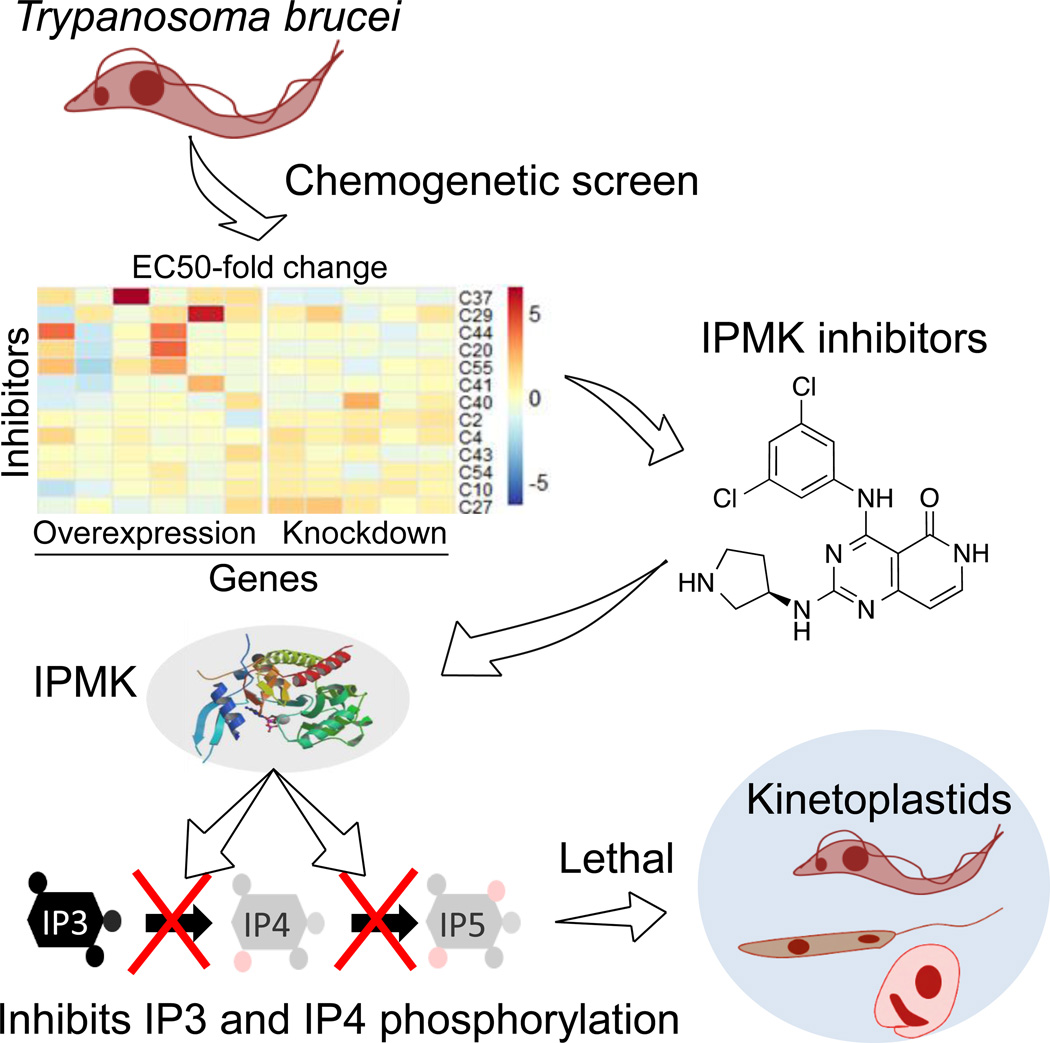
SUMMARY
Kinetoplastids cause Chagas disease, Human African Trypanosomiasis (HAT) and leishmaniases. Current treatments for these diseases are toxic and inefficient, and our limited knowledge of drug targets and inhibitors has dramatically hindered the development of new drugs. Here we used a chemogenetic approach to identify new kinetoplastid drug targets and inhibitors. We conditionally knocked down Trypanosoma brucei inositol phosphate (IP) pathway genes and showed that almost every pathway step is essential for parasite growth and infection. Using a genetic and chemical screen we identified inhibitors that target IP pathway enzymes and are selective against T. brucei. Two series of these inhibitors acted on T. brucei inositol polyphosphate multikinase (IPMK) preventing Ins(1,3,4)P3 and Ins(1,3,4,5)P4 phosphorylation. We show that IPMK is functionally conserved among kinetoplastids and its inhibition is also lethal for Trypanosoma cruzi. Hence, IP enzymes are viable drug targets in kinetoplastids and IPMK inhibitors may aid the development of new drugs.
Graphical Abstract
eTOC Blurb
Cestari et al. identified steps of the inositol phosphate pathway that are essential for Trypanosoma brucei infection. Using a chemical genetic approach they discovered small molecules that inhibit phosphorylation of Ins(1,4,5)P3 and Ins(1,3,4,5)P4 and that are lethal to T. brucei and other related parasites.
INTRODUCTION
Kinetoplastid parasites cause Chagas disease, Human African Trypanosomiasis and leishmaniases, which together affect over 2 million people annually worldwide (Stuart, et al., 2008). Chagas disease is caused by Trypanosoma cruzi and affects people in South and Central America, although it has also spread to the United States (Bern, 2015). Trypanosoma brucei sp. primarily affects sub-Saharan Africa and causes HAT (or Nagana in cattle) and Leishmania spp. occur in over 90 countries in the tropics, subtropics, and parts of Europe (Stuart, et al., 2008). Current drugs available for treating these diseases are inefficient, highly toxic and drug resistance is spreading (Renslo and McKerrow, 2006; Stuart, et al., 2008). Thus, there is an urgent unmet need for safe and effective drugs against these pathogens.
Typical drug discovery efforts against kinetoplastids have included untargeted inhibitor screens and repurposing approaches (Pena, et al., 2015; Planer, et al., 2014). Although these approaches have met with some success, the lack of ligand-target knowledge hinders chemical improvement of hits and questions of target specificity are also difficult to resolve. The development of target-based inhibitors is an attractive approach for addressing some of these concerns; however, few targets have been validated in T. cruzi or Leishmania due to their limited genetic tractability. Advances in T. brucei genetics have allowed genome-wide gene essentiality screens and the validation of new targets and chemotypes for the development of novel drugs (Alsford, et al., 2011; Cestari and Stuart, 2013; Kalidas, et al., 2014). Since the genomes of these parasites are highly conserved (~94% synteny and ~60% overall identity of orthologous genes) (El-Sayed, et al., 2005), the identification of new targets and discovery of T. brucei-specific inhibitors may be useful for chemical validation of orthologous enzymes in T. cruzi and Leishmania sp.
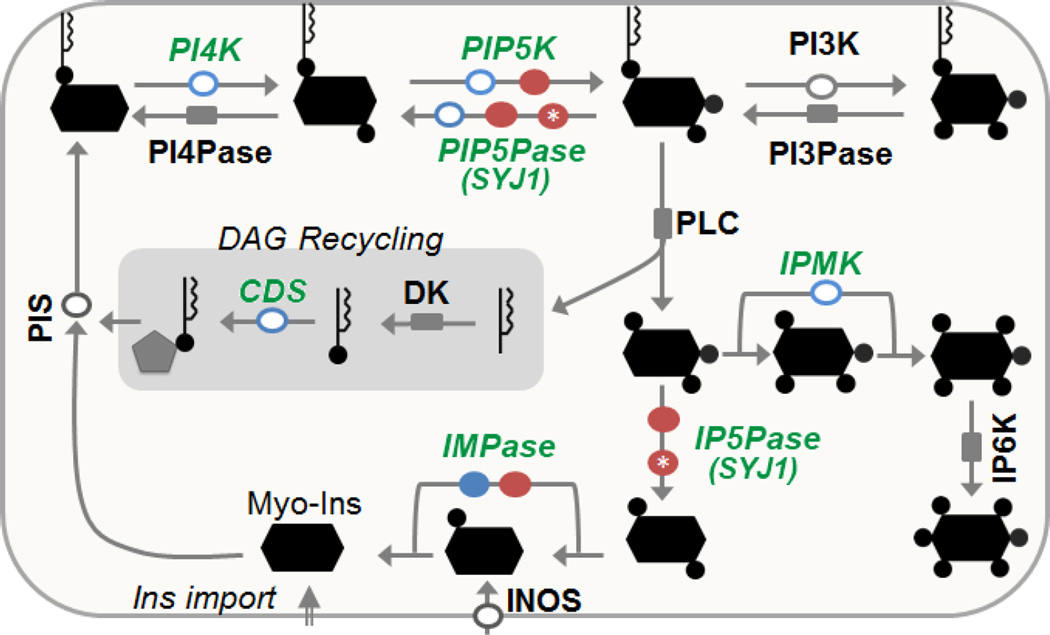
The IP pathway (Fig 1) plays important roles in regulating essential biological processes from yeast to humans, including mRNA transport from the nucleus to the cytoplasm (Wickramasinghe, et al., 2013), transcriptional regulation (Millard, et al., 2013; Steger, et al., 2003), embryogenesis (Seeds, et al., 2015), endocytosis (Yamaoka, et al., 2015), and ciliary function and signal transduction (Chavez, et al., 2015). We recently showed that this pathway controls transcription and allelic exclusion of variant surface glycoprotein (VSG) genes in T. brucei (Cestari and Stuart, 2015), allowing T. brucei to evade the host immune response. The IP pathway also regulates Ca2+ homeostasis (Huang, et al., 2013) and Golgi biogenesis (Hall, et al., 2006; Rodgers, et al., 2007) in T. brucei, differentiation and infectivity of T. cruzi (Hashimoto, et al., 2013), and its enzymes are also targets for malaria drugs (Mbengue, et al., 2015). IPs are also found on surface molecules, i.e. glycosylphosphatidylinositol (GPI), which anchors T. brucei VSGs, T. cruzi surface mucins and Leishmania surface molecules (Martin and Smith, 2006). Moreover, genes involved in the synthesis of IPs or phosphatidylinositol (PIs) are essential for parasite growth (Martin and Smith, 2005; Martin and Smith, 2006). We and others have also shown that some PI kinases and phosphatases are essential for bloodstream forms (BF) of T. brucei (Cestari and Stuart, 2015; Hall, et al., 2006; Rodgers, et al., 2007) (Fig 1). The essentiality of these enzymes is likely due to the variety of processes that their metabolic products control (Millard, et al., 2013; Watson, et al., 2012). Since their metabolic products often regulate protein function by direct interaction, IP pathway enzymes are attractive targets for antiparasitic drugs.
Fig 1. Outline of T. brucei IP pathway.
Arrows indicate enzyme reactions with enzymes manipulated in the present study indicated in green and italicized. Open circles, essential genes; closed circles, nonessential genes; blue, overexpressed genes; red, knockdown or nulls in chemogenetic experiments. A gray square indicates an enzyme whose activity has not been studied or a gene essentiality that has not been analyzed. The red circles with white asterisks indicate TbSYJ1 which is annotated as synaptojanin (N-terminal domain) and is predicted to be a PI/IP 5-phosphatase. At least two genes encode enzymes with TbPIP5K, TbPIP5Pase and TbIMPase predicted activity. Annotation is based on predicted or demonstrated function (Table 1).
Here, we show that almost every step of the T. brucei IP pathway contains a gene that is essential for growth and some are essential for infection. Using chemogenetics we identified inhibitors that target enzymes of this pathway, including two series that inhibit TbIPMK. This enzyme phosphorylates Ins(1,3,4)P3 and Ins(1,3,4,5)P4 and diverges substantially from its human ortholog. IPMK inhibitors were effective against related kinetoplastid IPMKs and inhibited T. cruzi amastigote proliferation. Thus, IP enzymes are potential drug targets and IPMK inhibitors may provide the basis for the development of new antiparasitic drugs.
RESULTS
T. brucei IP pathway genes are essential for infection of mice
We first sought to identify candidate drug targets in the T. brucei IP pathway. We knocked out various genes in the pathway and tested the ability of the resultant parasites to infect mice. We initially attempted to generate null cell lines by replacing both endogenous alleles of each target gene with drug resistance markers. In cases where null cells could not be obtained, we generated conditional null (CN) cells by replacing both endogenous alleles in cells that transcribe an ectopic copy of the target gene under tetracycline (tet) control. Using this approach we showed previously that the genes encoding TbPIP5K1, TbPIP5Pase1, TbIPMK and TbCDS enzymes were essential for T. brucei BF growth in vitro, whereas TbIMPase1 was not essential (Cestari and Stuart, 2015) (Table 1). We show here that genes encoding a predicted TbPIP5Pase2 and TbIP5Pase are not essential for growth of T. brucei BF, although knockdown of TbPIP5Pase2 slightly reduced parasite growth (Table 1 and Fig S1). We obtained null BF cells for genes encoding TbPIP5K2, TbSYJ1 (Synaptojanin, predicted IP/PI 5-phosphatase), and TbIMPase2 and found that these cells grew slightly slower (~7h doubling time) than the parental cell line (SM427, ~6h doubling time) (Fig S1). The genes encoding TbINOS, TbPIS, TbPI4K and TbPI3K are also essential for BF growth (Hall, et al., 2006; Martin and Smith, 2005; Martin and Smith, 2006; Rodgers, et al., 2007). Altogether, these results indicate that at least eight of the 14 studied genes encoding enzymes of the IP pathway are essential for growth of BF T. brucei.
Table 1.
T. brucei genes encoding IP pathway enzymes genetically analyzed for essentiality in vitro.
| Gene ID | Product name and abbreviation | Essentiality |
|---|---|---|
| Tb927.4.1620 | Phosphatidylinositol 4-phosphate 5-kinase (TbPIP5K1) | Yes (conditional null)1 |
| Tb927.10.4770 | Phosphatidylinositol 4-phosphate 5-kinase (TbPIP5K2) | No (null)a |
| Tb927.11.6270 | Phosphatidylinositol (4,5)/(3,4,5)-phosphate 5-phosphatase (TbPIP5Pase1) |
Yes (conditional null)1 |
| Tb927.9.5680 | Phosphatidylinositol 5-phosphatase (TbPIP5Pase2) | No (conditional null) a |
| Tb927.10.5510 | Inositol polyphosphate 5-phosphatase (TbIP5Pase) | No (conditional null) a |
| Tb927.9.10640 | Synaptojanin 1 (TbSYJ1, predicted IP/PI 5-phosphatase) | No (null) a |
| Tb927.8.6210 | Phosphatidylinositol 3-kinase (TbPI3K) | Yes (RNA interference)2 |
| Tb927.3.4020 | Phosphatidylinositol 4-kinase alpha (TbPI4K) | Yes (RNA interference)3 |
| Tb927.9.12470 | Inositol polypohosphate multikinase (TbIPMK) | Yes (conditional null)1 |
| Tb927.7.220 | CDP-diacylglycerol synthase (TbCDS) | Yes (conditional null)1 |
| Tb927.9.1610 | CDP-diacylglycerol inositol 3-phosphatidyltransferase (TbPIS) |
Yes (conditional null)4 |
| Tb927.10.7110 | Inositol-3-phosphate synthase (TbINOS) | Yes (conditional null)5 |
| Tb927.9.6350 | inositol-1(or 4)-monophosphatase 1 (TbIMPase1) | No (conditional null)1 |
| Tb927.5.2690 | Inositol-1(or 4)-monophosphatase 2 (TbIMPase2) | No (null) a |
This work (see Fig S1);
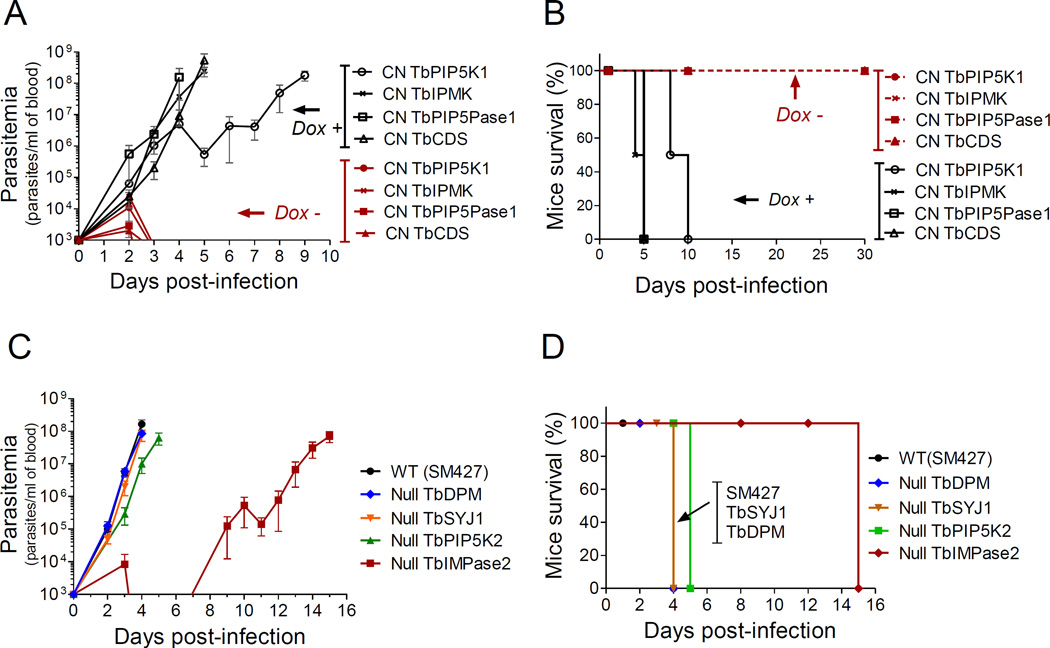
We infected mice with T. brucei BF CNs to test the in vivo essentiality of the genes encoding TbPIP5K1, TbPIP5Pase1, TbIPMK and TbCDS, all of which were essential in vitro. Doxycycline (dox, a stable tet analog) was added to the drinking water of one group of mice (dox-mice) 18h prior to infection to induce transcription of the ectopic target gene, while another group of mice had dox omitted from the drinking water (no dox-mice) to knockdown transcription of the target gene. All mice in both groups became infected, but parasitemia in the dox-mice reached lethal levels (>1.0×108 parasites/ml of blood) between four and five days post-infection (Fig 2A and B), whereas in no dox-mice parasitemia was cleared for all four cell lines indicating that these genes are all essential for infection.
Fig 2. T. brucei IP pathway genes are essential for infection of mice.
A) Parasitemia with CN cell lines. Mice were infected with 1.0×104 parasites and parasitemia was counted daily by tail prick. Dox = 200 µg/ml, n = 3 mice per group except for TbPIP5K in which n = 6 mice per group. B) Mouse survival after infection with CN cell lines from A. C) Mouse infection with SM427 or null cell lines. Experiments as in A except that mice did not receive dox, n = 3 per group. D) Mouse survival after SM427 or null cell lines infection from C. Mice were euthanized if parasitemia reached ≥ 1.0×108 parasites/ml. Data are represented as mean ± SEM.
We also infected mice with null cell lines for TbPIP5K2, TbSYJ1 and TbIMPase2 (Fig 2C and D), none of which was essential for growth in vitro. We compared their parasitemias with that of the SM427 parental cell line and to a null for dolichol phosphate α-1,2-mannosyltransferase (TbDPM) gene, which is involved in GPI glycosylation. Mice infected with null TbSYJ1 and TbDPM cells had parasitemias that were similar to those of the parental line; all mice died at day 4 post-infection. Mice infected with the TbPIP5K2 cell line survived slightly longer, although all were infected. However, mice infected with null TbIMPase2 cells initially had a low parasitemia that became undetectable until day 9 post-infection, after which parasitemia rose with 14h doubling time (2.5-fold slower than WT). All mice died by day 15 post-infection. Thus, although the TbIMPase2 null cells did not exhibit severe growth defects in vitro, they exhibited reduced infectivity in vivo. The reasons underlying this recrudescent parasitemia after a subpatent period are unknown, but it may be due to the IP pathway role in regulating VSG gene expression (Cestari and Stuart, 2015) or IP requirements for GPI synthesis (Martin and Smith, 2006). Overall, at least four of the eight IP pathway genes essential for T. brucei BF in vitro are also essential for infection of mice, whereas one nonessential gene (TbIMPase2) is important for infectivity. Thus, almost every step of the pathway has a gene essential for T. brucei; hence they are targets for drug development.
Identification of effective and selective inhibitors of trypanosome growth
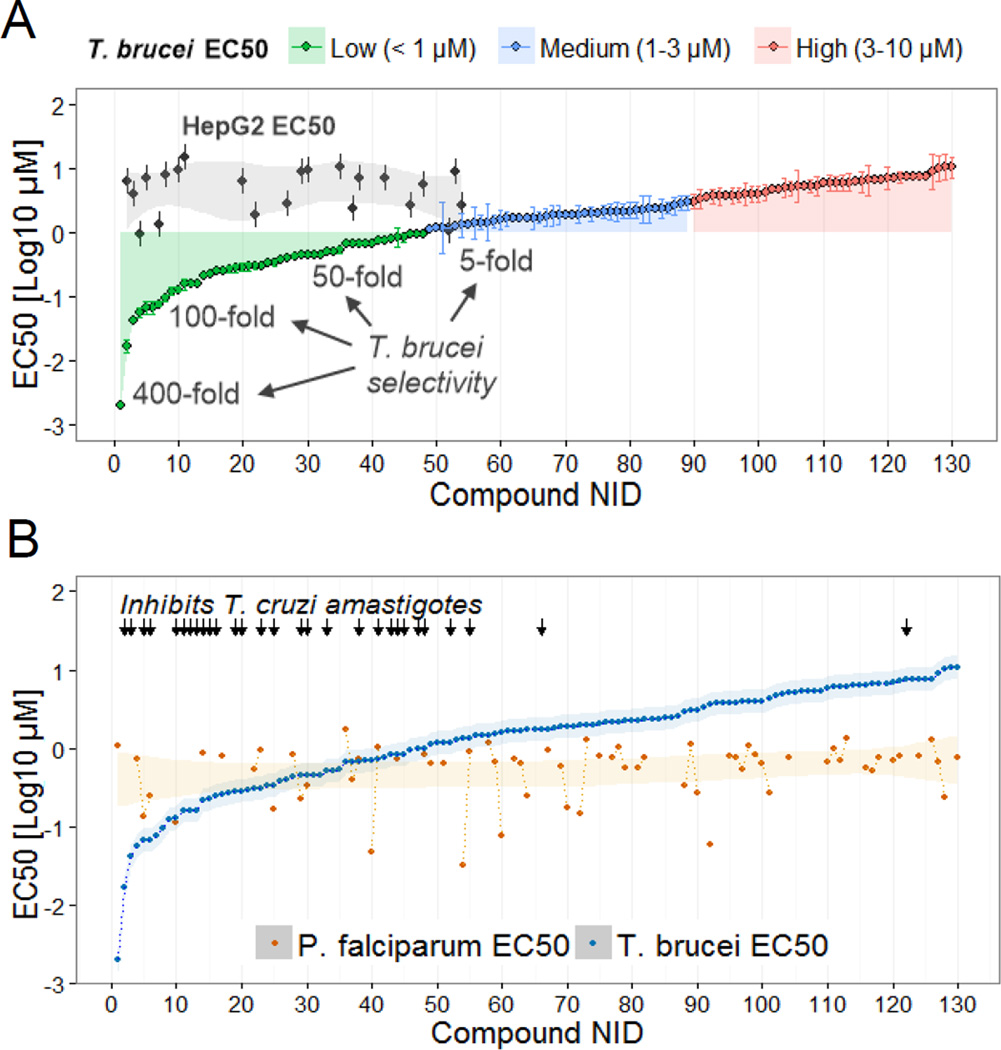
To identify inhibitors of the IP pathway enzymes, we initially screened a GlaxoSmithKline (GSK) library of 520 compounds predicted to act on protein kinases and PI kinases and that had not been previously tested against T. brucei BF. We identified 130 compounds with EC50s (effective concentrations that inhibit BF growth by 50%) ranging from 2 nM to 10 µM (Fig 3A and Table S1). Moreover, 48 compounds had an EC50 < 1 µM, 41 had an EC50 between 1–3 µM, and 41 had an EC50 between 3–10 µM. The remaining compounds were not inhibitory at concentrations up to 15 µM. Compound clustering by structural similarity revealed additional information on chemotypes (Table S1).
Fig 3. Effective and selective inhibitors of trypanosome growth.
A) EC50s of 130 compounds against T. brucei BFs. Compounds are grouped according to low (green), medium (blue) and high (red) EC50 values. Compounds have a numerical ID (NID) to facilitate identification (details in Table S1). The EC50s of a subset of compounds tested against HepG2 cells are shown (black dots in gray area) and the selectivity range is indicated. B) Plot of T. brucei and P. falciparum 3D7 EC50s. P. falciparum EC50s were obtained from (Gamo, et al., 2010). Compounds effective against T. cruzi intracellular amastigotes at 5 µM are indicated with black arrows. Shading shows EC50 trends. Data are represented as mean ± SEM.
We determined the EC50s of a subset of compounds (EC50 < ~1.5 µM) against HepG2 human hepatocytes. Of the 45 compounds that were tested, 23 did not affect HepG2 viability and growth at concentrations up to 15 µM. Most compounds with EC50s below 15 µM against HepG2 cells were still one to two orders of magnitude more selective against T. brucei, with some compounds being up to 400-fold more selective (Fig 3A). We tested the most potent and selective compounds against T. cruzi and found that 20 of these also significantly inhibited intracellular amastigote proliferation (Fig 3B and 6E, Table S1). Moreover, 67 of the 130 compounds were shown to be effective against the erythrocytic stage of P. falciparum 3D7 (Fig 3B) and 25 of these were also effective against the multidrug resistant P. falciparum strain Dd2 (Gamo, et al., 2010). Overall, we found compounds that are selective and effective against trypanosomes and potential chemotypes for the development of antiparasitic drugs.
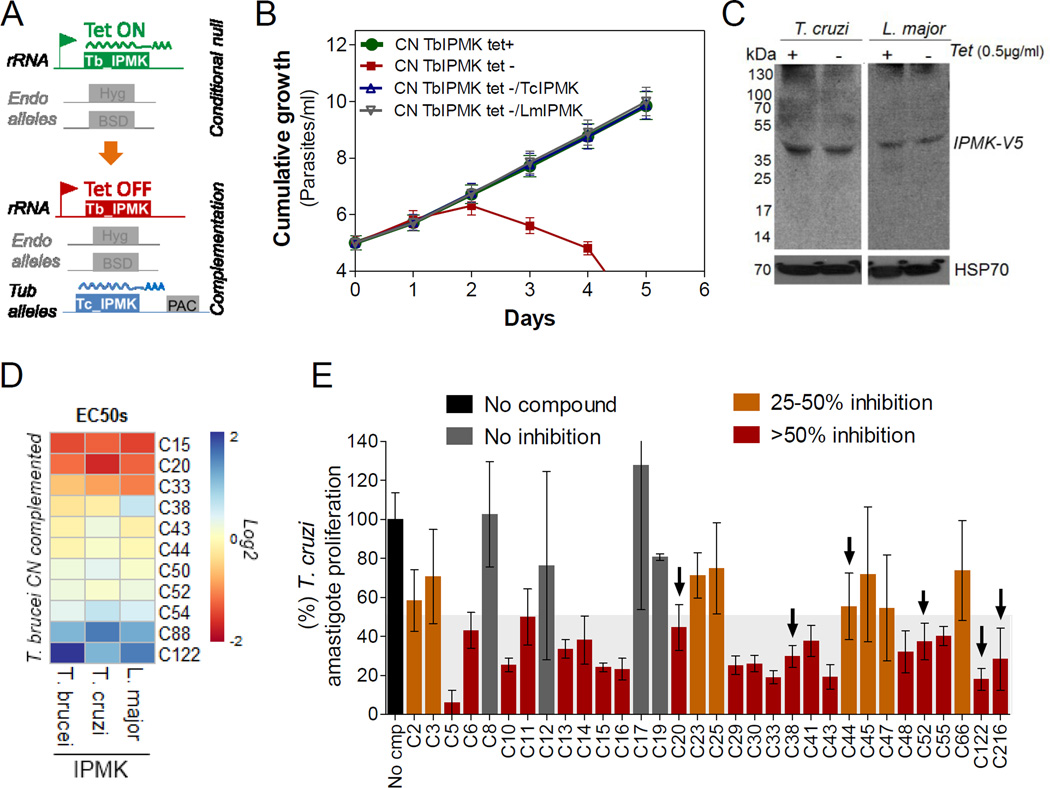
Fig 6. Chemical validation of T. cruzi and L. major IMPK.
A) Diagram showing T. brucei CN complemented with the orthologous IPMK gene from T. cruzi or L. major. In the absence of tet, T. brucei cells exclusively express the orthologous IPMK genes. B) Cumulative growth curve analysis of T. brucei CN TbIPMK complemented with T. cruzi or L. major genes. C) Western analysis of 3V5-tagged TcIPMK (left) or LmIMPK (right) in complemented T. brucei CN TbIPMK. D) Heatmap showing the EC50s of the TbIPMK inhibitors and analogs against T. brucei SM427 or T. brucei CN exclusively expressing T. cruzi or L. major IPMK genes. E) Effect of TbIPMK inhibitors against T. cruzi intracellular amastigotes (arrows). Other compounds are also shown for comparison. Compounds were used at 5 µM. Gray shaded area indicates values between 0–50%. Dark red bars (>50% inhibition) show compounds that significantly inhibited amastigote proliferation with p<0.05, t-test. Data are represented as mean ± SEM, except for heat map in D which shows means.
Chemogenetic identification of T. brucei IP pathway inhibitors
We next used a chemogenetic approach to identify compounds that target the IP pathway. We generated 11 cell lines in which expression of genes for specific steps in the pathway was either eliminated, knocked down or overexpressed by using null, conditional null or tet-regulatable overexpressor cells, respectively. These cells were screened with the T. brucei-selective compounds (EC50 < ~1.5 µM) to identify changes in cell sensitivity. Overexpression of a target enzyme is expected to modulate the compound inhibitory effect, i.e. increase the EC50. Elimination of a nonessential enzyme is predicted to generate cells dependent on the activity of a homologous enzyme and increase sensitivity to the inhibitor, i.e. lower EC50, or have other effects on drug sensitivity depending on the overall consequences of the loss of the function of the deleted gene.
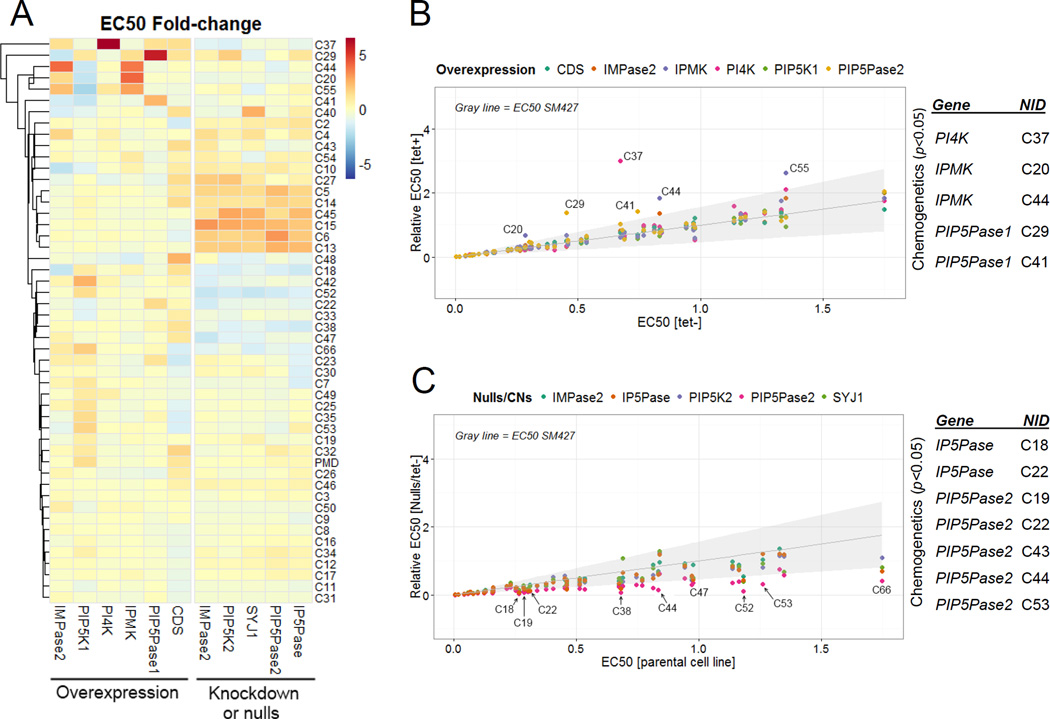
We performed a blind screen using 50 compounds against the transgenic cells and included pentamidine, a drug used to treat T. brucei infection, as a control for assay reproducibility (Fig 4A). Using a false discovery rate (FDR) of 1%, we identified seven compounds where gene overexpression increased the EC50 of the compound, compared to noninduced or parental cell lines, by about 2- to 5-fold (Fig 4B and Table S2). Analysis of variance (ANOVA) indicated that four of these changes were statistically significant. Overexpression of TbPI4K increased the EC50 to C37 (p<0.001), of TbIPMK to C20 (p<0.01) and C44 (p<0.01), and of TbPIP5Pase1 to C29 (p<0.001) and C41 (p<0.001). These results imply that the compounds primarily act on these enzymes.
Fig 4. Chemogenetic analysis.
A) Heatmap showing fold changes in compound sensitivity of overexpressing or null/CN cell lines compared to parental (SM427) or noninduced cells. The subset of compounds with EC50 <~1.5 µM was used for analysis. PMD, pentamidine. B–C) Fold EC50 changes for each compound in each overexpressing cell line (B) or CN/null cell line (C). A FDR of 1% was used as a cutoff (gray shading). NIDs are indicated for some compounds which EC50 changes. A list of compounds and genes showing significant changes by chemogenetic analysis with p ≤ 0.05 by ANOVA are shown on the right of each graph (see details in Table S2). Data are represented as mean.
Gene elimination or knockdown increased cell sensitivity to some compounds, resulting in as much as a 4.5-fold decrease in the EC50 compared to parental cells (Fig 4A and C). Conditional knockdown of TbIP5Pase decreased the EC50 to C18 (p<0.05) and C22 (p<0.05), and conditional knockdown of TbPIP5Pase2 decreased the EC50 to C19 (p<0.05), C22 (p<0.05), C43 (p<0.05), C44 (p<0.001) and C53 (p<0.05). The knockdowns of TbIP5Pase or TbPIP5Pase2 increased sensitivity to C22, suggesting that this compound acts against 5-phosphatases (Fig 1). On the other hand, knockdown of TbPIP5Pase2 or overexpression of TbIPMK altered sensitivity to C44, with the knockdown resulting in cells more being sensitive and the overexpression in cells being less sensitive to C44 (Fig 4A). The differential effect of C44 on these cell lines may be due to alterations of the pathway metabolic flux. Other compound treatments resulted in apparent changes in cell sensitivity, but not all of those were significant, possibly due to compound specificity. Thus, the chemogenetic analysis identified compounds that target enzymes of the IP pathway or processes related to the pathway function.
Enzymatic validation of T. brucei IPMK inhibitors
The chemogenetic data suggest that compounds C20 and C44 inhibit T. brucei IPMK. IPMKs are enzymes that phosphorylate soluble IPs downstream of phospholipase C and exhibit 6-/3-/5-kinase activity (Endo-Streeter, et al., 2012; Holmes and Jogl, 2006), with the exception of human IPMK, which also phosphorylates PI substrates (Maag, et al., 2011). T. brucei predicted IPMK was annotated as a hypothetical protein and has not been characterized. TbIPMK exhibits only 12% amino acid identity with human IPMK, 13% with yeast, 8% with Drosophila, 5% with P. falciparum and 45% and 35% with the predicted T. cruzi and L. major enzymes, respectively. Nevertheless, key amino acids of the catalytic site including the IP binding domain and the SSLL and IDF signatures are conserved among kinetoplastid IPMKs and in other organisms (Fig 5A). Its divergence from human enzymes and its essentiality for infection (Fig 2) make TbIPMK an attractive target candidate for drug development. We expressed and purified TbIPMK from E. coli and tested the activity of the recombinant TbIPMK (rTbIPMK) with IP and PI metabolites (Fig 5B–C). rTbIPMK phosphorylated soluble Ins(1,4,5)P3 and to a lesser extent Ins(1,3,4,5)P4 substrates, but not any of the other IP or PIs tested (Fig 5C). Enzymological analysis confirmed that rTbIPMK acts preferentially on Ins(1,4,5)P3 compared to Ins(1,3,4,5)P4, even at saturating substrate concentrations, with a rTbIPMK efficiency (Kcat/Km) about 6-fold higher for Ins(1,4,5)P3 than Ins(1,3,4,5)P4 (Fig 5C, inset and Table S3). It is important to note that reactions in the presence of Ins(1,4,5)P3 could sequentially phosphorylate two inositol sites, yielding Ins(1,3,4,5)P4 and Ins(1,3,4,5,6)P5. Hence, T. brucei IPMK catalyzes the phosphorylation of specific soluble IPs and lacks activity against PIs, in contrast to its human ortholog.
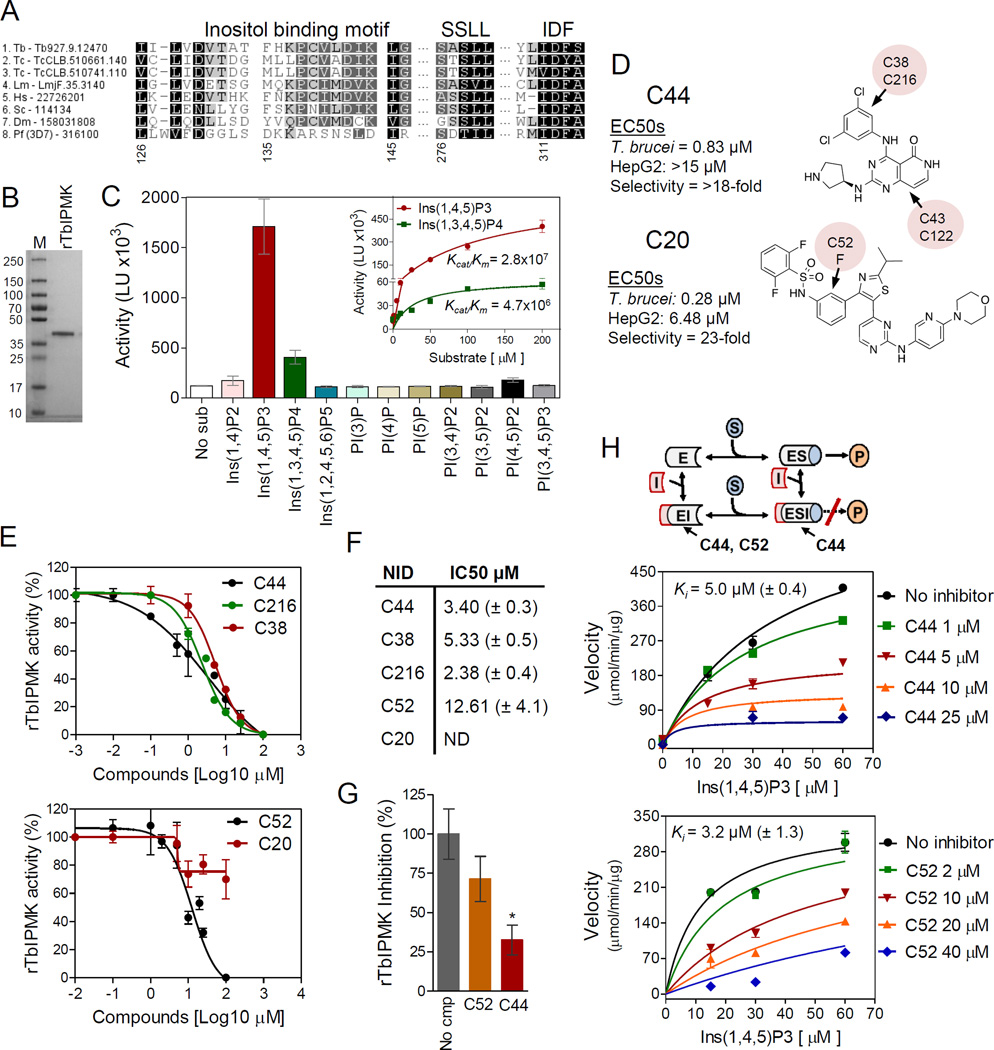
Fig 5. IPMK enzymology and inhibitor validation.
A) Alignment of catalytic sites of IPMKs of T. brucei (Tb), T. cruzi (Tc), L. major (Lm), P. falciparum (Pf), human (Hs), Saccharomyces cerevisiae (Sc) and Drosophila melanogaster (Dm). Gene IDs are shown after the species name abbreviations. B) Recombinant T. brucei IPMK (37 kDa) resolved on a 4–20% SDS/PAGE gel stained with Imperial Coomassie (Pierce). C) rTbIPMK activity assay with IP and PI substrates. Inset, comparison of rTbIPMK activity with Ins(1,4,5)P3 and Ins(1,2,4,5)P4 substrates. Enzyme efficiency (Kcat/Km) unit is S−1M−1 (see Table S3 for details). D) Structure and activity information for compounds C44 and C20. Arrows with pink circles indicate where modifications of analogous compounds are (see Fig S2 for detailed structure and activity of analogs). E) Inhibition assay of rTbIPMK with C44 and its close analogs (top) and C20 and C52 (bottom). F) IC50s of compounds tested in E. G) Inhibition assay of rTbIPMK with Ins(1,3,4,5)P4 substrate; cmp, compound. H) Mechanism of rTbIPMK inhibition by C44 and C52 compounds. Kinetic analyses show rTbIPMK inhibition by C44 (middle) and C52 (bottom) used to derive the type of inhibition. Inhibition constants (Ki) are shown in each graph. Diagram (top) summarizes steps of reaction inhibited by each compound. Enzyme (E), inhibitor (I), substrate (S) and product (P). C44 interacts with E or the ES complex (non-competitive inhibitor) whereas C52 interacts with E and competes for substrate binding (competitive inhibitor), both limiting product formation. Data are represented as mean ± SEM.
We performed rTbIPMK inhibition assays with C44, C20 and related compounds using primarily I(1,4,5)P3 as a substrate (Fig 5D–F). Compound C44 inhibited rTbIPMK activity with an IC50 (a concentration that inhibits enzyme activity to 50% of the maximum) of 3.4 µM. Compound structure similarity analysis identified C38, C43, C122 and C216 as related to C44 (Fig S2), and C38 and C216 each inhibited rTbIPMK with IC50s similar to that of C44. Although C38 and C216 have minor structural differences at the dichlorophenyl group (Fig 5D), the C38 EC50 against T. brucei cells was 0.51 µM, similar to that of C44 (0.83 µM), whereas C216 was not inhibitory at concentrations up to 15 µM. In contrast, C43 and C122, both of which differ from C44 at the pyridopyrimidine group, had almost no activity against rTbIPMK (Fig S2), suggesting that this chemical group may be important for the specificity of these inhibitors. Furthermore, C20 partially inhibited rTbIPMK activity, even at high concentrations (Fig 5E), but compound C52 which has a fluorine (F) at a phenyl group (Figs 5D and S2) completely inhibited rTbIPMK activity. Given the similarity of C20 and C52 it is likely that C20 activity against T. brucei IPMK in cell assays results from analogous products originating from C20 modifications in the cells. In addition, C44, and to a lesser extent C52, inhibited rTbIPMK activity in the presence of Ins(1,3,4,5)P4, the other TbIPMK substrate (Fig 5G).
Kinetic analysis showed that C44 inhibited rTbIPMK by decreasing its Vmax independently of substrate concentration with a Ki of 5.0 µM (Fig 5H), indicating that C44 acts as a noncompetitive inhibitor of rTbIPMK. However, C52 inhibited rTbIPMK with an Ki of 3.2 µM and resulted in an apparent increase of the Km but no significant alterations of Vmax, indicating that C52 acts as a competitive inhibitor of rTbIPMK with respect to Ins(1,4,5)P3. Due to differences in these compound mechanisms of inhibition, it is likely that they inhibit rTbIPMK by interacting with different enzyme sites with C52 competing with the Ins(1,4,5)P3 binding site. Overall, T. brucei IPMK functions in the synthesis of inositol phosphate metabolites and C44 and C20 represent two distinct classes of TbIPMK inhibitors.
Chemical validation of T. cruzi and L. major IPMK as drug targets
Due to the high genomic conservation between kinetoplastid parasites (El-Sayed, et al., 2005), we hypothesized that T. brucei IPMK inhibitors might also be effective against related kinetoplastid IPMKs. To test this hypothesis, we complemented the T. brucei IPMK CN cells with T. cruzi and L. major genes encoding for the orthologous predicted IPMKs. A C-terminally V5-tagged copy of the T. cruzi or L. major IPMK gene was inserted into the tubulin locus of T. brucei IPMK CN cells to enable constitutive expression (Fig 6A). Conditional knockdown of TbIPMK resulted in cells that exclusively expressed the T. cruzi or L. major IPMK gene and depended on this gene for survival and growth. The orthologous IPMK genes from both parasite species complemented T. brucei growth and the complemented cells grew with similar rates to that of T. brucei CN cells in the presence of tet (Fig 6B and C). Thus, the T. cruzi and L. major genes predicted to encode IPMK are functional in T. brucei and likely function as IPMK enzymes.
Parasites exclusively expressing either TcIPMK or LmIPMK were also sensitive to IPMK inhibitors and their analogs (Fig 6D). However, cell sensitivity differed from compound to compound and also among the IPMK genes expressed. For example, T. brucei cells exclusively expressing TcIPMK were more sensitive to C20, C44 and C52 and less sensitive to C50 and C88 compared to cells expressing TbIPMK, whereas the exclusive expression of LmIPMK resulted in cells more sensitive to C33 and less sensitive to C38 than cells expressing TbIPMK. These differences may result from IPMK amino acid sequence variations between the parasites.
Finally, we analyzed the effects of IPMK inhibitors and additional selective compounds against T. cruzi intracellular amastigotes. Importantly, IPMK mRNA has been shown to be upregulated in T. cruzi amastigotes and trypomastigotes compared to noninfectious stages (Aslett, et al., 2010); thus, if this gene is essential for T. cruzi amastigotes its inhibition would be lethal. We infected mammalian cells for 24h with T. cruzi Silvio X10/1 metacyclic trypomastigotes. At 72h post-infection we added various compounds at 5 µM and quantified the numbers of intracellular amastigotes and infected cells at 120h postinfection. Several compounds, including the C20 and C44 compound series, inhibited intracellular amastigote growth > 50% (Fig 6E). C44 only partially inhibited amastigote growth; however its analogs C38 and C216 strongly inhibited amastigote proliferation > 70%. In addition, both C20 and C52 also inhibited amastigote proliferation > 50%. These data imply that TcIPMK inhibition is lethal to amastigotes and chemically validates T. cruzi IPMK as a target for drug development. Moreover, compounds C52 and C44 are chemotypes for the development of clinical inhibitors of kinetoplastid IPMKs.
DISCUSSION
The development of new drugs against kinetoplastid parasites has been hindered by the lack of knowledge of drug targets, target-specific chemotypes and their modes of action. We show here that the IP pathway is a target for anti-kinetoplastid drug discovery. We found that eight genes of this pathway are essential for T. brucei BF growth, five of which are also important for infection. We identified 130 molecules that inhibited T. brucei growth, many of which were highly selective compared with mammalian cells. Using a chemogenetic approach that involved perturbation of IP pathway gene expression and compound screening we identified compounds that target IP enzymes. Two series of compounds were biochemically validated as inhibitors of T. brucei IPMK. This enzyme phosphorylates Ins(1,4,5)P3 and Ins(1,3,4,5)P4 and diverges from human IPMK. Genetic complementation experiments showed that IPMKs are functionally conserved among kinetoplastids, and that IPMK inhibitors were effective against T. cruzi and L. major IPMKs and lethal for intracellular T. cruzi amastigotes. Thus, our chemogenetic approach using T. brucei as an experimental surrogate of less manipulable parasites is valuable for identifying target-based inhibitors.
An attractive feature of the IP pathway as a drug target is that IP metabolites have multiple essential regulatory functions in eukaryotes. Hence, disruption of the pathway may have pleiotropic detrimental effects on the pathogen. This pathway has not been heavily studied in kinetoplastids, but is known to function in the control of VSG gene transcription and antigenic variation, acidocalcisome function, Golgi maintenance, and cytokinesis in T. brucei (Cestari and Stuart, 2015; Hall, et al., 2006; Huang, et al., 2013; Rodgers, et al., 2007) and likely in other processes. Our functional analyses using null and CN cell lines avoided the uncertainties related to insufficient gene knockdown in RNAi-based approaches. All four genes that were essential in vitro were also essential for infection in mice, but one of the four non-essential genes in vitro was important for infection. For example, the null TbIMPase2 cells were partially cleared before establishing lethal infections. In addition, infections with CN TbPIP5K cells in the presence of dox showed variable parasitemia levels before establishing lethal infections (Fig 2). Similar results were obtained with CN TbPIP5K in infections with various concentrations of dox, indicating that ample dox is present (not shown), and the in vitro growth rate of these cells in the presence of tet was the same as in WT cells (Cestari and Stuart, 2015). We did not explore the basis for these cases of decreased parasitemia levels followed by the establishment of infection and mouse lethality. However, we previously showed that TbPIP5K regulates monoallelic expression of VSG genes and antigenic variation (Cestari and Stuart, 2015), and TbIMPase2 is predicted to encode one of the two T. brucei enzymes that dephosphorylate Ins(1,4)P2. The product of this reaction may be utilized in the synthesis of GPIs, which anchor VSGs on the parasite surface (Martin and Smith, 2006). Because the in vivo growth profile of the CN TbPIP5K and null TbIMPase2 cells resembles a relapse due to antigenic switching, one possibility is that their antigenic switching is altered. Alternatively, perturbation of the IP pathway may have had other consequences on parasite-host interactions, given the numerous functions of these metabolites (Cestari and Stuart, 2015; Hall, et al., 2006; Huang, et al., 2013).
In contrast to target-based screening against recombinant proteins (Pedro-Rosa, et al., 2015) and compound repurposing strategies (Planer, et al., 2014), our chemogenetic approach took advantage of our knowledge of IP pathway enzyme essentiality and our ability to genetically manipulate T. brucei. Furthermore, potential target-based inhibitors were identified directly in T. brucei infectious stages. T. brucei sensitivity to 10 compounds changed after genetic perturbation of the IP pathway. While the shift in EC50 associated with the change in expression of a particular IP enzyme may reflect targeting of that enzyme by the compound, it may alternatively reflect alteration in IP metabolite flux or change in reliance on another, e.g. paralog, enzyme or process that is regulated by an IP metabolite. Since little is known about IP pathway metabolites and their flux and regulation, e.g. feedback loops and substrate-product conversion rates, we cannot pinpoint specific targets by this approach alone. However, chemogenetic data point the way for biochemical validation.
The chemogenetic results indicate that compounds C44 and C20 target T. brucei IPMK. Kinetoplastid parasites, which are diploids, have one IPMK gene. The likely two alleles in the T. cruzi Cl Brener strain, which is a hybrid, have >97% identity. T. brucei IPMK catalyzes the sequential phosphorylation of Ins(1,4,5)P3 into Ins(1,3,4,5)P4 and of this metabolite into Ins(1,3,4,5,6)P5. In contrast to human IPMKs (Maag, et al., 2011), this enzyme does not phosphorylate PI substrates. The sequence (<12% amino acid identity) and functional diversity from human IPMK and its essentiality for infection indicate that T. brucei, and perhaps the closely related kinetoplastid, IPMKs are useful targets for drug development. The precise function of IPMK in trypanosomes is unknown, but IPMK products regulate various processes in yeast and mammals, including nuclear mRNA export (Steger, et al., 2003; Wickramasinghe, et al., 2013) and the HDAC complex (Millard, et al., 2013; Watson, et al., 2012).
Kinetic analysis showed that C44 inhibits rTbIPMK in the presence of both Ins(1,4,5)P3 or Ins(1,3,4,5)P4 substrates and acts as a noncompetitive inhibitor, as may its analogs C38 and C216. C20 partially inhibited enzyme activity but its analog C52, from which it differs by a single F, completely inhibited rTbIPMK activity. C52 acted as a competitive inhibitor of Ins(1,4,5)P3 but poorly inhibited rTbIPMK in presence of Ins(1,3,4,5)P4. The differences in C52 inhibition with these substrates may be due to differences in IPMK requirements for substrate recognition, as suggested by structural and mutational analyses of orthologous IPMKs (Endo-Streeter, et al., 2012; Holmes and Jogl, 2006). Either type of inhibitor may be suitable for drug development, since both competitive and noncompetitive inhibitors have become drugs against infectious agents (Sarafianos, et al., 2009).
Our chemogenetic and biochemical data suggest that T. brucei IP enzymes may be the primary targets of these inhibitors, but they may also have secondary targets. The identification of analogs, such as the C52 and C20 or C44 series, also demonstrates the validity of the identified chemotypes. Some analogs were not identified as positive chemogenetic hits, which may be due to a combination of factors such as statistical stringency, compound transport and/or intracellular compound modifications. Limitations of gene overexpression levels in trypanosomes may also have influenced the identification of less specific compounds. The IPMK inhibitors also inhibited T. cruzi and L. major IPMKs, which we show by genetic complementation experiments to be functionally conserved among kinetoplastids. In addition, IPMK inhibition was also lethal for intracellular T. cruzi amastigotes, demonstrating the potential value of this drug target in T. cruzi. This extends the short list of validated targets for drug development in T. cruzi, which includes cytochrome b, cruzain, sirtuins and sterol 14α-demethylase (Doyle, et al., 2010; Khare, et al., 2015; McGrath, et al., 1995; Moretti, et al., 2015). Some IPMK inhibitors are also effective against P. falciparum, suggesting that these inhibitors may be broadly used to develop new antiparasitic drugs.
Overall, we identified kinetoplastid IP pathway enzymes that are druggable and two IPMK inhibitor chemotypes to aid the development of new drugs against kinetoplastids and perhaps other pathogens.
SIGNIFICANCE
Kinetoplastid parasites cause a spectrum of neglected tropical diseases that affect ~2 million people annually worldwide. Current drug treatments are inadequate and the development of new drugs has been hindered by a limited knowledge of drug targets and inhibitors, especially against T. cruzi and Leishmania sp. which are genetically less tractable. We used T. brucei as a surrogate system to identify new drug targets and employed a chemogenetic approach to identify target-based inhibitors. We focused on the IP pathway and genetically demonstrated that almost every step is essential for T. brucei. The pathway essentiality is likely due to various processes regulated by its metabolites, and highlights its biological importance and druggability. We identified inhibitors that target IP enzymes, specifically two chemotypes that inhibit IPMK with distinct mechanisms of action. We show that IPMK phosphorylates Ins(1,4,5)P3 and Ins(1,3,4,5)P4, well-known second messengers whose perturbation is anticipated to have pleiotropic effects. Moreover, IPMK inhibitors were selective to kinetoplastids compared with humans and also affected plasmodium. Thus, these inhibitors promise to aid the development of drugs against kinetoplastids and perhaps other pathogens. These inhibitors will be useful to dissect the IP pathway function in T. cruzi and Leishmania sp., and demonstrate that our chemogenetic approach is a powerful method to advance target-based drug discovery.
EXPERIMENTAL PROCEDURES
Cell culture
The T. brucei BF were maintained in HMI-9 medium supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2. SM427 and nulls were maintained with 2 µg/ml of G418, and CNs with also 2.5 µg/ml of phleomycin and 0.5 µg/ml of tet. T. cruzi was maintained as epimastigotes in LIT medium at 28°C and metacyclic trypomastigotes were obtained by DEAE-cellulose purification (Cestari and Ramirez, 2010). L. major LT252 (MHOM/IR/1983/IR) were cultivated in M199 medium supplemented with 10% fetal bovine serum, 100 uM adenine, 10 ug/mL heme, 40 mM HEPES pH 7.4, 50 units/mL penicillin and 50 ug of streptomycin. HepG2 and HeLa cells were maintained in DMEM supplemented with 10% FBS at 37°C in 5% CO2.
Transgenic cell lines and growth curves
Cell lines that conditionally express IP pathway genes, nulls or CN cell lines were generated as described (Cestari and Stuart, 2015). Gene IDs are shown in Table 1, except for TbPI4K and TbDPM which are Tb927.4.1140 and Tb927.6.1140, respectively. Cells that conditionally expressed a C-terminally V5 tagged protein were generated by introducing a tet dependent allele containing three V5 tag sequences at its 3’ terminus into the rRNA intergenic region of SM427 cells using pLEW100-3V5. T. brucei CN cells that exclusively expressed a T. cruzi (TcCLB.510741.110) or Leishmania major (LmjF.35.3140) IPMK gene were generated by transfection of pHD-1344-3V5, which contains a 3V5 tag at its 3’-terminus for constitutive expression by integration into one of the tubulin loci. All genes were amplified from genomic DNA of T. brucei strain 427, T. cruzi strain Silvio ×10/1 or L. major strain LT252 using specific primers (Table S4) and the DNeasy Blood & Tissue Kit (QIAGEN) following the manufacturer’s instructions. Gene replacements and insertions were confirmed by PCR and regulated RNA expression was determined by qRT-PCR analysis of cells grown in the presence or absence of 0.5 – 1.0 µg/ml tet. Tet-induced (1.0 µg/ml) conditional protein expression was assayed by western analysis of cell lysates with anti-V5 (Life Technologies) and anti-mouse IgG HRP-conjugated secondary antibodies (Pierce). Growth curves were obtained as previously described (Cestari and Stuart, 2015), except that no drugs were used for null cells. Parasites were counted by Neubauer chambers or a particle counter (Beckman Coulter).
Mouse experiments
T. brucei SM427, CN or null cell lines growing at mid-log phase were used to infect BALB/cAnNHsd mice (6–8 week-old males; Harlan Laboratories). Mice were injected intraperitoneally with 1.0×104 parasites in 200 µL of HMI-9 medium. Mice that were infected with CN cell lines were given drinking water 18h before infection containing 200 µg/ml doxycycline and 5% sucrose to induce expression of target genes, whereas gene knockdown was accomplished by giving the mice drinking water containing only 5% sucrose; water was replaced daily. Parasitemias were monitored daily by tail prick starting 2 days postinfection. Mice with a parasitemia of 1.0×108 parasites/ml of blood were sacrificed. All procedures were performed in the vivarium of the Center for Infectious Disease Research in compliance with all applicable laws and institutional guidelines (Institutional Animal Care and Use Committee protocol KS-01).
Compound assays
A library of 520 compounds diluted at 10 mM in dimethyl sulfoxide was provided by GSK. A screen was performed at 15 µM to identify compounds that inhibited T. brucei BF growth, and EC50s of 130 effective compounds were determined using two-fold serial dilutions ranging from 15 µM to 0.029 µM (Cestari and Stuart, 2013). For chemogenetic studies, survival assays were performed at the EC50 of each compound and fold-changes between genetic modified and parental cell lines were calculated as follows: (GMT ÷ PT) × (PNT ÷ GMNT), whereas GM is genetic modified cells (nulls/CNs or overexpression); P, parental cells (SM427 for nulls, noninduced for overexpression); T, compound treatment; NT, not treated with compounds, which correct for differences in cell lines growth rate. The relative EC50 was calculated by multiplying the EC50 of parental (SM427) by the survival fold-change. For HepG2 EC50s, 1.5×104 cells/well were plated in 96 well plates and assays performed as described (Cestari and Stuart, 2013). For T. cruzi drug assays, HeLa cells were incubated in 8-well chamber slides (Fisher Scientific) at 5.0×103 cells/well for 24h at 37°C in 5% CO2, then incubated with metacyclic trypomastigotes (10:1) for 24h. Extracellular parasites were removed by pipetting and mammalian cells incubated for an additional 48h to insure intracellular trypomastigote differentiation to amastigotes and initial replication. Next, compounds (5 µM) were added in 100 µl DMEM supplemented with 10% FBS, and the cells were incubated for 48h at 37°C in 5% CO2. The cells were washed, fixed in 100% cold methanol and stained with Giemsa. The number of intracellular amastigotes per cell and the number of infected cells per total cells were quantified. The percentage (%) of amastigote proliferation was calculated as follows: (ama ÷ inf) × (inf ÷ t), with ama = total intracellular amastigotes; inf = infected cells; and t = total cells. Infected cells not treated with compounds were set as 100%.
Enzyme kinetics
The T. brucei gene encoding IPMK was cloned into pET-29a (Novagen) and expressed in Escherichia coli DE3 pLysS Rosetta cells. Recombinant protein was purified using nickel-resin (Cestari and Stuart, 2015). Protein activity was assayed using the ADP luciferase assay (Promega). IPs and diC8 PIs were purchased from Echelon Biosciences. Assays were performed in 25 µl reactions in 96 well-plates using 20 mM HEPES, 150 mM NaCl, 2 mM MgCl2 at pH 7.5 at 37°C and 270 nM rTbIPMK. Substrate an alysis was performed for 60 min. with 50µM of each substrate, and dose-response assays were performed with 1–200 µM of Ins(1,4,5)P3 or Ins(1,2,4,5)P4 for 30 min. For inhibition assays all compounds were diluted in water and reaction mixes incubated for 30 min. Ins(1,4,5)P3, Ins(1,3,4,5)P4 and ATP were used at 30, 40 and 50 µM, respectively. For Ki analysis and mechanisms of enzyme inhibition, reactions were prepared with 10, 30 and 60 µM of Ins(1,4,5)P3 and C44 (0, 1, 5, 10 and 25 µM) or C52 (2, 10, 20 and 40 µM). Kinetic analyses were calculated by non-linear regression using GraphPad Prism for Windows 5.03 (GraphPad Software, Inc). Compound analogs were identified using an atom pair method and Tanimoto coefficients using ChemmineR (Cao, et al., 2008).
Data presentation and statistical analysis
Data are shown as means ± SEMs of at least three biological replicates. Comparisons among groups were made by two-tailed t test for repeated measures using GraphPad Prism. P values < 0.05 with a confidence interval of 95% were considered statistically significant. For chemogenetic interaction, FDR was calculated based on two-tailed normal distribution of combined data from SM427 and control cells (noninduced and nonknockdown). A 1% cutoff was used to define changes in fold-change after overexpression or knockdown or from null cells. Two-way ANOVA with Bonferroni correction was used to determine whether fold-changes were statistically significant with a P value < 0.05 and confidence interval of 95%. Graphs were prepared using GraphPad Prism or RStudio (Team, 2015).
Highlights.
Inositol phosphate pathway genes are essential for T. brucei infection
Chemical genetic screen used to identify inositol phosphate pathway inhibitors
Two series of molecules inhibit Ins(1,4,5)P3 and Ins(1,3,4,5)P4 phosphorylation
Inhibitors are lethal to T. brucei and for T. cruzi intracellular amastigotes
Acknowledgments
This work was supported by NIH grant R01AI078962 to KS and supplement R01AI014102-37S1 to KS and IC, and AHA fellowship 14POST18970046 to IC. SS and NSM received support from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, 2011/51973-3 to SS and 2012/09403-8 and 2013/20074-9 to NSM) and by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, 477143/2011-3 to SS). We thank GSK for compound library and Anna Sokolov for administrative support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, IC and KS; Methodology, IC; Investigation, IC, PH and NSM; Writing - Original Draft, IC; Writing – Review & Editing, IC, KS, SS, NSM; Funding Acquisition, KS, SS and IC.
REFERENCES
- Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21:915–924. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, et al. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38:D457–462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bern C. Chagas' Disease. N Engl J Med. 2015;373:1882. doi: 10.1056/NEJMc1510996. [DOI] [PubMed] [Google Scholar]
- Cao Y, Charisi A, Cheng LC, Jiang T, Girke T. ChemmineR: a compound mining framework for R. Bioinformatics. 2008;24:1733–1734. doi: 10.1093/bioinformatics/btn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestari I, Ramirez MI. Inefficient complement system clearance of Trypanosoma cruzi metacyclic trypomastigotes enables resistant strains to invade eukaryotic cells. PLoS One. 2010;5:e9721. doi: 10.1371/journal.pone.0009721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestari I, Stuart K. Inhibition of isoleucyl-tRNA synthetase as a potential treatment for human African Trypanosomiasis. J Biol Chem. 2013;288:14256–14263. doi: 10.1074/jbc.M112.447441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestari I, Stuart K. Inositol phosphate pathway controls transcription of telomeric expression sites in trypanosomes. Proc Natl Acad Sci U S A. 2015;112:E2803–E2812. doi: 10.1073/pnas.1501206112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez M, Ena S, Van Sande J, de Kerchove d'Exaerde A, Schurmans S, Schiffmann SN. Modulation of Ciliary Phosphoinositide Content Regulates Trafficking and Sonic Hedgehog Signaling Output. Dev Cell. 2015;34:338–350. doi: 10.1016/j.devcel.2015.06.016. [DOI] [PubMed] [Google Scholar]
- Doyle PS, Chen CK, Johnston JB, Hopkins SD, Leung SS, Jacobson MP, Engel JC, McKerrow JH, Podust LM. A nonazole CYP51 inhibitor cures Chagas' disease in a mouse model of acute infection. Antimicrob Agents Chemother. 2010;54:2480–2488. doi: 10.1128/AAC.00281-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed NM, Myler PJ, Blandin G, Berriman M, Crabtree J, Aggarwal G, Caler E, Renauld H, Worthey EA, Hertz-Fowler C, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309:404–409. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- Endo-Streeter S, Tsui MK, Odom AR, Block J, York JD. Structural studies and protein engineering of inositol phosphate multikinase. J Biol Chem. 2012;287:35360–35369. doi: 10.1074/jbc.M112.365031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamo FJ, Sanz LM, Vidal J, de Cozar C, Alvarez E, Lavandera JL, Vanderwall DE, Green DV, Kumar V, Hasan S, et al. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- Hall BS, Gabernet-Castello C, Voak A, Goulding D, Natesan SK, Field MC. TbVps34, the trypanosome orthologue of Vps34, is required for Golgi complex segregation. J Biol Chem. 2006;281:27600–27612. doi: 10.1074/jbc.M602183200. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Enomoto M, Morales J, Kurebayashi N, Sakurai T, Hashimoto T, Nara T, Mikoshiba K. Inositol 1,4,5-trisphosphate receptor regulates replication, differentiation, infectivity and virulence of the parasitic protist Trypanosoma cruzi. Mol Microbiol. 2013;87:1133–1150. doi: 10.1111/mmi.12155. [DOI] [PubMed] [Google Scholar]
- Holmes W, Jogl G. Crystal structure of inositol phosphate multikinase 2 and implications for substrate specificity. J Biol Chem. 2006;281:38109–38116. doi: 10.1074/jbc.M606883200. [DOI] [PubMed] [Google Scholar]
- Huang G, Bartlett PJ, Thomas AP, Moreno SN, Docampo R. Acidocalcisomes of Trypanosoma brucei have an inositol 1,4,5-trisphosphate receptor that is required for growth and infectivity. Proc Natl Acad Sci U S A. 2013;110:1887–1892. doi: 10.1073/pnas.1216955110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalidas S, Cestari I, Monnerat S, Li Q, Regmi S, Hasle N, Labaied M, Parsons M, Stuart K, Phillips MA. Genetic validation of aminoacyl-tRNA synthetases as drug targets in Trypanosoma brucei. Eukaryot Cell. 2014;13:504–516. doi: 10.1128/EC.00017-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S, Roach SL, Barnes SW, Hoepfner D, Walker JR, Chatterjee AK, Neitz RJ, Arkin MR, McNamara CW, Ballard J, et al. Utilizing Chemical Genomics to Identify Cytochrome b as a Novel Drug Target for Chagas Disease. PLoS Pathog. 2015;11:e1005058. doi: 10.1371/journal.ppat.1005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Maxwell MJ, Hardesty DA, Boucher KL, Choudhari N, Hanno AG, Ma JF, Snowman AS, Pietropaoli JW, Xu R, et al. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc Natl Acad Sci U S A. 2011;108:1391–1396. doi: 10.1073/pnas.1017831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KL, Smith TK. The glycosylphosphatidylinositol (GPI) biosynthetic pathway of bloodstream-form Trypanosoma brucei is dependent on the de novo synthesis of inositol. Mol Microbiol. 2006;61:89–105. doi: 10.1111/j.1365-2958.2006.05216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KL, Smith TK. The myo-inositol-1-phosphate synthase gene is essential in Trypanosoma brucei. Biochem Soc Trans. 2005;33:983–985. doi: 10.1042/BST0330983. [DOI] [PubMed] [Google Scholar]
- Martin KL, Smith TK. Phosphatidylinositol synthesis is essential in bloodstream form Trypanosoma brucei. Biochem J. 2006;396:287–295. doi: 10.1042/BJ20051825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue A, Bhattacharjee S, Pandharkar T, Liu H, Estiu G, Stahelin RV, Rizk SS, Njimoh DL, Ryan Y, Chotivanich K, et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520:683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath ME, Eakin AE, Engel JC, McKerrow JH, Craik CS, Fletterick RJ. The crystal structure of cruzain: a therapeutic target for Chagas' disease. J Mol Biol. 1995;247:251–259. doi: 10.1006/jmbi.1994.0137. [DOI] [PubMed] [Google Scholar]
- Millard CJ, Watson PJ, Celardo I, Gordiyenko Y, Cowley SM, Robinson CV, Fairall L, Schwabe JW. Class I HDACs share a common mechanism of regulation by inositol phosphates. Mol Cell. 2013;51:57–67. doi: 10.1016/j.molcel.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti NS, da Silva Augusto L, Clemente TM, Antunes RP, Yoshida N, Torrecilhas AC, Cano MI, Schenkman S. Characterization of Trypanosoma cruzi Sirtuins as Possible Drug Targets for Chagas Disease. Antimicrob Agents Chemother. 2015;59:4669–4679. doi: 10.1128/AAC.04694-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedro-Rosa L, Buckner FS, Ranade RM, Eberhart C, Madoux F, Gillespie JR, Koh CY, Brown S, Lohse J, Verlinde CL, et al. Identification of potent inhibitors of the Trypanosoma brucei methionyl-tRNA synthetase via high-throughput orthogonal screening. J Biomol Screen. 2015;20:122–130. doi: 10.1177/1087057114548832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena I, Pilar Manzano M, Cantizani J, Kessler A, Alonso-Padilla J, Bardera AI, Alvarez E, Colmenarejo G, Cotillo I, Roquero I, et al. New compound sets identified from high throughput phenotypic screening against three kinetoplastid parasites: an open resource. Sci Rep. 2015;5:8771. doi: 10.1038/srep08771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planer JD, Hulverson MA, Arif JA, Ranade RM, Don R, Buckner FS. Synergy testing of FDA-approved drugs identifies potent drug combinations against Trypanosoma cruzi. PLoS Negl Trop Dis. 2014;8:e2977. doi: 10.1371/journal.pntd.0002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renslo AR, McKerrow JH. Drug discovery and development for neglected parasitic diseases. Nat Chem Biol. 2006;2:701–710. doi: 10.1038/nchembio837. [DOI] [PubMed] [Google Scholar]
- Rodgers MJ, Albanesi JP, Phillips MA. Phosphatidylinositol 4-kinase III-beta is required for Golgi maintenance and cytokinesis in Trypanosoma brucei. Eukaryot Cell. 2007;6:1108–1118. doi: 10.1128/EC.00107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeds AM, Tsui MM, Sunu C, Spana EP, York JD. Inositol phosphate kinase 2 is required for imaginal disc development in Drosophila. Proc Natl Acad Sci U S A. 2015;112:15660–15665. doi: 10.1073/pnas.1514684112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Haswell ES, Miller AL, Wente SR, O'Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K, Brun R, Croft S, Fairlamb A, Gurtler RE, McKerrow J, Reed S, Tarleton R. Kinetoplastids: related protozoan pathogens, different diseases. J Clin Invest. 2008;118:1301–1310. doi: 10.1172/JCI33945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- Watson PJ, Fairall L, Santos GM, Schwabe JW. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481:335–340. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe VO, Savill JM, Chavali S, Jonsdottir AB, Rajendra E, Gruner T, Laskey RA, Babu MM, Venkitaraman AR. Human inositol polyphosphate multikinase regulates transcript-selective nuclear mRNA export to preserve genome integrity. Mol Cell. 2013;51:737–750. doi: 10.1016/j.molcel.2013.08.031. [DOI] [PubMed] [Google Scholar]
- Yamaoka M, Ando T, Terabayashi T, Okamoto M, Takei M, Nishioka T, Kaibuchi K, Matsunaga K, Ishizaki R, Izumi T, et al. PI3K regulates endocytosis after insulin secretion via signaling crosstalk between Arf6 and Rab27a. J Cell Sci. 2015 doi: 10.1242/jcs.180141. [DOI] [PubMed] [Google Scholar]