Abstract
Hypoxia induces a complex circuit of gene expression that drives tumor progression and increases drug resistance. Defining these changes allows for an understanding of how hypoxia alters tumor biology and informs design of lead therapeutics. We probed the role of microRNA-544 (miR-544), which silences mammalian target of rapamycin (mTOR), in a hypoxic breast cancer model by using a small molecule (1) that selectively impedes the microRNA's biogenesis. Application of 1 to hypoxic tumor cells selectively inhibited production of the mature microRNA, sensitized cells to 5-fluorouracil, and derepressed mRNAs affected by miR-544 in cellulo and in vivo, including boosting mTOR expression. Thus, small molecule inhibition of miR-544 reverses a tumor cell's physiological response to hypoxia. Importantly, 1 sensitized tumor cells to hypoxia-associated apoptosis at a 25-fold lower concentration than a 2′-O-methyl RNA antagomir and was as selective. Further, the apoptotic effect of 1 was suppressed by treatment of cell with rapamycin, a well-known inhibitor of the mTOR signaling pathway, illustrating the selectivity of the compound. Thus, RNA-directed chemical probes, which could also serve as lead therapeutics, enable interrogation of complex cellular networks in cells and animals.
A hallmark of tumorigenesis is the rapid, uncontrolled proliferation of incipient cancer cells due to aberrant cell cycle regulation.(1) Uncontrolled cell growth creates a hypoxic microenvironment with the emerging tumor, adaptation to which is essential for tumor progression. The transcription factor HIF-1α, which is induced in response to hypoxia, orchestrates cellular responses in tumor cells that promote survival in a low oxygen environment.(2) For example, HIF-1α induces expression of Ataxia telangiectasia mutated (ATM) protein and represses mammalian target of rapamycin complex 1 (mTORC1) activity, thereby restricting cell growth and metabolism in response to low oxygen.(3) HIF-1α also functions as a potent inducer of the epithelial-to-mesenchymal transition (EMT), which imparts tumor cells with invasive properties that allow escape (metastasis) from the low oxygen environment.(4) Importantly, emerging evidence suggests that a period of hypoxic stress occurs almost universally during tumorigenesis, and adaptation of tumor cells to hypoxia selects for drug resistant and metastatic phenotypes.(2, 5) Indeed, recent studies indicate that over 50% of locally advanced breast cancers have hypoxic regions, which reduces the overall effectiveness of chemotherapy and radiation.(6, 7) Consequently, probing hypoxia-responsive pathways provides a means to identify novel drug targets that can be exploited to disrupt pathways that drive metabolic adaptation of tumor cells to hypoxic stress, thus preventing progression of emerging tumors and their metastasis and relapse after therapy.
Previously, we identified a cluster of seven microRNAs (miRNAs; miRs-300, 382, 494, 495, 539, 543, and 544) within the imprinted DLK1-DIO3 locus that target proteins involved in tumor metastasis and showed that expression of this miRNA cluster is dysregulated in invasive ductal carcinomas.(8) To further interrogate the role of miR-544 in tumor cell growth under hypoxic and normoxic conditions, we designed a small molecule that inhibits miR-544 biogenesis using a strategy named Inforna.(9) Using this small molecule probe to interrogate miR-544 function, we demonstrate that miR-544 is strongly induced in tumor cells in response to hypoxia and directly represses expression of mTOR. Although HIF-1α, ATM, and miR-544 act synergistically to maintain a hypoxic state (see Figure 8), we show that inhibition of miR-544 processing with the small molecule (i) selectively induced apoptosis in triple negative breast cancer cells in response to hypoxic stress, (ii) sensitized these cells to 5-fluorouracil in vitro, and (iii) blocked their growth in vivo when transplanted to immunodeficient mice. Importantly, the small molecule targeting miR-544 elicited effects similar to that seen using a synthetic miR-544 antagomir but at a 25-fold lower concentration. Collectively, these studies illustrate that small molecule modulation of miRNA function can exert physiological responses in cancer and have broad applications in the design of RNA-targeting small molecule chemical probes and lead therapeutics.
Results and Discussion
MiR-544 Regulates mTOR expression
Previously, we reported that miR-544 modulates tumor cell growth by regulating the ATM pathway.(8) Both ATM and mTOR signaling pathways play critical roles in metabolic adaptation of tumor cells to hypoxic stress.(3, 10, 11) To demonstrate a role for miR-544 in this process, we quantified expressed levels of miR-544, ATM, mTOR, and HIF-1α in the breast adenocarcinoma cell lines MDA-MB-231 and MCF-7 and nontumorigenic mammary epithelial cell line MCF-10A under hypoxic (<1% O2) and normoxic (21% O2) conditions. Quantitative real time PCR (qRT-PCR) revealed significant induction of miR-544, HIF-1α, and ATM and significant down-regulation of mTOR transcripts in both tumor cell lines following exposure to hypoxia (Figure 1a). Additionally, Western blot analysis confirmed that hypoxic conditions induced accumulation of HIF-1α in cells (Supporting Information Figure 1). In contrast, no significant change in miR-544 or mTOR expression was observed in MCF-10A cells exposed to hypoxia as compared to normoxia, although HIF-1α and ATM were induced by low oxygen. A bioinformatics analysis of the 3′ untranslated region (UTR) of mTOR revealed two potential miR-544 binding sites (Figure 1b). Using a luciferase reporter fused to the 3′ UTR of mTOR, we confirmed that miR-544 represses mTOR expression (Figure 1c). These studies, together with our previous findings,(8) demonstrate that miR-544 modulates ATM and mTOR activity in response to hypoxia, confirming its role in metabolic adaptation of tumor cells to hypoxic stress. Based on these data, we hypothesized that inhibition of miR-544 biogenesis could short circuit this adaptive response, thereby inhibiting tumorigenesis.
Figure 1.
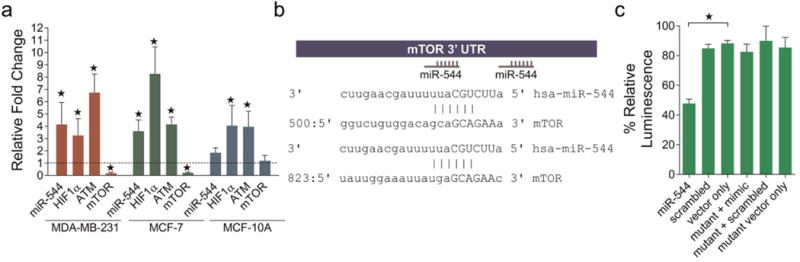
Induction of MiR-544 by hypoxia in tumor cells and direct repression of expression of mTOR. (a) Real-time qPCR analysis of the indicated transcripts in MDA-MB-231, MCF-7, and MCF-10A cell lines cultured under hypoxic conditions (1% O2). Data represent fold change compared to normoxic conditions (21% O2, dashed line). Errors bars represent 95% confidence interval. *, p < 0.05. (b) Two hexameric miR-544 binding sites in the 3′ UTR of mTOR were identified by using the miRanda microRNA target prediction algorithm. (c) Luciferase assay demonstrating inhibition of mTOR expression by miR-544. Mutant indicates base pair mutations in the 3′ UTR of mTOR. Error bars, SEM. *, p < 0.05.
Design of RNA-Targeting Small Molecules and Assessment of Lead Compound Activities Using a Novel Cell-based Screening Platform
The most common approach to targeting RNA is to use Watson–Crick base complementarity to design antisense or siRNA oligonucleotides. As an alternative, we employed a rational design-based approach(9) to identify small molecules that block miR-544 function by direct binding to the precursor miRNA. Briefly, the miR-544 hairpin precursor's secondary structure (Figure 2a) was obtained from miRbase, parsed into its composite motifs, then compared to a database of RNA motif–small molecule binding partners using a computational approach termed Inforna.(9) The hypothesis is that if compounds are identified that bind to these isolated motifs that they could bind to the biological RNA target and affect function.
Figure 2.
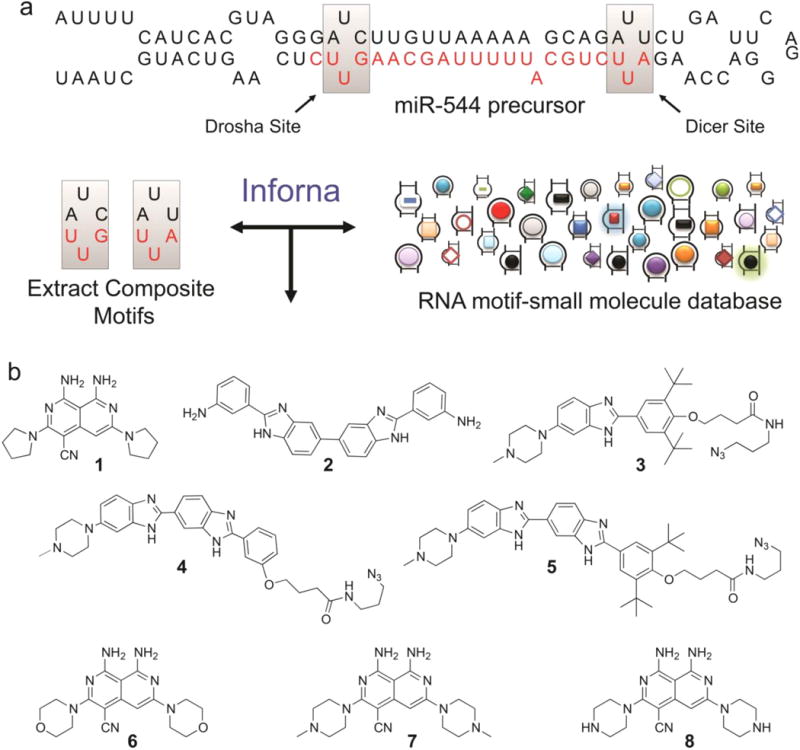
Identification of small molecules that impede miR-544 biogenesis via direct binding. (a) Hairpin structure of miR-544 precursor showing 1 × 1 nucleotide UU internal loops located in the Drosha and Dicer processing sites targeted by 1 and 2. (b) Compounds (1–5) identified from similarity searches between miR-544 precursor motifs and a fully characterized set of RNA motif small molecule interactions. Three scaffold modified derivatives of compound 1 (6–8) in which the pyrrolidine is substituted with morpholine, methylpiperazine, or piperazine.
In this application of Inforna, we queried over 900 small molecule–RNA motif binding partners to identify compounds that are predicted to bind the 1 × 1 nucleotide UU internal loops present in Dicer and Drosha processing sites within the miR-544 hairpin precursor. We previously showed that small molecule binding to processing sites can inhibit miRNA biogenesis;(9) thus binding of small molecules to the Dicer or Drosha processing sites in miR-544 could reduce levels of the mature miRNA and increase levels of its downstream targets such as mTOR by derepressing the effect of the microRNA on this target. No exact hits for either the Dicer of Drosha site motifs were identified when one includes both the 1 × 1 nucleotide internal loop and the internal loop's closing base pairs. Based on previous studies that showed the closing base pairs may or may not affect binding,(12, 13) we utilized Inforna to identify lead compounds that can target any RNA 1 × 1 nucleotide UU internal loop. These compounds would then be screened and tested for modulation of microRNA 544's biogenesis. Five potential heterocyclic lead small molecules (1–5; Figure 2b) were identified using this approach. Compounds 1 and 2 were previously shown to inhibit dysfunction of expanded r(CUG) repeats (r(CUG)exp), which fold into a hairpin structure with repeating 1 × 1 nucleotide internal loops and cause myotonic dystrophy type 1.(14, 15) Likewise, 4 was assembled into multivalent compounds that inhibit r(CUG)exp dysfunction.(16) It is important to note that although compounds 1, 2, and 4 do bind r(CUG)exp linked to myotonic dystrophy and have shown efficacy in treating the disease, these repeats are only found in individuals with myotonic dystrophy type 1(17) and thus do not represent a potential off-target. Further, our studies described below suggest that 1 selectively targets the miR-544 hairpin precursor, vide infra. Compound 3 binds a UU loop in miR-96 and inhibits its biogenesis.(9) Because heterocyclic, drug-like compounds were identified, there is the potential of being able to modify these compounds, if needed, to enhance binding or other pharmacological properties more easily than compared to, for example, aminoglycosides.
To assess the bioactivity and in cellulo selectivity of our lead small molecule for miR-544, we developed a novel fluorescent-based miRNA-drug interaction screening platform, pGRB-miR, capable of high-throughput application. The pGRB-miR vector expresses (i) the intergenic region coding for the miR-544 pre-miRNA sequence downstream of a GFP reporter (CMV promoter), (ii) the 3′ UTR from BMI1, a validated miR-544 target,(8) fused to an RFP reporter (PGK promoter), and (iii) a BFP reporter that serves as an internal transfection normalization control (SV40 promoter; Figure 3a; Supporting Information Figures 2 and 3). This configuration identifies miR-544 small molecule inhibitors or stabilizers based on changes in the RFP/BFP fluorescence ratio and serves as an effective and novel tool to identify selective miRNA modulators.
Figure 3.
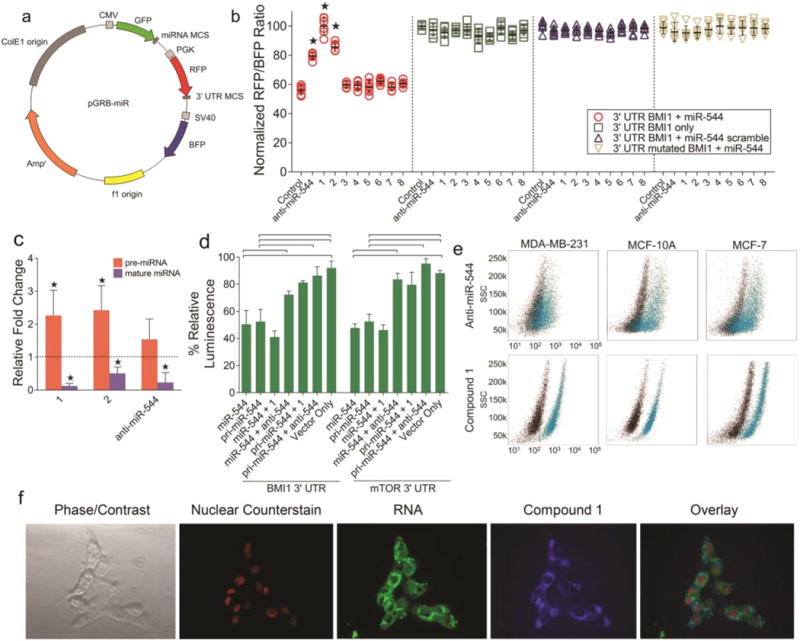
Cell-based screening platform to evaluate activities of lead small molecules. (a) Schematic representation of the pGRB-miR fluorescent drug screening vector system. (b) Effect of compounds 1–8 or a miR-544 antagomir on the relative RFP/BFP ratio expressed from pGRB-miR. Error bars, s.e.m. *, p < 0.05. (c) Real-time qPCR fold change analysis of precursor and mature miR-544 levels in MCF-10A cells treated with or without 1, 2, or a miR-544 antagomir. Errors bars represent 95% confidence interval. *, p < 0.05. (d) Luciferase-based secondary screens showing 1 inhibits precursor but not mature miR-544 binding to BMI1 (e) or mTOR (f) 3′-UTR. Errors bars, s.e.m. and brackets, p < 0.05. (e) Flow cytometric analysis of 1 (20 nM) or a FITC-labeled miR-544 antagomir (500 nM) uptake in MDA-MB-231, MCF-7, and MCF-10A cells. black, untreated; blue, drug treated or transfected cells. (f) Fluorescent microscopy of MDA-MB-231 cells treated with 20 nM of 1 demonstrating localization of 1 to the cytoplasmic space and nuclear exclusion.
We confirmed that 1 and 2 disrupt miR-544-mediated inhibition of BMI1 based on significant increases in the RFP/BFP ratio using this platform (Figure 3b). Moreover, they had no effect on fluorescent output from orthologous screening vectors lacking the pre-miR-544 or BMI1 3′ UTR sequences, indicating that the probes target the miRNA rather than the interaction between the miRNA and mRNA target sequence. A miR-544 antagomir also increased the RFP/BFP ratio of pGRB-miR but to a lesser extent than 1 and 2 and also had no effect on orthologous screening controls. To further assess the structure–activity relationships of 1, we made several derivatives of this structure. We synthesized three derivatives of 1 in which the pyrrolidine was substituted with morpholine, methylpiperazine, or piperazine (6–8, Figure 2b and Supporting Information Figure 4).(18, 19) Each of these compounds that have various ring substitutions were not as bioactive as the parent compound.
Consistent with these data, qRT-PCR analysis of MCF-10A cells treated with 1 or 2 resulted in accumulation of pre-miR-544 and a decrease in miR-544 levels (Figure 3c). In contrast, a synthetic miR-544 antagomir reduced levels of the mature miRNA but had no effect on precursor levels. Herein, 2 was less effective at reducing miR-544 levels in cellulo as compared to 1. Consequently, we employed a secondary luciferase-based screening assay to directly verify biogenesis inhibition of the mature miRNA by analyzing the effect of 1 on the activity of either the pri-miRNA or the mature forms of the miRNA using multiple downstream 3′ UTR target sequences. Indeed, 1 selectively inhibited the function of pri-miR-544 but not miR-544, preventing down-regulation of luciferase fused to either the 3′ UTR of BMI1 or mTOR, indicating the mechanism of action is a block in miRNA processing (Figure 3d). The siRNA inhibition of Dicer in the presence of 1 prevented pre-miR-544 down-regulation of mTOR, indicating that the mechanism of inhibition of 1 is Dicer-dependent (Supporting Information Figure 5).
One of the major hindrances to using RNAi as a mechanism to probe molecular pathways is poor cellular uptake and the requirement for cellular delivery of these compounds to be enabled by transfection agents.(20, 21) The issue is confounded in patients due to a variety of reasons. First, blood RNases, as presented by certain malignancies such as breast cancer, can rapidly degrade the oligonucleotide.(22, 23) Second, tumor microenvironments contain a complex mixture of cell types that can have very different transfection efficiencies. Therefore, we compared the uptake of 1, which is fluorescent, to a fluorescein-labeled anti-miR-544 antagomir. Flow cytometric analysis confirmed that the uptake of 1 into MDA-MB-231, MCF-7, and MCF-10A cells was highly efficient and exceeded that of the dye-labeled miR-544 antagomir, which was transfected into cells using lipofectamine (Figure 3e). The antagomir treated cell lines contained significant populations of cells that failed to take up the antisense oligonucleotide (33% of MCF-7 cells, 44% of MDA cells, and 21% of MCF-10A cells, respectively). Additionally, the uptake of 1 was confirmed by fluorescent microscopy, which showed general localization to the cytoplasmic space (Figure 3f) indicating inhibition of Dicer processing as the primary mechanism of miRNA biogenesis ablation.
Affinity and Selectivity of 1
Next, we measured the affinity of 1 for RNA constructs that contain Dicer or Drosha sites in the miR-544 hairpin precursor and to a control RNA in which the 1 × 1 nucleotide UU loop in the Dicer site is replaced with an AU base pair (Supporting Information Figure 6). These studies showed that 1 bound selectively to RNAs containing the 1 × 1 nucleotide internal loops with binding constants in the midnanomolar range (Figure 4a) but failed to bind to the control RNA constructs.
Figure 4.
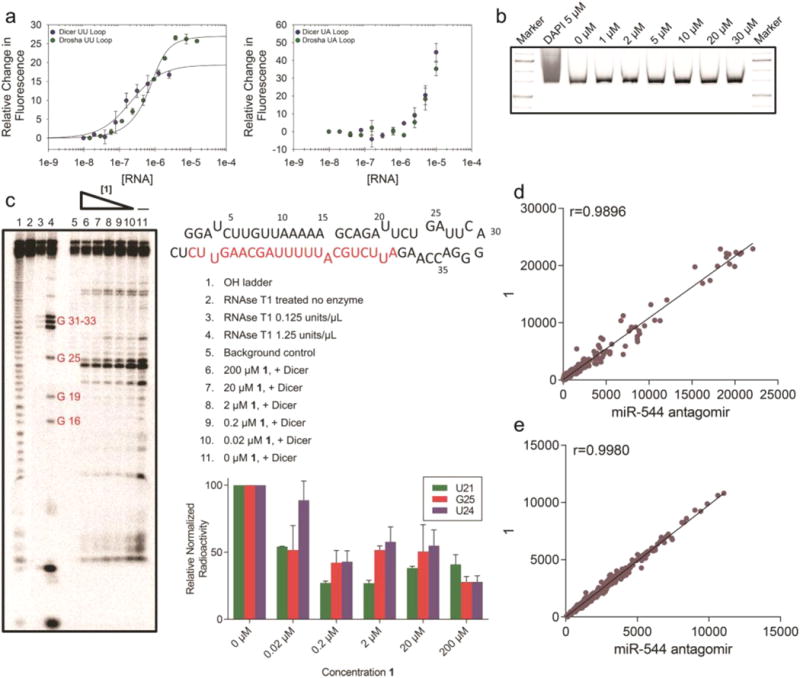
Characterization and specificity of 1. (a) Representative binding curves for the binding of 1 to Drosha and Dicer processing sites of miR-544 with the internal loops present (left) or ablated (right). (b) 1 at the indicated molar concentrations was incubated with a ∼600 bp fragment of PCR amplified DNA for a period of 2 h. The fragment was then subjected to PAGE analysis followed by SYBR-Gold staining. Unlike DAPI incubated DNA, 1 did not elicit a shift in the DNA fragment even at high molar concentrations. (c) Radioactive end-labeled pre-miR-544 was cleaved with Dicer in the presence of 1 at the indicated concentrations and subjected to PAGE analysis. Cleavage of pre-miR-544 at uracil 21 in the purported Dicer site was 50% protected in the presence of 20 nM of 1. (d) Dissimilarity measure (Pearson correlation coefficient) of miRNA microarray data for hypoxic MDA-MB-231 cells treated with 1 vs miR-544 antagomir. Dots represent gene counts (normalized intensity values) of at least 50. (e) Dissimilarity measure (Pearson correlation coefficient) of gene microarray data for hypoxic MDA-MB-231 cells treated with 1 vs miR-544 antagomir. Dots represent genes counts (normalized intensity values) of at least 50.
We also performed binding assays of compounds similar to 1 (6, 7, and 8) to study the effect of steric bulk toward selectivity of the miR-544 hairpin precursor. Compound 1 was most selective for the Dicer site (greater than 50-fold over the base paired control) followed by compound 8 (40-fold selectivity), while compound 7 exhibited the least selectivity (7-fold), and no binding was observed for compound 6 up to 10 μM of RNA. On the basis of these results, it is likely that compounds 7 and 8 have diminished selectivity relative to the starting compound due to the addition of a hydrogen bond donor or increased charge. The pKa of the amine on the substituted ring methyl-piperazine or piperazine is approximately 7–9 and could increase its nonspecific binding. Compound 6, the morpholino derivative, was inactive presumably because of the addition of steric bulk and partially negatively charged ring oxygen, which ablated binding to the target RNA. Collectively, these studies show that recognition of the RNA by the entire molecule and a balance between hydrogen bond donors, charge, and partial charge drives bioactivity and selective target recognition.
To confirm that the small molecule did not bind DNA, we performed gel shift assays with 1 and a 650 bp DNA fragment. No binding was observed even when 1 was added in 1500-fold excess (30 μM vs 20 nM; Figure 4b). The ability of 1 to inhibit processing of the miR-544 precursor in vitro by Dicer was also evaluated using an RNase protection assay. As shown in Figure 4c, compound 1 inhibited nuclease processing of the hairpin precursor at nanomolar concentrations in a dose-dependent manner. Collectively, these studies show that Inforna-designed small molecules can selectively bind to the miR-544 hairpin precursor and affect its biogenesis/maturation at low nanomolar concentrations.
Microarray analysis was subsequently used to analyze the selectivity of 1 for the miR-544 precursor in cellulo. In these studies, we compared the selectivity of 1 to a miR-544 antagomir, which allowed us to probe differences in target modulation, e.g. a small molecule that recognizes an RNA structure versus an oligonucleotide that recognizes sequence. The miRNA microarray was performed on hypoxic MDA-MB-231 cells treated with either a 500 nM miR-544 antagomir or 20 nM 1. Little dissimilarity in the miRNA expression profiles between the two treatment groups was observed based on the Pearson correlation coefficient (r = 0.9896) indicating that 1 is at least as selective as the miR-544 antagomir (Figure 4d). As seen in Supporting Information Figure 7, miR-544 exhibited the largest decrease in expression when treated with either miR-544 antagomir or 1 when compared to an untreated sample. A strong correlation (r = 0.9980) was also observed between treatment groups based on mRNA microarray (Figure 4e), revealing few if any off-target events that may complicate probing of the miR-544 signaling pathway in hypoxic cancer cells.
Small Molecule Inhibition of miR-544 Biogenesis Impairs Survival in Response to Hypoxia and Enhances Chemosensitivity of Tumor Cells
We completed a comparative systems biology approach to assess the ability of 1 to modulate miRNA function in order to produce a physiological response in cellulo and in vivo in hypoxic cancer models. We began by gauging the responsiveness of cancerous and noncancerous breast cell lines in either hypoxic or normoxic environments to interruption of miR-544 signaling by either antagomir or 1 treatment. As anticipated, treatment with 1 decreased miR-544, HIF-1α, and ATM transcripts and increased mTOR transcripts as determined by qRT-PCR analysis in MDA-MB-231 and MCF-7 cells under hypoxic conditions to a similar extent as a miR-544 antagomir but at a 25-fold lower concentration (Figure 5a). Compound 1 yielded similar affects in MCF-10A cells, but treatment with a miR-544 antagomir failed to significantly alter ATM and HIF-1α levels.
Figure 5.
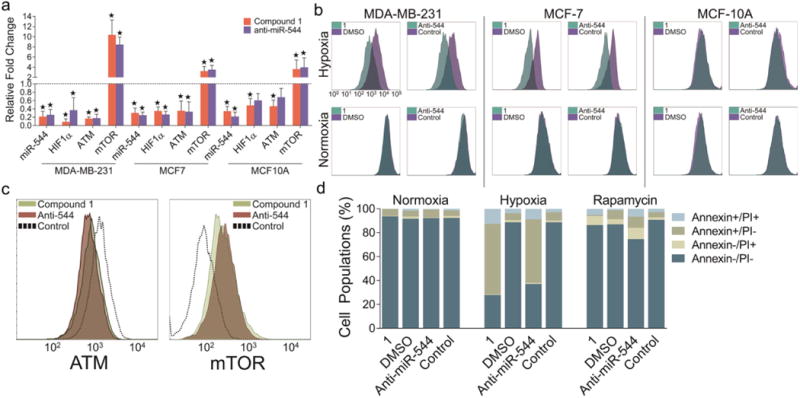
Compound 1 treatment impairing tumor cell growth and survival in response to hypoxia. (a) Real-time qPCR analysis of miR-544 and hypoxia-associated genes in MDA-MB-231, MCF-7, and MCF-10A cell lines treated with either 1 or miR-544 antagomir and cultured under hypoxic conditions. Data represent fold-change as compared to normoxic conditions (dashed line). Errors bars represent 95% confidence interval. *, p < 0.05. (b) CSFE-label retention in MDA-MB-231, MCF-7, and MCF-10A cell treated with 1 (20 nM) or a miR-544 antagomir (500 nM) and cultured for 72 h in hypoxic or normoxic conditions. (c) Flow cytometric analysis of MDA-MB-231 cells stained with anti-ATM (left) or anti-mTOR (right) antibodies following treatment with 1 (20 nM) or a miR-544 antagomir (500 nM) and cultured under hypoxic conditions. (d) Graphic representation of flow cytometric analysis of Annexin V and PI stained MDA-MB-231 cells treated with either 1 or miR-544 antagomir and cultured in normoxic vs hypoxic conditions in the presence or absence of rapamycin (100 nM).
Given that miR-544 overexpression in normoxic cancer cells induced G1-S phase arrest,(8) we examined if inhibition of miR-544 in a hypoxic microenvironment results in increased cell proliferation resulting in failure of cells to engage hypoxia-mediated survival mechanisms. Flow cytometric analysis of carboxyfluorescein succinimidyl ester (CFSE)-stained cells showed that 1- and antagomir-mediated inhibition of miR-544 prevented growth arrest of tumor cell lines under hypoxic conditions but had no effect on immortalized epithelial cells in normoxia or hypoxia (Figure 5b).
Reduction of ATM levels in tumor cells under hypoxic conditions is associated with an mTOR-mediated apoptosis response. As anticipated, ATM and HIF-1α protein levels were significantly down-regulated, and mTOR protein levels were significantly up-regulated in MDA-MB-231 cells treated with 1 or a miR-544 antagomir under hypoxic conditions (Figure 5c, Supporting Information Figure 8). We studied if disruption of the miR-544 signaling pathway was sufficient to induce a hypoxic apoptosis response in our model by flow cytometric analysis. Treatment with both 1 and a miR-544 antagomir induced apoptosis in MDA-MB-231 and MCF-7 cells cultured in hypoxic conditions as indicated by increased Annexin V staining but had no effect on cell survival in normoxic conditions (Figure 5d and Supporting Information Figure 9a). In contrast, miR-544 inhibition had no significant effect on the survival of MCF-10A cells in normoxia or hypoxia. To determine if the observed apoptotic effect was due to increased expression of mTOR, we attempted to block this effect by treating cells with rapamycin, a well-known inhibitor of the mTOR signaling pathway.(24) MDA-MB-231 cells cultured under hypoxic conditions were pretreated with rapamycin then exposed to the antagomir or 1. Both significantly suppressed miR-544 dependent hypoxia-induced apoptosis by approximately 5-fold (Figure 5d and Supporting Information Figure 9b), indicating that increased levels of mTOR drive the apoptotic effect.
Hypoxia also selects for tumor cells with drug resistant phenotypes, thereby reducing sensitivity to chemotherapeutic agents.(25) Our data demonstrate that inhibition of miR-544 function impairs the ability of tumor cells to adapt to hypoxic stress in vitro and, therefore, should enhance their chemosensitivity to agents such as 5-fluorouracil (5-FU). As shown in Figure 6, MCF-7 and MDA-MB-231 cells treated with 1 or a miR-544 antagomir exhibited significantly reduced chemoresistance to 5-FU under hypoxic conditions as examined by MTT assay. Collectively, these studies confirm a role for miR-544-dependent modulation of the ATM/mTOR axis in driving tumor cell adaptation to a low oxygen microenvironment and the selectivity of 1 observed in the rapamycin rescue experiment. Moreover, they validate the platforms described herein used to identify and evaluate the activity of RNA-targeting small molecules and demonstrate that these molecules effectively inhibit miR-544, thus altering cellular responses to hypoxia with high potency and selectivity.
Figure 6.
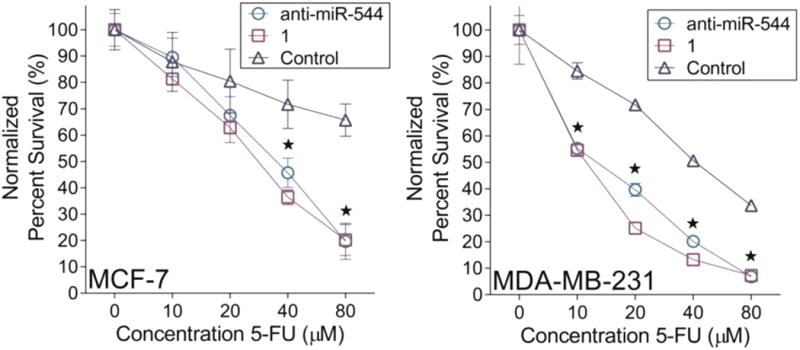
Compound 1 treatment enhances chemosensitivity. Viability of MCF-7 (left) and MDA-MB-231 (right) cells cultured in hypoxia following treatment with or without 1 (20 nM) or a miR-544 antagomir (500 nM) in the presence of varying doses of 5-fluoruracil. Error bars, SEM. *, p < 0.05 of both miR-544 antagomir and 1 compared to control.
Potency of Small Molecule- vs Antagomir-Mediated Inhibition of miR-544 Activity on Tumor Growth in Vivo
Our in vitro comparative analysis between miR-544 antagomir and 1 in hypoxic breast cancer cell lines led us to assess the potential of 1 as a cancer therapeutic in an in vivo breast cancer model. To determine if miR-544 inhibition prevents metabolic adaptation of tumor cells to hypoxic stress in vivo, we implanted MDA-MB-231-GFP-luc cells subcutaneously into immunodeficient NOD/Scid mice. MDA-MB-231-GFP-luc cells were pretreated with either 20 nM 1, transfected with 500 nM miR-544 antagomir, or left untreated prior to implantation. Tumor progression was monitored weekly by physical palpation of the tumor injection site until tumor masses were apparent in untreated control mice. As expected, MDA-MB-231-GFP-luc cells produced large tumors that extended well beyond the margins of the inoculation site when untreated, but pretreatment with 1 or transfection with a miR-544 antagomir significantly impeded tumor growth in vivo as evidenced by live animal bioluminescent imaging at 3 weeks post-transplant (Figure 7a).
Figure 7.
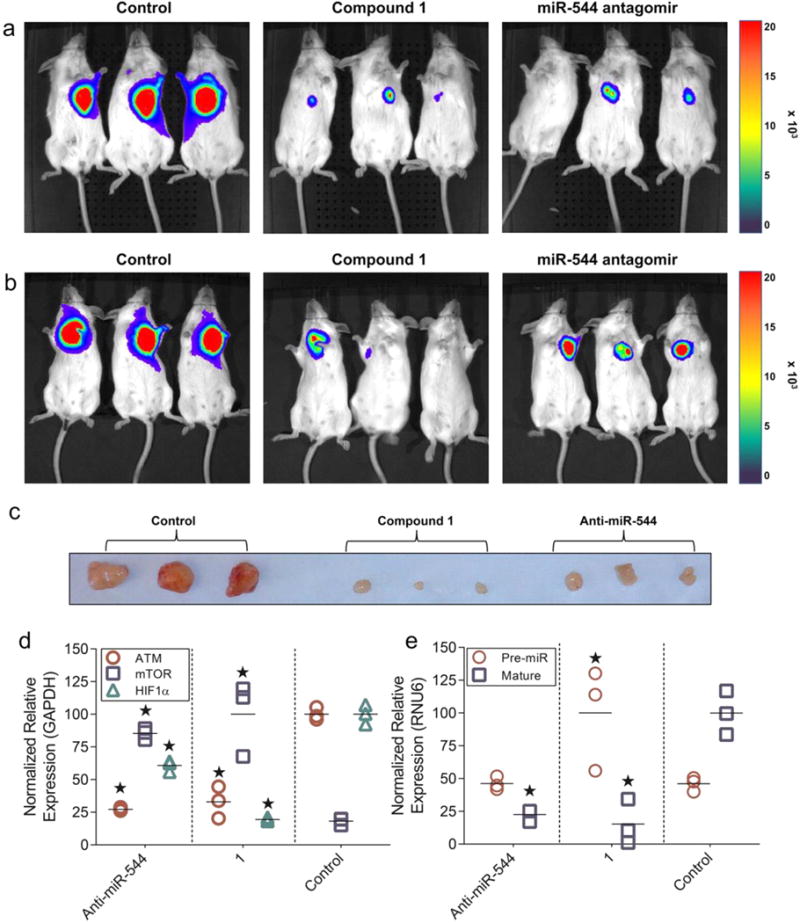
Compound 1 impeding tumor growth in vivo at a lower effective dose than a synthetic miR-544 antagomir. (a and b) Live bioluminescent imaging of NOD/Scid mice at 3 weeks postimplant of MDA-MB-231-GFP-luc cells. Images show mice administered GFP-labeled tumor cells that were pretreated with 1 or miR-544 antagomir in vitro prior to transplantation (a) or were administered an IP injection of 1 or antagomir 24 h postimplantation of tumor cells (b). (c) Images of MDA-MB-231-GFP-luc/Matrigel tumor masses resected from untreated, 1-treated, or miR-544 antagomir-treated mice 21 days after IP treatment injection. (d and e) Real-time qPCR analysis of hypoxia-associated gene transcripts (d) and miR-544 precursor and mature transcripts (e) in tumor samples resected from NOD/Scid mice from b. *, p < 0.05.
In order for RNA-targeting therapies against hypoxia to be efficacious, they must be able to reach the low oxygen microenvironments within the tumor masses that tend to form away from sites of angiogenesis initiation. Therefore, we compared the therapeutic potential of 1 and the miR-544 antagomir by implanting MDA-MB-231-GFP-luc cells into NOD/Scid mice and then dosing mice 24 h later with a single intraperitoneal (IP) injection (100 μL of 40 μM 1 or 100 μL of 200 μM antagomir). Once again, nontreated mice exhibited significant tumor growth into surrounding tissues by 3 weeks post-transplant, whereas mice receiving a single IP injection of miR-544 antagomir had significantly diminished tumor growth (Figure 7b). In contrast, two of three mice administered 1 had no detectable tumors by bioluminescent imaging. Importantly, IP injections of both 1 and miR-544 antagomir were generally well tolerated and produced no overt side effects. Established tumors were then resected and analyzed by qRT-PCR for expression of pre-miR-544, miR-544, ATM, mTOR, and HIF-1α. Macroscopic examination of the tumors revealed dramatic differences in size consistent with the bioimaging data (Figure 7c). In untreated mice, levels of miR-544, HIF-1α, and ATM were elevated in the primary tumor, whereas tumors treated with either a miR-544 antagomir or 1 had significantly decreased levels of miR-544, ATM, and HIF-1α and increased levels of mTOR (Figure 7d,e). Importantly, only tumors treated with 1 expressed increased levels of pre-miR-544, thereby confirming that 1 arrests tumor growth by impeding miR-544 biogenesis and altering expressed levels of miR-544 targets in vivo (Figure 7e). As a whole, these data demonstrate that 1 is a more potent modulator of miR-544 function in comparison to the miR-544 antagomir and that 1 is capable of altering signaling pathways that drive adaptation to hypoxic stress even within an in vivo three-dimensional environment. Indeed, a single IP injection of 1 was able to produce dramatic reductions in tumor mass with long-lived effects over the course of 3 weeks.
Over the past decade, significant strides have been made in the development of RNA-based cancer therapeutics, but their clinical intervention has been hampered by the lack of suitable in vivo delivery methods that ensure sufficient cellular uptake and protect against nuclease degradation of the RNA.(20) Moreover, a growing number of studies indicate that synthetic siRNAs are potent inducers of the inflammatory immune response.(26, 27) Therefore, administration of high dose RNA-based therapeutics could potentially elicit anti-RNA antibody responses as seen in patients with systemic lupus erythematosus.(28) In this study, we have put forth a novel drug discovery platform to rapidly identify, screen, and test small molecule inhibitors of cancer-associated miRNAs. Using this platform, we have identified a small molecule that selectively impedes the biogenesis of miR-544 via direct binding and elicits biological responses with few off-target effects at a potency that exceeds that of synthetic oligonucleotides. This small molecule exhibits rapid and efficient uptake in vivo as evidenced by changes in precursor and mature miR-544 levels and that of downstream miR-544 targets. Furthermore, our data demonstrate that targeting miRNAs that orchestrate metabolic adaptation to hypoxic stress provides a means to selectively sensitize tumor cells to hypoxia-induced killing and increase their sensitivity to chemotherapeutic drugs (Figure 8). These synergistic studies demonstrate that RNA targets are indeed druggable with small molecules, which have improved activities and selectivities relative to oligonucleotides in vivo and open up intriguing possibilities for developing small molecule therapeutics for other disease-associated RNAs.
Figure 8.
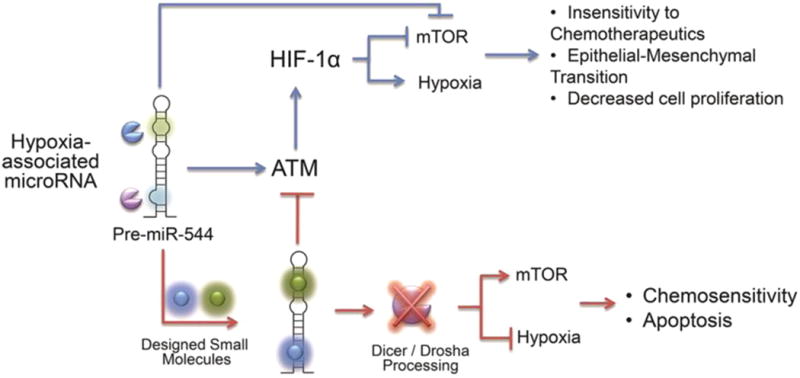
Schematic overview of miR-544/ATM/mTOR signaling axis. Overview of miR-544 signaling pathways under hypoxic conditions. Small molecule inhibition of miR-544 results in dysregulation of hypoxia-associated survival mechanisms leading to apoptosis through failure to adapt to low oxygen microenvironment.
Methods
Cell Culture
Cell lines were obtained from the Cell-Based Screening Facility at The Scripps Research Institute. MDA-MB-231, MDA-MB-231-GFP-luc, and MCF-7 cell lines were maintained in RPMI 1640 supplemented with 10% FBS and 5% Pen/Strep. MCF-10A cells were maintained in DMEM/F12 supplemented with 10% FBS, 20 ng/mL EGF, 0.5 mg mL−1 hydrocortisone, 100 ng/mL cholera toxin, 10 μg/mL insulin, and 5% Pen/Strep, and HEK-293 cells were maintained in DMEM with 10% FBS and 5% Pen/Strep. Cells cultured under hypoxic conditions were maintained at <1% O2 in a nitrogen filled hypoxic chamber with 5% CO2 at 37 °C (Billups-Rothenberg, Inc.) for a period of 5 days before conducting experimentation. Culture media were sampled for pH analysis to confirm similar pH conditions in cells exposed to hypoxia and normoxia and rule out effects of acidosis. Cells cultured under normoxic conditions were maintained in ambient air (∼21% O2) with 5% CO2 at 37 °C. The RNAiMax transfection reagent (Life Technologies) was used to transfect cells with a miR-544 antagomir (Qiagen) via the reverse transfection method and Lipofectamine 2000 (Life Technologies) was used to transfect pGRB-miR and luciferase constructs according to the manufacturer's instructions.
Mice
NOD/Scid mice (B6.CB17-Prkdcscid/Sz, Jackson Laboratories) were housed in the Scripps Florida vivarium, and all experiments employing live animals were approved by the Scripps Florida Institutional Animal Care and Use Committee. MDA-MB-231-GFP-luc cells (5 × 106) were pretreated with or without 1 (20 nM) or a miR-544 antagomir (500 nM) for 48 h. Cells were suspended in 100 μL of PBS/Matrigel (1:1 - BD Biosciences) and subcutaneously injected into mice. Tumor growth was monitored weekly, and after 21 days animals were imaged on an IVIS 200 system following IP injection of 200 μL d-luciferin (15 mg mL−1). Alternatively, mice were injected with MDA-MB-231-GFP-luc cells (5 × 106) and 24 h later administered an IP injection of 1 (100 μL of 40 μM) or a miR-544 antagomir (100 μL of 200 μM), and tumor growth was monitored as described. Where indicated, tumors were resected from mice following euthanasia and analyzed by real-time PCR.
Gene Expression Analysis
Total RNA was isolated using the Vantage Total RNA Purification Kit (Origene) according to the manufacturer's instructions. cDNA was generated from 1 μg of RNA using the miScript II Reverse Transcription Kit (Qiagen). Quantitative real time RT-PCR was performed as previously described(8) using the miScript Primer Assays (Qiagen) for miR-544 and pre-miR-544. The sequences for all qPCR primers used in this study can be found in Supporting Information Table 1. All mRNAs were normalized to GAPDH, and miRNAs were normalized to RNU6b (Qiagen). An unpaired Student's t test of the replicate 2(-ΔCt) values for each gene in the control group and treated group were used to determine statistical significance.
Fluorescence-based Drug Screening
The pGRB-miR screening system was first calibrated by transfecting HEK-293 cells with pGRB-miR using Lipofectamine 2000 according to manufacturer's instructions. Cells were visualized at 24 h post-transfection by fluorescent microscopy to ensure efficient transfection before being plated in 384-well plates. Two days after plating, cells were analyzed by fluorescence using a combination of excitation/emission filters on a Sapphire Infinite 200 Pro plate reader (Tecan) to determine the optimal excitation and emission spectra for each of the three fluorescent proteins. These values were used to determine the ability of 1 to inhibit miR-544 regulation of BMI1. To do so, HEK-293 cells were transfected with pGRB-miR, pGRB-miR lacking miR-544, or pGRB-miR encoding a 3′-UTR of BMI1 wherein the miR-544 binding site was mutated. Cells were replated 48 h post-transfection in 96-well plates in sextuplicates and treated with either small molecule inhibitors of miR-544 (20 nM) or a miR-544 antagomir (500 nM). After a 48 h incubation period, expression levels of GFP, RFP, and BFP were quantified using a Sapphire Infinite 200 Pro fluorescent plate reader using the previously derived emission/excitation spectra for GFP, RFP, and BFP. The RFP/BFP ratio was used to determine changes in miR-544 activity relative to the overall transfection efficiency. GFP expression was used to gauge miR-544 expression. Additionally, luciferase secondary screens were performed to confirm the reproducibility of our pGRB-miR system. Briefly, HEK293 cells were cotransfected with either a pri-miR-544 or miR-544 mimic (500 nM) and a pmiRGlo (Promega) dual luciferase expression vector containing human genomic DNA-derived 3′-UTRs of either BMI1 or mTOR cloned downstream of firefly luciferase. Cells were treated at 48 h post-transfection with either 1 (20 nM) or a miR-544 antagomir (500 nM). After a 48 h incubation, luciferase assays were performed using the Dual Glo Luciferase Assay Kit (Promega) according to the manufacturer's instructions. An unpaired Student's t test of the replicate relative luminescence values was used to determine significance.
Cell Proliferation, Apoptosis, and Drug Uptake
MDA-MB-231, MCF-7, and MCF-10A were labeled with CFSE (0.5 μM) at 37 °C for 15 min, washed three times with PBS, plated in six-well plates, treated with or without 1 (20 nM) or a miR-544 antagomir (500 nM), and then incubated in hypoxic or normoxic conditions for 5 days. Cell growth was determined by flow cytometric analysis using an LSRII (BD Biosciences). Alternatively, 1 or antagomir treated cells were dissociated from plates using 100 μL of Accumax, rinsed two times with PBS, and stained using the Annexin V APC Apoptosis Detection Kit (eBiosciences) according to the manufacturer's instructions. Cells were then incubated in propidium iodide (1 μg/mL) for 5 min and analyzed by flow cytometry. Where indicated, rapamycin (Mp Biomedicals Inc.) was added to the cell culture medium at a concentration of 100 nM. In a separate experiment, MDA-MB-231, MCF-7, and MCF-10A cells were transfected with a FITC-labeled miR-544 antagomir (500 nM) or treated with 1 (20 nM), incubated for 72 h under normoxic conditions, and analyzed by flow cytometry at 488 nm excitation/530 nm emission or 405 nm excitation/440 nm emission, respectively. Data analysis was performed using FlowJo Software (TreeStar).
DNA Constructs
The CMV promoter region was PCR-amplified from pCDNA3.1+ and ligated to eGFP, which was PCR-amplified from pCWD. The PGK promoter region was PCR-amplified from pmiRGlo (Promega) and ligated with DsRed (RFP) from pCAG-RFP-int (Addgene). The SV40 promoter region was amplified from psiChek2 (Promega) and ligated to BFP, which was PCR-amplified from pDONR223_BFP (Addgene). The CMV-GFP product was cloned into pBluescript II SK+ after restriction digest with KpnI and XhoI. After sequence confirmation, SV40-BFP was cloned into the NotI/SacII site of pBluescript-CMV-GFP, and PGK-RFP was cloned into the EcoRI/BamHI site of pBluescript-CMV-GFP-SV40-BFP to generate pGRB-miR. MiR-544 was cloned downstream of the CMV-GFP site, and the 3′-UTR region of BMI1 was cloned downstream of the PGK-RFP site. Additional constructs lacking miR-544 and with a mutated miR-544 binding site in the 3′-UTR of BMI1 were generated in a similar manner.
Microarray Analysis
MDA-MB-231 cells were transfected with a miR-544 antagomir (500 nM) or treated with 1 (20 nM) and placed under hypoxic conditions for a period of 5 days. Total RNA was isolated as described above, converted to cDNA using the Affymetrix cDNA Synthesis Kit (Affymetrix), and transcribed in vitro using the IVT labeling kit. Sample cleanup was carried out using the GeneChip Sample Cleanup Module (Affymetrix). Biotinylated cRNA (20 μg) was fragmented and hybridized to either an Affymetrix GeneChip miRNA 3.0 Array or Affymetrix GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix) overnight. Data were collected using the GeneChip Scanner 3000 (Affymetrix) and then analyzed by the Genechip Operating software package. Microarray data can be accessed via Gene Expression Omnibus (GEO, accession number GSE64437).
Supplementary Material
Acknowledgments
We thank J. Childs-Disney for helpful discussions and comments on the manuscript. The research was funded by a grant from the National Institutes of Health to M.D.D. (1R01GM097455 and 1R01GM097455-S1). C.L.H. is supported by the TSRI Stem Cell Postdoctoral Fellowship provided by the estate of Burt and Virginia Polin.
Footnotes
Author Contributions: C.L.H. designed the study, cloned constructs, collected and analyzed data, and wrote the paper. S.P.V. performed RNA protection studies. J.R.S. collected data. W-Y.Y. synthesized molecules. M.D.D. and D.G.P. designed the study, analyzed data, and wrote the paper. All authors discussed results and commented on the manuscript.
The authors declare no competing financial interest.
Supporting Information: Additional figures, tables, and materials and methods. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.5b00265.
Contributor Information
Matthew D. Disney, Email: Disney@scripps.edu.
Donald G. Phinney, Email: DPhinney@scripps.edu.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 3.Cam H, Easton JB, High A, Houghton PJ. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1α. Mol Cell. 2010;40:509–520. doi: 10.1016/j.molcel.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoo YG, Christensen J, Gu J, Huang LE. HIF-1α mediates tumor hypoxia to confer a perpetual mesenchymal phenotype for malignant progression. Sci Signaling. 2011;4:pt4. doi: 10.1126/scisignal.2002072. [CrossRef], [PubMed] [DOI] [PubMed] [Google Scholar]
- 5.Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012;33:207–214. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosse JP, Michiels C. Tumour hypoxia affects the responsiveness of cancer cells to chemotherapy and promotes cancer progression. Anti-Cancer Agents Med Chem. 2008;8:790–797. doi: 10.2174/187152008785914798. [DOI] [PubMed] [Google Scholar]
- 7.Rohwer N, Cramer T. Hypoxia-mediated drug resistance: novel insights on the functional interaction of HIFs and cell death pathways. Drug Resist Updates. 2011;14:191–201. doi: 10.1016/j.drup.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Haga CL, Phinney DG. MicroRNAs in the imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal transition by targeting the TWIST1 protein signaling network. J Biol Chem. 2012;287:42695–42707. doi: 10.1074/jbc.M112.387761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velagapudi SP, Gallo SM, Disney MD. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat Chem Biol. 2014;10:291–297. doi: 10.1038/nchembio.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ditch S, Paull TT. The ATM protein kinase and cellular redox signaling: beyond the DNA damage response. Trends Biochem Sci. 2012;37:15–22. doi: 10.1016/j.tibs.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menendez JA, Cufi S, Oliveras-Ferraros C, Martin-Castillo B, Joven J, Vellon L, Vazquez-Martin A. Metformin and the ATM DNA damage response (DDR): accelerating the onset of stress-induced senescence to boost protection against cancer. Aging. 2011;3:1063–1077. doi: 10.18632/aging.100407. [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran T, Disney MD. Molecular Recognition of 6′-N-5-Hexynoate Kanamyin A and RNA 1 × 1 Internal Loops Containing CA Mismatches. Biochemistry. 2011;50:962–969. doi: 10.1021/bi101724h. [ACS Full Text ACS Full Text], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo Y, Disney MD. Bottom-up Design of Small Molecules that Stimulate Exon 10 Skipping in Mutant MAPT Pre-mRNA. ChemBioChem. 2014;15:2041–2044. doi: 10.1002/cbic.201402069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Childs-Disney JL, Stepniak-Konieczna E, Tran T, Yildirim I, Park H, Chen CZ, Hoskins J, Southall N, Marugan JJ, Patnaik S, Zheng W, Austin CP, Schatz GC, Sobczak K, Thornton CA, Disney MD. Induction and reversal of myotonic dystrophy type 1 pre-mRNA splicing defects by small molecules. Nat Commun. 2013;4:2044. doi: 10.1038/ncomms3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkesh R, Childs-Disney JL, Nakamori M, Kumar A, Wang E, Wang T, Hoskins J, Tran T, Housman D, Thornton CA, Disney MD. Design of a Bioactive Small Molecule That Targets the Myotonic Dystrophy Type 1 RNA via an RNA Motif–Ligand Database and Chemical Similarity Searching. J Am Chem Soc. 2012;134:4731–4742. doi: 10.1021/ja210088v. [ACS Full Text ACS Full Text], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Childs-Disney JL, Hoskins J, Rzuczek SG, Thornton CA, Disney MD. Rationally designed small molecules targeting the RNA that causes myotonic dystrophy type 1 are potently bioactive. ACS Chem Biol. 2012;7:856–862. doi: 10.1021/cb200408a. [ACS Full Text ACS Full Text], [PubMed], [CAS] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, Sohn R, Zemelman B, Snell RG, Rundle SA, Crow S, Davies J, Shelbourne P, Buxton J, Jones C, Juvonen V, Johnson K, Harper PS, Shaw DJ, Housman DE. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 18.Atkinson JD, Johnson MC. Syntheses of some highly substituted pyridines, 2,7-naphthyridines and 1H-pyrimido[4,5,6-i,j][2,7]naphthyridines. J Chem Soc C. 1968:1252–1258. doi: 10.1039/j39680001252. [CrossRef], [CAS] [DOI] [Google Scholar]
- 19.Kelly RB, Slomp G, Caron EL. 2-Cyanomethyl-1,1,3,3-tetracyanopropene, a Self-Condensation Product of Malononitrile. J Org Chem. 1965;30:1036–1038. doi: 10.1021/jo01015a020. [ACS Full Text ACS Full Text], [CAS] [DOI] [Google Scholar]
- 20.Gavrilov K, Saltzman WM. Therapeutic siRNA: principles, challenges, and strategies. Yale J Biol Med. 2012;85:187–200. [CAS] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Lu Z, Wientjes MG, Au JS. Delivery of siRNA Therapeutics: Barriers and Carriers. AAPS J. 2010;12:492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haupenthal J, Baehr C, Kiermayer S, Zeuzem S, Piiper A. Inhibition of RNAse A family enzymes prevents degradation and loss of silencing activity of siRNAs in serum. Biochem Pharmacol. 2006;71:702–710. doi: 10.1016/j.bcp.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Haupenthal J, Baehr C, Zeuzem S, Piiper A. RNAse A-like enzymes in serum inhibit the anti-neoplastic activity of siRNA targeting polo-like kinase 1 Int. J Cancer. 2007;121:206–210. doi: 10.1002/ijc.22665. [DOI] [PubMed] [Google Scholar]
- 24.Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan R, Paré GC, Frederiksen LJ, Semenza GL, Graham CH. Hypoxia-induced resistance to anticancer drugs is associated with decreased senescence and requires hypoxia-inducible factor-1 activity. Mol Cancer Ther. 2008;7:1961–1973. doi: 10.1158/1535-7163.MCT-08-0198. [DOI] [PubMed] [Google Scholar]
- 26.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 27.Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 28.Blanco F, Kalsi J, Isenberg DA. Analysis of antibodies to RNA in patients with systemic lupus erythematosus and other autoimmune rheumatic diseases. Clin Exp Immunol. 1991;86:66–70. doi: 10.1111/j.1365-2249.1991.tb05775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


