Abstract
Autologous CD34+ cells are widely used for vascular repair; however, in individuals with diabetes and microvascular disease these cells are dysfunctional. In this study, we examine expression of the clock genes Clock, Bmal, Per1, Per2, Cry1, and Cry2 in CD34+ cells of diabetic and nondiabetic origin and determine the small encoding RNA (miRNA) profile of these cells. The degree of diabetic retinopathy (DR) was assessed. As CD34+ cells acquired mature endothelial markers, they exhibit robust oscillations of clock genes. siRNA treatment of CD34+ cells revealed Per2 as the only clock gene necessary to maintain the undifferentiated state of CD34+ cells. Twenty-five miRNAs targeting clock genes were identified. Three of the miRNAs (miR-18b, miR-16, and miR-34c) were found only in diabetic progenitors. The expression of the Per2-regulatory miRNA, miR-92a, was markedly reduced in CD34+ cells from individuals with DR compared with control subjects and patients with diabetes with no DR. Restoration of miR-92a levels in CD34+ cells from patients with diabetes with DR reduced the inflammatory phenotype of these cells and the diabetes-induced propensity toward myeloid differentiation. Our studies suggest that restoring levels of miR-92a could enhance the usefulness of CD34+ cells in autologous cell therapy.
Introduction
The endogenous circadian system drives the rhythm of many physiological and behavioral processes. Approximately 10% of the transcriptome in all tissues is under the control of clock genes (1–3). Abnormal clock gene expression in endothelial cells is implicated in the pathogenesis of both macrovascular (2) and microvascular (3) complications. In addition, clock gene dysregulation poses a risk for metabolic syndrome, obesity, premature aging, and abnormal sleep cycle (4).
While the suprachiasmatic nucleus drives the central rhythm, the control of the clock at the molecular level involves self-regulatory transcriptional feedback loops composed of positive (Clock and Bmal1) and negative (Per1, Per2, Cry1, Cry2, and Rev-Erbα) sets of genes (5). Previously, we reported that in type 2 diabetic (T2D) rats, defects in clock gene expression of bone marrow progenitor cells (BMPCs) contributed to development of diabetic retinopathy (6). Using a mouse model lacking the clock gene Per2, we showed that these mice develop the histological features of diabetic retinopathy (DR) without hyperglycemia and also develop diabetes-like BMPC dysfunction (3). Per2 mutant mice display a range of endothelial defects, including reduced tube formation, decreased endothelium-dependent vasodilatation, and increased senescence (7). Mice lacking Per2 in BMPCs undergo autoamputations, largely due to the reduced function of BMPCs that hinders vascular repair in these mice (8). Mice deficient in Bmal1 also exhibit endothelial defects with increased oxidative stress and reduced endothelial nitric oxide synthase (9). Together, these studies support an important role of clock genes in both normal endothelial function and vascular disease pathogenesis.
Small noncoding RNAs (miRNAs), 19–25 nucleotides in length, are involved in repressing mRNA translation or cleaving target mRNA and have been implicated in the regulation of clock gene expressions in angiogenesis (10). The miR-17-92 cluster is highly expressed in endothelial cells, and in particular, miR-92a is reported to have a role in angiogenesis by targeting mRNAs of proangiogenic proteins such as integrin α5 (11). Strikingly, CD34+ cells show a 5- to 10-fold increase in the expression of miR-92a (12) that potentially targets the clock gene Per2 (13).
Differentiation of progenitor cells is a complex process. A variety of transcription factors (e.g., Ets, Forkhead, GATA, and Kruppel-like families) (14), posttranscriptional regulators including miRNA-mediated repression (15), and the microenvironment (16) together define the characteristics of a particular progenitor population and their tendency to differentiate toward the endothelial-like linage. However, while only a limited number of CD34+ cells and early endothelial progenitor cells (eEPCs) (CD34+CD133+vascular endothelial growth factor [VEGF] receptor 2+) differentiate into endothelial cells, the cells transition through reduced levels of primitiveness, and this transition dramatically influences their secretome. The secretome of CD34+ cells and eEPCs is paramount to their mode of action, which is that of providing paracrine support to the injured vasculature (17). In contrast, endothelial colony–forming cells (ECFCs) (CD144+VEGFR2+CD133−) serve as building blocks by directly participating in blood vessel formation (18). Interestingly, ECFCs can undergo numerous population doublings, while the proliferative potential of CD34+ cells and eEPCs is limited (19). ECFCs possess mature endothelial markers like CD31 and CD144, which support their role in vessel formation (18,20). The paracrine function of eEPCs, like CD34+ cells, is mainly mediated via secretion of a variety of potent stem cell growth factors including stem cell factor (SCF), hepatocyte growth factor (HGF), and thrombopoietin (TPO) as well as cytokines such as interleukins (IL), chemokine (C-C motif) ligand 2, and granulocyte colony–stimulating factor. Diabetes causes defects in ex vivo expansion of ECFCs (success rate 15%) and in the paracrine function of eEPCs resulting in a decrease in SCF, HGF, and TPO (21).
In the current study, we hypothesized that the differentiation of vascular progenitors is under the regulation of clock genes with specific miRNAs regulating clock gene oscillations. We further hypothesized that diabetes interferes with the expression of these specific miRNAs leading to altered differentiation and paracrine function of these cells.
Research Design and Methods
Isolation of CD34 Cells
Mononuclear cell fractions from healthy individuals or patients with diabetes were negatively selected for lineage-negative (Lin−) population using a human progenitor cell enrichment kit (Stem Cell Technologies, Vancouver, BC, Canada). Lin− cells were stained with a Pacific Blue CD34 and PE/Cy7 CD45 antibodies (BioLegend, San Diego, CA) and sorted for Lin−CD34highCD45dim population using a FACSAria II cell sorter (BD Biosciences, San Jose, CA).
Culture of CD34+ Cells and Analysis for Surface Expression
Isolated CD34+ cells were propagated in round-bottom 96-well culture plates with no more than 5,000 cells per well under two different culturing conditions for 4 days: 1) undifferentiated, Stem Span Media supplemented with cytokine cocktail containing IL-3, IL-6, Flt, and SCF (Stem Cell Technologies) and 2) differentiated, EBM-2 media supplemented with EGM-2MV growth supplements (Lonza, Allendale, NJ) containing human epidermal growth factor, hydrocortisone, human fibroblast growth factor beta, VEGF, recombinant analog of insulin-like growth factor, ascorbic acid, heparin, and FBS.
At 4 days, the cells were stained with a cocktail of antibodies containing Pacific Blue CD34, PE/Cy7 CD45, FITC CD144, and PE CD133 (BioLegend). Cell-surface expression was analyzed using a BD LSR II Flow Cytometer, and the analysis was performed using FCS express (De Novo Software, Los Angeles, CA).
Quantitative RT-PCR for Clock Gene Expression
mRNA was extracted using TRIzol (Life Technologies, Grand Island, NY) and reverse transcribed using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Transcript levels of clock, bmal1, Per1, Per2, Cry1, and Cry2 were determined using TaqMan Gene Expression assays (Life Technologies) and an ABI-7500 Fast Real-Time PCR system.
miRNA Microarrays and Analysis
Total RNA was extracted with the miRNeasy Serum/Plasma extraction kit (Qiagen, Redwood City, CA) and purified using RNeasy MinElute Spin Columns. A total of 250 ng RNA was then reverse transcribed with a miScript II RT kit (Qiagen). miRNA expression profile was then measured using Qiagen’s Human Serum & Plasma miRNA PCR Array. Quantitative PCR was performed using a StepOnePlus machine (Applied Biosystems) per the manufacturer’s instructions. miRNA array data were analyzed using Ingenuity pathway analysis software (Qiagen) and DIANA-mirPath.
Statistics
Data were expressed as means ± SEM, and each experiment was repeated at least in triplicate unless otherwise specified. Statistical analysis was conducted using either Prism (GraphPad, La Jolla, CA) or SigmaPlot (Systat Software, San Jose, CA). The data were analyzed using one-way ANOVA followed by the Tukey-Karmer post hoc test.
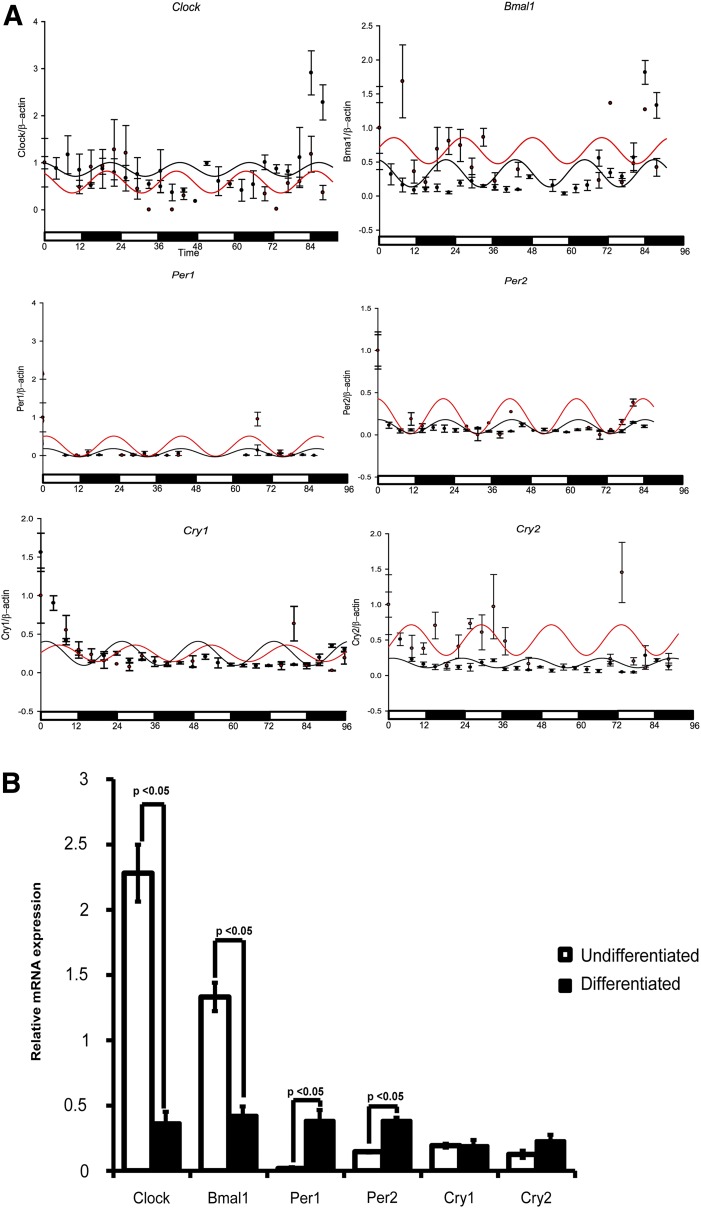
For the CD34+ cell differentiation study (Fig. 3A), a smooth curve for the expression of each gene was fitted by single cosine function of f(t)=A*cos(ωt+Φ)+M, where f(t) is the gene expression level at time t, A is amplitude, ω is angular frequency, Φ is acrophase, and M is mesor (22). The data were considered to be statistically significant when the P value was <0.05 using a zero-amplitude test.
Figure 3.
Endothelial differentiation of CD34+ cells alters the expression pattern of clock genes. A: Lin−CD34highCD45mid cells were cultured in hematopoietic progenitor–supporting (stem cell maintenance media [black lines]) or endothelial differentiation conditions (differentiation [red lines]) and harvested every 4 h to analyze mRNA expression of clock genes (Clock, Bmal, Per1, Per2, Cry1, and Cry2). Each data point represents the experimentally observed mRNA expressions for clock genes, while the solid line shows simultaneous data set fitted for the cosine model (see research design and methods) over 96 h. The cosine model was used to predict the biological rhythm of individual clock genes. Clock constituting “positive arm” of circadian cycle showed suppressed expression with endothelial differentiation, while Bmal1, Per2, and Cry2 exhibited rapid oscillatory pattern in endothelial supporting conditions. There was no significant change in the expression pattern for Per1 and Cry1. B: Bar chart showing relative mRNA expression at day 4 for individual clock gene expression. n = 4.
Justification for Online Supplemental Material
In order to address our hypothesis, we preformed key experiments which are included in the Supplementary Data online. We believe this additional information, which is not part of the original article, will help readers understand our study design and conclusions.
Results
Differentiation of CD34+ Cells Leads to a Decrease in CD133 Expression
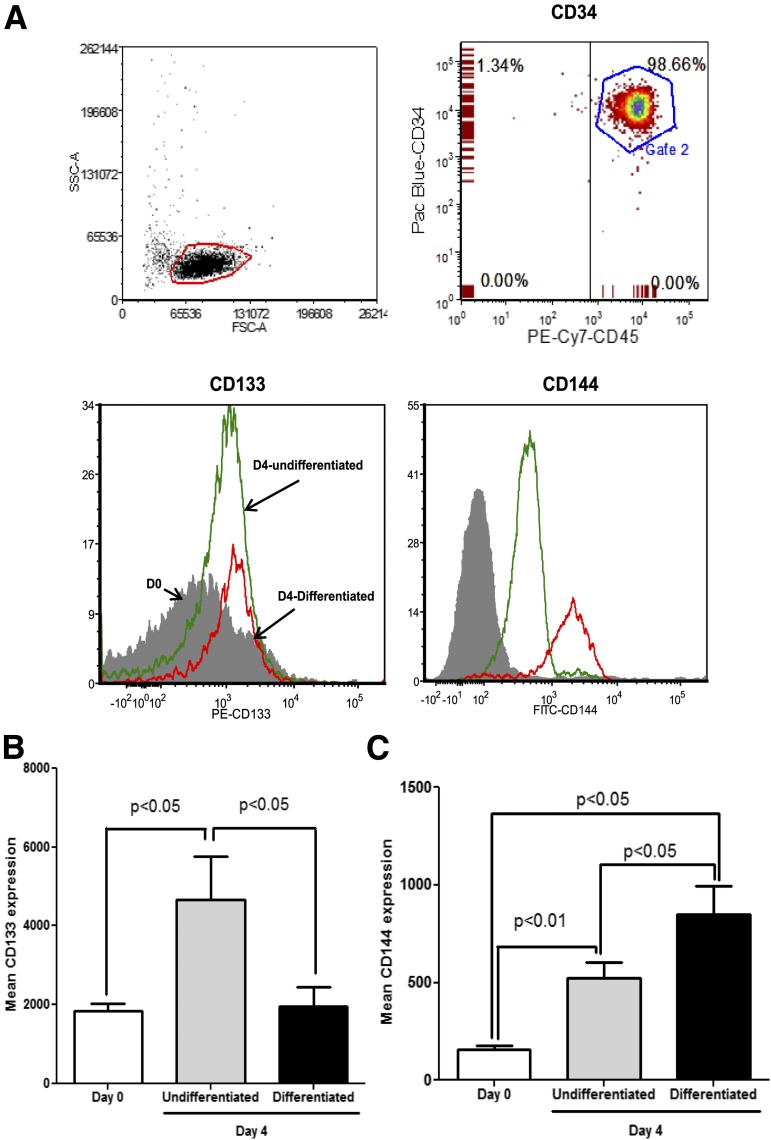
We examined the differentiating effect of specific growth factors on highly purified Lin−CD34+ cells (>97% purity) (Fig. 1A, top panel) using media that either maintained CD34+ cells in the undifferentiated state (cells supplemented with SCF, IL-3, IL-6, and Flt-1) or promoted the differentiated state (cells supplemented with VEGF, IGF-1, fibroblast growth factor, and 5% serum). CD34+ cells maintained under undifferentiated conditions showed a rapid expression of the primitive stem cell marker CD133 (P < 0.05) by day 4 (23). In contrast, endothelial-differentiating conditions resulted in a significant decrease in CD133 expression (P < 0.05) compared with undifferentiating conditions (Fig. 1B). CD34+ cells maintained in either undifferentiating or differentiating media increased expression of CD144 (P < 0.05), a cell junction protein specifically expressed by mature endothelial cells. CD144 expression was significantly higher (P < 0.05) in cells maintained in differentiating media compared with undifferentiating media (Fig. 1C) (P < 0.05).
Figure 1.
CD133 and CD144 expression in CD34+ cells under hematopoietic and endothelial culture conditions. Lin−CD34highCD45dim cells were analyzed for surface expression of CD133 and CD144 under hematopoietic and endothelial differentiating culturing conditions at day 0 and day 4. A: Representative flow cytometry dot plots showing highly purified fraction of cells exhibiting Lin−CD34highCD45dim population expressing the above markers under conditions for differentiation or for maintenance of the stem cell characteristics. At day 4, endothelial differentiation medium reduced the expression of stem cell marker CD133 and increased expression of the mature endothelial marker CD144. B and C: Bar chart showing mean increase in expression of CD133 and CD144 from freshly isolated cells (day 0) in hematopoietic or endothelial-culturing conditions. n = 7.
Per2 Expression Is Required to Maintain Undifferentiated Stem Cell Characteristics in CD34+ Cells
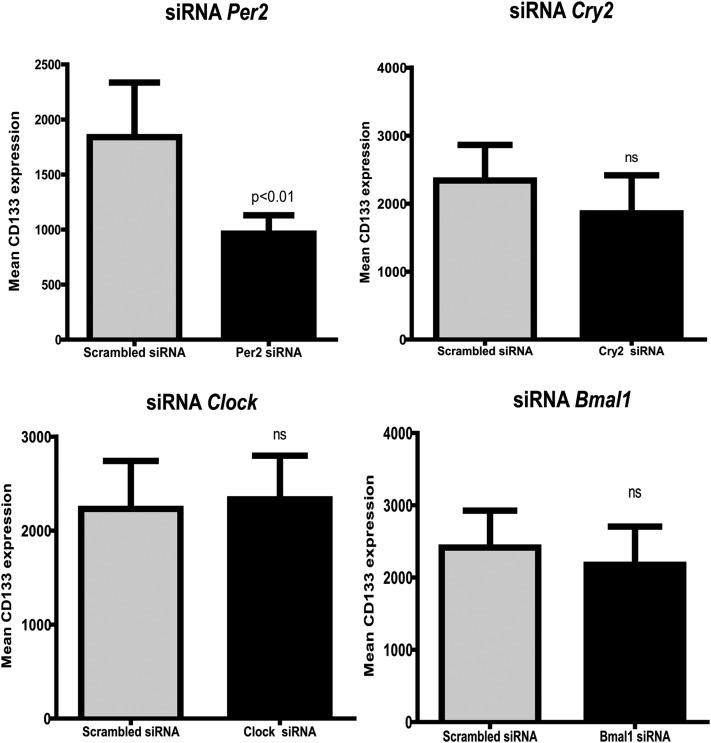
Clinical reports support a dampened clock gene response in metabolic (24) disorders; however, there are no reports describing the involvement of clock genes in regulating differentiation of CD34+ cells. To gain insights into this question, we placed CD34+ cells with small interfering RNAs (siRNAs) for the clock genes, Clock, Bmal1, Per2, and Cry2. siRNA for Clock, Bmal1, and Cry2 did not have any significant effect on expression of CD133. Only the silencing of Per2 had an effect on CD34+ cell differentiation, leading to a decrease in CD133 expression (P < 0.05) compared with treatment of CD34+ cells with control siRNA (Fig. 2). This suggested an important role of Per2 in maintaining the stem cell characteristics of CD34+ cells.
Figure 2.
Per2 is required to maintain expression of CD133 stem cell marker. Lin−CD34highCD45mid cells were treated with siRNAs specific to the clock genes clock, Bmal1, Per2, and Cry2 or a scrambled/control siRNA, and surface expression for CD133 was determined using flow cytometry. Bar chart showing that only Per2 is necessary to maintain primitive stem cell characteristics compared with the other clock genes Clock, Bmal1, and Cry2. n = 8. ns, not significant.
We next tested the role of clock genes in controlling endothelial differentiation by examining changes in CD144 expression. Silencing of the clock genes Clock, Bmal1, and Cry2 did not cause a change in CD144 expression (Supplementary Fig. 1).
Differentiation Toward an Endothelial Phenotype Alters the Clock Gene Oscillatory Pattern of CD34+ Cells
Previous studies indicate that differentiation of neural progenitor cells toward neurons was associated with clock gene oscillation (25). To determine whether in vitro differentiation influenced clock gene expression patterns in CD34+ cells, we propagated CD34+ cells in either undifferentiating or differentiating conditions. Cells were harvested every 4 h for 4 days, and mRNA expression for clock genes was determined using real-time quantitative RT-PCR (qRT-PCR). A rapid change in the clock gene Clock, Bmal1, Per1, Per2, Cry1, and Cry2 expression was observed. The cosine model, which uses a least squares method to fit a cosine wave to a time series study, was used to interpret clock gene expression (Fig. 3A). Each data point represents the experimentally observed mRNA expression patterns for clock genes, while the solid line shows the simultaneous fit for mRNA expression over 96 h. Per1, Per2, Cry1, and Cry2 (negative arm) exhibited rapid oscillatory patterns under endothelial-differentiating conditions (P < 0.05); however, the positive arm of clock gene expression represented by Clock and Bmal1 showed an overall decrease at the end of the 4-day time course (Fig. 3B).
Diabetic CD34+ Cells Fail to Maintain Stem Cell Characteristics or to Differentiate in Endothelial-Supporting Conditions
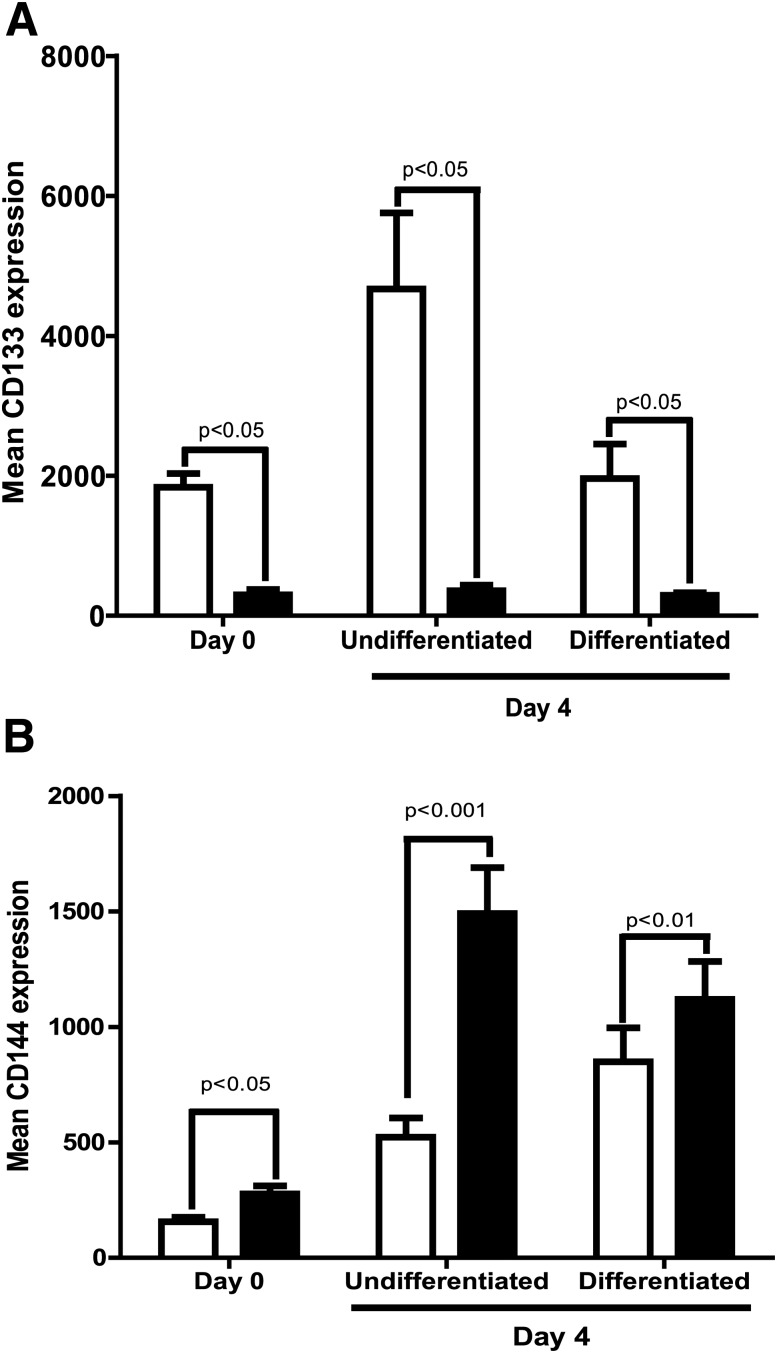
To test the relevance of our findings that clock genes influence the differentiation of CD34+ cells, we examined the differentiation pattern of diabetic CD34+ cells under the same experimental conditions described above for control (nondiabetic) cells. Diabetic CD34+ cells showed a significant reduction (P < 0.05) in baseline levels of CD133 and also failed to respond to conditions required to maintain the undifferentiated state (Fig. 4A). Exposure of diabetic CD34+ cells to promote differentiation failed to show a characteristic gain in differentiation marker CD144 under endothelial-supporting conditions (Fig. 4B), suggesting defects in the ability both to maintain “stemness” and to differentiate toward endothelial cells.
Figure 4.
Decrease in “stemness” and endothelial differentiation in diabetic CD34+ cells. Bar chart showing CD133 (A) and CD144 (B) surface expression of Lin−CD34highCD45mid population when first isolated and then when placed in vitro under conditions to maintain the undifferentiated state or the differentiated state (n = 4). □, control; ■, diabetes.
Differential Expression of miRNA Signature in Nondiabetic and Diabetic Progenitor Populations
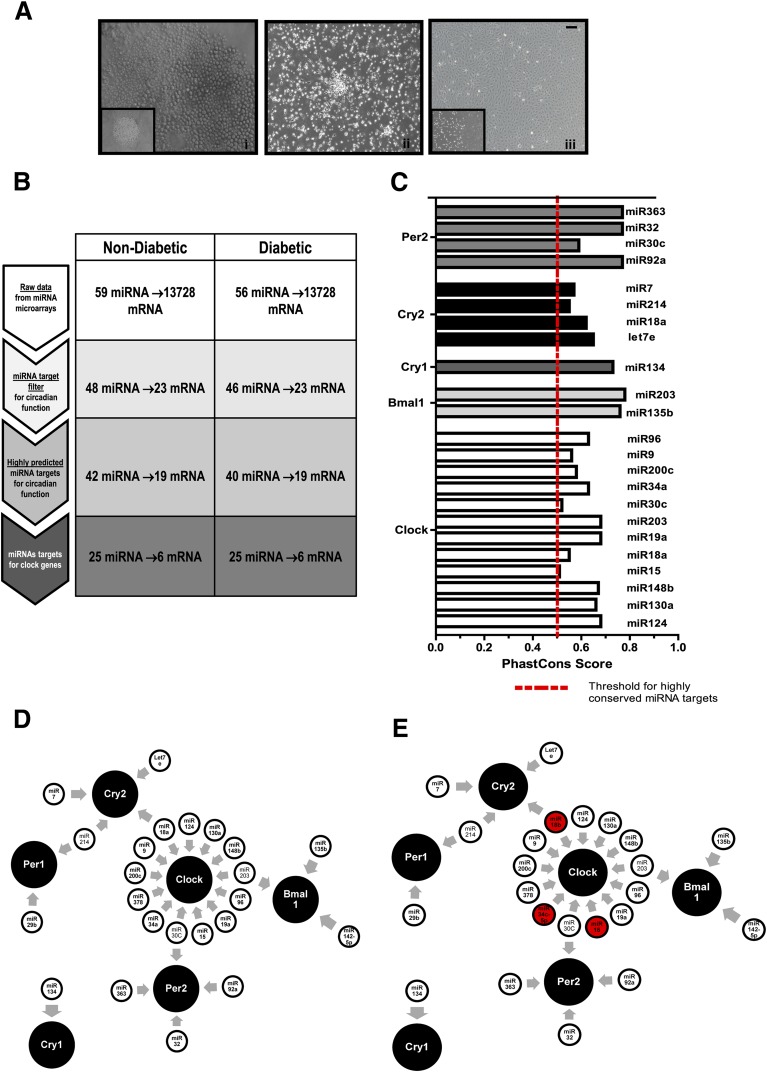
Emerging studies suggest a role for miRNA in regulation of physiological clock function (26). CD34+ cells obtained from peripheral blood and bone marrow as well as during development show a differential pattern of miRNA expression (27). To study the influence of miRNA expression and clock gene function on progenitor cell differentiation, we selected three interrelated but distinct populations of endothelial progenitors that exhibit clear differences in their propensities to differentiate to endothelial cells, CD34+ cells, eEPCs, or ECFCs (Fig. 5A). These three populations were generated from individuals with diabetes and age- and sex-matched individuals without diabetes. Microarrays for miRNAs were performed on these three populations using the Qiagen Human Serum & Plasma miRNA PCR Array. Data were analyzed using Ingenuity Pathway Analysis software. From both the control subjects and patients with diabetes, 59 miRNAs were mapped in the three different progenitor populations. With the use of the miRNA target filter for circadian function, 48 miRNAs targeting 23 mRNAs associated with circadian function were selected, and our database was filtered to enrich for only highly predicted miRNA targets associated with circadian function. Finally, 25 miRNAs were identified that were only associated with clock gene function. These miRNA specifically targeted six clock gene mRNAs (i.e., Clock, Bmal1, Per1, Per2, Cry1, and Cry2) (Fig. 5B). We further determined phastCons scores for individual miRNAs using a genome database (28). Only the miRNAs that crossed a threshold of 0.5 or 50% were considered highly predicted targets for respective clock gene mRNAs. Of the 25 filtered miRNAs, 20 miRNA passed this threshold with the exception of miR-15 and miR-30c; however, the phastCons score for miR-127-5p and miR-29b could not be determined owing to the unavailability of sufficient information in database (Fig. 5C). The miRNAs mapped by the three populations of progenitors are depicted in Fig. 5D along with their respective mRNA targets (Supplementary Table 1). Several unique miRNAs only target clock genes in diabetic progenitor populations and not in any of the nondiabetic populations. These were miR-18b (miR-17 family), miR-16 (miR-15 family), and miR-34c (miR-34 family).
Figure 5.
miRNA microarrays identify unique clock gene regulatory miRNAs in control and diabetic cells in three distinct stages of endothelial differentiation. A: Differentiation pattern and clock gene expression in freshly isolated CD34+ cells (i), cultured eEPCs (ii), and ECFCs (iii). B: Analysis scheme for miRNA microarray data using Ingenuity miRNA target analysis to predict highly significant targets for clock genes. C: Bar chart showing highly conserved miRNA targets expressed in terms of their phastCons score determined using a combination of miRNA target filter and mRNA expression pairing. D: Schematic showing highly predicted miRNAs for circadian target genes mapped by three progenitor populations with distinct capabilities to differentiate to endothelial cells. miRNAs identified only in the diabetic population are marked in red. Scale bar, 100 μm. The array plates were run at minimum in sextuplicate, with at least 6 unique samples for each sample category.
Differential Expression of miRNA in Nondiabetic and Diabetic Endothelial Progenitor Populations
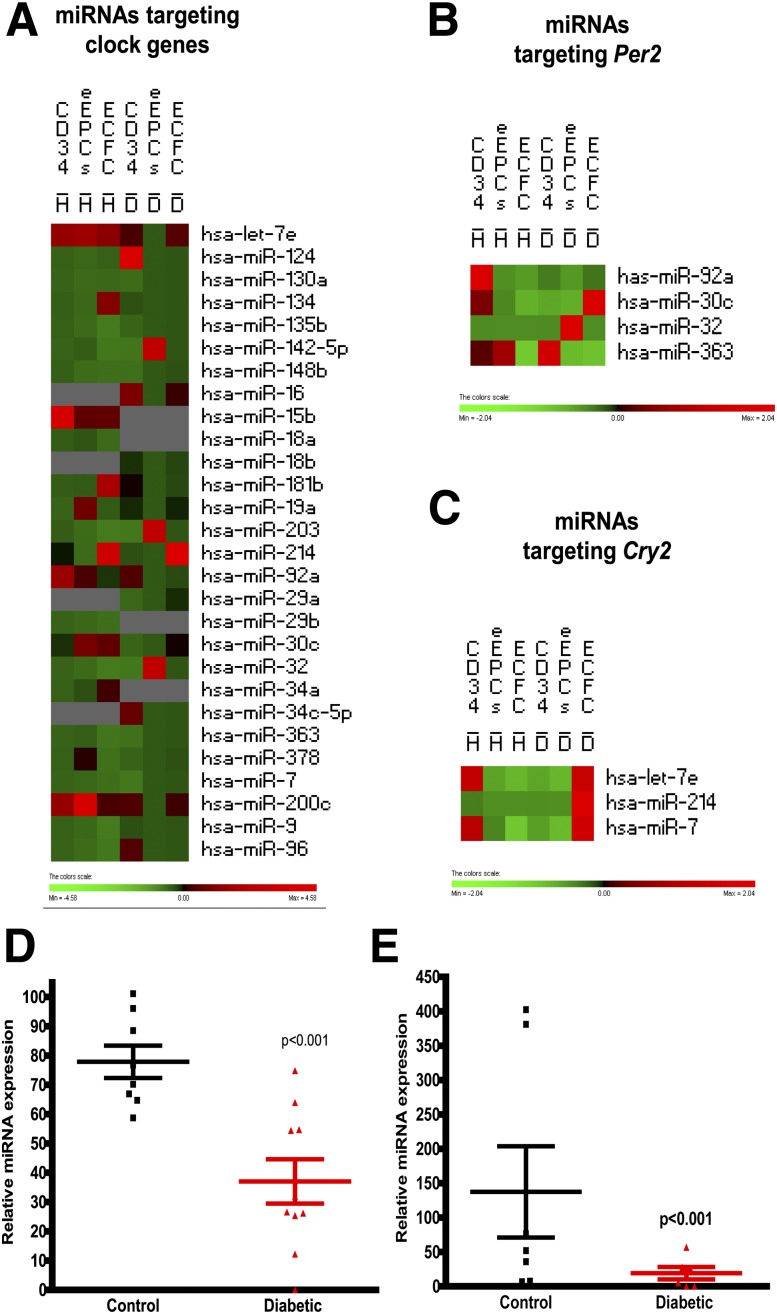
The enriched data set mapped by the three progenitor populations of diabetic or nondiabetic origin was further analyzed using PermutMatrix cluster analysis software. The change in expression pattern of miRNAs due to differentiation toward endothelial linage or due to diabetes was examined (29). Clustering identified candidate miRNAs that changed dramatically among these distinct progenitor populations, e.g., let-7e, miR-92a, miR-30c, and miR-200c (Fig. 6A).
Figure 6.
Differential expression of miRNA signatures in populations with diabetes and populations without diabetes with different propensities for endothelial differentiation. A: Heat map showing individual expression of miRNAs that are known to target clock genes in CD34 cells, eEPCs, and ECFCs. Highly altered miRNAs were further separated to identify unique miRNAs targeting Per2 (B) and Cry2 (C) mRNA. n = 3. D: qRT-PCR validating miRNA expression of miR-92a and let-7e. n = 8, control subjects without diabetes; n = 10, diabetes.
With the use of the combination of TargetScan (www.targetscan.org [30]) and cluster analysis, miRNAs for the mRNA targets Per2 (Fig. 6B) and Cry2 (Fig. 6C) were selected, and qRT-PCR was performed to validate the miRNA changes. For these studies, CD34+ cells were selected, as these cells are the most feasible for use in autologous cell therapy and to date the use of autologous CD34+ cells in patients with diabetes has been severely limited by the dysfunction of these cells. Using qRT-PCR, we showed a significant decrease in miR-92a (Fig. 6C) (P < 0.001), which is known to target Per2. We also observed a decrease in let-7e (Fig. 6D) (P < 0.001), which previously had been shown to possess circadian oscillations, predicted to repress Cry2 and known to be involved in glucose homeostasis and insulin sensitivity (31). We also validated miR-9, miR-96, miR-124, miR-135, and miR-363, which are known to regulate the clock genes Clock and Bmal1 (Supplementary Fig. 2). On the basis of the studies supporting a role of miR-92a in angiogenesis (11), we selected miR-92a as the candidate miRNA for additional examination.
Expression of miR-92a in CD34+ Cells of Long-term Patients With Diabetes With DR
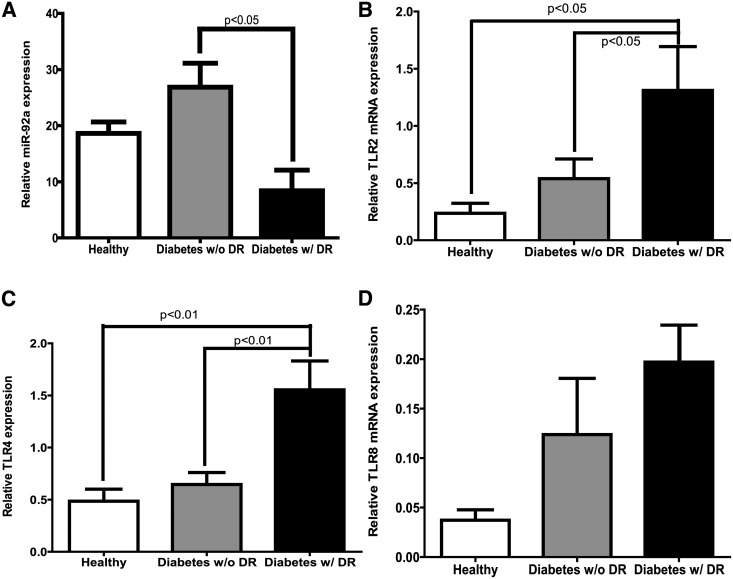
In order to understand the role of miR-92a in CD34+ cells, we selected a unique cohort of individuals who, despite long-standing poorly controlled diabetes of >40 years’ duration, remained free of DR (Diabetes w/o DR). We compared CD34+ cells from the “protected” patients with diabetes with individuals matched for age, sex, and duration of diabetes who developed DR (Diabetes w/DR) and with age- and sex-matched control subjects without diabetes. The evaluation of miR-92a expression showed a significant upregulation (P < 0.05) of miR-92a in Diabetes w/o DR compared with the patients that developed retinopathy (Fig. 7A). miR-92a is a negative regulator of inflammation modulating Toll-like receptors (TLR) to reduce inflammatory gene expression (32). Therefore, we next examined the miR-92a–TLR target gene network in the three cohorts and analyzed the data using Ingenuity Pathway Analysis. Our microarray studies have previously been published (33). The pathway analysis for TLR receptors showed downregulation in TLR2, TLR4, TLR5, TLR7, and TLR8 (Supplementary Fig. 3A), known targets of IL-1. We observed similar changes in TLR2 (Fig. 7B), TLR4 (Fig. 7C), and TLR8 (Fig. 7D) gene expression when the microarray results were validated using qRT-PCR. We also observed a profound increase in clock genes Bmal (Arntl), Per1, Per2, Cry1, and Cry2 in individuals protected from DR (Supplementary Fig. 3B).
Figure 7.
Patients with diabetes protected from DR show an upregulation in miR-92a and a decrease in TLR expression. A: Bar chart showing increase in miR-92a expression in Diabetes w/o DR compared with Diabetes w/DR. mRNA expression determined using real-time quantitative PCR showing decrease in expression of TLR2 (B), TLR4 (C), and TLR8 (D) in the Diabetes w/o DR group. n = 5, Diabetes w/o DR; n = 3, Diabetes w/DR; n = 5, control subjects without diabetes.
Overexpression of miR-92a Corrects Inflammation in Diabetic CD34+ Cells
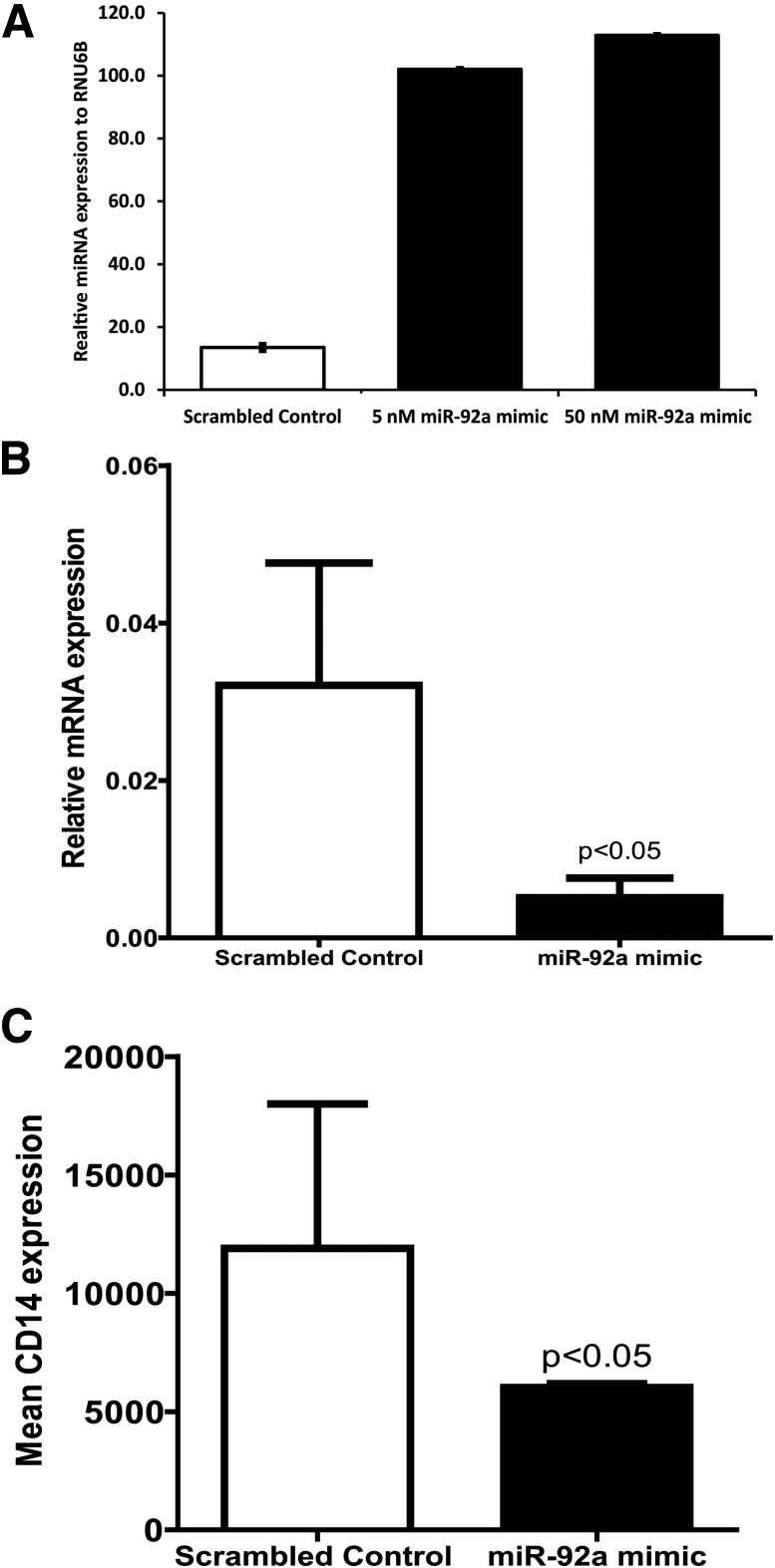
Overexpressing miR-92a in diabetic Lin−CD34+CD45+ cells was accomplished using an miR-92a mimic, and expression of miR-92a was determined using qRT-PCR. A significant increase in the concentration of miR-92a was observed at two concentrations (5 nmol/L and 50 nmol/L) of the miR-92a mimic tested (Fig. 8A). All additional studies were performed using the lower concentration of the miR-92a mimic. Next, we examined whether restoring optimum levels of miR-92a in diabetic CD34+ cells could correct known dysfunction, specifically, defects in proliferation and differentiation and changes in inflammatory gene expression in these cells. For this purpose, we treated diabetic CD34+ cells with miR-92a mimic and propagated the cells in methylcellulose semisolid media. At 10 days after culture, the numbers of colonies were enumerated using a light microscope. The morphology of the colonies was similar in both the control miRNA and the miR-92a mimic-treated groups (Supplementary Fig. 4A and B). Similarly, no effect of miR-92a overexpression on diabetic CD34+ cell proliferation was observed as determined by enumeration of colonies. Next, we tested whether differentiation of diabetic CD34+ cells was affected by miR-92a overexpression using the protocol described in Fig. 1A. Overexpression of miR-92a failed to show the expected decrease in CD133 expression under endothelial-differentiating conditions (Supplementary Fig. 5A). Treatment with the miR-92a mimic also did not cause a change in CD144 expression compared with control miRNA (Supplementary Fig. 5B).
Figure 8.
miR-92a overexpression downregulates inflammation and myeloid differentiation CD34+ cells obtained from patients with diabetes. A: miR-92a mimic (5 nmol/L) was overexpressed in CD34+ cells using Optimem and Lipofectamine 2000, and the miR-92a expression was evaluated using qRT-CPR. Bar chart showing upregulation in expression of miR-92a after treatment with miR-92a mimic. B: miR-92a mimic treatment showing downregulation in IL-1β mRNA expression. C: Bar chart showing surface expression of CD14 after treatment with miR-92a mimic determined using flow cytometry. n = 4.
Diabetic CD34+ cells exhibit paracrine dysfunction and are proinflammatory (34). We next studied the effect of miR-92a mimic treatment on the expression of inflammatory genes in these cells; we measured mRNA expression of a variety of cytokines in these cells. We could not detect IL-6 or IL-15 by qRT-PCR, while IL-8 and tumor necrosis factor (TNF)-α were detected in only 25% of samples. There was a significant decrease in expression of IL-1β after treatment with miR-92a mimic (Fig. 8B). Flow cytometry analysis revealed a significant decrease in surface expression of CD14 (Fig. 8C), while CD11a and CD13 expression remained unchanged (Supplementary Fig. 6).
Discussion
The salient findings of our study are that only Per2 is required for maintaining the undifferentiated state of CD34+ cells, while robust oscillations of clock genes are needed for differentiation toward the endothelial linage. Patients with diabetes who are protected from the development of DR maintain higher levels of miR-92a expression in their CD34+ cells compared with Diabetes w/DR. Importantly, restoration of miR-92a levels in CD34+ cells from patients with Diabetes w/DR reduces the inflammatory phenotype of these cells and the diabetes-induced propensity toward myeloid differentiation.
Several major regulators of differentiation include the Sox, Ets, Forkhead, GATA, and Kruppel-like families. Of these, the Ets family members are expressed in hematopoietic stem cells and have been shown to be under clock control. The nuclear binding of Ets is necessary to maintain the undifferentiated state in progenitor cells (35). Interestingly, the CLOCK/BMAL and CRY2 complex displaces Ets in the promoter region (36), suggesting that nuclear binding of the clock gene complex may promote progenitor differentiation by suppression of Ets transcription. However, studying the potential role of clock genes in endothelial transcription will require a robust experimental design. Specifically, the studies would need to ascertain molecular regulators of the respective clock genes and screen for a series of endothelial transcription factors followed by validation in animal studies.
A key determinant of clock gene function is posttranscriptional and posttranslational regulation. Seminal studies show that posttranslational modifications of PER2 determine the duration of the clock (5). In addition, emerging studies document that miRNA regulation is critical in defining clock gene expression. A variety of miRNAs such as miR-181d (clock, bmal) and miR-192/194 (Per1, Per2, and Per3) have previously been implicated in clock gene regulation (37). Our study highlights 25 key miRNAs targeting mRNAs of clock gene transcripts that influence the three populations of progenitors examined. We identify miR-18b, miR-16, and miR-34c-5p as unique to the diabetic cell populations. miR-18b has been identified as an important marker of cell proliferation and adhesion and as a diagnostic marker in individuals with hepatocellular carcinoma (38). Our study demonstrates that miR-18b, similar to miR-92a, may serve as an indicator of CD34+ cell dysfunction in diabetes. miR-16 plays a critical role in angiogenesis by targeting VEGF (39) and in cell-cycle control by targeting regulators of cell-cycle progression such as cyclin D1, cyclin E1, and cylcin-dependent kinase 1 (40). Interestingly, miR-16 is reduced in skeletal muscles of individuals with diabetes (41) and is involved in insulin resistance (42). Optimum restoration of miR-16 may correct diabetes-induced dysfunction in CD34+ cells; however, such experiments are beyond the scope of this study.
Another main finding of our study is the reduction of miR-92a, a Per2-regulatory miRNA, in CD34+ cells of individuals with DR. Previously, we reported that Per2 mutant mice show decreased colony-forming ability (3) suggesting the important role of Per2 in maintaining the stem cell nature of BMPCs. It would be of interest to determine whether miR-92a overexpression in Per2 mutant mice could help correct BMPC dysfunction and prevent retinopathy. Our study provides a basis for future studies in this direction.
miR-92a is activated in cancer cells including solid tumors and neuroblastomas but also is highly expressed in endothelial cells, acting as an endogenous repressor of angiogenesis by regulating mRNAs of proangiogenic protein integrin subunit α5 (3,11). miR-92a targets TLR, and forced overexpression of miR-92a using a mimic leads to a decrease in expression of inflammatory cytokines, IL-6 and TNF, in macrophages. This response of miR-92a is likely mediated via a c-Jun NH2-terminal kinase (JNK/c-Jun) pathway, which results in a decrease of TLR4 expression (32). In CD34+ cells of individuals with diabetes protected from DR, we observed a profound decrease in TLR2, TLR4, and TLR8 that was associated with an increase in miR-92a expression. We speculate that this unique cohort of protected individuals maintains high levels of miR-92a throughout the course of their disease, which prevents the TLR-mediated inflammatory response typically seen in the CD34+ cells of patients with diabetes with DR. Previously, we reported that CD34+ cells from patients with diabetes protected from DR exhibit other unique phenotypic characteristics including a reduction of transforming growth factor-β1 (33), modulation of the renin angiotensin system (43), and protection of their long-term repopulation ability (under revision). Together, these likely represent the mechanisms for the sustained vascular repair capacity observed in the CD34+ cells isolated from the individuals with diabetes protected from DR.
CD34+ cells of diabetic origin express a proinflammatory secretome compared with cells from healthy control subjects (21). Diabetic CD34+ cells show a dramatic decrease in proangiogenic growth factors (e.g., SCF, HGF, TPO), which are known to be involved in proliferation, differentiation, and migration of CD34+ cells (21). Interestingly, inflammatory molecules such as TNF-α and IL-1β possess a diurnal rhythm, and sleep deprivation results in an increase of these cytokines (44). We speculate that the autocrine production of inflammatory factors such as TNF-α and IL-1β by diabetic CD34+ cells may be responsible for the defective differentiation observed in these cells.
In CD34+ cells of individuals with diabetes protected from DR, we observed a profound decrease in TLR2, TLR4, and TLR8 that was associated with an increase in miR-92a expression. CD34+ cells widely express TLR4, -7, and -8 receptors. Activation of these receptors results in biased differentiation toward more committed monocytic and macrophagic cells characterized by an increase in expression of CD13, CD11c, and CD14 (45). Normally, there is a perfect balance between myeloid, lymphoid, and stem cell numbers; however, with diabetes this balance shifts toward more myeloid and fewer lymphoid cells (46). Our results would suggest that miR-92a downregulation is linked to this imbalance. Restoration of optimum miR-92a would not only correct myeloid bias but also reduce inflammation; thus, we believe that the protective effect of miR-92a in diabetes is possibly mediated via reduced activation of TLR receptors.
Our miRNA validation and subsequent miR-92a overexpression studies focused on diabetic CD34+ cells because CD34+ cells possess the greatest translational potential due to their ease of isolation, which requires minimum manipulation, unlike eEPCs or ECFCs, which require ex vivo expansion. CD34+ cells serve as “reporter” cells that can gauge diabetes-induced dysfunction (47). In these cells, this dysfunction represents multiple defects including the shift to a myeloid phenotype characterized by a rapid expression of dendritic markers such as CD11c and CD14 (48,49).
Taken together, we consider that the shift of healthy nondiabetic CD34+ progenitor cells toward a more differentiated state is a dynamic process in which clock genes play an integral role. At the nuclear level, clock genes promote transcription of endothelial differentiation genes, thus reducing stem cell characteristics, while at a posttranscriptional level a variety of miRNAs regulating clock genes may augment this endothelial differentiation phenotype. Unanswered questions remain, including the nature of the transcription factors that bind to the clock genes that orchestrate these events and whether overexpression of miR-92a can correct endothelial dysfunction in Per2 mutant mice or miR-16 can correct the dysfunction of CD34+ cells isolated from patients with diabetes with vascular complications.
While acknowledging the limitations of this study, we have shown an important role of clock genes in the phenotypic shift of progenitors toward a more differentiated phenotype and that unique miRNAs can regulate this process. In an era that is bringing stem cell therapies to the clinics, use of miR-92a mimics may restore diabetic CD34+ cells toward functional progenitors with enhanced reparative capacity. Furthermore, our study has identified TLR as a potential target regulated by miR-92a. miR-92a may serve to correct diabetes-associated inflammation as well as restore normal circadian function in CD34+ cells, enhancing their therapeutic potential.
Article Information
Acknowledgments. The authors thank Mrs. Amy Davis, University of Florida, for her support as clinical coordinator for this study.
Funding. This work was supported by an American Heart Association Postdoctoral Fellowship (09POST2380125), and the Ralph and Grace Showalter Trust Fund (to A.D.B.); National Institutes of Health (NIH), National Eye Institute (NEI) 1RC1EY020341-01 and 1R43EY020030-01 (to S.B.) and EY022091-01 (to B.C.); and NIH, National Heart, Lung, and Blood Institute, HL-110170-03, NEI R01EY007739-23 and R01EY012601-15, and NIH R01-DK-090730-04 to M.B.G. This work is also supported by an unrestricted award from the Research to Prevent Blindness Foundation to the Department of Ophthalmology, Indiana University.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. A.D.B. researched data and wrote the manuscript. Y.Y. helped with the cosine model. V.S., S.H., and M.K. generated data. S.B. reviewed the manuscript. B.C. performed miRNA microarrays and contributed to the discussion. M.B.G. contributed to the discussion and reviewed and edited the manuscript. A.D.B. and M.B.G. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db15-0521/-/DC1.
References
- 1.Miller BH, McDearmon EL, Panda S, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A 2007;104:3342–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeda N, Maemura K, Horie S, et al. Thrombomodulin is a clock-controlled gene in vascular endothelial cells. J Biol Chem 2007;282:32561–32567 [DOI] [PubMed] [Google Scholar]
- 3.Bhatwadekar AD, Yan Y, Qi X, et al. Per2 mutation recapitulates the vascular phenotype of diabetes in the retina and bone marrow. Diabetes 2013;62:273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcheva B, Ramsey KM, Buhr ED, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 2010;466:627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 2001;107:855–867 [DOI] [PubMed] [Google Scholar]
- 6.Busik JV, Tikhonenko M, Bhatwadekar A, et al. Diabetic retinopathy is associated with bone marrow neuropathy and a depressed peripheral clock. J Exp Med 2009;206:2897–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viswambharan H, Carvas JM, Antic V, et al. Mutation of the circadian clock gene Per2 alters vascular endothelial function. Circulation 2007;115:2188–2195 [DOI] [PubMed] [Google Scholar]
- 8.Wang CY, Wen MS, Wang HW, et al. Increased vascular senescence and impaired endothelial progenitor cell function mediated by mutation of circadian gene Per2. Circulation 2008;118:2166–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anea CB, Cheng B, Sharma S, et al. Increased superoxide and endothelial NO synthase uncoupling in blood vessels of Bmal1-knockout mice. Circ Res 2012;111:1157–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suárez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res 2009;104:442–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonauer A, Carmona G, Iwasaki M, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 2009;324:1710–1713 [DOI] [PubMed] [Google Scholar]
- 12.Ohyashiki JH, Umezu T, Kobayashi C, et al. Impact on cell to plasma ratio of miR-92a in patients with acute leukemia: in vivo assessment of cell to plasma ratio of miR-92a. BMC Res Notes 2010;3:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res 2008;36:D149–D153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Val S, Black BL. Transcriptional control of endothelial cell development. Dev Cell 2009;16:180–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCall MN, Kent OA, Yu J, Fox-Talbot K, Zaiman AL, Halushka MK. MicroRNA profiling of diverse endothelial cell types. BMC Med Genomics 2011;4:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Falco E, Porcelli D, Torella AR, et al. SDF-1 involvement in endothelial phenotype and ischemia-induced recruitment of bone marrow progenitor cells. Blood 2004;104:3472–3482 [DOI] [PubMed] [Google Scholar]
- 17.Ramos AL, Darabi R, Akbarloo N, et al. Clonal analysis reveals a common progenitor for endothelial, myeloid, and lymphoid precursors in umbilical cord blood. Circ Res 2010;107:1460–1469 [DOI] [PubMed] [Google Scholar]
- 18.Yoder MC, Mead LE, Prater D, et al. Redefining endothelial progenitor cells via clonal analysis and hematopoietic stem/progenitor cell principals. Blood 2007;109:1801–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingram DA, Mead LE, Tanaka H, et al. Identification of a novel hierarchy of endothelial progenitor cells using human peripheral and umbilical cord blood. Blood 2004;104:2752–2760 [DOI] [PubMed] [Google Scholar]
- 20.Crosby CV, Fleming PA, Argraves WS, et al. VE-cadherin is not required for the formation of nascent blood vessels but acts to prevent their disassembly. Blood 2005;105:2771–2776 [DOI] [PubMed] [Google Scholar]
- 21.Jarajapu YP, Hazra S, Segal M, et al. Vasoreparative dysfunction of CD34+ cells in diabetic individuals involves hypoxic desensitization and impaired autocrine/paracrine mechanisms. PLoS One 2014;9:e93965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson W, Tong YL, Lee JK, Halberg F. Methods for cosinor-rhythmometry. Chronobiologia 1979;6:305–323 [PubMed] [Google Scholar]
- 23.Yin AH, Miraglia S, Zanjani ED, et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood 1997;90:5002–5012 [PubMed] [Google Scholar]
- 24.Ando H, Takamura T, Matsuzawa-Nagata N, et al. Clock gene expression in peripheral leucocytes of patients with type 2 diabetes. Diabetologia 2009;52:329–335 [DOI] [PubMed] [Google Scholar]
- 25.Kimiwada T, Sakurai M, Ohashi H, Aoki S, Tominaga T, Wada K. Clock genes regulate neurogenic transcription factors, including NeuroD1, and the neuronal differentiation of adult neural stem/progenitor cells. Neurochem Int 2009;54:277–285 [DOI] [PubMed] [Google Scholar]
- 26.Chen R, D’Alessandro M, Lee C. miRNAs are required for generating a time delay critical for the circadian oscillator. Curr Biol 2013;23:1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin P, Wang E, Ren J, et al. Differentiation of two types of mobilized peripheral blood stem cells by microRNA and cDNA expression analysis. J Transl Med 2008;6:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 2005;15:1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caraux G, Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics 2005;21:1280–1281 [DOI] [PubMed] [Google Scholar]
- 30.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15–20 [DOI] [PubMed] [Google Scholar]
- 31.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A 2011;108:21075–21080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai L, Song Y, Liu Y, et al. MicroRNA-92a negatively regulates Toll-like receptor (TLR)-triggered inflammatory response in macrophages by targeting MKK4 kinase. J Biol Chem 2013;288:7956–7967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hazra S, Stepps V, Bhatwadekar AD, et al. Enhancing the function of CD34(+) cells by targeting plasminogen activator inhibitor-1. PLoS One 2013;8:e79067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarajapu YP, Grant MB. The promise of cell-based therapies for diabetic complications: challenges and solutions. Circ Res 2010;106:854–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seeger FH, Chen L, Spyridopoulos I, Altschmied J, Aicher A, Haendeler J. Downregulation of ETS rescues diabetes-induced reduction of endothelial progenitor cells. PLoS One 2009;4:e4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kondratov RV, Shamanna RK, Kondratova AA, Gorbacheva VY, Antoch MP. Dual role of the CLOCK/BMAL1 circadian complex in transcriptional regulation. FASEB J 2006;20:530–532 [DOI] [PubMed]
- 37.Mehta N, Cheng HY. Micro-managing the circadian clock: The role of microRNAs in biological timekeeping. J Mol Biol 2013;425:3609–3624 [DOI] [PubMed] [Google Scholar]
- 38.Murakami Y, Tamori A, Itami S, et al. The expression level of miR-18b in hepatocellular carcinoma is associated with the grade of malignancy and prognosis. BMC Cancer 2013;13:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun CY, She XM, Qin Y, et al. miR-15a and miR-16 affect the angiogenesis of multiple myeloma by targeting VEGF. Carcinogenesis 2013;34:426–435 [DOI] [PubMed] [Google Scholar]
- 40.Goretti E, Rolland-Turner M, Léonard F, Zhang L, Wagner DR, Devaux Y. MicroRNA-16 affects key functions of human endothelial progenitor cells. J Leukoc Biol 2013;93:645–655 [DOI] [PubMed] [Google Scholar]
- 41.Bork-Jensen J, Scheele C, Christophersen DV, et al. Glucose tolerance is associated with differential expression of microRNAs in skeletal muscle: results from studies of twins with and without type 2 diabetes. Diabetologia 2015;58:363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye EA, Steinle JJ. miR-15b/16 protects primary human retinal microvascular endothelial cells against hyperglycemia-induced increases in tumor necrosis factor alpha and suppressor of cytokine signaling 3. J Neuroinflammation 2015;12:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jarajapu YP, Bhatwadekar AD, Caballero S, et al. Activation of the ACE2/angiotensin-(1-7)/Mas receptor axis enhances the reparative function of dysfunctional diabetic endothelial progenitors. Diabetes 2013;62:1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krueger JM, Fang J, Taishi P, Chen Z, Kushikata T, Gardi J. Sleep. A physiologic role for IL-1 beta and TNF-alpha. Ann N Y Acad Sci 1998;856:148–159 [DOI] [PubMed] [Google Scholar]
- 45.Sioud M, Fløisand Y, Forfang L, Lund-Johansen F. Signaling through toll-like receptor 7/8 induces the differentiation of human bone marrow CD34+ progenitor cells along the myeloid lineage. J Mol Biol 2006;364:945–954 [DOI] [PubMed] [Google Scholar]
- 46.Muller-Sieburg C, Sieburg HB. Stem cell aging: survival of the laziest? Cell Cycle 2008;7:3798–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fadini GP, Avogaro A. Diabetes impairs mobilization of stem cells for the treatment of cardiovascular disease: a meta-regression analysis. Int J Cardiol 2013;168:892–897 [DOI] [PubMed] [Google Scholar]
- 48.Rondelli D, Lemoli RM, Ratta M, et al. Rapid induction of CD40 on a subset of granulocyte colony-stimulating factor-mobilized CD34(+) blood cells identifies myeloid committed progenitors and permits selection of nonimmunogenic CD40(-) progenitor cells. Blood 1999;94:2293–2300 [PubMed] [Google Scholar]
- 49.Ferrero E, Bondanza A, Leone BE, Manici S, Poggi A, Zocchi MR. CD14+ CD34+ peripheral blood mononuclear cells migrate across endothelium and give rise to immunostimulatory dendritic cells. J Immunol 1998;160:2675–2683 [PubMed] [Google Scholar]