SUMMARY
Influenza A virus hemagglutinin (HA) changes conformation and drives membrane fusion of viral and endosomal membrane at low pH. Membrane fusion proceeds through an intermediate called hemifusion1,2. For viral fusion the hemifusion structures are not determined3. Here, influenza virus-like particles (VLP)4 carrying wild-type (WT) HA or HA hemifusion mutant G1S5 and liposome mixtures were studied at low pH by Volta phase plate (VPP) cryo-electron tomography (cET) which improves signal-to-noise ratio close to focus. We determined two distinct hemifusion structures: a hemifusion diaphragm (HD) and a novel structure termed lipidic junction. Liposomes with lipidic junctions were ruptured with membrane edges stabilized by HA. The rupture frequency and HD diameter were not affected by G1S mutation, but decreased when the cholesterol level in the liposomes was close to physiological concentrations. We propose that HA induces merger between the viral and a target membrane by one of two independent pathways: rupture-insertion pathway leading to lipidic junction and hemifusion-stalk pathway leading to fusion pore. The latter is relevant under the conditions of influenza virus infection of cells. Cholesterol concentration functions as a pathway switch due to its negative spontaneous curvature in the target bilayer as determined by continuum analysis.
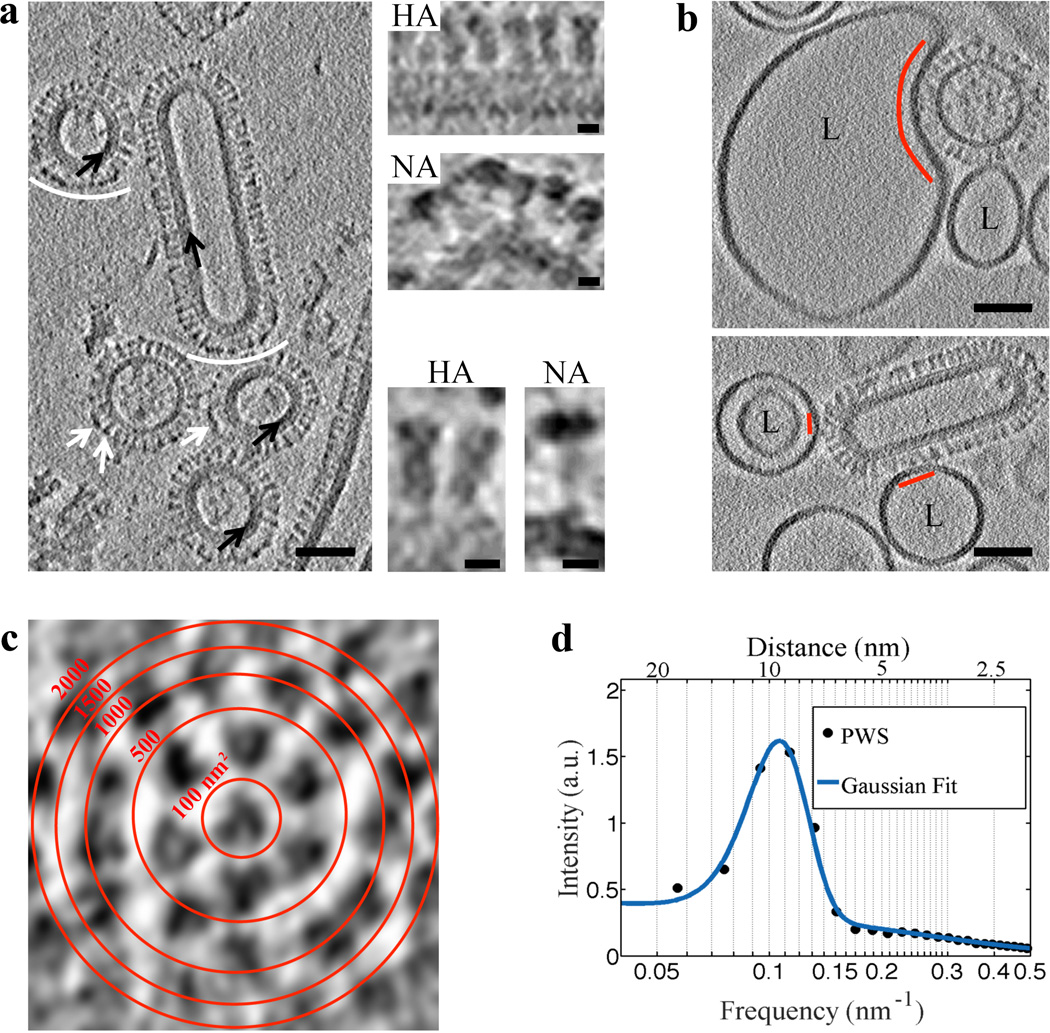
To study WT and G1S hemagglutinin-induced membrane fusion intermediates, we used influenza VLP that are composed of both HA and neuraminidase (NA) glycoproteins, matrix 1 (M1) layer and an ion channel M2. These VLP are structurally similar to influenza virus6 (Fig. 1a). VLP morphology was either filamentous with an M1 layer or spherical with or without an M1 layer underlying the membrane (Fig. 1a). The VLP glycoprotein spacing was 9.5±1.8 nm (mean ± standard deviation (SD)) (Fig 1c and 1d), similar to that found on influenza X31 viruses (~11 nm)7. Thus, VLP glycoprotein density is ~11 glycoproteins per 1000 nm2.
Figure 1. VLP structural features are resolved using VPP and indicate that influenza VLP carrying G1S mutated HA engage 1–7 HA glycoproteins on binding to liposomes.
a, A tomogram slice (3 nm thick) of WT VLP acquired using VPP and calculated by the weighted-back projection, capturing spherical particles without or with matrix layer (black arrows), and a filamentous particle. NA spikes (marked by white arrows or arcs) are characterized by a large globular domain and thin stalk as opposed to HA which has a cylindrical shape7. NA clusters (white arcs) observed in VLP are also found in filamentous influenza virus27. The tomographic slice in a is representative of VLP found in 7 tomograms collected on the same grid. Scale bars: 50 nm. Magnified views of HA and NA spikes. Scale bars: 5 nm. b, VLP carrying the G1S mutation in HA, when mixed with liposomes (labeled L) containing 16 mol % cholesterol and incubated for 20 minutes at neutral pH and subsequently subjected to pH 5 at 37°C for approximately 2 minutes demonstrate binding (binding area indicated by red arc and lines). VPP-cET tomogram slices (3 nm thick) calculated by the weighted-back projection method, showing VLP that are bound to liposomes but did not undergo conformational change. Scale bars: 50 nm. c, Tomogram slice of a G1S VLP surface showing transverse cross-sections of glycoproteins. Red concentric rings mark area boundaries within a 50×50 nm image. d, Radially averaged power spectrum of glycoprotein cross-sections shows an average 9.5±1.8 nm spacing (centers of the glycoproteins) on the surface of the G1S VLP. The tomographic slice b, is representative of 21 VLP-liposome binding events found in 12 tomograms collected on 2 independent grids as a technical replica of the sample.
To characterize the VLP based fusion system, we first compared the low pH formed liposomal fusion products of intact influenza virus (X31) to those of WT VLP. Virus or VLP were incubated at neutral pH with liposomes (66 mol % phosphatidylcholine (PC), 13 mol % phosphatidylethanolamine (PE), 16 mol % cholesterol and 5 mol % gangliosides as receptor) and then exposed to low pH for two minutes at 37°C prior to vitrification and cryo-electron microscopy (cEM). Observed events were classified as binding, fusion intermediate and complete fusion (Material and Methods). In the binding class, the liposomes bound to VLP retained their spherical shape or were compressed by adhesive forces to HA glycoproteins. The binding areas between liposome and VLP were typically distributed between 100 and 600 nm2, corresponding to occupancy of 1–7 glycoproteins, respectively (Fig. 1b and 1c). In contrast, the fusion intermediate class showed liposomes with teardrop shapes pointing towards VLP where HA glycoproteins were disorganized. WT VLP had similar numbers of binding and fusion intermediates and lower numbers of complete fusion events compared to X31 virus (Supplementary Fig. 2). For G1S VLP-liposome mixtures, binding and fusion intermediates were observed, but not complete fusion, as expected from this mutation’s phenotype in cell-cell fusion (Supplementary Fig. 1). G1S HA supports lipid mixing but not fusion pore opening or content mixing, conditions necessary for observable hemifusion (Supplementary Fig. 1)5 Structurally, influenza virus and VLP form similar fusion intermediates with liposomes. However, unlike virus, we can produce VLP carrying mutations in the HA fusion peptide that prevent viral replication.
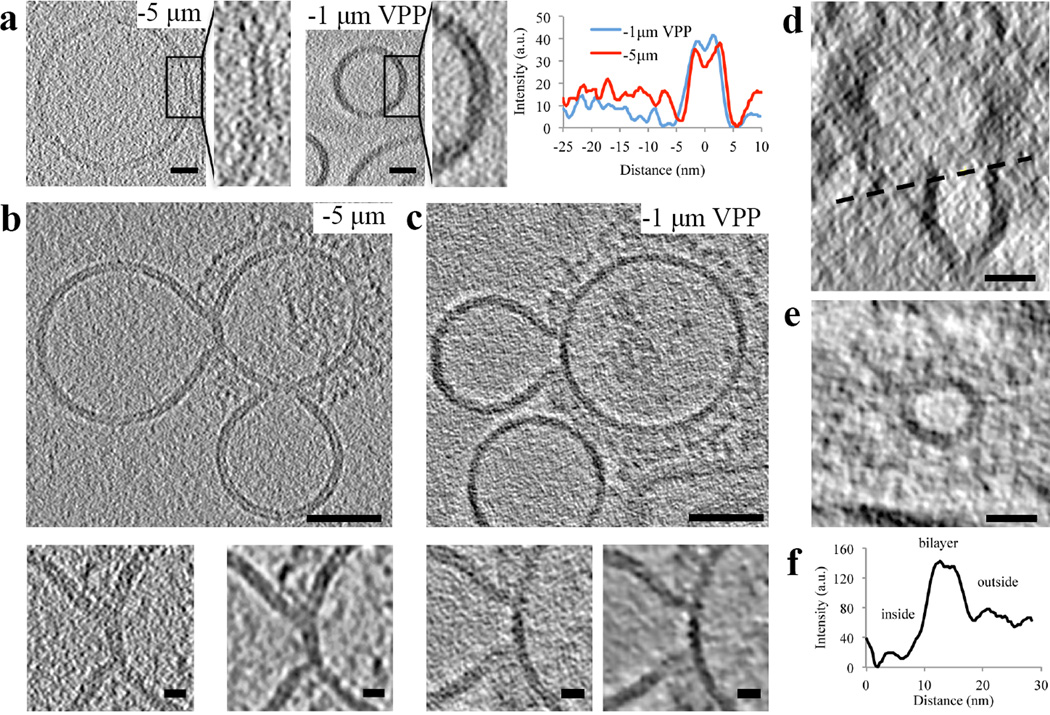
cET has been successfully applied to study membrane fusion8–10. However, cET relies on phase contrast achieved by defocusing at the expense of image distortion and poorer resolution which limits the interpretation of molecular details, e.g. membrane-protein interactions11. Therefore, we used the recently developed VPP-cET which enables close-to-focus imaging with higher signal-to-noise ratios12. We compared liposomal membrane bilayers obtained by either VPP-cET at defocus −1 µm or cET at defocus −5 µm (Fig. 2a). While bilayers appeared more evident without VPP at defocus −5 µm, presumably due to fringes appearing on both sides of the two monolayers, the signal-to-noise ratio was approximately half compared to the VPP tomogram (Fig. 2a).
Figure 2. Defocus-phase contrast cET and VPP-cET of liposomal membrane, “Y” shaped lipidic junctions and hemifusion diaphragms.
a, Tomogram slices (0.74 nm thick), calculated by the weighted-back projection method, from a tilt series acquired with an electron dose 80 e−/Å2 either at defocus −5 µm without VPP or at defocus −1 µm with VPP showing liposomal membrane. The radially averaged density profile plots are shown on the right. Scale bars: 10 nm. b and c, G1S VLP when mixed with liposomes containing 16 mol % cholesterol and incubated for 20 minutes at neutral pH and subsequently subjected to pH 5 at 37°C for approximately 2 minutes show junctions and hemifusion. b, Tomogram slice (3 nm thick) acquired at defocus −5 µm capturing two “Y” shaped junctions of the hemifusion diaphragm (HD). Magnified HD regions below before (left) and after (right) nonlinear anisotropic diffusion (NAD, k=50, 10 iterations) are shown below. The bilayer thickness, ~6 nm at the “Y” shaped lipidic junction, did not increase when compared with liposome or VLP. c, Tomogram slices (3 nm thick) acquired at defocus −1 µm with Volta phase plate (VPP) capturing “Y” shaped junctions of the hemifusion diaphragm (HD). Magnified HD regions below before (left) and after (right) nonlinear anisotropic diffusion (NAD, k=10, 10 iterations) are shown below. d, Transverse (X-Z) tomogram slice of a filamentous VLP and the hemifused liposome forming a complete HD. e, Tomogram slice (3 nm thick) of the circular junction connecting the liposome and the filamentous VLP indicated by the black dashed line shown in d. e, The radially averaged density profile plot of the circular junction shown in e (zero on the x-axis marks the center of the hemifusion diaphragm interior). Scale bars: 50 nm (b and c), 20 nm (d and e), 10 nm (Magnified views). The tomographic slices are representative of 40 and 16 VLP-liposome interactions found in 12 and 10 tomograms collected with and without VPP on 2 independent grids of two independent samples, respectively.
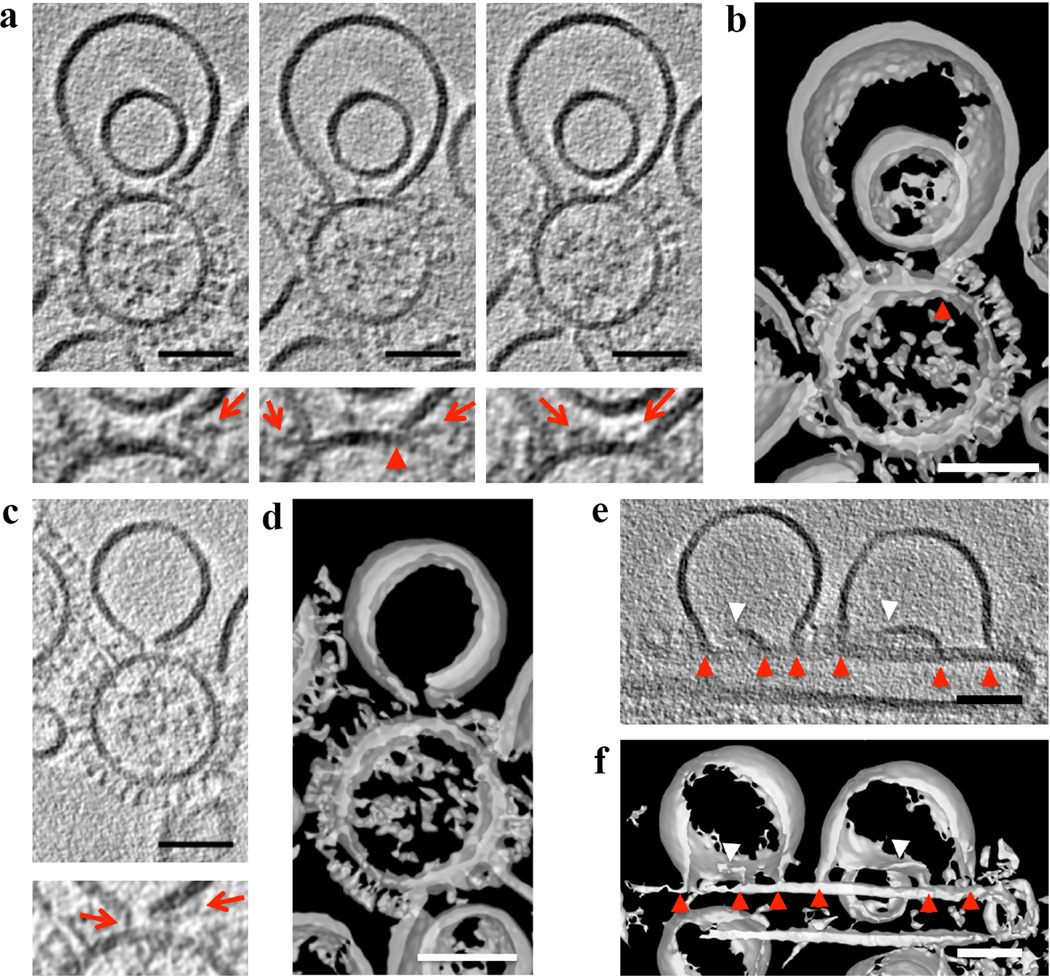
When analyzing the fusion intermediate class, we resolved liposomal membrane inserted into VLP membrane forming characteristic “Y” shaped junctions using both defocus phase contrast cET and VPP-cET (Fig. 2b and 2c, Supplementary Video 1 and 2, respectively). The membrane thickness at the “Y” junction (~6 nm) did not differ significantly compared to liposomal bilayer, consistent with “Y” junctions where the phospholipid acyl chains are tilted to prevent energetically forbidden bilayer voids as theoretically predicted13. There were neither structural features nor any increase in electron density over that of a lipid bilayer in the hydrophobic center of the “Y” junction (Supplementary Fig. 3). Because the “Y” junction appeared lipidic (i.e. there were no obvious barriers that would inhibit lipid mixing) we termed it a ‘lipidic junction’. All analyzed G1S VLP fusion intermediates showed lipidic junctions. Three-dimensional analysis revealed that the liposome was often ruptured with arc-shaped membrane sheets connected to VLP (53%, n=34) (Fig. 3, Supplementary Video 2 and 4). Ruptures were even more apparent in liposomes showing multiple lipidic junctions on the surface of filamentous G1S VLP (Fig. 3e and 3f, Supplementary Video 5 and 6). These tomographic slices show an architectural aspect to the leaky membrane conductance measured during influenza-mediated fusion14. About half (47%, n=34) of the lipidic junctions completely encircled a discoid membrane bilayer (Fig. 2d–f, Supplementary Video 3). This discoid membrane separated the aqueous contents of the liposome and VLP and is consistent with the definition of a hemifusion diaphragm (HD). The combined average of the HD diameters and lipidic junction chords formed by G1S VLP was ~ 20 nm. HD and lipidic junctions were found independently of the M1 layer (Supplementary Fig. 4, Supplementary results).
Figure 3. HA spikes in close proximity to lipidic junctions and ruptured membranes.
a and c, Tomogram slices (3 nm thick), calculated by weighted-back projection method, from a tilt series acquired at defocus −1 µm with VPP and after NAD and Gaussian filtering, capturing a liposome containing 16 mol % cholesterol in proximity to G1S VLP at low pH. a (left panel) and c, Ruptured liposome membrane in close proximity to glycoproteins, a, Central and right panel show lipidic junction of membrane sheet inserted into the VLP membrane (red arrowhead) and right panel shows glycoproteins inside the HD. Corresponding magnified views shown below. Red arrows mark glycoproteins in the proximity to HD or open liposomal membrane. b and d, Isosurfaces of the NAD-filtered tomographic volume calculated by the weighted simultaneous iterative reconstruction technique shown in a and c, respectively, also show material within the lipidic junctions, and a lack of HD completion. e, A tomogram slice (3 nm thick), calculated by weighted-back projection method, from a tilt series acquired at defocus −1 µm with VPP showing fusion intermediates between liposomes and filamentous G1S VLP. Red arrowheads mark lipidic junctions and white arrowheads mark free membrane edges. f, Isosurface of the NAD-filtered tomographic volume calculated by weighted simultaneous iterative reconstruction technique shown in e further illustrate the lipidic junctions and free membrane edges. Scale bars: 50 nm. The tomographic slices are representative of 40 and 16 VLP-liposome interactions found in 12 and 10 tomograms collected with and without VPP on 2 independent grids of two independent samples, respectively.
HA spikes in proximity to lipidic junctions were thinner than at neutral pH, often bent and attached to the edge of ruptured liposomal membrane (Fig. 3a and 3c). Occasionally, HA spikes were tilted parallel with the HD, suggesting an HA post-insertion folding stabilizing lipidic junctions (Fig. 3a). Although the majority of the HD interiors were free of glycoproteins, a number of HD interiors (11%, n=37) showed spike-like densities (Fig. 3a and 3b). Since spikes cannot be retained in an HD formed from hemifusion stalk expansion, this result demonstrates that the HD formed from lipidic junctions result from a different pathway.
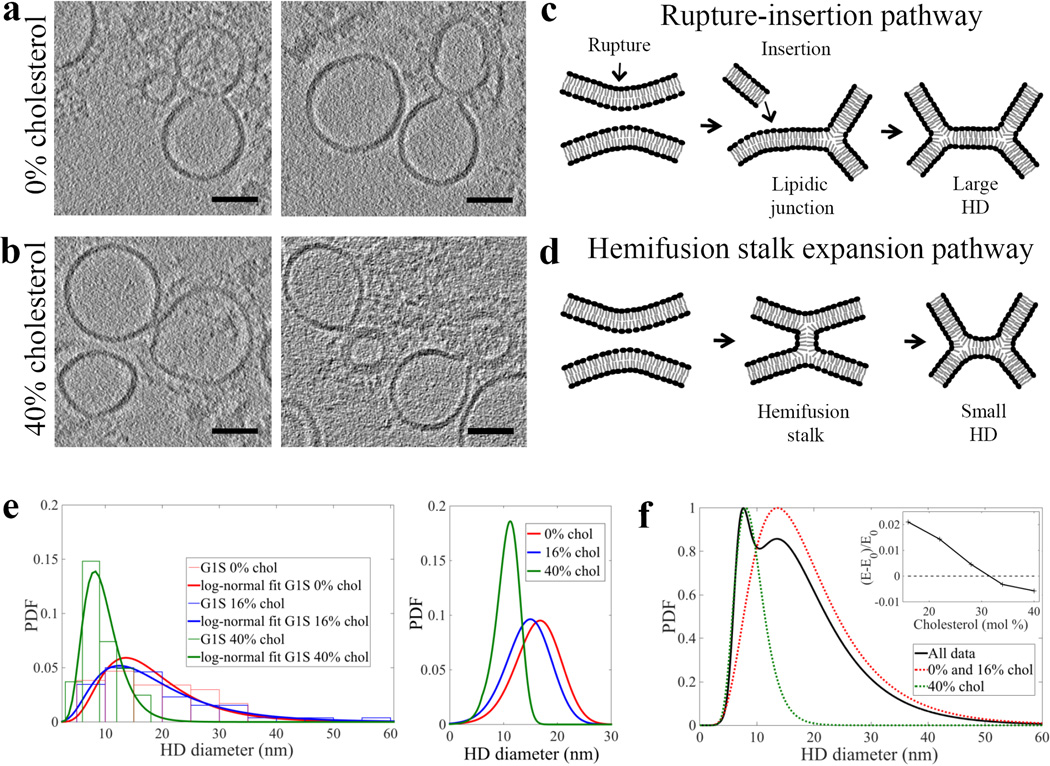
Since negative spontaneous curvature, conferred by lipids such as cholesterol and PE, is known to play a role in membrane fusion15 we analyzed G1S VLP fusion intermediates with liposomes containing different lipid compositions: (i) 95 mol % PC and 5 mol % gangliosides and (ii) 42 mol % PC, 13 mol % PE, 40 mol % cholesterol and 5 mol % gangliosides, in addition to the liposome composition described above (66 mol % PC, 13 mol % PE, 16 mol % cholesterol and 5 mol % gangliosides). Observed HD structure was independent of the cholesterol concentration or the absence of PE in the G1S VLP (Fig. 4). Samples obtained using 0 mol % cholesterol liposomes showed no differences from 16 mol % cholesterol liposomes in rupture frequency and HD diameter (Fig. 4). However, in samples with liposomes containing 40 mol % cholesterol, we detected smaller HD diameters (~9 nm) and both lipidic junctions and ruptures were rare (10%, n=29) (Fig. 4e and Supplementary Table 1).
Figure 4. Two independent, cholesterol concentrations dependent, pathways lead to HD of different diameter.
Two different classes of HD – “large” and “small” – were produced when G1S VLP were mixed with liposomes containing either 0 mol % (a) or 40 mol % (b) cholesterol, respectively, incubated for 20 minutes at neutral pH, and then subjected to low pH for 2 min at 37°C. Tomogram slices (3 nm thick), calculated by weighted-back projection method from a tilt series acquired with VPP, capturing hemifusion events formed between liposomes and VLP. White arrow marks an edge of a ruptured liposome. Scale bars: 50 nm. c) Schema of rupture-insertion pathway that leads to formation of larger HD, predominant at low cholesterol. d) Schema of canonical, hemifusion stalk expansion pathway leading to formation of smaller HD. e) Histograms and probability density functions (PDF) of the measured diameters and chords for liposomes containing 0, 16 and 40 mol % cholesterol (left). PDF calculated using continuum analysis for different monolayer spontaneous curvatures at cholesterol concentrations 0, 16 and 40 mol % (right), as described in supplementary information. f) PDF of experimentally observed hemifusion diameters and chords for combined WT and G1S at different cholesterol concentrations (black; peak normalized to 1); the combined data were fit by a sum of two lognormal distributions. The relative heights of the two peaks (black curve) reflect the amount of data represented in the study and not the relative probability. With fixed lognormal distribution parameters, the relative probability was calculated from the fitted proportions in each of the individual data sets (see Supplementary Figure 9). The red and green dash PDF curves, peak normalized to 1, represent HD populations with small and large diameter, respectively. Using the continuum analysis for stalk formation (and hence for choosing the stalk expansion trajectory, see supplementary information), the relative energy changes between the fused stalk shape and two spherical shapes were calculated to decrease with increasing cholesterol and become negative at approximately 31% cholesterol (inset). The tomographic slices are representative of 46 and 27 VLP-liposome interactions detected in 12 tomograms of each sample using either liposomes with 0 and 40 mol % cholesterol, respectively.
To elucidate the effect of G1S mutation we analyzed WT VLP-liposome mixtures at low pH. Small fusion pores were never detected, presumably due to their short lifetime. Instead, WT VLP-liposome fusion products demonstrated the same lipidic junctions and HD as found in G1S VLP. Moreover, HD diameters, lipid junction chords, and rupture frequency found in WT VLP exhibited the same dependence on liposome lipid composition (Supplementary Fig. 5, Supplementary Table 1). Interestingly, differences between WT and G1S were revealed when the presence and absence of M1 layer was taken into consideration. Distributional properties of WT (HD size and energy) in the presence of M1 was dependent on cholesterol and the differences between WT with and without M1 was much greater than for G1S (Supplementary Fig. 6 and 10 and Supplementary information).
Our data support two independent pathways, via two different structures, leading to hemifusion: (i) formation of large HD (~20 nm), which are products of a liposome rupture-insertion pathway, (Fig. 4c) and (ii) formation of small HD (~9 nm) at high cholesterol concentrations, which are likely a result of hemifusion stalk expansion rather than rupture (Fig. 4d). When all HD diameters are combined, the resulting density function revealed two distinct peaks and is described by a sum of two lognormal distributions representing the two populations of HD diameters. This result supports the hypothesis that two different pathways lead to two distinct HD diameter populations (Fig. 4f).
The rupture-insertion structure, predominating at low cholesterol, is confirmed by the presence of ruptured liposomal membrane edges and spikes inside the HD. Also, a cryo-ET study using defocus phase contrast imaged a variety of intermediates but critically lacked the HA receptor in the target liposome (here provided by ganglioside) and lacked sufficient resolving power9. The HA-stabilized membrane edges and lipidic junctions of variable sizes indicate that HD formation could proceed through multiple steps of liposomal membrane insertion into VLP and lateral merging of the resulting lipidic junctions. We propose that the rupture-insertion pathway directly depends on the initial binding area and its complement of HA. Consistent with this idea, distributions of binding area are similar to HD areas (Supplementary Fig. 7). Based on the average HD diameter of WT HA (21±8 nm) and VLP glycoprotein spacing (9.5 nm) prior to HA conformational changes, we calculated that 4±1 HA trimers were present in binding areas involved in the formation of HD, consistent with previous studies16,17. Perhaps HA fusion peptides stabilize the otherwise thermodynamically unstable bilayer edges of ruptured liposomes.
In the hemifusion stalk expansion pathway, prevalent at high cholesterol content, we rarely observed either rupture or lipidic junctions, suggesting that HD is formed by expansion of a hemifusion stalk; the HD lack any evidence of proteinaceous materials. Consistent with this view, liposomes with higher cholesterol (40 mol %), having a more negative intrinsic monolayer curvature (−0.24 nm−1), could spontaneously expand stalks into HD with the diameter range we measured18. HD diameter distributions calculated by continuum analysis based on a Helfrich Hamiltonian are in agreement with distributional analysis of experimental data (Fig. 4e and Supplementary Fig. 8). The energy of forming a hemifusion stalk decreases with increasing cholesterol and becomes negative at approximately 31 mol % cholesterol concentration (Fig. 4f inset). Therefore, at concentrations higher than 31 mol %, the hemifusion stalk expansion pathway is favored since energy is gained as a result of the membrane curvatures matching the more negative spontaneous curvature. Decreased rupture frequency might be explained by an increased elastic area expansion moduli Ka (mN/m) in the presence of high cholesterol (~40 mol %)19 as well as by other physical effects of cholesterol such as lipid diffusional terms, including membrane fluidity20.
The cholesterol content in the late endosomes, a site of influenza A virus membrane fusion, is not known precisely. The cholesterol concentration in the endosomal recycling compartment was estimated to be ~35%,21 indicating that late endosomal cholesterol concentration is around 30%22. Other lipids with negative spontaneous curvature such as lysobisphosphatidic acid, known to be enriched in the late endosomes, may serve the same role as increased cholesterol and influence which pathway HA follows23,24. Since we observe HD over a wide range of cholesterol concentrations we conclude that HD is also common in late endosomal membrane. Although the rupture-insertion pathway leading to large HD may not be an energetically favorable pathway to a completed fusion product, not all WT VLP remained arrested as hemifusion products even at low cholesterol concentrations (Supplementary Fig. 2, Supplementary Video 7). The density of detected hemifusion events was approximately twice lower using liposomes containing 40 mol % compared to 0 mol % cholesterol. Assuming no changes in binding, the decrease in hemifusion is consistent with an increase in complete membrane fusion supporting the hypothesis that the hemifusion-stalk pathway is more efficient in producing complete membrane fusion. The variation in the small and large HD fractions as a function of cholesterol and the fraction differences between WT and G1S with and without M1 suggest that compensatory mechanisms may exist for handling differences in the endosomal cholesterol concentration and/or negative spontaneous curvature depending on the infected host cell (Supplementary Fig. 9).
Cholesterol, influencing both membrane elasticity and spontaneous monolayer curvature, is thus expected to be pivotal in all membrane fusion beyond hemagglutinin-induced fusion. Consistent with this expectation, large stable HD were observed in systems where SNAREs mediate liposome fusion,25 and low membrane cholesterol content prolongs a hemifusion intermediate which only rarely develops into a fusion pore26. We have shown that at low cholesterol, the rupture/insertion pathway of membrane merger is more favored than the stalk pathway. The reverse is true at high cholesterol content. We suggest that the stalk pathway is more likely to evolve into a fusion pore and is likely more relevant to cellular infection. Continuum membrane mechanics offers a quantitative physical explanation for the phenomenon through the intrinsic curvature of the target membrane.
MATERIALS AND METHODS
Plasmids
HA (A/Aichi/2/68), NA (A/Singapore/1/57) DNA sequences cloned into eukaryotic expression vector pCAGGS28 were a kind gift from H.D. Klenk, University of Marburg. Sequences of M1 (A/Hong Kong/8/68) and M2 (synthetic) were cloned into separate pCAGGS plasmids. Nucleotide mutations resulting in G1S mutant in HA (A/Aichi/2/68), was introduced by QuikChange II XL site-directed mutagenesis kit (Agilent).
Cells
HEK-293T (ATCC CRL-1573) human embryonic kidney cells and Cos7 (ATCC CRL-1651) African green monkey fibroblast cells were authenticated visually by light microscopy, tested negative for Mycoplasma contamination, and aliquoted in frozen stocks at the time of purchase from ATCC (Manassas, VA). Cells were maintained at 37°C and 5% CO2 in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), L-glutamine, and primocin. We have chosen HEK-293T cells for their high transfection efficiency and high production of the influenza A VLP. Since HEK-293T cells are listed in the databases of commonly misidentified cell lines maintained by ICLAC and NCBI Biosample, we purchased the HEK-293T directly from ATCC to avoid using a cross-contaminated cell line.
VLP preparation
293T cells were seeded in a 10 cm diameter tissue culture dish (3.2×106 cells/dish) and grown at 37°C and 5% CO2 in DMEM-10% FCS supplemented with L-glutamine and primocin. 24 h later, cells were transfected with the appropriate plasmids using Trans-IT LT1 (Mirus) according to the manufacturer's instructions. VLP were produced using the following quantities: 2.5 µg pCAGGS-HA(wt) or HA(G1S), 0.5 µg pCAGGS-NA, 1 µg pCAGGS-M1, and 0.25 µg of pCAGGS-M2. At 5 h post-transfection, the medium was replaced with serum-free SFM4MegaVir and exogenous bacterial neuraminidase from Clostridium perfrigens (Sigma-Aldrich) was added at a final concentration of 25 mU/ml. At 48 h TPCK-treated trypsin (5 µg/ml) was directly added to the medium in the culture dish and incubated at 37°C for 15 min. Trypsin was inactivated by the addition of soybean trypsin inhibitor (0.1 mg/ml); supernatant was collected and clarified at 4°C by centrifugation at 1000 × g for 10 min. Subsequently, the supernatant was layered onto a 3 ml 30% (wt/vol) sucrose-KHE (100 mM KCl, 10 mM HEPES [pH 7.4], 1 mM EDTA) cushion and centrifuged at 200,000 × g for 2 h at 4°C in a Beckman Optima centrifuge using a 50.2Ti rotor (Beckman Coulter, Fullerton). The pellet was re-suspended in KHE buffer and centrifuged again at 130,000 × g for 30 min at 4°C using a TLA 100.3 rotor. The pellet was re-suspended in KHE buffer.
Red blood cells (RBC) labeling with hydrophilic and hydrophobic fluorescent dyes
Human red blood cells (RBC) were obtained from NIH Blood Bank from healthy, normal donors who consented to participate in the IRB approved Research Donor Program at the NIH/FDA laboratories. RBC were spun at 700 × g for 8 min at 4°C and subsequently washed 5× with PBS without Ca2+ and Mg2+ to remove serum. The RBC pellet was hemolyzed with hemolysis buffer (PBS without Ca2+ and Mg2+: H2O, 1:1) and incubated on ice for 2–15 min with aqueous fluorescent dye (5-(and 6-)carboxy-2’,7’-dichloro-fluorescein; Molecular Probes), FITC-anionic dextran 10 kDa, or FITC- anionic dextran 500 kDa (Invitrogen). The RBC membrane was sealed according to Ellens (1989)29 and spun at 300 × g for 4 min at 4°C to remove any unincorporated fluorescent dye. The RBC membrane was labeled with PKH26 (Sigma).
Cell-RBC fusion assay
Cos7 cells were seeded in a 2 well Lab-Tek chambered coverglass plate (Nunc) at density 3.5×104 cells per well and grown at 37°C and 5% CO2 in DMEM-10% FCS supplemented with L-glutamine and primocin. 24 h later, cells were transfected with 2.5 µg pCAGGS-HA(wt) or HA(G1S) using Trans-IT LT1 (Mirus) according to the manufacturer's instructions. One day post-transfection the cells were washed twice with PBS (pH 7.4) and treated for 4 min at RT with 10 µg/ml TPCK-treated trypsin and 0.2 mg/ml exogenous bacterial neuraminidase from Clostridium perfrigens (Sigma-Aldrich). Subsequently, the RBC labeled with content and lipid fluorescent dye were added to transfected Cos7 cells and incubated for 20 min at RT. The Cos7 RBC cell-pairs were washed 3 times with PBS with Ca2+ and Mg2+ (pH 7.4), incubated for 2 min with acidified PBS (pH 4.9) at 37°C and an additional 20 min with PBS with Ca2+ and Mg2+ (pH 7.4) at 37°C. Cell-pairs were imaged using an Axiovert 25 (Zeiss) inverted microscope equipped with 20× Zeiss objective (NA 0.8) and filter sets 45 and 42 (Zeiss). Cell-RBC pairs showing content and lipid dye mixing were counted. Fusion was evaluated using the number of cell-pairs exhibiting both content dye and lipid dye mixing and presented as a number ratio.
Liposome preparation
Phospholipids were selected based on phospholipids typically found in endosomal membrane except for charged phospholipids22. POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine), POPE (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine), cholesterol, and total gangliosides (GA) (Avanti Polar Lipids) were dissolved in chloroform-methanol (2:1) at a final concentration of 10 mM. Liposomes with 3 different lipid compositions were prepared: A) 95 mol % POPC and 5 mol % GA; B) 66 mol % POPC, 13 mol % POPE, 16 mol % cholesterol and 5 mol % GA; C) 42 mol % POPC, 13 mol % POPE, 40 mol % cholesterol and 5 mol % GA. The solvent was removed by evaporation; first using a stream of argon gas and second, high vacuum for 1 hour. The lipid film was re-suspended in KHE buffer, processed using 21 freeze-thaw cycles, and extruded at room temperature through 100 nm PC membrane (Whatman) using a mini-extruder set (Avanti Polar Lipids). Liposomes were stored at 4°C and used within 1 to 5 days.
cEM of VLP and liposome fusion products and quantification
For cEM, 5 µl of VLP and 1µl of liposomes (10 mM) were incubated for 20 min at 37°C. Membrane fusion was triggered by the addition of 0.9 µl of 17 mM citric acid in 20 mM HEPES to achieve a final pH of 5.0 and incubated at 37°C for 2–5 min. VLP-liposome mixtures containing 10 nm Protein A gold were applied to C-flat grids CF-2/1-2C-50 (Protochips) maintained at 25°C, 100% relative humidity and vitrified by plunging into liquid ethane using a Vitrobot (FEI). Cryo-electron micrographs of vitrified samples were collected using a transmission electron microscope T20 (FEI) equipped with a K2 base camera (Gatan) at defocus −3 to −5 µm. We counted three different stages of fusion: (i) binding, (ii) fusion intermediate and (iii) complete fusion based on the following criteria. Bound liposomes retained their spherical shape or were compressed and glycoprotein spikes were clearly discernable. In contrast, fusion intermediates showed a teardrop shape pointing towards the disorganized layer of HA glycoproteins. Complete fusion was characterized by complete expansion of the fusion pore and a disorganized matrix layer.
cET and VPP-cET of VLP and liposome fusion products
Samples were prepared as for cEM described above. Tilt series were collected using a transmission electron microscope Titan Krios at 300kV equipped with Falcon II direct detector (FEI, Netherlands). Tilt series were acquired at a nominal magnification of 22,500× with angular range from −65° to +65°, an angular increment of 2° using an −5 µm defocus, 70 µm objective aperture, 0.368 nm/pixel, and a total dose of 80 e−/Å2. Tilt series with Volta-induced phase plate were acquired as above substituting the 70 µm objective aperture with a phase plate and at −1 µm defocus12. The phase plate was aligned to obtain cross-over at the back-focal plane in diffraction mode and beam-shift pivot points30. Each tilt series was collected on a different phase plate location and in most of the cases was preconditioned for 300–500 sec to induce phase shift. Tomograms were reconstructed using the weighted back projection technique in the IMOD software suite31 following nonlinear anisotropic diffusion (NAD) de-noising32 if indicated. Isosurfaces were calculated from tomograms reconstructed by simultaneous iterative technique (10 iterations) and de-noised with NAD using k value set to 10, and 10 iterations in IMOD.
Measurements and statistical analysis of HD geometry
HD diameter was measured as the farthest distance between fully formed HD “Y” junctions or as chords of arcs in partial HD found in the three-dimensional volume using Imod slicer window in the tomograms binned to 0.736 nm/pixel. Measurement of HD diameter was done from the centers of membrane bilayers. HD angles were measured using FIJI on both side of the “Y” junction. PDF and CDF fits were calculated in MATLAB. The Kolmogorov-Smirnov test was performed using an online tool: http://www.physics.csbsju.edu/.
Radially averaged power spectrum calculation
3 nm slices from a single tomogram showing glycoproteins cross-sections at the surface of four different VLP (2 filamentous and 2 spherical) were extracted from a tomogram and converted to a tiff using mrc2tif in IMOD. Ten (53×53 nm) images were cropped from the tomogram slices and tapered in the MATLAB image-processing toolbox. The calculated power spectra of each image were averaged in MATLAB. A radially averaged power spectrum was calculated from the averaged power spectrum using the MATLAB function rotavg written by Bruno Olshausen (University of California, Berkeley) and available online at https://redwood.berkeley.edu/bruno/npb261b/lab2/rotavg.m.
Radially averaged bilayer profile and signal to noise ratio comparison
Slices (0.74 nm thick) containing a liposome were cropped after binning by 2 either from a tomogram collected at defocus −5 µm without phase plate or at defocus −1 µm with Volta-induced phase plate. Radially averaged bilayer profiles were obtained using the FIJI radial profile plugin written by Paul Baggethun. The tomogram thicknesses were estimated in IMOD during tomogram reconstruction. Signal to noise ratios were calculated as a ratio of the intensity across the bilayer and the intensity across the interior of the liposome.
Continuum Energy Calculation
The energy of hemifusion between the liposome membrane and the viral membrane was calculated numerically as a function of the diameter of the HD, the portion of the membrane that continues to separate virus and liposome aqueous interiors. The total energy of the VLP-liposome merger product was determined from a Helfrich Hamiltonian33;
The Hamiltonian accounts for: two dimensional splay of the lipid director d (div d; twice the negative mean curvature when d = n; the unit surface normal) with splay modulus KC and spontaneous curvature k0; saddle splay (det ∇d) with Gaussian curvature modulus KG, tilt (d × n, also called twist) with tilt modulus Kθ; and local stretch/compression to the area per lipid (a) from the resting area (a0) for area modulus KA. The monolayer surfaces Σ of the VLP-liposome merger product were partitioned into inner/outer VLP monolayers and inner/outer liposome monolayers. Interstices (i.e. voids) were prevented by requiring opposing monolayer acyl chains to terminate at the same point. To determine area per lipid, we used volume incompressibility, and an equilibrium acyl chain length l0 = 1.7 nm for both liposome and VLP. For a head group diameter 0.8 nm, this yields a bilayer thickness 5.0 nm34. The elastic constants were: KC = 4 kT, KG = −3.2 kT and Kθ = 4 kT nm−2 in the liposomal bilayer and KC = 12 kT, KG =−9.6 kT and Kθ = 12 kT nm−2 in the VLP monolayers34–36. The liposome was thus roughly three times more elastic than the VLP37. For DOPC lipids, the splay modulus is insensitive to cholesterol concentration38, but the spontaneous curvature and area modulus are sensitive. For 0, 16, 40 mol % cholesterol, we used KA = 14, 16, 30 kT nm−2 for the area modulus and k0 = −0.05, −0.13, −0.24 nm−1 for the spontaneous curvature respectively (Table S1).
In order to conserve membrane, the liposome and VLP inner monolayer areas were both constrained to 31,600 nm2 (radius 50 nm prior to merger). They represent size distributions when both particles in the VLP-liposome merged product have diameter roughly 100 nm. To model the cET results, the merged VLP and liposome monolayers were discretized by piecewise linear, cylindrical finite elements (N = 600 grid points in total), with mesh preserving equally spaced distal monolayer grid points. The HD radius was confined to the range 3 nm – 30 nm (endpoints corresponding to small stalk-like HD and a large HD). Bilayer shape was deformed in the direction of steepest gradient descent - ∇E using a fully implicit backward Euler scheme. An adaptive time stepping strategy mitigated the problem of numerical stiffness and greatly reduced the computer time of calculation; short range forces near the junction where three bilayers meet equilibrated rapidly (the angles of the “Y” varied to minimum energy), after tens of iterations with time step Δt ~10−5 s; long range curvature forces away from the “Y” were driven to equilibrium after hundreds of iterations with Δt ~1 s. Iterations were terminated once a decrease in numerical energy was within 0.1% relative error of minimal energy.
Supplementary Material
Acknowledgments
We are grateful to Vinod Nair for assistance with the Krios transmission electron microscope at Rocky Mountain Laboratories Microscopy Unit, NIAID, NIH. We thank Lars-Anders Carlson, Leonid Chernomordik, Ivonne Morales and Tom Reese for critical reading of the manuscript. This work was supported by the Division of Intramural Research (DIR) of the Intramural Program of the NIH (IRP).
Footnotes
STATEMENT OF INDIVIDUAL CONTRIBUTIONS
Project was planned by P.C., P.S.B. and J.Z.; Experimental work was done P.C. and E.M.; H.W. generated pCAGGS-M1 and pCAGGS-M2 constructs; Cryo-electron microscopy and tomography was done by: P.C., C.L.S. and E.R.F.; Statistical analysis and image processing was done by P.C. and P.S.B.; Continuum analysis was calculated by R.J.R. and F.S.C.; Manuscript was written by P.C., R.J.R., F.S.C., P.S.B. and J.Z. All authors assisted editing the manuscript and contributed to the data analysis.
COMPETING INTERESTS
The authors declare no competing financial interests.
ACCESSION NUMBERS
Tomograms for Fig. 2 have been deposited to EMBD database under accession number EMD-8088 and EMD-8089.
REFERENCES
- 1.Chernomordik LV, Zimmerberg J, Kozlov MM. Membranes of the world unite! J Cell Biol. 2006;175:201–207. doi: 10.1083/jcb.200607083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kemble GW, Danieli T, White JM. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 3.Harrison SC. Viral membrane fusion. Virology. 2015;479–480C:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen BJ, Leser GP, Morita E, Lamb RA. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. Journal of virology. 2007;81:7111–7123. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiao H, Armstrong RT, Melikyan GB, Cohen FS, White JM. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol Biol Cell. 1999;10:2759–2769. doi: 10.1091/mbc.10.8.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chlanda P, et al. Structural analysis of the roles of influenza A virus membrane-associated proteins in assembly and morphology. Journal of virology. 2015 doi: 10.1128/JVI.00592-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris A, et al. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnafous P, et al. Treatment of influenza virus with beta-propiolactone alters viral membrane fusion. Biochimica et biophysica acta. 2014;1838:355–363. doi: 10.1016/j.bbamem.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 9.Lee KK. Architecture of a nascent viral fusion pore. The EMBO journal. 2010;29:1299–1311. doi: 10.1038/emboj.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurer UE, Sodeik B, Grunewald K. Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10559–10564. doi: 10.1073/pnas.0801674105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erickson HP, Klug A. Measurement and Compensation of Defocusing and Aberrations by Fourier Processing of Electron Micrographs. Philos Trans R Soc Lond B Biol Sci. 1971;261:105–118. [Google Scholar]
- 12.Danev R, Buijsse B, Khoshouei M, Plitzko JM, Baumeister W. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:15635–15640. doi: 10.1073/pnas.1418377111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozlovsky Y, Kozlov MM. Stalk model of membrane fusion: solution of energy crisis. Biophysical journal. 2002;82:882–895. doi: 10.1016/S0006-3495(02)75450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frolov VA, Dunina-Barkovskaya AY, Samsonov AV, Zimmerberg J. Membrane permeability changes at early stages of influenza hemagglutinin-mediated fusion. Biophysical journal. 2003;85:1725–1733. doi: 10.1016/S0006-3495(03)74602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nat Struct Mol Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanovic T, Choi JL, Whelan SP, van Oijen AM, Harrison SC. Influenza-virus membrane fusion by cooperative fold-back of stochastically induced hemagglutinin intermediates. eLife. 2013;2:e00333. doi: 10.7554/eLife.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danieli T, Pelletier SL, Henis YI, White JM. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozlovsky Y, Chernomordik LV, Kozlov MM. Lipid intermediates in membrane fusion: formation, structure, and decay of hemifusion diaphragm. Biophysical journal. 2002;83:2634–2651. doi: 10.1016/S0006-3495(02)75274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Needham D, Nunn RS. Elastic deformation and failure of lipid bilayer membranes containing cholesterol. Biophysical journal. 1990;58:997–1009. doi: 10.1016/S0006-3495(90)82444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmerberg J, Gawrisch K. The physical chemistry of biological membranes. Nat Chem Biol. 2006;2:564–567. doi: 10.1038/nchembio1106-564. [DOI] [PubMed] [Google Scholar]
- 21.Hao M, et al. Vesicular and non-vesicular sterol transport in living cells. The endocytic recycling compartment is a major sterol storage organelle. J Biol Chem. 2002;277:609–617. doi: 10.1074/jbc.M108861200. [DOI] [PubMed] [Google Scholar]
- 22.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi T, et al. Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat Cell Biol. 1999;1:113–118. doi: 10.1038/10084. [DOI] [PubMed] [Google Scholar]
- 24.Yang ST, Zaitseva E, Chernomordik LV, Melikov K. Cell-penetrating peptide induces leaky fusion of liposomes containing late endosome-specific anionic lipid. Biophysical journal. 2010;99:2525–2533. doi: 10.1016/j.bpj.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diao J, et al. Synaptic proteins promote calcium-triggered fast transition from point contact to full fusion. eLife. 2012;1:e00109. doi: 10.7554/eLife.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreutzberger AJ, Kiessling V, Tamm LK. High cholesterol obviates a prolonged hemifusion intermediate in fast SNARE-mediated membrane fusion. Biophysical journal. 2015;109:319–329. doi: 10.1016/j.bpj.2015.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. Structural organization of a filamentous influenza A virus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10685–10690. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 29.Ellens H, Doxsey S, Glenn JS, White JM. Delivery of macromolecules into cells expressing a viral membrane fusion protein. Methods in cell biology. 1989;31:155–178. doi: 10.1016/s0091-679x(08)61607-6. [DOI] [PubMed] [Google Scholar]
- 30.Fukuda Y, Laugks U, Lucic V, Baumeister W, Danev R. Electron cryotomography of vitrified cells with a Volta phase plate. J Struct Biol. 2015 doi: 10.1016/j.jsb.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [pii] 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 32.Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- 33.Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z Naturforsch C. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 34.Nagle JF, Tristram-Nagle S. Structure of lipid bilayers. Biochim Biophys Acta. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamm M, Kozlov MM. Elastic energy of tilt and bending of fluid membranes. Eur. Phys. J. 2000;3:323–335. [Google Scholar]
- 36.Siegel DP, Kozlov MM. The gaussian curvature elastic modulus of N-monomethylated dioleoylphosphatidylethanolamine: relevance to membrane fusion and lipid phase behavior. Biophysical journal. 2004;87:366–374. doi: 10.1529/biophysj.104.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S, et al. pH-Controlled two-step uncoating of influenza virus. Biophysical journal. 2014;106:1447–1456. doi: 10.1016/j.bpj.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan J, Tristram-Nagle S, Nagle JF. Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;80:021931. doi: 10.1103/PhysRevE.80.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.