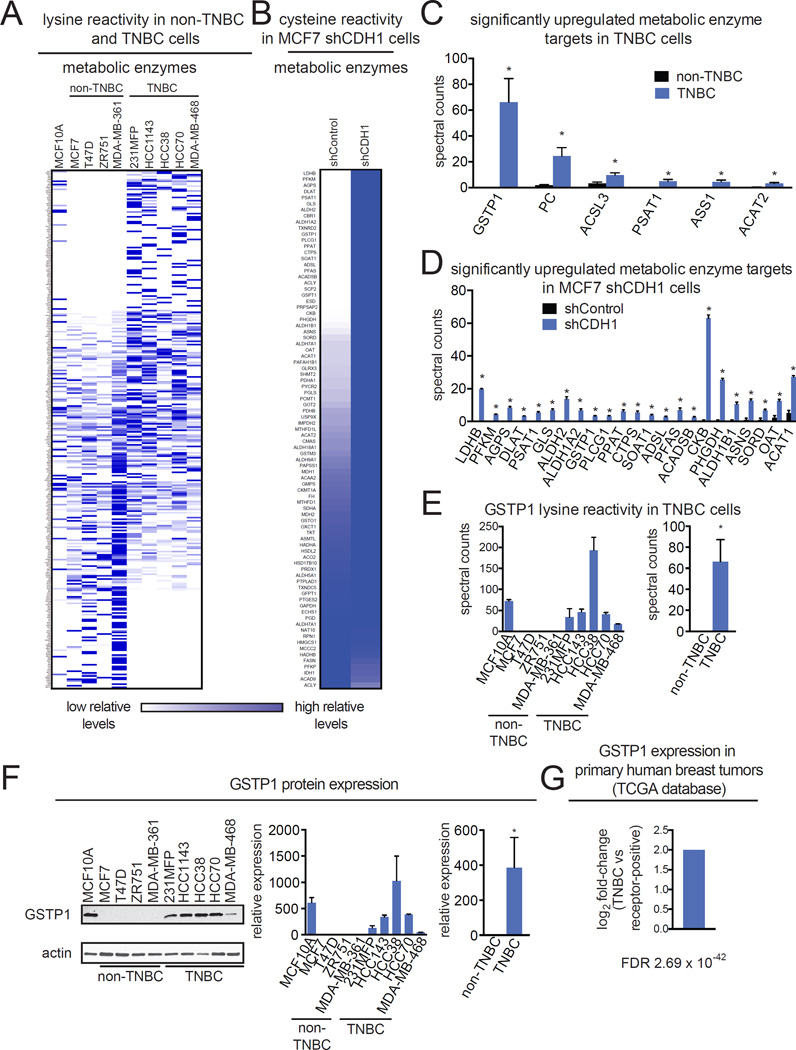
Figure 1. Profiling dysregulated metabolic enzyme targets in TNBC cells and CDH1 knockdown breast cancer cells.
(A, B) Chemoproteomic profiling of a panel of MCF10A non-transformed mammary epithelial cells, non-TNBC, and TNBC cell lines (A) and shControl and shCDH1 MCF7 breast cancer cells (B) with the lysine-reactive dichlorotriazine-alkyne and cysteine-reactive iodoacetamide-alkyne probes, respectively. Probe-labeled proteins were avidin-enriched and enriched proteins were trypsinized and analyzed by LC-MS/MS and quantified by spectral counting. Heatmaps represent relative levels for each protein where dark and light blue indicate higher and lower enrichment of the protein, respectively. (C,D) Significantly upregulated metabolic enzyme targets in TNBC cells (C) and shCDH1 MCF7 cells (D). (E) GSTP1 lysine reactivity in TNBC cells from each individual cell line and combined averaged spectral counts from non-TNBC and TNBC cells. (F) GSTP1 expression was measured across a panel of MCF10A non-transformed mammary epithelial cells, non-TNBC, and TNBC cell lines by Western blotting. Western blotting image shown is a representative image from n=3/group. GSTP1 expression was normalized to actin loading control and quantified by densitometry. (G) GSTP1 expression in receptor-positive and TNBC primary human breast tumors from The Cancer Genome Atlas (TCGA) database. FDR refers to false-discovery rate. Data in (A) is presented as mean ± SEM, n=3–5/group. Significance is presented as *p<0.05 compared to non-TNBC or shControl cells. Detailed data from the chemoproteomic study in (A–D) can be found in Table S1. Characterization of shCDH1 cells is shown in Fig. S1.
We labeled cell lysates from these two breast cancer models with previously validated lysine-reactive dichorotriazine-alkyne or cysteine-reactive iodoacetamide-alkyne reactivity-based probes to tag proteins bearing functional lysines or cysteines, respectively, for subsequent attachment of a biotin handle by click-chemistry, enrichment, and analysis by mass-spectrometry, using an adapted method established by Weerapana et al (Fig. 1A, B; Table S1) (Shannon et al., 2014; Weerapana et al., 2010). We chose to profile cysteine and lysine-reactivity in this study since these amino acids are important mediators of enzyme activity or function through nucleophilic and redox catalysis, allosteric regulation, metal binding, and structural stabilization across a wide range of protein classes (Pace and Weerapana, 2013; Shannon and Weerapana, 2015). Upon proteomic analysis of probe-enriched protein targets, we filtered these proteins for metabolic enzyme targets that were significantly upregulated (p<0.01, >5-fold) in TNBC cells or in shCDH1 MCF7 cells compared to non-TNBC or shControl counterparts, respectively (Fig. 1C, 1D; Table S1). GSTP1 was the most significantly upregulated target in TNBC cells that was also significantly heightened in shCDH1 cells so we focused on investigating the pathogenic role of this target in breast cancer (Fig. 1C, 1D).
Many human cancers, including breast, lung, colon, and ovarian cancers have been shown to express high levels of GSTP1 and its expression has been correlated with both disease progression and resistance to chemotherapeutic drugs (Laborde, 2010). GSTP1 inhibitors have been studied in the context of cancer for many years as adjuvant therapies with chemotherapy agents to prevent their detoxification by cancer cells, towards improving chemotherapy efficacy (Tew and Townsend, 2011; Townsend and Tew, 2003). Yet, the majority of chemotherapeutic drugs have a relatively weak affinity for GSTP1, compared to other GST enzymes. This discrepancy between expression of GSTP1 and its correlation with the development of multidrug-resistance suggests additional roles for GSTP1 on influencing metabolic and signaling pathways in cancer cells (Laborde, 2010). GSTP1 inhibitors have not yet been investigated for their potential to be stand-alone therapies towards thwarting breast cancer. We thus focused our attention on further investigating the role of GSTP1 in driving breast cancer pathogenicity.
GSTP1 is a TNBC-Specific Target
We next investigated the expression pattern of GSTP1 in different types of breast cancer cell lines and primary human breast tumors. Interestingly, we show that GSTP1 is not expressed in any of the non-TNBC cells (MCF7, T47D, ZR751, and MDA-MB-361), but is highly expressed across all of the TNBC cells (231MFP, HCC1143, HCC38, HCC70, and MDA-MB-468) (Fig. 1E, F), shown both by dichlorotriazine-alkyne reactivity and Western blotting. We also show that GSTP1 expression is significantly heightened in TNBC compared to receptor-positive primary human breast tumors (Fig. 1G). Overall, our data indicate that GSTP1 may represent a TNBC-specific target.
Genetic or Pharmacological Inactivation of GSTP1 Impairs TNBC Pathogenicity
To investigate the role of GSTP1 in TNBC pathogenicity, we generated two knockdown lines of GSTP1 in TNBC 231MFP cells (shGSTP1-1 and shGSTP1-2) (Fig. 2A). We showed that GSTP1 knockdown impairs serum-free cell survival in 231MFP cells without affecting cell proliferation (Fig. 2B). GSTP1 knockdown also impaired in vivo 231MFP breast tumor xenograft growth in immune-deficient mice (Fig. 2C).
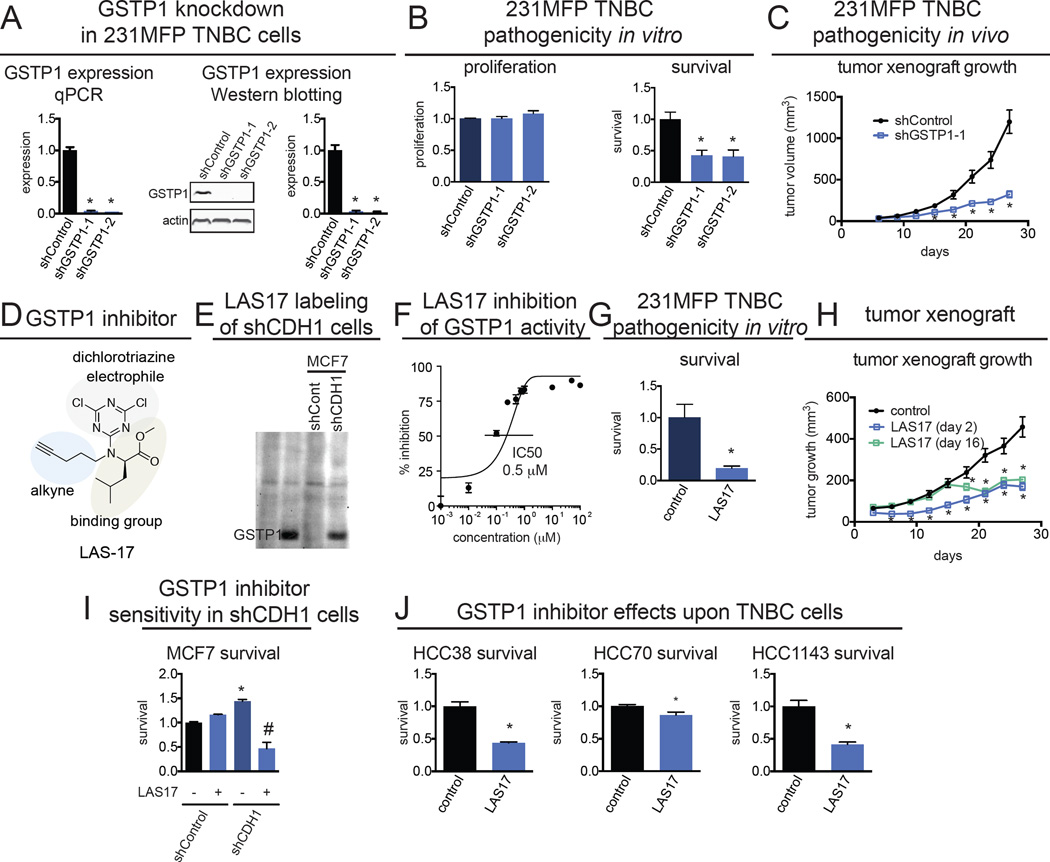
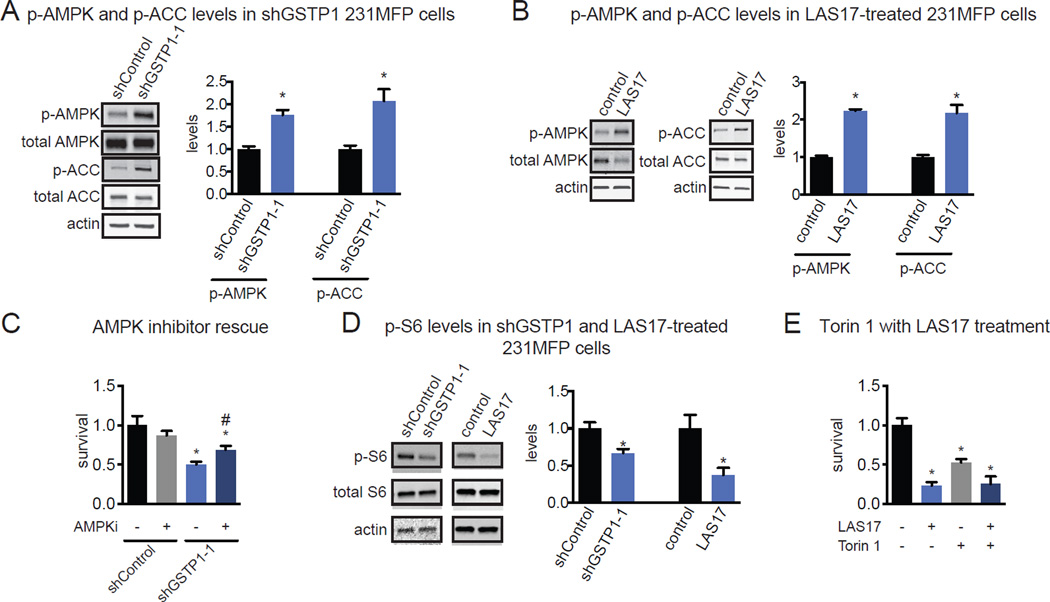
Figure 2. The effect of genetic and pharmacological inactivation of GSTP1 on breast cancer pathogenicity.
(A) GSTP1 was knocked down in 231MFP TNBC cells using two independent shRNA and expression was confirmed by both qPCR and Western blotting. Western blotting image is a representative image from n=3/group and protein expression was quantified by densitometry. (B) GSTP1 knockdown in 231MFP cells shows impaired serum-free cell survival, with no change in cell proliferation 48 h after seeding. Survival and proliferation were assessed using the Hoechst stain. (C) shGSTP1 231MFP cells show impaired tumor growth in SCID mice compared to shControl cells. (D) Selective and irreversible dichlorotriazine GSTP1 inhibitor LAS17 bearing a dichlorotriazine electrophile, GSTP1 binding group, and an alkyne handle for subsequent click chemistry. (E) LAS17 labeling of shControl and shCDH1 MCF7 cells showing heightened GSTP1 in shCDH1 cells. Cell lysates were labeled with LAS17 for 30 min prior to click chemistry with rhodamine-azide. Proteomes were separated on SDS/PAGE and visualized by in-gel fluorescence. (F) LAS17 inhibits GSTP1 activity with a 50 % inhibitory concentration (IC50) of 0.5 µM. GSTP1 activity was assessed by pre-incubating vehicle DMSO or LAS17 with pure GSTP1 protein for 30 min prior to measuring the rate of conjugation of glutathione to CDNB and quantifying the rate of increase in the absorption of the reaction product, glutathione-DNB conjugate. (G) Serum-free survival 48 h after seeding in 231MFP cells treated with DMSO vehicle or LAS17 (10 µM), determined by the Hoechst staining. (H) 231MFP tumor xenograft growth in SCID mice upon once-per-day daily treatment with vehicle (18:1:1 PBS:ethanol:PEG40) or LAS17 (20 mg/kg, ip) either initiated 2 or 16 days after subcutaneous injection of 2 × 106 231MFP cells. (I) Serum-free cell survival in shControl and shCDH1 MCF7 cells treated with DMSO vehicle or LAS17 treatment (10 µM) in shControl and shCDH1 MCF7 cells, determined by WST-1 assay. (J) Serum-free cell survival 48 h after seeding in HCC38, HCC70, and HCC1143 cells treated with DMSO vehicle or LAS17 (10 µM), determined by Hoechst staining. Data in (A, B, C, F, G, H, I, and J) are presented as mean ± SEM, n=3/group for (A), n=5/group in (B), n=8/group in (C), n=3/group in (F), n=5/group in (G), n=8/group in (H), and n=5/group in (I, J). Significance is presented as *p<0.05 compared to shControl or vehicle-treated controls. Body weights and further effects of GSTP1 inhibition upon TNBC pathogenicity are shown in Fig. S2.
Intrigued by these results, we next tested whether pharmacological inhibition of GSTP1 with a highly selective GSTP1 inhibitor LAS17 also impaired breast cancer pathogenicity. LAS17 was recently developed as a highly potent, selective, and irreversible dichlorotriazine GSTP1 inhibitor that targets an active-site tyrosine (Fig. 2D) (Crawford, L.A., Weerapana, E.). In addition to its property as a GSTP1 inhibitor, LAS17 also bears a bioorthogonal alkyne handle, which can be coupled with copper-catalyzed click chemistry (Martell and Weerapana, 2014) to append a fluorophore-azide or biotin-azide analytical handle for subsequent fluorescent or proteomics-based detection of GSTP1, respectively (Fig. 2D, 2E). LAS17 inhibits GSTP1 activity in vitro with a 50 % inhibitory concentration (IC50) value of 0.5 µM (Fig. 2F).
LAS17 treatment in 231MFP breast cancer cells recapitulated the serum-free cell survival impairments observed with genetic inactivation of GSTP1 (Fig. 2G). Daily administration of LAS17 (20 mg/kg ip, once per day) significantly impaired 231MFP breast tumor xenograft growth in immune-deficient mice when treatment was initiated 2 days after subcutaneous injection of cells, and LAS17 even slowed tumor growth when initiated 16 days after tumor implantation (Fig. 2H), with no observable toxicity and no weight-change (Fig. S2A). LAS17 had no effect on serum-free survival of shControl MCF7 non-TNBC cells that do not express GSTP1, but significantly impaired the survival of shCDH1 MCF7 breast cancer cells (Fig. 2I). While GSTP1 inhibition selectively impaired survival in shCDH1 cells, LAS17 treatment in shCDH1 cells did not rescue vimentin expression or impair proliferation, indicating that GSTP1 is not a critical driver of the EMT process (data not shown). LAS17 treatment also impaired serum-free cell survival in additional TNBC lines HCC38, HCC70, and HCC1143 cells (Fig. 2J). We also show that GSTP1 knockdown in HCC38 cells also impairs cell survival (Fig. S2B, S2C). We thus show that pharmacological GSTP1 inactivation impairs breast cancer pathogenicity in culture and in vivo in a seemingly TNBC-specific manner, suggesting that GSTP1 inhibitors may be promising therapeutics for treating TNBCs.
Functional Metabolomic Profiling Reveals GSTP1 Control Over Glycolytic Metabolism, Energetics, and Macromolecular Building Blocks
We next wanted to understand the mechanisms through which GSTP1 was controlling breast cancer pathogenicity. Previous studies have shown that GSTP1 may directly affect cancer pathogenicity through protein-protein interactions with the MAPK family protein JNK, leading to JNK phosphorylation and activation (Tew and Townsend, 2011; Townsend and Tew, 2003). However, GSTP1 genetic or pharmacological inactivation did not lead to JNK activation compared to treatment with the JNK activator anisomycin in 231MFP cells (Fig. S3A). We also postulated that GSTP1 could modulate glutathione levels and oxidative stress to cause cell survival impairments. However, genetic or pharmacological inactivation of GSTP1 did not change oxidative stress levels or reduced to oxidized glutathione (GSH/GSSG) ratios under both basal and menadione-induced oxidative stress conditions compared to controls (Fig. S3B–S3D).
To identify alternate mechanisms, we next performed functional metabolomic profiling to comprehensively map metabolic alterations conferred by genetic or pharmacological inactivation of GSTP1 in 231MFP breast cancer cells (Fig. 3A–3C, Fig. S4A–S4F, Table S2). We used a targeted single-reaction monitoring (SRM)-based liquid chromatography-mass-spectrometry (LC-MS/MS) approach to comparatively measure the levels of ~200 metabolites encompassing glycolytic, TCA cycle, amino acid, nucleotide, and lipid metabolism. We also used an untargeted LC-MS approach in which we collected mass spectra from mass-to-charge ratios (m/z) of 100–1200 and analyzed the resulting features by XCMSOnline (Gowda et al., 2014) to identify metabolites that were altered upon GSTP1 inactivation. We subsequently analyzed any changing m/z by METLIN to identify candidate metabolites, used targeted SRM-based LC-MS/MS to quantify the levels of these metabolites, and added these data to our total targeted analyses. Through these approaches, we identified several metabolites that were commonly changing between both shGSTP1 231MFP lines, including lowered levels of lactic acid, ATP, nucleotides, diacylated phospholipids, and alkylacyl ether lipids and increased levels of acyl carnitines (ACs), ceramides, and lysophospholipids (Fig. 3A–3C, Table S2). LAS17 treatment in 231MFP cells also showed reduced levels of ATP, lactic acid, purine nucleotides, and diacylated phospholipids and alkylacyl ether lipids and increased levels of ACs, ceramides, lysophospholipids (Fig. S4B–S4F, Table S2). While there were many metabolomic changes that were unique to either shGSTP1 or LAS17 treatment, we attributed these differences to metabolic alterations that may manifest under chronic and stable knockdown versus acute inhibition of GSTP1. Nonetheless, between the common changes and even the uncommon changes in metabolite levels, we were able to identify overlapping metabolic pathways that were altered upon GSTP1 inhibition.
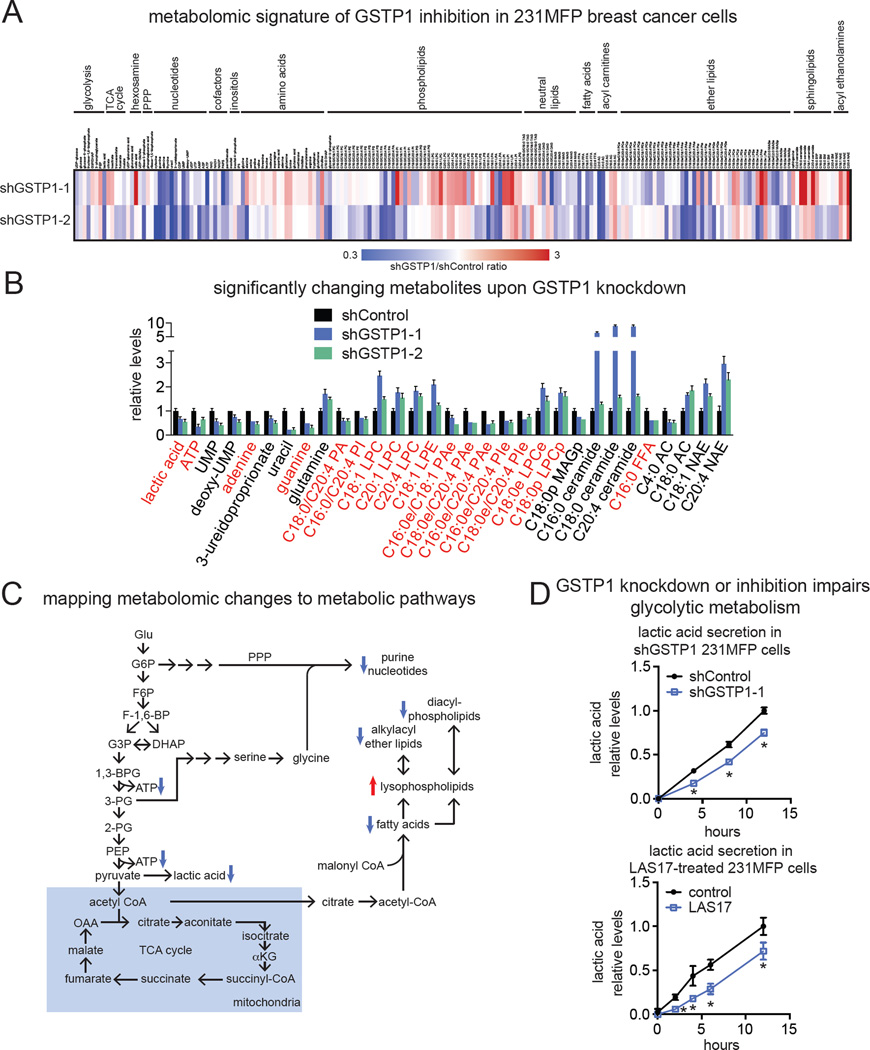
Figure 3. Functional metabolomic profiling and pathway mapping of GSTP1 inactivation in TNBC cells.
(A) We performed untargeted and targeted SRM-based metabolomic profiling of shControl and shGSTP1 231MFP cancer cells. Shown is a heatmap of shGSTP1/shControl ratios of all the metabolites that were measured by SRM analysis. (B) Shown are metabolites that were significantly (p<0.05) changing in the same direction in both shGSTP1-1 and shGSTP1-2 cells compared to shControl cells. Highlighted in red are the metabolite changes that were mapped to metabolic pathways in (C). Abbreviations: ATP, adenosine triphosphate; UMP, uridine monophosphate; PA, phosphatidic acid; PI, phosphatidyl inositol; LPC, lysophosphatidyl choline; PAe, phosphatidic acid-ether; PIe, phosphatidyl inositol-ether; LPCe, lysophosphatidylcholine-ether; LPCp, lysophosphatidylcholine-plasmalogen; MAGp, monoalkylglycerol-plasmalogen; FFA, free fatty acid; AC, acyl carnitine; NAE, N-acyl ethanolamine. (C) Pathway mapping of many of the metabolic changes observed in shGSTP1 231MFP cells indicates a primary impairment in glycolysis with resulting secondary impairments in nucleotide and lipid metabolism. (D) Lactic acid secretion into the media in shControl and shGSTP1-1 231MFP cells or DMSO vehicle-treated or LAS17-treated (10 µM) 231MFP cells. Lactic acid was measured using a lactic acid measurement kit. Data in (B and D) are presented as mean ± SEM, n=5/group. Significance in (D) is presented as *p<0.05 compared to shControl or DMSO vehicle-treated controls. Detailed data, explanation of abbreviations, and SRM transitions are in Table S2. Effect of GSTP1 inhibition on JNK signaling and oxidative stress is shown in Fig. S3, and metabolic characterization of LAS17-treated 231MFP cells are shown in Fig. S4.
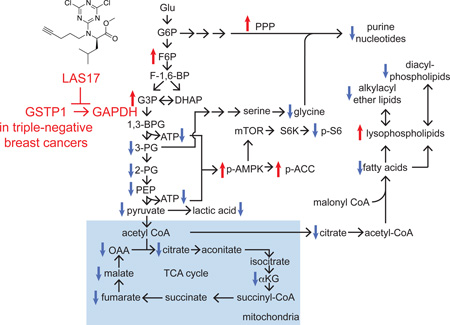
Upon mapping the observed metabolomic changes to metabolic pathway maps, our data suggested that GSTP1 inactivation led to impairments in glycolytic metabolism, leading to reduced lactic acid and ATP levels as well as reductions in the levels of macromolecular building blocks, including purine nucleotides, fatty acids, diacylphospholipids, and alkylacyl ether lipids (Fig. 3C). Consistent with this premise, we observed significant and time-dependent reductions in lactic acid secretion or glucose consumption upon GSTP1 knockdown in shGSTP1 cells or upon LAS17 treatment in 231MFP cells, compared to shControl or vehicle-treated controls, respectively (Fig. 3D, Fig. S4E), indicating reduced glycolytic metabolism.
GSTP1 Inhibition Impairs Oncogenic Signaling Pathways
The reduced levels of ATP and increased levels of ACs (C18:0 AC), a product of carnitine palmitoyltransferase 1 (CPT1), suggested heightened phosphorylation and activity of AMP kinase (AMPK) and also phosphorylation and inhibition of the downstream substrate acetyl CoA carboxylase (ACC). Inhibited ACC would presumably lead to decreased malonyl CoA levels and derepression of CPT1, and thus the observed increase in AC levels. We show that GSTP1 knockdown in both shGSTP1 cells and LAS17 treatment in 231MFP cells results in increased levels of phosphorylated AMPK and ACC (Fig. 4A, 4B).
Figure 4. GSTP1 Inhibition Impairs Oncogenic Signaling Pathways.
(A, B) shGSTP1 (A) and LAS17-treated (B) 231MFP cells show increased levels of phosphorylated AMPK and ACC compared to shControl and DMSO vehicle-treated controls, respectively, as assessed by Western blotting. (C) Serum-free cell survival impairments 48 h post-seeding in shControl and shGSTP1 231MFP cells treated with DMSO vehicle or AMPK inhibitor dorsomorphin (10 µM). (D) Phosphorylated S6 levels, a readout of mTOR activity, in shControl and shGSTP1 or DMSO vehicle or LAS17-treated (10 µM) 231MFP cells. (E) Serum-free cell survival 48 h post-seeding in 231MFP cells treated with DMSO vehicle or LAS17 (10 µM) and DMSO vehicle or mTOR inhibitor Torin 1 (250 nM). Western blotting images in (A, B, and D) are representative images from n=3/group. Quantification was performed by normalizing to actin or total S6 loading controls by densitometry. Data in (A–E) are presented as mean ± SEM, n=3–5/group. Significance is presented as *p<0.05 compared to shControl or DMSO vehicle-treated controls, respectively in (A–E) and #p<0.05 compared to DMSO vehicle-treated shGSTP1 in (C).
Activation of AMPK has been previously shown to impair breast cancer pathogenicity, partly by inhibiting mTOR activity and downstream protein synthesis (Dowling et al., 2007). Consistent with the role of AMPK in driving the GSTP1 inhibition-mediated breast cancer pathogenicity impairments, we show that the cell survival impairments conferred by GSTP1 knockdown are partially rescued upon treatment with the AMPK inhibitor dorsomorphin (Fig. 4C). We also show that the levels of phosphorylated S6, downstream of mTOR and S6 kinase, are lower in shGSTP1 and LAS17-treated 231MFP breast cancer cells compared to shControl or vehicle-treated controls cells, respectively (Fig. 4D). Consistent with the role of impaired mTOR signaling in GSTP1 inhibition-mediated survival defects, treatment of 231MFP cells with mTOR inhibitor Torin 1 alone causes survival impairments, but co-treatment of 231MFP cells with Torin 1 and LAS17 does not cause additional survival impairments beyond those observed with LAS17 treatment alone (Fig. 4E).
GSTP1 Interacts with and Activates GAPDH Activity to Influence Glycolytic Metabolism
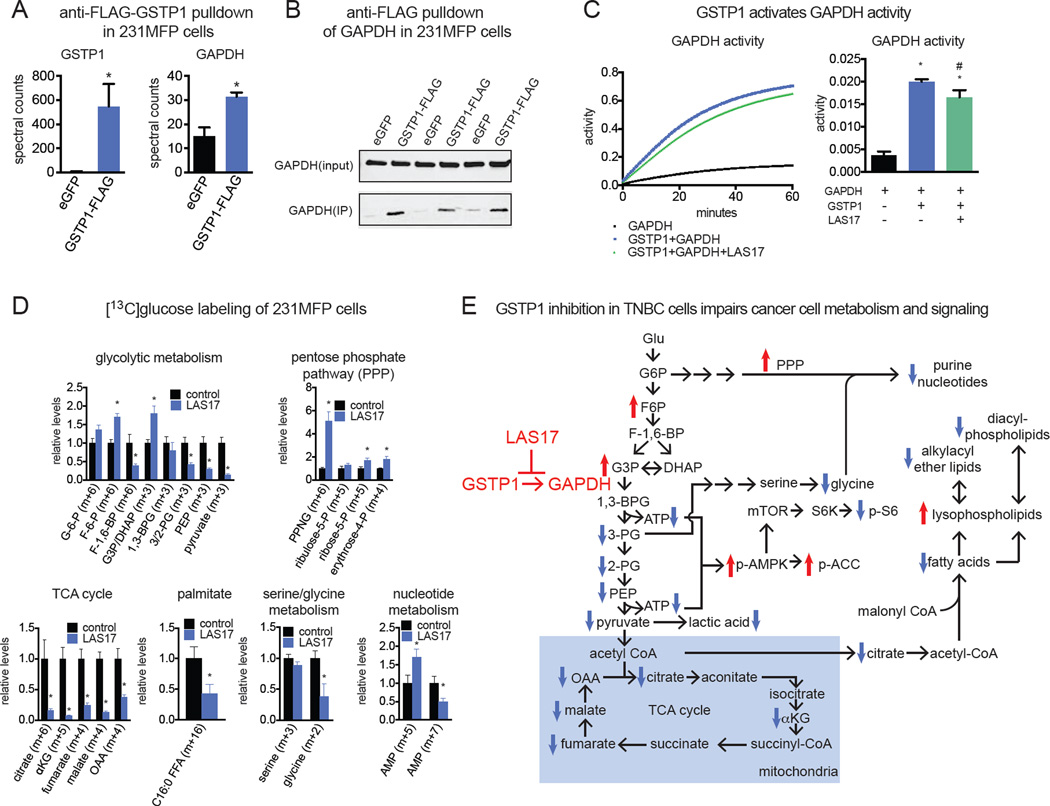
We next wanted to investigate the mechanism through which GSTP1 controlled glycolytic metabolism. GSTP1 has been previously shown to control signaling pathways through protein-protein interactions. We thus generated a 231MFP cell line stably overexpressing a FLAG-tagged GSTP1 and performed a pull-down study to identify GSTP1 interacting proteins through proteomic profiling (Fig. S5A–S5C). We found that anti-FLAG pulldown in GSTP1-FLAG-overexpressing cell lysates significantly enriched 7 proteins compared to mock-infected control cell lysates (Fig. S5C). Among these 7 proteins was glyceraldehyde-3-phosphate dehydrogenase (GAPDH) that was significantly enriched in GSTP1-FLAG-expressing cells compared to mock-infected control 231MFP cells (Fig. 5A). While we could not confirm this GSTP1 interaction with endogenous GAPDH, we showed that addition of exogenous pure and active GAPDH enzyme could lead to enrichment of GAPDH upon anti-FLAG pulldown in GSTP1-FLAG-overexpressing cell lysates compared to mock-infected control lysates (Fig. 5B).
Figure 5. GSTP1 Interacts with and Activates GAPDH Activity to Influence Glycolytic Metabolism.
(A) We generated stable GFP-expressing control and GSTP1-FLAG overexpressing 231MFP cells to identify GSTP1 interaction partners. Upon anti-FLAG pulldown of GFP-expressing control (eGFP) or GSTP1-FLAG-overexpressing 231MFP lysates and subsequent proteomic analysis of pulled-down proteins, we found enrichment of GSTP1 and GAPDH. (B) This GSTP1-GAPDH interaction was confirmed by incubation of pure and active GAPDH enzyme with GFP-expressing or GSTP1-FLAG overexpressing 231MFP cell lysates, subsequent anti-FLAG pulldown, and Western blotting of GAPDH. Shown are GAPDH blots for input and IP fractions. (C) Effect of coincubating pure and active GSTP1 and GAPDH proteins individually or together on GAPDH activity, assessed by a GAPDH activity assay measuring NADH, a byproduct of the conversion of glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate. GAPDH was treated with DMSO vehicle and GAPDH + GSTP1 was treated with DMSO vehicle or LAS17 (10 µM) 30 min prior to assessment of GAPDH activity. (D) Isotopic tracing of [13C]glucose into metabolic pathways in 231MFP cells. Cells were pre-treated with DMSO vehicle or LAS17 (10 µM) 1h prior to labeling of 231MFP cells with [13C]glucose (10 mM, 8 h). Cell metabolomes were analyzed by SRM-based LC-MS/MS. (E) Model of mechanisms underlying GSTP1 control over breast cancer pathogenicity, based on steady-state metabolomics, isotopic tracing, and Western blotting data. Data in (A and D) are presented as mean ± SEM, n=3–5/group. Significance is presented as *p<0.05 compared to eGFP or DMSO vehicle-treated controls, respectively. Data in (C) are presented as mean ± SEM, n=3/group. Significance in (C) is presented as *p<0.05 compared to GAPDH only and #p<0.05 compared to the GAPDH and GSTP1 group. Further characterization of GSTP1 protein interactions and the role of GSTP1 in MCF10A cells are described in Fig. S5.
We thus postulated that GSTP1 may interact with GAPDH to influence its activity. Consistent with this hypothesis, we show that GSTP1 greatly activates GAPDH activity in vitro (Fig. 5C). This activation in GAPDH activity was independent of glutathione (GSH) or oxidized glutathione (GSSG), indicating that this activity was not due to glutathione conjugation of a small-molecule metabolite or glutathionylation of GAPDH (Fig. S5D). LAS17 itself does not inhibit GAPDH activity (Fig. S5E). This GSTP1-induced GAPDH activity was, however, partially suppressed by LAS17 pre-treatment, indicating that LAS17 binding to GSTP1 at least partially disrupts the GSTP1-GAPDH protein-protein interaction (Fig. 5C).
To complement our steady-state metabolomic profiling data and further confirm our proposed mechanism that GSTP1 inhibition indirectly inhibits glycolysis through impairing GAPDH activity, we performed [13C]glucose isotopic tracing analysis in 231MFP cells to follow 13C-carbon into relevant metabolic pathways using SRM-based LC-MS/MS methods (Fig. 5D, 5E). GSTP1 inhibition with LAS17 led to an accumulation of the GAPDH substrate [13C]glyceraldehyde-3-phosphate/dihydroxyacetone phosphate (G3P/DHAP) and lowered [13C]glycolytic intermediates downstream of GAPDH, which represent the ATP-generating steps of glycolysis (Fig. 5D, 5E). We also show an accumulation in the levels of [13C]glycolytic and [13C]pentose phosphate pathway (PPP) metabolites upstream of GAPDH (Fig. 5D, 5E). Overall, these data are consistent with an inhibition of GAPDH activity from GSTP1 inhibition. [13C]fructose-1,6-bisphosphate (F-1,6-BP) levels were reduced indicating that GSTP1 may potentially also affect additional nodes in glycolysis. We also show that the impaired glycolytic metabolism downstream of GAPDH also affects other related downstream pathways, including lowered levels of [13C]TCA cycle and [13C]glycine metabolites. This reduction of carbon into TCA cycle and particularly into [13C]citrate is also consistent with reduced de novo lipogenic pools of [13C]palmitate levels (Fig. 5D, 5E), and the reductions in the levels of phospholipid and ether lipid levels (Fig. 3B, 3C, 5E). Our steady-state metabolomic data also showed reduced levels of purine nucleotides such as adenine and guanine, which is counter-indicative to heightened PPP observed from our tracing studies. However, detailed tracing analysis shows that the levels of [13C]adenine mononucleotide phosphate (AMP) (m+7), derived from isotopic incorporation of glucose into PPP metabolites (m+5) and glycine (m+2), are lowered, whereas levels of [13C]AMP (m+5), derived only from glucose incorporation into PPP metabolites, are elevated (Fig. 5D). These data indicate that the observed reductions in nucleotide levels are likely due to reduction in glycine necessary for de novo synthesis of purines (Fig. 3B, 3C, 5D, 5E).
While GSTP1 expression is heightened in TNBC compared with non-TNBC cells, we also showed that GSTP1 is expressed in non-transformed MCF10A mammary epithelial cells. We find that GSTP1 inhibition in MCF10A cells does not impair cell survival compared to significantly impaired survival in 231MFP cells (Fig. S5F). Interestingly, LAS17 treatment in MCF10A cells also impairs lactic acid secretion, indicating that glycolytic metabolism may also be impaired in these cells (Fig. S5G). We postulated that TNBC cells may be more addicted to glycolytic metabolism compared to MCF10A cells. Consistent with this premise, 231MFP cell survival is significantly impaired by the glycolytic hexokinase inhibitor 2-deoxyglucose (2DG) to a comparable degree as LAS17, but MCF10A cells show no survival impairment upon 2DG treatment (Fig. S5H).
Discussion
Taken together, our data point to GSTP1 as a novel TNBC target that, when inactivated, impairs glycolytic metabolism through a unique mechanism of disrupting GSTP1-induced activation of GAPDH, leading to lower levels of macromolecular building blocks (e.g. lipids and nucleotides) and ATP levels. This reduction in energy levels in-turn also leads to impaired oncogenic signaling through activation of AMPK and inhibition of mTOR signaling (Fig. 5E).
While our interpretations that we present here are consistent with the metabolomic and signaling changes that we observe with GSTP1 inactivation in breast cancer cells, we expect that there are also additional mechanisms involved that may arise from the metabolomic changes that we observed. For example, we showed increased levels of ACs with GSTP1 inhibition, which we presume to be downstream of ACC inhibition and de-repression of CPT1. These results could indicate that fatty acid beta oxidation pathways may be activated upon GSTP1 inactivation. While we did not observe rescue of cell survival impairments with the CPT1 inhibitor etomoxir (data not shown), the observed increase in AC levels may play a role in other aspects of GSTP1-mediated effects (Carracedo et al., 2013).
GSTP1 has also been linked to many other functions in cancer and other human pathologies and even in drug addiction. Beyond glutathonylation and detofixication functions, GSTP1 has been shown to possess chaperone functions, regulation of nitric oxide pathways, control over various kinase signaling pathways (Zhang et al., 2014). For example, GSTP1 inhibits JNK signaling and prevents downstream transcriptional activation of cell stress pathways. Under cellular stress conditions where reactive oxygen stress is heightened, GSTP1 has been shown to dimerize into larger aggregates and preclude binding to JNK, enabling JNK activation. In the context of hematopoiesis, GSTP1 inhibition has been shown to play a cytoprotective role in both erythroid and lymphoid cells, and the GSTP1 inhibitor Ezatiostat has been shown to be clinically effective for myelodisplastic syndrome (Mahadevan and Sutton, 2015; Zhang et al., 2014). While we report here that GSTP1 inhibition does not activate JNK signaling in our TNBC cells, GSTP1 may still directly regulate other signaling pathways through protein interactions or glutathionylation-mediated pathways (Zhang et al., 2014).
We also acknowledge that the apparent activation of GAPDH activity by GSTP1 is only modestly reduced upon LAS17 treatment indicating that there may be additional complexities involved in the mechanism underlying the GSTP1-induction of GAPDH activity. While our isotopic tracing data clearly indicate that GSTP1 inhibition leads to an impairment in the ATP-generating steps of glycolysis downstream of GAPDH, we do not yet understand the mechanism through which this occurs. GAPDH activity is dependent on a highly reactive catalytic cysteine that coordinates the interconversion between 1,3-bisphosphoglycerate into glyceraldehyde-3-phosphate and is particularly sensitive to oxidation by agents such as hydrogen peroxide or other oxidants which can inhibit GAPDH activity (Hildebrandt et al., 2015). Reports have also shown that GAPDH can even be inhibited by its reactive 1,3-bisphosphoglycerate product on a hyper-reactive and functional lysine to inhibit its activity (Moellering and Cravatt, 2013). It would be of future interest to investigate whether GSTP1 interacts with GAPDH to protect hyper-reactive sites from being adducted and inhibited. GSTP1 has also been shown to glutathionylate cysteines on proteins to post-translationally regulate protein structure and function and protect proteins from degradation from sulfhydryl overoxidation or proteolysis (Grek et al., 2013). While we were not able to detect GAPDH glutathionylation by GSTP1 and the activation of GAPDH by GSTP1 was independent of GSSG and GSH, it may still be possible that GAPDH activity and glycolytic metabolism may be regulated by GSTP1-mediated glutathionylation. Furthermore, while GAPDH is not generally considered to be a rate-limiting step of glycolysis, a recent study has shown that GAPDH is a rate-limiting in cancer cells that possess aerobic glycolysis (Hildebrandt et al., 2015; Moellering and Cravatt, 2013; Shestov et al., 2014). Our studies with GSTP1 provide additional support for how GAPDH may act as a major regulatory hub for TNBC glycolytic activity.
We show that pharmacological inactivation of GSTP1 over a sustained period does not show any observable toxicity and not only prevents breast tumor growth, but even slows established breast tumor growth in mice. A highly potent GSTP1 inhibitor Ezatiostat developed by Telik, Inc. has passed Phase II clinical trials in human patients for treatment of myelodysplastic syndrome, indicating that GSTP1 inhibitors are likely to be well-tolerated in human patients (Mahadevan and Sutton, 2015). Beyond the many previously reported biochemical and therapeutic roles of GSTP1 (Grek et al., 2013; Tew and Townsend, 2011; Townsend and Tew, 2003), our study suggests that GSTP1 inhibitors may also be promising stand-alone therapeutics for TNBCs. Our study also underscores the utility of using reactivity-based chemoproteomic platforms coupled with functional metabolomic approaches to identify novel metabolic drivers and pathways underlying breast cancer malignancy.
Significance
TNBCs and stem-like breast cancers that have undergone EMT have been linked to breast cancer malignancy and poor prognosis. While these breast cancer cell types are particularly dangerous, there are little to no therapeutic strategies that specifically target these types of breast cancer cells. Here, we have used chemoproteomic profiling platforms to identify GSTP1 as a novel potential therapeutic target for TNBCs. We show that GSTP1 interacts with GAPDH to activate its activity and that GSTP1 inactivation impairs glycolytic metabolism after the GAPDH step in glycolysis to impair ATP generation, nucleotide and fatty acid metabolism, and oncogenic signaling pathways. We show that GSTP1 inhibitors show selective killing of TNBC cells over non-TNBC or non-transformed mammary epithelial cells and that long-term GSTP1 inhibition in mice does not cause overt toxicity or weight loss. Taken together, we show that GSTP1 inhibitors may potentially be novel therapeutic agents to specifically target TNBCs.
Experimental Procedures
Chemicals
The AMPK inhibitor, dorsomorphin dihydrochloride and the mTOR inhibitor, Torin 1 were obtained from Tocris. LAS17 was synthesized as described previously (Crawford, L.A., Weerapana, E.). Menadione was obtained from Spectrum Chemical.
Cell Culture
The 231MFP cells were obtained from Professor Benjamin Cravatt and were generated from explanted tumor xenografts of MDA-MB-231 cells. MCF7, MCF10A, T47D, ZR751, MDA-MB-361, HCC1143, HCC38, HCC70, MDA-MB-468, and HEK293T cells were obtained from ATCC. HEK293T cells were cultured in DMEM containing 10% (vol/vol) fetal bovine serum (FBS) and maintained at 37°C with 5% CO2 231MFP, MDA-MB-361, and MDA-MB 468 cells were cultured in L15 media containing 10% FBS and maintained at 37°C with 0% CO2. MCF10A cells were cultured in DMEM/F12K media containing 5% horse serum, 20 ng/ml epidermal growth factor, 100ng/ml cholera toxin, 10 ng/ml insulin, and 500 ng/ml hydrocortisone and maintained at 37°C with 5% CO2. MCF7, T47D, ZR751, HCC1143, HCC38, and HCC70 cells were cultured in RPMI media containing 10% FBS and maintained at 37°C with 5% CO 2.
Gene Expression by qPCR
qPCR was performed using the manufacturer’s protocol for Fisher Maxima SYBRgreen with 10 µM primer concentrations or for BioRad SsoAdvanced Universal Probes Supermix. Primer sequences for Fisher Maxima SYBRgreen were derived from Primer Bank. Primer sequences for BioRad SsoAdvanced Universal Probes Supermix were designed with Primer 3 Plus.
Constructing Knockdown Cell Lines
We used two independent short-hairpin oligonucleotides to knockdown the expression of GSTP1 and one short-hairpin oligonucleotide to knockdown the expression of CDH1 using previously described methods (Benjamin et al., 2013). For generation of stable shRNA lines, lentiviral plasmids in the pLKO.1 backbone containing shRNA (Sigma) against human GSTP1 were transfected into HEK293T cells using Fugene (Roche).
Lentivirus was collected from filtered cultured media and used to infect target cancer cell line with polybrene. Target cells were selected over 3 days with 1 µg/mL puromycin. The short-hairpin sequences used for generation of the GSTP1 knockdown lines were:
shGSTP-1:
CCGGCGCTGACTACAACCTGCTGGACTCGAGTCCAGCAGGTTGTAGTCAG
CGTTTTTG and shGSTP-2:
CCGGCCTCACCCTGTACCAGTCCAACTCGAGTTGGACTGGTACAGGGTGAGGTTTTTG.
The short-hairpin sequence used for generation of the CDH1 knockdown line was CCGGAGAAGGGTCTGTTCACGTATTCTCGAGAATACGTGAACAGACCCTTCTTTTTTG.
The control shRNA was targeted against GFP with the target sequence GCAAGCTGACCCTGAAGTTCAT. Knockdown was confirmed by qPCR or Western blotting.
Constructing FLAG-tagged GSTP1 Cells
GSTP1 cDNA (Dharmacon) was subcloned into the pENTR4-FLAG vector (Addgene). This entry vector was recombined via an attL-attR (LR) reaction into a pLenti CMV puro DEST vector (Addgene). For generation of the FLAG-tagged GSTP1 line, the lentiviral plasmid containing FLAG-GSTP1 was transfected into HEK293T cells using Fugene (Roche). Lentivirus was collected from filtered cultured media and used to infect target cancer cell line with polybrene. Target cells were selected over 3 days with 1 µg/mL puromycin.
Cellular Phenotype Studies
Cell proliferation, serum-free cell survival, and migration assays were performed in a similar manner to previously described assays (Benjamin et al., 2013, 2015). More details can be found in Supplemental Methods.
Tumor Xenograft Studies
Human tumor xenografts were established by transplanting cancer cells ectopically into the flank of C.B17 severe combined immunodeficiency (SCID) mice (Taconic Farms) as previously described (Nomura et al., 2010). Briefly, cells were washed twice with PBS, trypsinzied, and harvested in serum-containing medium. Harvested cells were washed twice with serum-free medium and resuspended at a concentration of 2.0 × 104cells/µl and 100 µl was injected. Tumors were measured every 2 days with calipers.
Metabolomic Profiling of Cancer Cells
Metabolomic analyses were conducted using previously described methods (Benjamin et al., 2013, 2014,2015; Long et al., 2011; Mulvihill et al., 2014). More details can be found in Supplemental Methods.
Lactic Acid Secretion
Lactic acid secretion from L-15 media was measured by collecting media and performing a colorimetic lactic acid assay using a kit purchased from Abcam per the manufacturer’s protocol.
Glucose Consumption
Glucose consumption from RPMI media was measured by collecting media and performing a colorimetric glucose assay using a kit purchased from Abcam per the manufacturer’s protocol.
Western blotting
E-Cadherin antibody was obtained from BD Biosciences. Vimentin antibody was obtained from Sigma. Antibodies to cyclophilin, GSTP1, β-actin, phospho-AMPK α (Thr172), AMPK α, phospho-ACC (Ser79), ACC, phospho-S6, total S6, phospho-JNK (Thr183/Tyr185), JNK, and GAPDH were obtained from Cell Signaling Technology. FLAG antibody was obtained from Cayman Chemicals.
Cells were lysed in lysis buffer (CST) containing both protease and phosphatase inhibitors. Proteins were resolved by electrophoresis on 4–15% Tris-glycine precast Mini-PROTEAN TGX gel (BioRad Laboratories) and transferred to nitrocellulose membranes using the iBlot system (Invitrogen). Blots were blocked with 5% nonfat milk in Tris-buffered saline containing Tween-20 (TBST) solution for 1 hour at room temperature, washed in TBST, and probed with primary antibody diluted in recommended diluent per manufacturer overnight at 4°C. Following washes wit h TBST, the blots were incubated in the dark with a IR-linked secondary antibody at room temperature for 1 hour. Blots were visualized using an Odyssey Li-Cor scanner after additional washes.
Anti-FLAG Pulldown Studies
Pulldown studies were performed used Anti-FLAG M2 Magnetic beads (Sigma) according to the manufacturer’s protocol. FLAG-tagged GSTP1-overexpressing and GFP-overexpressing control cells were lysed in lysis buffer (CST) and 500 µg of lysate were incubated with 32 µl of anti-FLAG magnetic beads for 2 hours at 4°C. B eads were collected and washed three times with TBS before elution with 4% SDS (w/v) in 120 mM Tris-HCI. Samples were heated to 95°C for 3 minutes. Eluent was subsequently prepared for proteomic profiling with a shotgun proteomic analysis protocol as described below.
Shotgun Proteomic Profiling of Anti-FLAG Pulldown
Pulldown products were TCA precipitated in 20%TCA at −80°C overnight and centrifuged at 10,000 x g at 4°C for 10 minutes to pellet protein. Pelleted p roteins were washed three times with 8 M urea in PBS. After solubilization, 30 µl of 0.2% Protease Max Surfactant (Promega) was added and the resulting mixture was vortexed followed by the addition of 40 µl of 100 mM ammonium bicarbonate and 10 mM tris(2-carboxyethyl)phosphine. After 30 minutes, 12.5 mM iodoacetamide was added and allowed to react for 30 minutes in the dark before adding 120 µl PBS and 1.2 µl 1% Protease Max Surfactant. The protein solution was vortexed and 0.5 µg/µl sequencing grade trypsin (Promega) was added and allowed to react overnight at 37°C. The peptide solution was then centrifuged at 10,000 × g before the supernatant was subsequently analyzed by LC-MS/MS.
GAPDH Activity Assay
GAPDH activity was measured using a colorimetric kit purchased from BioVision and performed according to the manufacturer’s protocol. Active human GSTP1 and active human GAPDH full-length proteins were purchased from Abeam. Proteins were co-incubated at 37°C for 1 hour before GAPDH activity was measured using kit.
Oxidative Stress
Oxidative stress was measured using CellROX Green Reagent (Invitrogen) per the manufacturer’s protocol.
Cysteine and Lysine Reactivity Profiling
Cysteine and lysine reactivity profiling was performed based on an adapted method that has been previously described (Medina-Cleghorn et al., 2015; Weerapana et al., 2010). Tryptic digests for proteomic profiling were analyzed using a Thermo LTQ-XL and quantified by spectral counting using previously described methods (Nomura et al., 2010). More details can be found in Supplemental Methods.
GSTP1 activity assay
The 50% inhibitory concentration (IC50) of LAS17 was determined using a GSTP1 activity assay. CDNB (1-chloro-2,4-dinitrobenzene) was incubated with 200 mM of reduced glutathione and 1 µg of active GSTP1 protein with 100, 10, 1, 0.1, 0.01, 0.001, and 0 µM of LAS17. GSTP1 catalyzes the conjugation of glutathione to CDNB, generating the reaction product glutathione-DNB conjugate, which absorbs at 340 nm. The rate of increase in the absorption of the product is proportional to GSTP1 activity and was used to measure GSTP1 activity.
In vitro labeling of GSTP1 with LAS17
50 µg of protein was incubated with 5 µM of LAS17 for 30 minutes at room temperature. Following probe labeling, 25 µM rhodamine-azide, 1 mM TCEP, 100 µM TBTA, and 1mM Cu(II)SO4 were added and incubated for 1 hour at room temperature. Laemmli sample buffer was added, heated to 95°C for 5 minut es, and then cooled to room temperature. Samples were loaded onto a SDS-PAGE gel and imaged by in-gel fluorescence using a Typhoon flatbed scanner.
Supplementary Material
Highlights.
We used chemoproteomics to profile metabolic drivers of breast cancer
GSTP1 is a novel triple-negative breast cancer-specific target
GSTP1 inhibition impairs triple-negative breast cancer pathogenicity
GSTP1 inhibition impairs GAPDH activity to affect metabolism and signaling
Acknowledgments
We thank the members of the Nomura Research Group for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (R01CA172667 for DKN, SL, EG, LD and R01CA170447 for AG), American Cancer Society Research Scholar Award (RSG14-242-01-TBE for DKN, SL, EG, LD), DOD Breakthroughs Award (CDMRP W81XWH-15-1-0050 for DKN, SL, EG, LD), DOD Era of Hope Award (CDMRP W81XWH-12-1-0272 for AG), the Searle Scholar Foundation (DKN, SL, EG, LD), the National Science Foundation Graduate Research Fellowship (SL), Damon Runyon-Rachleff Innovator Award (DRR-18-12 for EW, LAC), the Smith Family Foundation (EW, LAC), Boston College (EW, LAC), and the Diabetes, Endocrinology and Metabolism training grant (T32 DK007418 for RC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
DKN and SML conceived of the project, designed and performed experiments, analyzed and interpreted data, and wrote the paper. EAG designed and performed experiments, analyzed and interpreted data. LAC performed synthetic efforts and provided GSTP1 pure protein; also provided reagents and designed and performed experiments. LD designed and performed experiments, analyzed and interpreted data. RC performed data analysis and provided data. TRH and DKM performed synthetic efforts for probes used in the study and performed experiments. AG performed data analysis and provided data. EW designed and performed experiments, wrote the paper, provided reagents.
References
- Benjamin DI, Cravatt BF, Nomura DK. Global profiling strategies for mapping dysregulated metabolic pathways in cancer. Cell Metab. 2012;16:565–577. doi: 10.1016/j.cmet.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin DI, Cozzo A, Ji X, Roberts LS, Louie SM, Mulvihill MM, Luo K, Nomura DK. Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14912–14917. doi: 10.1073/pnas.1310894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin DI, Louie SM, Mulvihill MM, Kohnz RA, Li DS, Chan LG, Sorrentino A, Bandyopadhyay S, Cozzo A, Ohiri A, et al. Inositol phosphate recycling regulates glycolytic and lipid metabolism that drives cancer aggressiveness. ACS Chem. Biol. 2014;9:1340–1350. doi: 10.1021/cb5001907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin DI, Li DS, Lowe W, Heuer T, Kemble G, Nomura DK. Diacylglycerol Metabolism and Signaling Is a Driving Force Underlying FASN Inhibitor Sensitivity in Cancer Cells. ACS Chem. Biol. 2015;10:1616–1623. doi: 10.1021/acschembio.5b00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat. Rev. Cancer. 2013;13:227–232. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford LA, Weerapana E. A Tyrosine-Reactive Irreversible Inhibitor for Glutathione S-Transferase Pi (GSTP1) doi: 10.1039/c6mb00250a. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson SJ, Provenzano E, Caldas C. Triple negative breast cancers: clinical and prognostic implications. Eur. J. Cancer Oxf. Engl. 2009;1990(45 Suppl. 1):27–40. doi: 10.1016/S0959-8049(09)70013-9. [DOI] [PubMed] [Google Scholar]
- Dietze EC, Sistrunk C, Miranda-Carboni G, O’Regan R, Seewaldt VL. Triple-negative breast cancer in African-American women: disparities versus biology. Nat. Rev. Cancer. 2015;15:248–254. doi: 10.1038/nrc3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling RJO, Zakikhani M, Fantus IG, Pollak M, Sonenberg N. Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res. 2007;67:10804–10812. doi: 10.1158/0008-5472.CAN-07-2310. [DOI] [PubMed] [Google Scholar]
- Gowda H, Ivanisevic J, Johnson CH, Kurczy ME, Benton HP, Rinehart D, Nguyen T, Ray J, Kuehl J, Arevalo B, et al. Interactive XCMS Online: simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal. Chem. 2014;86:6931–6939. doi: 10.1021/ac500734c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grek CL, Zhang J, Manevich Y, Townsend DM, Tew KD. Causes and consequences of cysteine S-glutathionylation. J. Biol. Chem. 2013;288:26497–26504. doi: 10.1074/jbc.R113.461368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt T, Knuesting J, Berndt C, Morgan B, Scheibe R. Cytosolic thiol switches regulating basic cellular functions: GAPDH as an information hub? Biol. Chem. 2015;396:523–537. doi: 10.1515/hsz-2014-0295. [DOI] [PubMed] [Google Scholar]
- Laborde E. Glutathione transferases as mediators of signaling pathways involved in cell proliferation and cell death. Cell Death Differ. 2010;17:1373–1380. doi: 10.1038/cdd.2010.80. [DOI] [PubMed] [Google Scholar]
- Long JZ, Cisar JS, Milliken D, Niessen S, Wang C, Trauger SA, Siuzdak G, Cravatt BF. Metabolomics annotates ABHD3 as a physiologic regulator of medium-chain phospholipids. Nat. Chem. Biol. 2011;7:763–765. doi: 10.1038/nchembio.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan D, Sutton GR. Ezatiostat hydrochloride for the treatment of myelodysplastic syndromes. Expert Opin. Investig. Drugs. 2015;24:725–733. doi: 10.1517/13543784.2015.1021003. [DOI] [PubMed] [Google Scholar]
- Martell J, Weerapana E. Applications of copper-catalyzed click chemistry in activity-based protein profiling. Mol. Basel Switz. 2014;19:1378–1393. doi: 10.3390/molecules19021378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Cleghorn D, Bateman LA, Ford B, Heslin A, Fisher KJ, Dalvie ED, Nomura DK. Mapping Proteome-Wide Targets of Environmental Chemicals Using Reactivity-Based Chemoproteomic Platforms. Chem. Biol. 2015;22:1394–1405. doi: 10.1016/j.chembiol.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering RE, Cravatt BF. Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science. 2013;341:549–553. doi: 10.1126/science.1238327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill MM, Benjamin DI, Ji X, Le Scolan E, Louie SM, Shieh A, Green M, Narasimhalu T, Morris PJ, Luo K, et al. Metabolic Profiling Reveals PAFAH1B3 as a Critical Driver of Breast Cancer Pathogenicity. Chem. Biol. 2014;21:831–840. doi: 10.1016/j.chembiol.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Long JZ, Niessen S, Hoover HS, Ng S-W, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NJ, Weerapana E. Diverse functional roles of reactive cysteines. ACS Chem. Biol. 2013;8:283–296. doi: 10.1021/cb3005269. [DOI] [PubMed] [Google Scholar]
- Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- Shannon DA, Weerapana E. Covalent protein modification: the current landscape of residue-specific electrophiles. Curr. Opin. Chem. Biol. 2015;24:18–26. doi: 10.1016/j.cbpa.2014.10.021. [DOI] [PubMed] [Google Scholar]
- Shannon DA, Banerjee R, Webster ER, Bak DW, Wang C, Weerapana E. Investigating the proteome reactivity and selectivity of aryl halides. J. Am. Chem. Soc. 2014;136:3330–3333. doi: 10.1021/ja4116204. [DOI] [PubMed] [Google Scholar]
- Shestov AA, Liu X, Ser Z, Cluntun AA, Hung YP, Huang L, Kim D, Le A, Yellen G, Albeck JG, et al. Quantitative determinants of aerobic glycolysis identify flux through the enzyme GAPDH as a limiting step. eLife. 2014:3. doi: 10.7554/eLife.03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tew KD, Townsend DM. Regulatory functions of glutathione S-transferase P1-1 unrelated to detoxification. Drug Metab. Rev. 2011;43:179–193. doi: 10.3109/03602532.2011.552912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend DM, Tew KD. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. 2003;22:7369–7375. doi: 10.1038/sj.onc.1206940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MBD, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Grek C, Ye Z-W, Manevich Y, Tew KD, Townsend DM. Chapter Four - Pleiotropic Functions of Glutathione S-Transferase P. In: Tew KD, editor. Advances in Cancer Research. Academic Press; 2014. pp. 143–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.