Abstract
Receptors for the Fc portion (FCR) of Ig have been extensively characterized and are known to regulate humoral responses, but members of the closely related FCR-like (FCRL) family have not been found to bind Ig and to date no ligand has been identified for any FCRL. Using a cell-based GFP reporter system and a recombinant Fc chimeric protein, we show that human FCRL6, a receptor selectively expressed by cytotoxic T and NK cells, directly binds HLA-DR, a major histocompatibility complex (MHC) class II molecule. Given the similarity among constant regions of Ig and MHC molecules, these findings suggest that representatives of the FCR and FCRL multigene families may have independently evolved to engage two ancestral elements fundamental to adaptive immunity. This discovery may offer new insight into the interaction between cytotoxic lymphocytes and antigen presenting cells and may have important implications for better understanding HLA disease susceptibility and pathogenesis.
INTRODUCTION
The characterization in recent years of multiple receptor-gene families with activating or inhibitory potential has disclosed that the immune system is integrated with a remarkable number of regulatory mechanisms to balance effector responses. The classical Fc receptors (FCR) for IgG and IgE are located on human chromosome 1q21–23 and play fundamental roles in both positive and negative immune regulation (1–3). The discovery of an extended family of FCR-like (FCRL1-6) genes positioned in the same human chromosomal region has uncovered unexpected diversity at this locus (4–6). In addition to their genomic linkage and similar genetic organization, FCRL1-6 share many features with the FCRs including related extracellular Ig-like domains and cytoplasmic tyrosine-based signaling capability of their encoded type I transmembrane protein products (6). Of these, human FCRL6 is distinctly expressed by cytotoxic T and NK cells, is upregulated on expanded populations of terminally differentiated CD8+ T cells in HIV and B cell chronic lymphocytic leukemia (CLL) patients, and possesses a cytoplasmic immunoreceptor tyrosine-based inhibition motif (ITIM) that is capable of being phosphorylated and recruiting the src homology 2-domain containing phosphatase 2 (SHP-2) (7, 8).
Despite the many similarities between the FCR and FCRL families, no FCRL has been shown to bind Ig and thus ligands for these receptors remain unknown. In this study we report that the MHC class II molecule HLA-DR is a ligand for human FCRL6. Using a cell-based reporter system, FCRL6 ligand reactivity was found to be restricted to antigen presenting cells and the development of a panel of blocking antibodies facilitated the identification of HLA-DR as the interacting partner. This association was further confirmed using HLA-DR transductants for FCRL6-specific induction assays and selective binding of a soluble FCRL6-Fc chimeric molecule.
MATERIALS AND METHODS
Cells
43-1 FCRL6ζ cells were generated as previously described (9). Briefly, the FCRL6 extracellular region was PCR amplified from full-length cDNA with the KOD polymerase (Novagen) using the following SfiI-flanked primers: forward 5’-TAT AGG CCA TTA TGG CCG AAC TGT CTG GCT GTA C-3’ and reverse 5’-TAT AGG CCA CCG CGG CCA GTG AAC AAG ACT TG-3’, and inserted into the pEF.CD3ζ backbone to generate a lentiviral vector plasmid termed pEF.FCRL6ζ. Vector packaging, transduction, and transductant sorting was performed as previously described (9).
Full-length HLA-DRα and HLA-DRβ1 cDNAs were amplified from human PBL cells and the SUDHL6 cell line, respectively. The DRβ3 (DRB3*01010201), DRβ4 (DRB4*01030101), and DRβ5 (DRB5*010101) allele cDNAs were purchased from Open Biosystems. HLA-DRα (DRA*0101), HLA-DRβ1 (DRB1*040101), and DRβ3-5 cDNAs were subcloned into the pMX-PIE retroviral vector, which contains the GFP gene, and used to transduce BW5147 mouse T cells as described previously (10). Doubly-transduced HLA-DRα+β1 and β3-5 cells were sorted with a PE-labeled anti-HLA-DR mAb (Sigma); singly transduced HLA-DRα and HLA-DRβ1 lines were sorted for GFP.
Human tonsil and spleen samples were obtained from the UAB Tissue Procurement program. Blood specimens were obtained from healthy adult volunteers with IRB approval following informed consent according to the Declaration of Helsinki. Mononuclear cells were isolated as detailed previously (8). Dendritic cells were generated from FACS sorted monocytes as described (11).
43-1 experiments
For antibody stimulation experiments, Immulon 96-well high binding plates (Thermo Scientific) were coated with antibodies for 1 h at RT and plated with 3×104 43-1 cells for 18 h. For co-culture experiments, primary cells were added at a 5:1 ratio and dendritic cells and cell lines at a 2:1 ratio with 43-1 reporter cell lines. In blocking experiments, 30µL of hybridoma supernatant or purified antibody diluted in culture media was added to stimulator cells prior to plating the 43-1 cells. After co-culture, cells were stained with anti-mouse CD5 PE (SBA) to delineate the 43-1 fraction and then analyzed by flow cytometry. To distinguish between the 43-1 and BW5147 cell lines, BW5147 cells were fluorescently labeled with the PKH26 cell membrane labeling kit (Sigma) prior to co-culture.
Antibodies
FCRL6-ligand mAbs were generated by immunizing BALB/c mice with the SUDHL6 and Mino cell lines according to established techniques (12). All procedures were approved by the UAB IACUC. Hybridoma supernatants were screened for reactivity with SUDHL6 and their ability to block 43-1 FCRL6ζ cell GFP induction by SUDHL6. Hybridomas were subcloned by limiting dilution, and isotyped using the SBA clonotyping system. The resulting mAbs are moIgG1κ (L2-1E2), moIgG3κ (L2-3F7, L2-3H2), and moIgMκ (L2-1A8, L2-2A6, L2-2B1, L2-3C5, L2-3C6, L2-4C2) isotypes. The anti-FCRL6 specific mAbs 7B7 and 1D8 (both moIgG1κ) were generated as previously described (8) and Fab digests were prepared using the ImmunoPure IgG1 Fab Preparation Kit (Pierce). The following additional Abs were used: control hamster IgG (anti-KLH) (BD Bioscience), HLA-DRβ clone DA2 (Santa Cruz), control mouse IgG Fab fragments (Rockland Immunochemicals), and the hamster anti-mouse CD3 clone 145-2C11 (a kind gift from Dr. Chandar Raman, UAB).
Fc chimeric proteins
Fc soluble recombinant proteins were generated by modifying the pRB1-Fcmut plasmid (a kind gift of Dr. Roberto Biassoni; Genoa, Italy) which contains a cDNA encoding the CH2-3 Fc portion of the human IgG1 Hc (IGHG1) with three amino acid residues mutated (L234A, L235A, G237A) to abrogate Fc receptor binding. The FCRL6-Fc and control-Fc constructs were generated by inserting the endogenous FCRL6 leader and ectodomain sequence or a murine Ig kappa (IGK) leader sequence, respectively, upstream of the CH2 portion of IGHG1.
Final cDNA constructs were subcloned into the pShuttle-CMV adenovirus transfer vector (Quantum Biotechnologies) for transfection and expression in 293T cells. Soluble protein containing supernatants were purified over Protein A Sepharose columns (GE Healthcare) before SDS-PAGE and analysis by Coomassie staining (BioRad) and Western Blot.
For staining, Fc proteins were incubated with Alexa-Fluor 647 labeled Protein A (Invitrogen) at a 2.5:1 molar ratio for 30 min at RT then diluted and used to stain BW5147 transductants at a 200nM concentration for 1 h at 37°C.
RESULTS AND DISCUSSION
Development of a GFP-inducible system to detect FCRL6 engagement
In order to assay functional engagement between FCRL6 and potential ligand sources, a reporter cell line was engineered. A chimeric molecule termed FCRL6ζ, which possesses the three FCRL6 Ig-like extracellular domains in frame with an uncharged hydrophobic transmembrane region and the immunoreceptor tyrosine-based activation motif (ITAM)-containing cytoplasmic portion of mouse CD3ζ, was transduced into a murine T cell hybridoma, 43-1, which harbors an inducible GFP gene downstream of three tandem nuclear factor of activated T cells (NFAT) transcription factor binding sites (9, 13). The rationale behind this approach was that an encounter between FCRL6 and its ligand should initiate a CD3ζ-driven activation cascade resulting in NFAT nuclear translocation and GFP expression.
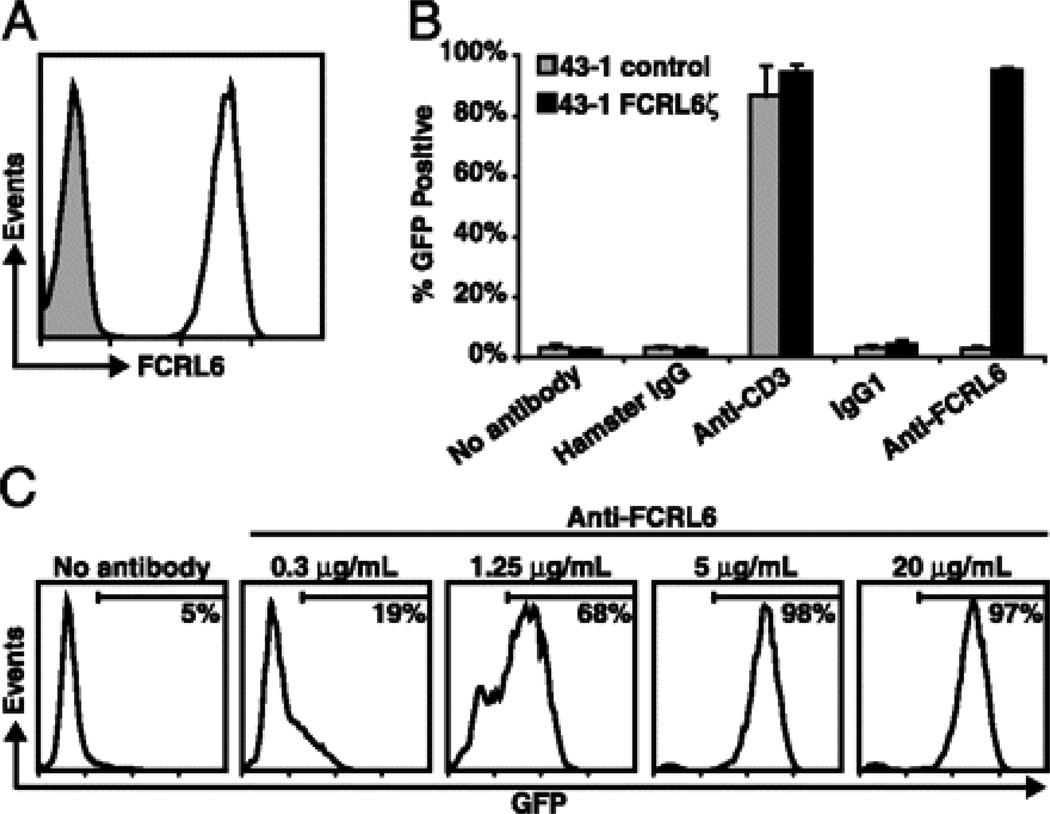
Following transduction of the chimeric construct into 43-1 cells, FCRL6ζ surface expression was verified by staining with an FCRL6-specific antibody (Fig. 1A). The 43-1 cells have an intact T cell receptor complex (13), therefore stimulation of parental or FCRL6ζ cells with monoclonal antibodies (mAb) to mouse CD3 resulted in potent GFP induction by both cell lines, whereas stimulation with an FCRL6-specific mAb (7B7) only induced GFP expression in FCRL6ζ cells (Fig. 1B). The response to mAb stimulation was dose dependent, such that a larger percentage of 43-1 cells expressed GFP when exposed to higher concentrations of stimulating antibody (Fig. 1C).
FIGURE 1.
Development of a GFP-inducible system to detect FCRL6 engagement. A 43-1 cells transduced with the FCRL6ζ reporter construct were stained with the anti-FCRL6 7B7 mAb (black line) or with an isotype-matched control (grey shade). B Untransduced 43-1 control (grey columns) or 43-1 FCRL6ζ cells (black columns) were stimulated at 5µg/mL with the indicated plate-bound antibodies for 18 h and GFP induction was assayed by flow cytometry. Columns represent the mean ± s.d.; n=3. C FCRL6ζ cells were stimulated as in panel (B) with different concentrations of plate-bound anti-FCRL6 mAb. The percentage of GFP+ 43-1 cells is indicated in each histogram and demarcated by horizontal bars.
FCRL6 interacts with a surface molecule on antigen presenting cells
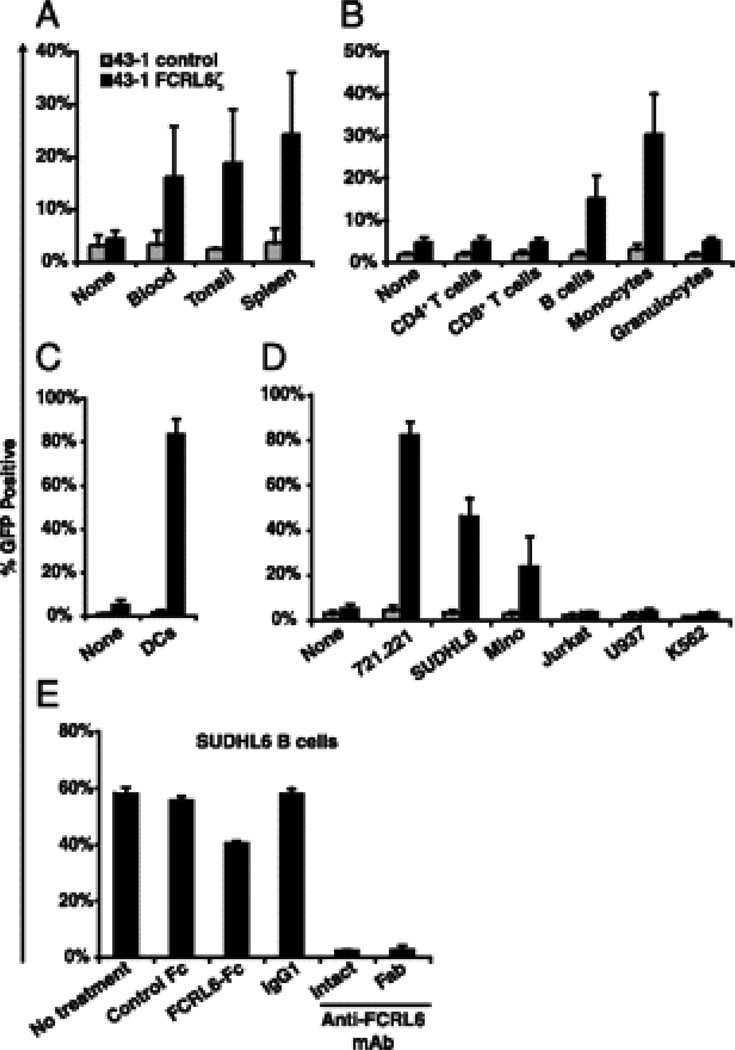
Having established a functional readout for FCRL6 engagement, we began investigating different lymphoid tissues to explore the cellular distribution of the FCRL6 ligand(s). In co-culture experiments, mononuclear cells isolated from blood, tonsil, and spleen were found to induce GFP expression in FCRL6ζ cells but not in control 43-1 cells (Fig. 2A). To ascertain which cell type(s) was responsible for this reactivity, distinct leukocyte populations were sorted from blood samples and assayed for FCRL6-dependent GFP induction. The putative FCRL6 ligand was found only on cells with antigen presenting capability, namely, B cells and monocytes, but not on resting CD4+ T cells, CD8+ T cells, or granulocytes (Fig. 2B). Monocyte-derived dendritic cells were also found to evoke a robust response (Fig. 2C).
FIGURE 2.
FCRL6 interacts with a molecule expressed by antigen presenting cells. FCRL6ζ (black columns) or untransduced 43-1 control cells (grey columns) were co-cultured with (A) mononuclear cells isolated from the indicated tissues; (B) FACS sorted blood CD4+ T cells (CD3+CD4+), CD8+ T cells (CD3+CD8+), B cells (CD19+), monocytes, or granulocytes (both defined by light scatter); (C) monocyte-derived dendritic cells; or (D) the indicated human cell lines. After 18 h, 43-1 cell GFP expression was analyzed by flow cytometry. 43-1 control cells cultured alone (“none”) were employed to determine background GFP levels. E FCRL6ζ cells were co-cultured with the SUDHL6 B cell line either alone (“no treatment”) or in the presence of the indicated blocking reagents and assayed for GFP expression. Columns represent the mean ± s.d.; n≥3.
To further define FCRL6 ligand expression and facilitate its eventual cloning, representative human cell lines were analyzed. Co-culture with EBV-immortalized B lymphoblastoid (721.221), diffuse large B lymphoma (SUDHL6), and mantle B cell lymphoma-derived (Mino) cell lines activated FCRL6ζ cells whereas no detectable response was observed following co-culture with Jurkat T cells or the U937 and K562 myeloid cell lines (Fig. 2D). Conditioned media from SUDHL6 cultured cells failed to stimulate GFP, suggesting that the interacting counterpart is a cell surface-associated molecule (data not shown). The FCRL6-specificity in this reporter system was supported by several lines of evidence. Induction by SUDHL6 in co-culture assays cells was blocked by both intact and monomeric Fab fragments of an FCRL6-specific mAb (1D8), as well as by the addition of a soluble chimeric FCRL6-Fc fusion protein but not a control Fc-only recombinant molecule consisting of the CH2-CH3 domains of human IgG1 (Fig. 2E). Furthermore, a mouse FCRL1ζ 43-1 transductant that only differs from human FCRL6ζ cells in its extracellular region failed to respond following co-culture with 721.221 or SUDHL6 B cell lines (Supplemental Fig. 1).
A panel of mAbs obstructs binding between FCRL6 and its ligand
As a tool to molecularly characterize the unknown FCRL6 ligand, we developed a panel of ligand-reactive mAbs. Hybridoma supernatants from mice immunized with the SUDHL6 and Mino B cell lines were tested for mAbs that would both react with SUDHL6 cells by flow cytometry and suppress SUDHL6-induced GFP expression by FCRL6ζ cells. This strategy led to the isolation of nine mAb-secreting clones that fulfilled both criteria (Supplemental Fig. 2A, 2B). However, prior to utilizing these mAbs in approaches to identify the FCRL6 ligand biochemically, we were struck by several compelling evolutionary observations that provided useful clues for predicting its possible nature.
HLA-DR is an FCRL6 ligand
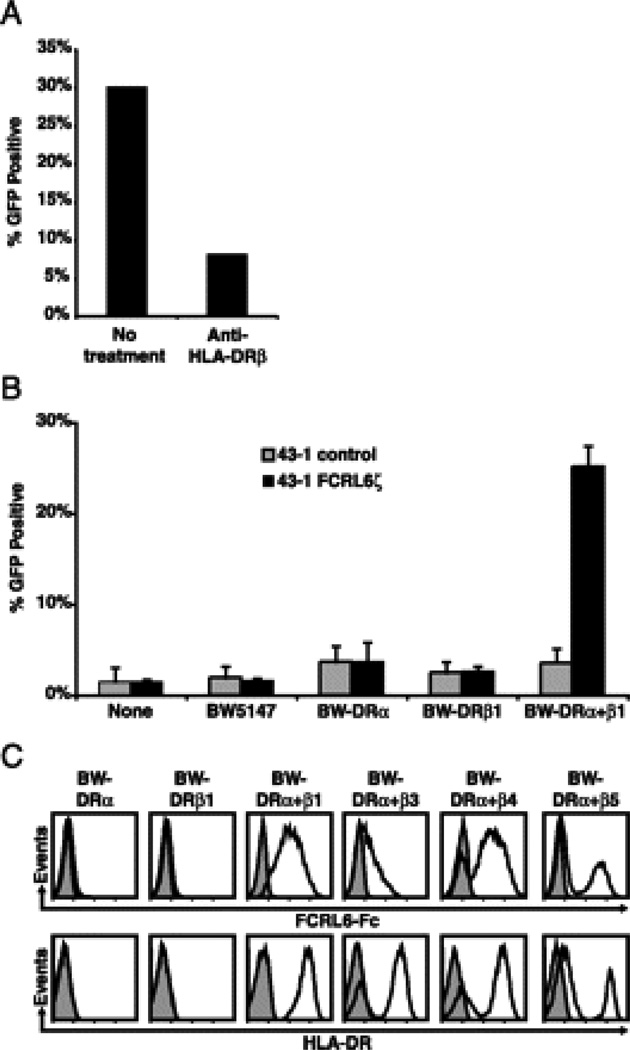
Striking phylogenetic relationships are evident between Ig and MHC constant regions (14–19). Thus, the finding that an FCR/FCRL family member recognized a surface determinant common to antigen presenting cells prompted us to consider that the ligand for FCRL6 could be MHC class II. To test this hypothesis we first performed competition assays with 43-1 FCRL6ζ cells utilizing an anti-HLA-DRβ mAb (clone DA2). Addition of this reagent in co-culture experiments with SUDHL6 cells indeed abrogated FCRL6ζ GFP induction (Fig. 3A). A series of BW5147 murine thymoma cell transductants expressing HLA-DRα and/or HLA-DRβ1 was then generated and employed in co-culture reporter assays. Surprisingly, HLA-DRα+β1 BW cells (surface HLA-DR+), but not parental or singly transduced control cells (surface HLA-DR−), triggered FCRL6ζ GFP expression (Fig. 3B). Importantly, the panel of FCRL6 ligand-reactive mAbs all selectively stained the HLA-DRα+β1 transductants and all but the L2-1E2 clone were capable of blocking FCRL6ζ activation when added to co-culture experiments with the HLA-DRα+β1 BW cells (Supplemental Fig. 3A, 3B).
FIGURE 3.
FCRL6 directly binds to HLA-DR. A FCRL6ζ cells were co-cultured for 18 h with SUDHL6 B cells either alone (“no treatment”) or in the presence of an HLA-DRβ-specific mAb (clone DA2) and assayed for GFP expression. B FCRL6ζ (black columns) or untransduced 43-1 control cells (grey columns) were cultured for 18 h either alone (“none”) or in the presence of parental BW5147 cells, BW cells singly transduced with HLA-DRα or HLA-DRβ1 (both surface HLA-DR−), or doubly-transduced with HLA-DRα and β1 (surface HLA-DR+) and analyzed for GFP expression. Columns represent the mean ± s.d.; n=3. C Recombinant FCRL6-Fc (black line) or a control Fc-only protein (grey shade) was complexed with Alexa-Fluor 647-labeled Protein A and used to stain the indicated BW5147 cells transduced with the following HLA-DR alleles (DRA*0101, DRB1*040101, DRB3*01010201, DRB4*01030101, and DRB5*010101). Histograms are representative of three independent experiments.
MHC class II is both extremely polymorphic and polygenic. Therefore we explored the potential of FCRL6 to bind HLA-DR heterodimers composed of different DRβ chains. HLA-DRβ3, β4, and β5 were each stably transduced into BW HLA-DRα cells and utilized for co-culture experiments with 43-1 cells. Interestingly, while HLA-DRα+β1, HLA-DRα+β4, and HLA-DRα+β5 cells invoked moderate to high levels of activation in the 43-1 system, HLA-DRα+β3 cells induced much lower levels of GFP expression (Supplemental Fig. 4). To independently verify the FCRL6/HLA-DR interaction, we then tested the ability of a recombinant soluble FCRL6-Fc fusion protein to stain the panel of HLA-DR transductants. Multimeric FCRL6-Fc selectively bound to HLA-DRα+β1, HLA-DRα+β4, HLA-DRα+β5 BW cells and to a lesser extent HLA-DRα+β3 transductants, but not to control cells (Fig. 3C). These results demonstrate that FCRL6 is capable of binding MHC class II molecules and suggest that it may have differential affinity for HLA-DR that varies according to its HLA-DRβ chain composition.
Concluding remarks
In summary, these findings clearly indicate that MHC class II is an FCRL6 ligand. Phylogenetic analyses suggest that the non-polymorphic constant region domains of Ig heavy chain isotypes and the membrane-proximal MHC class I α3 and MHC class II α2 and β2 domains, as well as β2 microglobulin, arose from a common ancestral C1-set Ig-like domain (14–19). Given the strong likelihood that the FCR and FCRL families arose from a common ancestor, it is quite remarkable that individual members have evolved to bind structurally and phylogenetically related ligands. Even more noteworthy are the parallels with members of the leukocyte receptor complex (LRC), an expanded monophyletic family of Ig-like receptor-genes in a region of human chromosome 19q13 (20, 21). This dense locus encompasses the Fc receptor for IgA (FCAR) as well as the multigene killer cell Ig-like receptor (KIR) and leukocyte Ig-like receptor (LILR) families that interact with classical and non-classical MHC class I molecules (22). Despite their separate genomic locations both the FCR/FCRL and LRC Ig-like multigene families encode activating and/or inhibitory receptors that bind either Ig or MHC and have expanded or contracted in a species-specific fashion (21, 23).
In addition to highlighting an interesting evolutionary relationship, the finding that human FCRL6 is a receptor for HLA-DR may have important implications for understanding the interactions between cytotoxic lymphocytes and antigen presenting cells. Dendritic cells can activate resting NK cells and conversely NK cells can induce dendritic cell activation and maturation (24, 25). Additionally, NK cells and CD8+ T cells can kill autologous dendritic cells; a mechanism proposed to regulate adaptive immune responses (24–27). What influence, if any, the FCRL6 – MHC class II interaction may have in this cross-talk is subject to further investigation. FCRL6 contains a cytoplasmic domain with a consensus ITIM motif that is capable of phosphorylation and SHP-2 recruitment (7, 8). It is therefore tempting to speculate that FCRL6 may function as an inhibitory receptor for MHC class II. However, studies using mAbs to ligate FCRL6 have failed to demonstrate convincing inhibitory function (7, 8) and definitive functional studies for this receptor await further exploration.
The discovery of this interaction raises many questions. For example, do other FCRL family members bind MHC class II molecules and is FCRL6 capable of recognizing other MHC class II isotypes? The data shown here demonstrate that FCRL6 is capable of binding HLA-DR molecules composed of distinct DRβ subunits; future studies addressing whether FCRL6 differentially binds MHC class II allelic variants may provide insight into the molecular mechanisms underlying HLA-based disease associations.
Supplementary Material
Acknowledgments
The authors would like to thank G. Larry Gartland for excellent assistance with cell sorting; Michael O. Alberti for help in generating 43-1 transductants; Takashi Saito, Kerry S. Campbell, Louis M. Staudt, Josef T. Prchal, and Eric Vivier, for donating cell lines; and Paul A. Goepfert, Olusimidele T. Akinsiku, Steffanie Sabaj, Peter D. Burrows, Allan J. Zajac, and Gary W. Litman for discussion and critical reading of the manuscript.
ABBREVIATIONS
- FCRL
Fc receptor-like
- LRC
leukocyte receptor complex
Footnotes
This study was supported in part by National Institutes of Health grants AI55638, AI067467, and CA131656, the Dana Foundation Program in Human Immunology, and the Cancer Research Institute (R.S.D.). D.M.S. received support from National Institutes of Health training grant AI007051 and the UAB MSTP T32 GM008361.
REFERENCES
- 1.Ravetch JV, Kinet JP. Fc receptors. Annu Rev Immunol. 1991;9:457–492. doi: 10.1146/annurev.iy.09.040191.002325. [DOI] [PubMed] [Google Scholar]
- 2.Daeron M. Fc receptor biology. Annu Rev Immunol. 1997;15:203–234. doi: 10.1146/annurev.immunol.15.1.203. [DOI] [PubMed] [Google Scholar]
- 3.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 4.Hatzivassiliou G, Miller I, Takizawa J, Palanisamy N, Rao PH, Iida S, Tagawa S, Taniwaki M, Russo J, Neri A, Cattoretti G, Clynes R, Mendelsohn C, Chaganti RS, Dalla-Favera R. IRTA1 and IRTA2, novel immunoglobulin superfamily receptors expressed in B cells and involved in chromosome 1q21 abnormalities in B cell malignancy. Immunity. 2001;14:277–289. doi: 10.1016/s1074-7613(01)00109-1. [DOI] [PubMed] [Google Scholar]
- 5.Davis RS, Wang YH, Kubagawa H, Cooper MD. Identification of a family of Fc receptor homologs with preferential B cell expression. Proc Natl Acad Sci U S A. 2001;98:9772–9777. doi: 10.1073/pnas.171308498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis RS. Fc receptor-like molecules. Annu Rev Immunol. 2007;25:525–560. doi: 10.1146/annurev.immunol.25.022106.141541. [DOI] [PubMed] [Google Scholar]
- 7.Wilson TJ, Presti RM, Tassi I, Overton ET, Cella M, Colonna M. FcRL6, a new ITIM-bearing receptor on cytolytic cells, is broadly expressed by lymphocytes following HIV-1 infection. Blood. 2007;109:3786–3793. doi: 10.1182/blood-2006-06-030023. [DOI] [PubMed] [Google Scholar]
- 8.Schreeder DM, Pan J, Li FJ, Vivier E, Davis RS. FCRL6 distinguishes mature cytotoxic lymphocytes and is upregulated in patients with B-cell chronic lymphocytic leukemia. Eur J Immunol. 2008;38:3159–3166. doi: 10.1002/eji.200838516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cannon JP, Haire RN, Magis AT, Eason DD, Winfrey KN, Hernandez Prada JA, Bailey KM, Jakoncic J, Litman GW, Ostrov DA. A bony fish immunological receptor of the NITR multigene family mediates allogeneic recognition. Immunity. 2008;29:228–237. doi: 10.1016/j.immuni.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrhardt GR, Davis RS, Hsu JT, Leu CM, Ehrhardt A, Cooper MD. The inhibitory potential of Fc receptor homolog 4 on memory B cells. Proc Natl Acad Sci U S A. 2003;100:13489–13494. doi: 10.1073/pnas.1935944100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou LJ, Tedder TF. CD14+ blood monocytes can differentiate into functionally mature CD83+ dendritic cells. Proc Natl Acad Sci U S A. 1996;93:2588–2592. doi: 10.1073/pnas.93.6.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kearney JF. Hybridomas and monoclonal antibodies. In: Paul WE, editor. Fundamental Immunology. New York: Raven Press; 1984. pp. 751–766. [Google Scholar]
- 13.Ohtsuka M, Arase H, Takeuchi A, Yamasaki S, Shiina R, Suenaga T, Sakurai D, Yokosuka T, Arase N, Iwashima M, Kitamura T, Moriya H, Saito T. NFAM1, an immunoreceptor tyrosine-based activation motif-bearing molecule that regulates B cell development and signaling. Proc Natl Acad Sci U S A. 2004;101:8126–8131. doi: 10.1073/pnas.0401119101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson PA, Cunningham BA, Berggard I, Edelman GM. 2 - Microglobulin--a free immunoglobulin domain. Proc Natl Acad Sci U S A. 1972;69:1697–1701. doi: 10.1073/pnas.69.7.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orr HT, Lancet D, Robb RJ, Lopez de Castro JA, Strominger JL. The heavy chain of human histocompatibility antigen HLA-B7 contains an immunoglobulin-like region. Nature. 1979;282:266–270. doi: 10.1038/282266a0. [DOI] [PubMed] [Google Scholar]
- 16.Larhammar D, Wiman K, Schenning L, Claesson L, Gustafsson K, Peterson PA, Rask L. Evolutionary relationship between HLA-DR antigen beta-chains, HLA-A, B, C antigen subunits and immunoglobulin chains. Scand J Immunol. 1981;14:617–622. doi: 10.1111/j.1365-3083.1981.tb00603.x. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman JF, Strominger JL. HLA-DR light chain has a polymorphic N-terminal region and a conserved immunoglobulin-like C-terminal region. Nature. 1982;297:694–697. doi: 10.1038/297694a0. [DOI] [PubMed] [Google Scholar]
- 18.Williams AF, Barclay AN. The immunoglobulin superfamily--domains for cell surface recognition. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 19.DuPasquier L, Chretien I. CTX, a new lymphocyte receptor in Xenopus, and the early evolution of Ig domains. Res Immunol. 1996;147:218–226. doi: 10.1016/0923-2494(96)87224-5. [DOI] [PubMed] [Google Scholar]
- 20.Wende H, Colonna M, Ziegler A, Volz A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome. 1999;10:154–160. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- 21.Martin AM, Kulski JK, Witt C, Pontarotti P, Christiansen FT. Leukocyte Ig-like receptor complex (LRC) in mice and men. Trends Immunol. 2002;23:81–88. doi: 10.1016/s1471-4906(01)02155-x. [DOI] [PubMed] [Google Scholar]
- 22.Barrow AD, Trowsdale J. The extended human leukocyte receptor complex: diverse ways of modulating immune responses. Immunol Rev. 2008;224:98–123. doi: 10.1111/j.1600-065X.2008.00653.x. [DOI] [PubMed] [Google Scholar]
- 23.Fayngerts SA, Najakshin AM, Taranin AV. Species-specific evolution of the FcR family in endothermic vertebrates. Immunogenetics. 2007;59:493–506. doi: 10.1007/s00251-007-0208-8. [DOI] [PubMed] [Google Scholar]
- 24.Degli-Esposti MA, Smyth MJ. Close encounters of different kinds: dendritic cells and NK cells take centre stage. Nat Rev Immunol. 2005;5:112–124. doi: 10.1038/nri1549. [DOI] [PubMed] [Google Scholar]
- 25.Walzer T, Dalod M, Robbins SH, Zitvogel L, Vivier E. Natural-killer cells and dendritic cells: "l'union fait la force". Blood. 2005;106:2252–2258. doi: 10.1182/blood-2005-03-1154. [DOI] [PubMed] [Google Scholar]
- 26.Ronchese F, Hermans IF. Killing of dendritic cells: a life cut short or a purposeful death? J Exp Med. 2001;194:F23–F26. doi: 10.1084/jem.194.5.f23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guarda G, Hons M, Soriano SF, Huang AY, Polley R, Martin-Fontecha A, Stein JV, Germain RN, Lanzavecchia A, Sallusto F. L-selectin-negative CCR7- effector and memory CD8+ T cells enter reactive lymph nodes and kill dendritic cells. Nat Immunol. 2007;8:743–752. doi: 10.1038/ni1469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.