Abstract
Background
Measures of neocortical amyloid burden (NAB) identify individuals who are at substantially greater risk of developing Alzheimer's disease (AD). Blood-based biomarkers predicting NAB would have great utility for the enrichment of AD clinical trials, including large-scale prevention trials.
Methods
Nontargeted proteomic discovery was applied to 78 subjects from the Australian Imaging, Biomarkers and Lifestyle Flagship Study of Ageing with a range of NAB values. Technical and independent replications were performed by immunoassay.
Results
Seventeen discovery candidates were selected for technical replication. α2-Macroglobulin, fibrinogen γ-chain (FGG), and complement factor H-related protein 1 were confirmed to be associated with NAB. In an independent cohort, FGG plasma levels combined with age predicted NAB had a sensitivity of 59% and specificity of 78%.
Conclusion
A single blood protein, FGG, combined with age, was shown to relate to NAB and therefore could have potential for enrichment of clinical trial populations.
Keywords: Plasma, β amyloid, Proteomics, Alzheimer's disease, Biomarker, Fibrinogen γ-chain, Clinical trials
1. Background
The diagnosis of Alzheimer's disease (AD) can only be confirmed, with certainty, by histologic examination of the brain tissue at autopsy. This inspection should demonstrate considerable evidence of the classic pathologic hallmarks of AD: extracellular amyloid β (Aβ) plaques and intracellular neurofibrillary tangles predominantly composed of hyperphosphorylated tau [1]. Although an age-related disease usually affecting people older than age 65, it is believed the accumulation of Aβ plaques begins 15 to 20 years before clinical presentation [2] and reaches a plateau when cognitive, functional, and behavioral decline occurs [3]. The existing treatments of AD are only capable of temporary symptomatic relief in a subset of patients [4]. Because elevated brain Aβ is an important risk factor for eventual AD, it has become critical to identify individuals at the early stages of Aβ deposition to recruit into clinical trials of potentially disease-modifying therapeutic agents. Three prevention trials of asymptomatic individuals at the early stages of Aβ deposition have recently begun [5].
At present, neuroimaging and cerebrospinal fluid biomarkers are the accepted standards used to provide evidence of ongoing AD pathophysiology related to Aβ plaques [6]. 11C-Pittsburgh compound B (PiB), coupled with positron emission tomography (PET), is widely used in research in measuring in vivo Aβ deposition, because its uptake in AD correlates with Aβ plaques measured neuropathologically in the same brains [7]. The availability of longer lived [18]F- labeled Aβ PET tracers, such as flutemetamol [8] and florbetapir [9] could foster wider usage in the clinic [10]. Early “proof of concept” PiB-PET studies demonstrated an increase of Aβ deposition in most individuals clinically diagnosed with AD, as judged by visual assessment [11] or quantification of tracer uptake [6], [12]. Two large studies, from Victoria (Australia) and the University of California, San Francisco, Memory and Aging Center (UCSF), have shown that PiB-PET could discriminate between AD and non-Aβ dementia [6], [13]. Some, but not all [14], [15], studies have also shown that amyloid deposition as measured using PiB-PET either predicts a decline in cognitive measures or tracks with such declines [2], [16].
Many disease-modifying therapeutic agents being developed target amyloid generation, deposition, or clearance [17]. Recent phase III trials targeting amyloid have reported that approximately 20% of trial participants actually had little or no Aβ when studied later using such PET imaging (suspected nonamyloid pathologic findings) [18]. This is a very serious problem for such trials—success is hard to find in the field of neurodegeneration but likely to be significantly more difficult when a large minority of trial subjects fail to have the primary target pathologic entity.
A solution is to use amyloid-PET scans (∼$3000 per scan) to ensure the presence of the primary target pathologic entity. The first study to use this will be the Anti Amyloid in Asymptomatic AD (A4; n = 1000) prevention trial. In A4, the screen failure rate is anticipated to be even greater (∼66%) owing to the use of asymptomatic subjects. The great expense of the anticipated ∼20% and ∼66% amyloid-PET screen failure rates for clinical and prevention anti-amyloid trials means that a blood test with even relatively low predictive accuracy for neocortical amyloid burden (NAB) has the potential to greatly reduce costs. This would work by applying the blood tests to large numbers of potentially eligible subjects and only performing PET scans on those whose blood test results are positive. This would reduce the screen failure rates and save money if the blood test was comparatively inexpensive. Therefore, a blood-based measure that correlates with the NAB would be of considerable value as an enrichment filter for clinical trials.
The obvious blood candidate biomarker of brain Aβ pathology would be Aβ itself. A systematic review of the published data and meta-analysis by Koyama et al. [19] of 10,303 subjects found that lower plasma Aβ42/Aβ40 ratios were significantly associated with the development of AD. However, the estimates had wide confidence intervals because of the high interstudy differences. As such, plasma Aβ42/Aβ40 ratios are unlikely to be useful by themselves for the prediction of NAB. The same study found that the individual Aβ42 and Aβ40 levels in blood were not significantly associated with AD. Clearly, novel biomarkers are needed that reflect the brain amyloid pathologic features in the blood.
There has been considerable effort in the search for AD blood-based biomarkers. Most studies have used a case-control design, with a clinical diagnosis of AD, determined by the medical history, cognitive assessment findings, and clinical examination results. This classic, case versus age-matched controls approach has identified a large number of putative plasma biomarkers [20], [21], [22]. However, such approaches are intrinsically flawed in the context of AD, because a considerable proportion of cognitively unimpaired controls will be in the prodromal phase of AD (e.g., asymptomatic but with elevated NAB).
An approach to overcome this is to use a nonapparent measure of disease activity (endophenotype paradigm). The endophenotype approach is increasingly being adopted, for example, to study blood-based biomarkers of cognitive decline [23], [24], apolipoprotein E ε4 (APOE ε4) carrier status [25], brain atrophy [26], [27], and hippocampal metabolism [28]. More recently, blood-based biomarkers of NAB, as measured by PiB-PET, have been reported [29], [30], [31]. Both Kiddle et al. [30] and Burnham et al. [31] used the Rules Based Medicine panel of 190 analytes to discover plasma proteins related to NAB and proposed a 13- and 5-analyte model, respectively. These models both contained the protein pancreatic polypeptide.
In a different approach, Thambisetty et al. [29] used two-dimensional gel electrophoresis (2D-GE), coupled with mass spectrometry (MS), to identify protein spots associating with NAB in an unbiased fashion. That study identified 6 proteins for spots associated with NAB, including APOE and complement C3, which were independently replicated in the study by Kiddle et al. [30]. 2D-GE is a well-established technique for blood biomarker research and offers many advantages. However, it is restricted by the lengthy procedure with poor reproducibility that can only identify a small number of “candidate spots” in limited sample sets.
In the present study, we used a method that combines the unbiased approach of gel-based proteomics with high-throughput multiplex technology and the latest in MS instrumentation. This has enabled the identification and quantification of several hundred proteins, comparable to some panel-based arrays, without losing the key advantages of unbiased gel-based discovery. This is the first application of this approach to identify blood-based biomarkers of NAB and was applied to a subset of patients from the Australian Imaging, Biomarkers and Lifestyle Flagship Study of Ageing (AIBL) cohort with either high or low NAB. Promising markers were then replicated using immunoassays, first in the same cohort and then in an independent cohort [13].
2. Material and methods
2.1. Australian Imaging, Biomarkers and Lifestyle Flagship Study of Ageing
The AIBL study is a longitudinal study of ageing, neuroimaging, biomarkers, lifestyle, and clinical and neuropsychological analysis, with a focus on early detection and lifestyle intervention (available at: http://www.aibl.csiro.au/). Additional specifics regarding subject recruitment, diagnosis, and study design have been previously described [32].
2.2. Discovery cohort: assessments, blood collection, and processing
We examined the plasma samples from a subset of 78 subjects from the AIBL study, who had undergone PiB-PET scans. A standardized uptake value ratio (SUVR) cutoff of 1.3 was used to classify subjects as belonging to the PiB+ and PiB− groups. To increase the statistical power, the subjects were selected to be enriched for clear cases of PiB negativity and positivity. The standardized clinical assessments included the Mini-Mental State Examination, and APOE genotypes were available.
The details of blood collection and sample processing have been previously reported [31]. Plasma proteomic analysis and immunoassay measures were undertaken at King's College London.
2.3. AIBL PiB-PET methods
The PiB imaging method of the AIBL study has been previously reported [33]. SUVRs were generated using the cerebellar gray matter as the reference region, as described in the study by Burnham et al. [31]. NAB was expressed as the average SUVR of the mean of the frontal, superior parietal, lateral temporal, lateral occipital, and anterior and posterior cingulate regions.
2.4. Tandem mass tag protein labeling, enzymatic digestion, and peptide extraction
Each sample was randomly assigned and labeled with an amine-reactive tandem mass tag (TMT) reagent (TMT127-TMT131; catalog no. 90064, Thermo Scientific), with TMT126 used to label the study reference, an equal pool of the plasma obtained from all 78 subjects. A complete TMT6Plex combined five labeled plasma samples with a labeled study reference. In general, sample preparation and TMT labeling was performed as previously described [23], [34], with some minor modifications (see Supplemental Methods 1, available online). Each TMT6Plex underwent one-dimensional gel electrophoresis and excised into 10 fractions (see Supplemental Methods 2, available online). The gel pieces were then destained and digested and the peptides extracted and lyophilized to completion before MS analysis (Supplemental Methods 3, available online). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) data separated with one-dimensional GE can show a single protein in multiple fractions. Therefore, identical protein identifications observed in different fractions were considered as separate entities defined as protein molecular weight (MW) isoforms (see Supplemental Methods 5b, available online).
2.5. LC-MS/MS data acquisition
The samples were analyzed using an LTQ Orbitrap Velos instrument (Thermo Scientific) coupled to a Proxeon EASY-nLC II system (Thermo Scientific). Additional details on chromatographic separation and MS data acquisition are outlined in Supplemental Methods 4 (available online).
2.6. Preprocessing of LC-MS/MS data
Raw data files produced in Excalibur software (Thermo Scientific) were processed using Proteome Discoverer, version 1.3 (Thermo Scientific) to determine peptide identification; the subsequent Mascot (version 2.3; available at: http://www.matrixscience.com) output file was used for additional preprocessing and analysis (see Supplemental Methods 5a, available online). A script was written in R to complete the preprocessing, taking into account the experimental setup described (available at: http://github.com/KHP-Informatics/PRQ). We named the script Preprocessing for Relative Quantification of LC-MS/MS data (PRQ; see Supplemental Methods–PRQ, available online). PRQ (1) performs median ratio normalization [35], (2) calculates ratios for each peptide, (3) derives protein level data from peptide scores, and (4) collects the protein scores across all TMT6Plexs.
2.7. UCSF Memory and Aging Center cohort
2.7.1. Replication cohort: assessments, blood collection, and processing
The replication cohort consisted of samples from 79 participants enrolled in the UCSF Alzheimer's Disease Research Center (Table 1). All subjects underwent APOE genotyping, neurologic and cognitive assessments [13], and plasma collection and storage [36], as previously described. AD, frontotemporal dementia, and mild cognitive impairment were diagnosed clinically by consensus applying standard research criteria [37], [38], [39]. All subjects underwent PiB-PET at Lawrence Berkeley National Laboratory on a Siemens ECAT EXACT HR PET (n = 69) or Biograph Truepoint 6 PET/computed tomography (n = 10) [13]. The scans were visually rated as PiB+ or PiB− by an experienced single rater, who was unaware of the clinical and plasma data [13]. The mean 50- to 70-minute SUVR values were extracted from the frontal, parietal, cingulate, and lateral temporal cortex, using the mean activity in the cerebellar gray matter as the reference tissue (for details of image processing, see Lehmann et al. [40]).
Table 1.
Demographics of selected subjects from AIBL and UCSF cohorts
| Variable | AIBL discovery cohort |
UCSF replication cohort |
||||
|---|---|---|---|---|---|---|
| Low neocortical SUVR (PiB−) | High neocortical SUVR (PiB+) | P value | Low neocortical visual PiB read (PiB−) | High neocortical visual PiB read (PiB+) | P value | |
| Subjects (n) | 38 | 40 | 47 | 32 | ||
| SUVR (missing) | 1.11 ± 0.06 | 2.34 ± 0.33 | 2.4 × 10−25 | 1.2 ± 0.12 (1) | 2.2 ± 0.35 (2) | 4.2 × 10−16 |
| Female gender | 18 (47) | 20 (50) | .83 | 18 (38) | 14 (44) | .65 |
| Age (y) | 75.8 ± 6.53 | 80.9 ± 8.22 | .0035 | 65 ± 8.8 | 64 ± 8.4 | .61 |
| Clinical diagnosis | .0037 | 1.9 × 10−10 | ||||
| HC | 13 (34) | 6 (15) | 2 (4.3) | 1 (3.1) | ||
| SMC | 18 (47) | 13 (40) | 1 (2.1) | 1 (3.1) | ||
| MCI | 7 (19) | 16 (30) | 0 (0) | 0 (0) | ||
| AD | 0 (0) | 6 (15) | 2 (4.3) | 23 (72) | ||
| FTD | 0 (0) | 0 (0) | 42 (89.3) | 7 (21.8) | ||
| APOE ε4 carrier | 14 (37) | 25 (63) | .36 | 8 (17) | 13 (41) | .036 |
| MMSE | 28.3 ± 1.8 | 26.8 ± 4.1 | .038 | 26 ± 4.3 | 21 ± 6.9 | .0011 |
Abbreviations: AD, Alzheimer's disease; AIBL, Australian Imaging, Biomarkers and Lifestyle Flagship Study of Ageing; APOE, apolipoprotein E; FTD, frontotemporal dementia; HC, healthy control; MCI, mild cognitive impairment; MMSE, Mini-Mental State Examination; PiB, Pittsburgh compound B; SUVR, standardized uptake value ratio; UCSF, University of California San Francisco.
NOTE. Data presented as mean ± standard deviation or n (%).
2.7.2. Immunoassay–enzyme-linked immunosorbent assay
Single analyte sandwich enzyme-linked immunosorbent assay (ELISA) was used to quantify the candidate proteins and were performed as per the manufacturer's instructions (see Supplemental Methods 6, available online).
2.8. Statistical analysis
All statistical analyses were performed in R (see Supplemental Methods 7, available online). For logistic and linear regression analysis, age, gender, and the presence of an APOE ε4 allele were used as covariates. For the ELISA analysis, data outliers were excluded (±3 standard deviations) and a fourth covariate, batch, was added. The PET scanner type was added as a covariate for the UCSF data. Benjamini-Hochberg q values were calculated as a multiple testing correction. Details of the pathway, regression, and classification analyses are given in Supplemental Methods 7 (available online).
3. Results
3.1. LC-MS/MS performed on AIBL subjects
LC-MS/MS was performed on plasma samples from 78 AIBL subjects, whose demographic data are listed in Table 1. Combining the data from all MS/MS runs, we identified 4518 unique peptides sequences that corresponded to 789 unique protein groups. PRQ was able to extract 2319 unique TMT peptides, 1139 MW isoforms, and 379 unique protein groups (see Supplemental Results 1a, available online), which was reduced to 116 confidently annotated unique protein groups after post-PRQ data clean up. This consisted of 381 protein MW isoforms (see Supplemental Results 1b, available online).
3.2. Plasma protein markers of global PiB-PET
Each protein MW isoform underwent Mann-Whitney U test and logistic regression analysis to compare PiB+ and PiB− groups and the Spearman rank correlation and linear regression to associate the protein MW isoform levels against PiB retention as a continuous measure. This was completed for the mean and median protein roll-up methods separately, giving a total of eight statistical tests per protein. One protein MW isoform, complement C4a, passed all eight statistical tests. A total of 69 protein MW isoforms passed at least one statistical test (uncorrected P < .05; see Supplemental Results 1c, available online). Pathway analysis (see Supplemental Results 2, available online) revealed that these protein groups were overrepresented for involvement in the complement and coagulation cascades (P = 3.7 × 10−22, q = 3.3 × 10−21), systemic lupus erythematosus (P = 2.65 × 10−4, q = 0.15), and prion diseases (P = 5.9 × 10−3, q = 0.051). Three albumin and 15 immunoglobulin MW isoforms were removed, leaving 51 protein MW isoforms associated with PiB-PET retention (Table 2).
Table 2.
LC-MS/MS data: Protein MW isoforms significantly associated with NAB
| UniProt ID | Protein name | 1D-GE fraction | Protein level data (roll up method indicated; i.e., mean or median) |
Tests with P < .05 (n) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean for all peptides mapping to protein |
Median over all peptides mapping to protein |
||||||||||||||||||
| Logistic regression |
Mann-Whitney U test |
Linear regression |
Spearman's rank correlation |
Logistic regression |
Mann-Whitney U test |
Linear regression |
Spearman's rank correlation |
||||||||||||
| β | P value | Median difference | P value | β | P value | ρ | P value | β | P value | Median difference | P value | β | P value | ρ | P value | ||||
| P0C0L4 | Complement C4a | 7 | −0.656 | .038∗ | −0.235 | .027∗ | −0.162 | .019∗ | −0.302 | .007∗ | −0.592 | .049∗ | −0.224 | .037∗ | −0.143 | .042∗ | −0.229 | .044∗ | 8 |
| P00738 | Haptoglobin | 8 | −0.932 | .015∗ | −0.331 | .011∗ | −0.170 | .026∗ | −0.248 | .047∗ | −1.048 | .018∗ | −0.190 | .030∗ | −0.157 | .043∗ | −0.226 | .070 | 7 |
| P02647 | Apolipoprotein A-I | 10 | −0.877 | .014∗ | −0.173 | .070 | −0.211 | .013 | −0.204 | .136 | −0.872 | .025∗ | −0.087 | .087 | −0.183 | .035∗ | −0.266 | .049∗ | 5 |
| O14791 | Apolipoprotein L-1 | 5 | 0.829 | .048∗ | 0.731 | .004∗ | 0.165 | .035∗ | 0.308 | .014∗ | 0.578 | .091 | 0.795 | .007∗ | 0.120 | .145 | 0.238 | .061 | 5 |
| P00738 | Haptoglobin | 5 | −0.728 | .023∗ | −0.521 | .017∗ | −0.198 | .005∗ | −0.280 | .013∗ | −0.446 | .109 | −0.367 | .134 | −0.149 | .041∗ | −0.181 | .112 | 5 |
| Q03591 | FHR-1 | 8 | 1.667 | .120 | 0.313 | .008∗ | 0.155 | .178 | 0.483 | .007∗ | 1.414 | .187 | 0.186 | .019∗ | 0.144 | .217 | 0.444 | .014∗ | 4 |
| P25311 | Zinc-α2-glycoprotein | 7 | 0.140 | .681 | 0.327 | .043∗ | 0.076 | .461 | 0.327 | .021∗ | 0.140 | .681 | 0.327 | .043∗ | 0.076 | .461 | 0.327 | .021∗ | 4 |
| O43866 | CD5 molecule like | 5 | −0.929 | .110 | −0.398 | .013∗ | −0.140 | .088 | −0.288 | .028∗ | −0.596 | .080 | −0.307 | .015∗ | −0.165 | .042∗ | −0.235 | .076 | 4 |
| P04196 | Histidine rich glycoprotein | 5 | −0.996 | .031∗ | −0.093 | .150 | −0.228 | .011∗ | −0.238 | .096 | −0.996 | .031∗ | −0.093 | .150 | −0.228 | .011∗ | −0.238 | .096 | 4 |
| P02747 | C1q subcomponent subunit | 2 | −0.746 | .043∗ | −0.460 | .095 | −0.201 | .024∗ | −0.210 | .165 | −0.746 | .043∗ | −0.460 | .095 | −0.201 | .024∗ | −0.210 | .165 | 4 |
| P00450 | Ceruloplasmin | 6 | −2.088 | .032∗ | −0.222 | .019∗ | −0.105 | .169 | −0.154 | .247 | −2.088 | .032∗ | −0.222 | .019∗ | −0.105 | .169 | −0.154 | .247 | 4 |
| P04196 | Histidine rich glycoprotein | 3 | 1.205 | .039∗ | 0.334 | .034∗ | 0.120 | .169 | 0.106 | .449 | 1.149 | .024∗ | 0.611 | .030∗ | 0.114 | .199 | 0.084 | .548 | 4 |
| P00751 | Complement factor B | 1 | 0.341 | .419 | 0.065 | .657 | 0.164 | .081 | 0.310 | .038∗ | 1.239 | .371 | 0.058 | .188 | 0.189 | .042∗ | 0.363 | .014∗ | 3 |
| P04003 | C4b-binding protein α chain | 4 | −0.405 | .189 | −0.108 | .174 | −0.127 | .076 | −0.248 | .035 | −0.466 | .106 | −0.473 | .038∗ | −0.105 | .154 | −0.275 | .018∗ | 3 |
| P02647 | Apolipoprotein A-I | 6 | −0.487 | .080 | −0.074 | .310 | −0.157 | .023∗ | −0.189 | .098 | −0.403 | .137 | −0.156 | .109 | −0.143 | .038∗ | −0.236 | .038∗ | 3 |
| P02679 | FGG | 6 | 0.108 | .789 | −0.038 | .081 | 0.044 | .613 | −0.331 | .021∗ | −0.671 | .085 | −0.206 | .164 | −0.191 | .021∗ | −0.298 | .039∗ | 3 |
| Q92620 | DEAH box protein 38 | 2 | −0.383 | .351 | 0.046 | .044∗ | −0.095 | .207 | 0.223 | .067 | −0.380 | .351 | 0.046 | .031∗ | −0.095 | .208 | 0.243 | .046∗ | 3 |
| Q06033 | ITI heavy chain H3 | 2 | 1.525 | .027∗ | 0.297 | .099 | 0.148 | .144 | 0.170 | .265 | 1.267 | .019∗ | 0.422 | .013∗ | 0.187 | .061 | 0.225 | .137 | 3 |
| P06727 | Apolipoprotein A-IV | 6 | −1.837 | .206 | −0.100 | .087 | −0.144 | .115 | −0.389 | .010∗ | −1.432 | .271 | −0.093 | .107 | −0.139 | .128 | −0.387 | .010∗ | 2 |
| O43866 | CD5 molecule like | 6 | 0.556 | .275 | 0.277 | .147 | 0.077 | .400 | 0.347 | .022∗ | 0.260 | .484 | 0.623 | .201 | 0.090 | .313 | 0.369 | .015∗ | 2 |
| P08603 | Complement factor H | 6 | 0.031 | .917 | 0.147 | .409 | −0.001 | .989 | 0.128 | .299 | 0.190 | .490 | 0.243 | .041∗ | −0.001 | .987 | 0.286 | .018∗ | 2 |
| P01023 | α2m | 3 | 0.046 | .853 | 0.331 | .130 | 0.048 | .497 | 0.290 | .010∗ | 0.034 | .893 | 0.351 | .223 | 0.042 | .559 | 0.254 | .025∗ | 2 |
| P07357 | Complement C8 α chain | 5 | −0.303 | .346 | −0.294 | .111 | −0.105 | .229 | −0.197 | .137 | −0.539 | .104 | −0.474 | .032∗ | −0.153 | .073 | −0.261 | .048∗ | 2 |
| P00739 | Haptoglobin-related protein | 5 | 0.960 | .076 | 0.190 | .072 | 0.209 | .016∗ | 0.232 | .104 | 0.960 | .076 | 0.190 | .072 | 0.209 | .016∗ | 0.232 | .104 | 2 |
| P08519 | Apolipoprotein(a) | 1 | 0.929 | .062 | 0.315 | .030∗ | 0.115 | .212 | 0.237 | .105 | 0.869 | .064 | 0.222 | .045∗ | 0.111 | .226 | 0.221 | .131 | 2 |
| P04003 | C4b-binding protein α chain | 7 | −0.602 | .064 | −0.198 | .238 | −0.153 | .028∗ | −0.156 | .173 | −0.648 | .052 | −0.199 | .151 | −0.158 | .023∗ | −0.150 | .189 | 2 |
| P19823 | ITI heavy chain H2 | 4 | −0.762 | .067 | −0.294 | .026∗ | −0.159 | .061 | −0.267 | .039∗ | −0.510 | .105 | −0.412 | .125 | −0.134 | .112 | −0.158 | .227 | 2 |
| P01024 | Complement C3 | 6 | −0.374 | .171 | 0.034 | .783 | −0.148 | .035∗ | −0.107 | .351 | −0.445 | .116 | −0.115 | .475 | −0.156 | .025∗ | −0.119 | .300 | 2 |
| Q92620 | DEAH box protein 38 | 5 | 0.449 | .199 | 0.123 | .576 | 0.197 | .038∗ | 0.144 | .318 | 0.449 | .199 | 0.123 | .576 | 0.197 | .038∗ | 0.144 | .318 | 2 |
| Q14624 | ITI heavy chain H4 | 1 | 1.244 | .041∗ | 0.187 | .118 | 0.251 | .013∗ | 0.222 | .142 | 0.651 | .220 | 0.127 | .281 | 0.152 | .139 | 0.136 | .373 | 2 |
| P00747 | Plasminogen | 1 | 0.901 | .026∗ | 0.610 | .078 | 0.151 | .112 | 0.154 | .295 | 0.786 | .049∗ | 0.409 | .102 | 0.128 | .182 | 0.119 | .420 | 2 |
| P06396 | Gelsolin | 5 | 1.018 | .251 | 0.174 | .033∗ | 0.095 | .275 | 0.171 | .198 | 1.149 | .202 | 0.151 | .042∗ | 0.095 | .275 | 0.088 | .509 | 2 |
| P01023 | α2m | 9 | −0.749 | .035∗ | −0.246 | .112 | −0.196 | .037∗ | −0.092 | .482 | −0.555 | .078 | −0.165 | .230 | −0.139 | .113 | −0.044 | .737 | 2 |
| O75636 | Ficolin-3 | 6 | −0.372 | .249 | 0.043 | .629 | −0.152 | .044∗ | 0.034 | .798 | −0.372 | .249 | 0.043 | .629 | −0.152 | .044∗ | 0.034 | .798 | 2 |
| P19827 | ITI heavy chain H4 | 8 | −0.396 | .253 | −0.252 | .301 | −0.155 | .115 | −0.265 | .069 | −0.337 | .373 | −0.196 | .282 | −0.164 | .113 | −0.313 | .030∗ | 1 |
| P01023 | α2m | 2 | −0.044 | .861 | −0.190 | .823 | 0.000 | .996 | 0.013 | .912 | 0.108 | .659 | 0.259 | .171 | 0.048 | .495 | 0.237 | .037 | 1 |
| P02790 | Hemopexin | 5 | 0.255 | .341 | 0.018 | .368 | 0.080 | .252 | 0.152 | .184 | 0.424 | .158 | 0.164 | .123 | 0.105 | .134 | 0.228 | .045∗ | 1 |
| P01023 | α2m | 1 | 0.325 | .196 | 0.198 | .133 | 0.057 | .420 | 0.118 | .303 | 0.340 | .181 | 0.469 | .046∗ | 0.063 | .380 | 0.184 | .107 | 1 |
| P13671 | Complement C6 | 2 | −0.454 | .132 | −0.338 | .064 | −0.089 | .288 | −0.167 | .190 | −0.430 | .145 | −0.662 | .048∗ | −0.085 | .310 | −0.203 | .110 | 1 |
| P02675 | Fibrinogen β chain | 6 | −0.441 | .241 | 0.042 | .869 | −0.113 | .117 | −0.082 | .504 | −0.539 | .090 | −0.213 | .341 | −0.172 | .016∗ | −0.188 | .125 | 1 |
| P02790 | Hemopexin | 6 | 0.587 | .049∗ | 0.216 | .053 | 0.090 | .203 | 0.230 | .059 | 0.429 | .147 | 0.234 | .082 | 0.041 | .562 | 0.166 | .176 | 1 |
| P01024 | Complement C3 | 8 | −0.569 | .058 | −0.480 | .030∗ | −0.129 | .101 | −0.166 | .161 | −0.422 | .132 | −0.401 | .081 | −0.091 | .249 | −0.135 | .255 | 1 |
| P02787 | Serotransferrin | 1 | 0.123 | .658 | 0.205 | .170 | 0.046 | .544 | 0.093 | .450 | 0.249 | .372 | 0.352 | .040∗ | 0.068 | .370 | 0.133 | .281 | 1 |
| P02647 | Apolipoprotein A-I | 5 | −0.458 | .221 | −0.293 | .126 | −0.101 | .229 | −0.263 | .046∗ | −0.237 | .484 | −0.099 | .333 | −0.026 | .762 | −0.129 | .335 | 1 |
| P0C0L4 | Complement C4a | 1 | 0.735 | .117 | 0.058 | .937 | 0.241 | .009∗ | 0.160 | .293 | 0.156 | .631 | −0.004 | .847 | 0.064 | .513 | 0.124 | .419 | 1 |
| P02647 | Apolipoprotein A-I | 9 | −0.599 | .039∗ | −0.061 | .255 | −0.134 | .060 | −0.033 | .780 | −0.414 | .134 | 0.036 | .812 | −0.090 | .211 | 0.067 | .566 | 1 |
| P00450 | Ceruloplasmin | 2 | 0.543 | .136 | −0.097 | .638 | 0.167 | .029∗ | 0.000 | .998 | 0.177 | .524 | 0.038 | .946 | 0.082 | .302 | 0.058 | .639 | 1 |
| P00734 | Prothrombin | 6 | 1.356 | .402 | 0.004 | .409 | 0.190 | .027∗ | 0.208 | .135 | −0.053 | .879 | −0.001 | .908 | −0.002 | .983 | 0.056 | .689 | 1 |
| P10909 | Clusterin | 6 | −0.480 | .134 | 0.036 | .422 | −0.159 | .037∗ | −0.102 | .408 | −0.434 | .179 | −0.150 | .646 | −0.140 | .068 | −0.033 | .789 | 1 |
| P02671 | Fibrinogen α chain | 4 | −0.698 | .122 | −0.175 | .107 | −0.142 | .052 | −0.284 | .012∗ | −0.054 | .835 | 0.065 | .700 | −0.054 | .472 | −0.028 | .809 | 1 |
| P04196 | Histidine rich glycoprotein | 6 | −0.459 | .343 | 0.005 | .447 | −0.185 | .049∗ | −0.194 | .214 | −0.315 | .485 | 0.251 | .895 | −0.163 | .083 | −0.021 | .895 | 1 |
Abbreviations: 1D-GE, one-dimensional gel electrophoresis; α2m, α2-macroglobulin; FGG, fibrinogen γ-chain; FHR-1, factor H-related protein 1; ID, identification; ITI, inter-α-trypsin inhibitor; LC-MS/MS, liquid chromatography tandem mass spectrometry; UniProt, Universal Protein Resource.
NOTE. All multiple testing corrected q values were >0.75; for regression analysis, age, gender, and presence of apolipoprotein E ε4 were used as covariates.
∗Statistically significant.
Subsequently, 17 proteins were selected for technical replication (Fig. 1). In addition to the statistical evidence, we also considered the candidate's relationship with the amyloid and/or AD Genome-Wide Association Studies results (see Supplemental Results 3, available online). We also chose to replicate histidine-rich glycoprotein, the protein most associated with NAB, but that had no previous evidence for a relationship with Aβ.
Fig. 1.
Flow diagram to select liquid chromatography tandem mass spectrometry plasma neocortical amyloid burden (NAB) candidate markers for technical replication. ∗Two protein molecular weight (MW) isoforms associated with NAB; ∗∗three protein MW isoforms associated with NAB; ∗∗∗four protein MW isoforms. Abbreviations: Aβ, amyloid β; α2m, α2-macrohpage; FGG, fibrinogen γ-chain; FHR-1, factor H-related protein 1.
3.3. Technical replication
We sought to translate our discovery findings to a simple-to-use commercially available ELISA format. The 17 protein candidates from MS were measured in plasma samples from the 78 AIBL subjects in the discovery cohort. Using linear regression models (including age, gender, APOE ε4, and ELISA plate as covariates), we found that two proteins, α2-macroglobulin (α2m) (q = 0.076) and fibrinogen γ-chain (FGG) (q = 0.076), replicated our findings from the LC-MS/MS discovery study (Table 3). In the discovery study, factor H-related protein 1 (FHR-1) was increased in the PiB+ group. Although FHR-1 (q = 0.076) was associated with NAB at the 0.1 q value in the ELISA technical replication, an opposite trend was observed. Apolipoprotein A-IV, gelsolin, histidine-rich glycoprotein, haptoglobin, and apolipoprotein(a) all showed the same directional change as in the LC-MS/MS discovery.
Table 3.
Technical replication of plasma protein candidates discovered by LC-MS/MS
| UniProt ID | Protein name | Outliers excluded (n) | Logistic regression with SUVR >1.3 |
Linear regression with SUVR |
||||
|---|---|---|---|---|---|---|---|---|
| β | P value | q value | β | P value | q value | |||
| P01023 | α2m | 10 | 1 | 8.9 × 10−3 | 0.076 | 0.2 | 7.9 × 10−3 | 0.068 |
| Q03591 | FHR-1 | 11 | −1 | 4.6 × 10−3 | 0.076 | −0.22 | 5.5 × 10−3 | 0.068 |
| P02679 | FGG | 0 | −0.7 | .041 | 0.23 | −0.2 | .014 | 0.081 |
| P08519 | Apolipoprotein(a) | 21 | 0.48 | .13 | 0.34 | 0.18 | .042 | 0.18 |
| P06396 | Gelsolin | 2 | −0.48 | .11 | 0.34 | −0.14 | .068 | 0.19 |
| P00738 | Haptoglobin | 2 | −0.38 | .18 | 0.39 | −0.13 | .089 | 0.19 |
| P04196 | Histidine rich glycoprotein | 2 | 0.48 | .14 | 0.34 | 0.14 | .081 | 0.19 |
| P06727 | Apolipoprotein A-IV | 2 | −0.63 | .083 | 0.34 | −0.17 | .067 | 0.19 |
| P01024 | Complement C3 | 0 | −0.61 | .25 | 0.47 | −0.21 | .13 | 0.25 |
| P0C0L4 | Complement C4a | 0 | −0.55 | .51 | 0.66 | −0.27 | .22 | 0.38 |
| P10909 | Clusterin | 0 | −0.27 | .36 | 0.51 | −0.091 | .27 | 0.41 |
| P02647 | Apolipoprotein A-I | 0 | 0.34 | .29 | 0.47 | 0.088 | .32 | 0.46 |
| P02671 | Fibrinogen α chain | 6 | −0.28 | .3 | 0.47 | −0.064 | .39 | 0.52 |
| P02787 | Serotransferrin | 1 | −0.013 | .96 | 0.96 | −0.041 | .6 | 0.73 |
| O14791 | Apolipoprotein L-1 | 0 | −0.09 | .74 | 0.89 | −0.026 | .73 | 0.77 |
| P08603 | Complement factor H | 3 | 0.066 | .8 | 0.89 | 0.027 | .7 | 0.77 |
| P00751 | Complement factor B | 2 | 0.053 | .84 | 0.89 | 0.018 | .81 | 0.81 |
Abbreviations: 1D-GE, one-dimensional gel electrophoresis; α2m, α2-macroglobulin; FGG, fibrinogen γ-chain; FHR-1, factor H-related protein 1; ID, identification; ITI, inter-α-trypsin inhibitor; LC-MS/MS, liquid chromatography tandem mass spectrometry; UniProt, Universal Protein Resource.
3.4. Independent replication
To verify the results from the AIBL samples, we measured the levels of the three proteins significantly associated with NAB (α2m, FHR-1, and FGG) using samples from an independent cohort. These proteins were measured using ELISA in 79 samples from the UCSF cohort (Table 1). FGG was significantly associated with PiB positivity, as determined both by visual examination of PiB-PET scans (q = 5.9 × 10−3) and by applying a threshold of 1.3 to SUVRs (q = 0.051; Table 4). Despite not being significantly associated with NAB, α2m correlated with SUVR positivity in the same direction as in the discovery study.
Table 4.
Independent replication of plasma protein candidates discovered by LC-MS/MS and technically replicated
| UniProt ID | Protein name | Logistic regression to visual read |
Logistic regression to SUVR >1.3 |
Linear regression to SUVR |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β | P value | q value | β | P value | q value | β | P value | q value | ||
| P01023 | α2m | −0.013 | .96 | 0.96 | 0.27 | .29 | 0.44 | 0.075 | .22 | 0.33 |
| P02679 | FGG | −1.0 | 2.0 × 10−3 | 5.9 × 10−3 | −0.74 | .017 | 0.051 | −0.21 | 4.1 × 10−4 | 1.2 × 10−3 |
| Q03591 | FHR-1 | −0.066 | .79 | 0.96 | 0.011 | .97 | 0.97 | 1.5 × 10−3 | .98 | 0.98 |
Abbreviations: α2m, α2-macroglobulin; FGG, fibrinogen γ-chain; FHR-1, factor H-related protein 1; ID, identification; LC-MS/MS, liquid chromatography-tandem mass spectrometry; SUVR, standardized uptake value ratio; UniProt, Universal Protein Resource.
NOTE. Only one outlier (>3 standard deviations from mean) was excluded, which was detected for FGG; for regression analysis, age, gender, presence of apolipoprotein E ε4, enzyme-linked immunosorbent assay plate, and scanner type were used as covariates.
3.5. Multivariate analysis
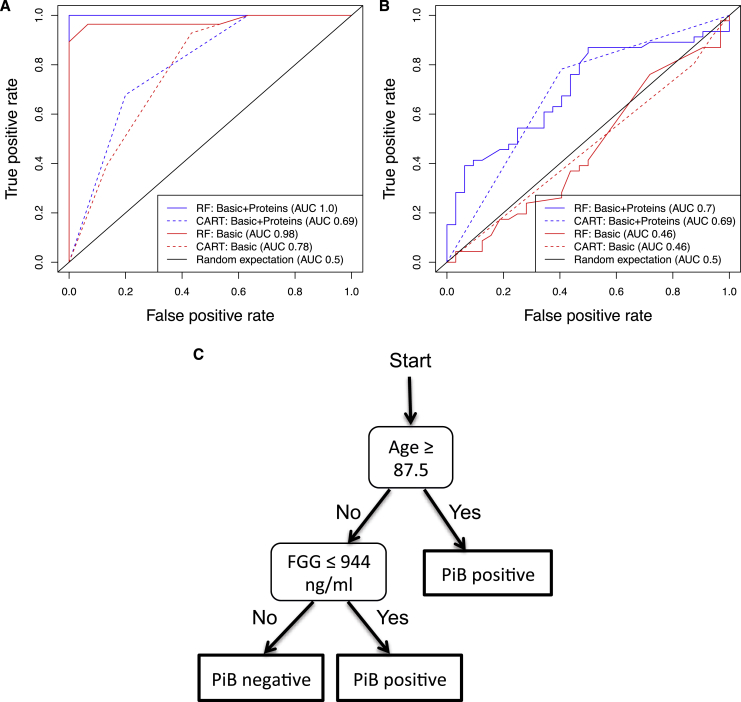
Subjects with any missing covariates or protein measurements were excluded from the multivariate analysis, leaving 58 subjects from AIBL (28 PiB−, 30 PiB+ using an SUVR >1.3) and 78 subjects from UCSF (46 PiB−, 32 PiB+ determined from visual inspection). Classification models were trained in the AIBL ELISA data to predict SUVR positivity (>1.3) and tested in the UCSF ELISA data to predict PiB positivity, determined by visual inspection (more robust across multiple scanners). A “basic” model (age, gender, APOE ε4) was compared with a “basic + protein” model, which also used the plasma concentration of FGG, α2m, and FHR-1. Fig. 2A and 2B shows a receiver operating characteristic analysis, in which area under the curve (AUC) was shown to be greater for the “basic + protein” model than for the “basic” model in the test data sets. The highest test AUC was found using the random forest approach, in which the basic + protein model (AUC 0.70) outperformed the basic model (AUC = 0.46) in the test data set. The random forest basic + protein model gave a test set sensitivity of 50% and specificity of 85%. Additionally, a classification tree was fitted to the basic + protein model to provide a simpler alternative with clear thresholds. The resulting classification tree used just two variables (age/plasma FGG level; Fig. 2C) and achieved a comparable AUC to that of the random forest model (AUC 0.69, sensitivity 59%, specificity 78%). In the UCSF cohort, 23 of 25 AD subjects were PiB+, and two PiB− subjects had plasma FGG levels greater than the threshold (see Supplemental Results 4, available online).
Fig. 2.
Receiver operating characteristic (ROC) curves for the prediction of Pittsburgh compound B (PiB) positivity. A “basic” model (age, gender, APOE ε4 presence) was compared with a “basic + protein” model that also included the plasma levels of fibrinogen γ-chain (FGG), α2-macrophage, and factor H-related protein 1. Random forest (RF) and classification and regression trees (CARTs) were used to fit models in classification and regression training using default parameters. The area under the curve (AUC) is given for each model. ROC curves are shown comparing the predictive accuracy of the models in (A) the training data set (Australian Imaging, Biomarkers and Lifestyle Flagship Study of Ageing [AIBL]) and (B) the test data set (University of California, San Francisco). Classification tree trained on AIBL enzyme-linked immunosorbent assay data to predict neocortical amyloid burden positivity and estimated cutoff (C).
4. Discussion
With the failure of serial amyloid-based therapeutic agents in clinical trials compromised by the inclusion of substantial numbers of participants without the target pathologic entity [18], and with the prospect of very large trials in presymptomatic AD such as the A4 trial and others [5], the need for blood-based markers of NAB has never been greater. Blood-based biomarkers could be used to screen large numbers of potential participants, and only those predicted to have abnormally high NAB would be retested using cerebrospinal fluid assays or PET scans, reducing the screen failure rates. This could reduce recruitment time and costs and allow eligible subjects to be identified more readily (e.g., from biobanks with permission for recontact).
The present study has demonstrated that a simple blood test consisting of FGG plasma levels and patient age could have some potential for predicting NAB, achieving a test set sensitivity, specificity, and AUC of 59%, 78%, and 69% respectively, highlighting its potential use in stratifying patients for anti-amyloid trials. This independent replication was performed in a mixed dementia cohort (UCSF), suggesting that FGG and age could also have utility for distinguishing between amyloid and nonamyloid dementias. Additionally, because the classification model was trained in a subset of the AIBL cohort containing very few AD subjects, it is more likely that FGG will be able to predict PiB positivity in non-AD subjects. However, because the UCSF cohort contained only two cognitively normal individuals, additional work is needed to determine the sensitivity and specificity in those who are cognitively normal. These measures will determine the cost-saving potential of this blood test for prevention trials. Preliminary data generated from a cognitively normal cohort in our laboratory supports this (data not shown). Previously, Burnham et al. [31] reported a blood test that achieved 79% sensitivity and 76% specificity in an independent test set. Although our sensitivity was slightly lower, this was achieved by measuring a single plasma protein compared with the 6 plasma proteins used in the Burnham model.
Although the sensitivity and specificity of these markers for predicting NAB was not high enough to use clinically, they would be useful for enrichment of clinical trials if they performed at this level in relevant populations. The strongest case can be made for prevention trials in asymptomatic subjects because of the large expected screen failure rate (∼66% or higher) when searching for individuals with elevated NAB. Because of the relatively high cost of amyloid-PET scans (∼$3000) versus blood protein ELISAs, even a blood test without clinical utility could theoretically save millions of dollars for studies the size of A4 (n = 1000).
The APOE ε4 allele is a substantial risk factor for AD [41] and amyloid [30], [42]. Although we considered APOE ε4 during our analyses, we were not surprised to find that the APOE genotype markers did not improve our classification model, because the study was designed to be independent of this effect. However, in a general population sample, the APOE genotype is likely to contribute to the prediction of NAB.
It was interesting that FGG, and to a lesser extent complement C3 and fibrinogen α-chain, were associated with NAB in our study, which has been previously found [29], [30]. However, in the study by Burnham et al. [31], total fibrinogen was not associated with NAB. In contrast, Kiddle et al. [30] showed that it was negatively associated with NAB. Furthermore, decreased levels of plasma FGG have been shown to be associated with a smaller whole brain volume in AD subjects [26] and measures of whole fibrinogen in plasma have shown an increase [43], [44]. Discrepancies in these findings might have resulted from the platform used to measure total fibrinogen or might highlight the importance of studying specific fibrinogen chains.
FGG is normally rejected from the brain by the blood-brain barrier (BBB), yet has still been detected in mice and human brain tissue [45], [46]. This could result from the reported dysfunction of the BBB in mice [47] and humans in AD [48]. However, the movement of fibrinogen across a defected BBB seems to be molecule-specific, because smaller molecules are not BBB permeable in AD [49]. Fibrinogen has been shown to accumulate over time as the AD pathology progresses [46] and codeposits with Aβ in brain tissue [50]. Ahn et al. [51] demonstrated that fibrinogen binds to Aβ, which enhances aggregation and increases Aβ fibrillization. It is possible that the decreased FGG levels associated with high NAB in our study resulted from movement of fibrinogen across a compromised BBB in the subjects with AD pathologic features.
After FGG, α2m was the second most promising candidate, shown for the first time to associate with NAB. This is noteworthy, because α2m has been found to be one of the most replicable markers of other AD-related phenotypes, including diagnosis, hippocampal metabolism, and response to treatment with divalproex sodium [20]. Future studies should aim to replicate all previously discovered markers of NAB and investigate which combination of analytes would achieve greater sensitivity and specificity.
To our knowledge, this is the first study to apply an unbiased and nontargeted quantitative LC-MS/MS discovery approach, combining LC-MS/MS with TMT labeling, for the investigation of plasma proteins related to NAB. Furthermore, this method will allow the unprecedented exploration of plasma peptide and modified proteins as markers of NAB. We have also described a novel and automated bioinformatic pipeline, PRQ, to accurately preprocess the TMT-MS data. PRQ not only conducts rigorous normalization of MS data [35] but also automates the calculation of peptide/protein ratios against the study reference.
Subsequently, technical replication was performed to reduce the number of false-positive results and to ensure translation of LC/MS-MS findings using a platform more applicable to the clinical setting. Using commercially available immunoassays, we confirmed that α2m, FGG, and FHR-1 significantly predicted NAB, with a 0.1 q-value significance level. All except FHR-1 displayed a similar direction of association between discovery and replication. Immunoassays cannot always distinguish between sequence variants, proteins modified with different post-translational modification, or different truncated forms of a same protein seen by LC/MS-MS. This could also explain the differences seen in the association trend between discovery and replication in some cases (e.g., FHR-1); therefore, these candidates should not necessarily be discounted. The discrepancies observed between the two platforms point to the need for investigation of protein modifications as potential biomarkers in future studies.
The discrepancies between the findings from the AIBL and UCSF studies could have resulted from the low statistical power, differences in disease stage, or differences in preanalytical factors. The major difference in the preanalytical factors is the centrifugation step of plasma collection. AIBL used a two-step centrifugation (200g, remove supernatant, then 800g), and UCSF used a single centrifugation step (1300–1800g). This highlights the importance of standardization of blood collection and preparation for biomarker studies.
Although many have agreed that Aβ deposition is the earliest event in AD pathogenesis, one group has shown changes in episodic memory preceding the changes in Aβ levels [52]. If confirmed in other cohorts, it would be interesting to compare the ability of episodic memory and our blood test to predict NAB in asymptomatic individuals.
In conclusion, the present study reports a potential blood test, consisting of measuring FGG, which, along with age, showed some ability to predict NAB in an independent sample. To ensure the robustness and relevance of these findings, this test must be replicated in larger cohorts that are more representative of the relevant clinical trial populations. Our results provide additional evidence that differences in the plasma proteome in relation to AD and its pathology do exist. Therefore, such changes could be used to stratify patients for anti-amyloid treatment trials. This could lower barriers to the development of an effective treatment to combat the increasing concern of dementia.
Research in context.
-
1.
Systematic review: We searched PubMed up to June 2014 using the keywords, Alzheimer's disease (AD), plasma biomarkers, pathology, fibrinogen gamma chain (FGG), beta amyloid.
-
2.
Interpretation: Previous targeted studies have demonstrated that the combination of several blood proteins had the potential to predict the NAB. Our study is the first to apply an unbiased and nontargeted quantitative LC-MS/MS discovery approach, combined with isobaric labeling, to investigate the plasma proteins related to NAB. Our findings showed a prediction of NAB, with comparable accuracy to previous studies, which used a single plasma protein. This highlights the potential use of a simple blood-based screen to stratify patients for anti-amyloid trials.
-
3.
Future directions: To ensure the robustness and reliability of these findings, this test should be replicated in larger cohorts more representative of relevant clinical trial populations.
Acknowledgments
Steven Kiddle is supported by an MRC Career Development Award in Biostatistics (MR/L011859/1). We are grateful for grant funding from Alzheimer's Research UK, the Alzheimer's Society (to S.W.), and the National Institute for Health Research (NIHR) Biomedical Research Centre for Mental Health and Biomedical Research Unit for Dementia at the South London and Maudsley NHS Foundation Trust and Kings College London, and a joint infrastructure grant from Guy's and St Thomas' Charity and the Maudsley Charity. This report presents independent research funded in part by the National Institute for Health Research. A portion of this work was funded by GE Healthcare and Janssen Research & Development. The views expressed are those of the authors and not necessarily those of the NHS, NIHR, Department of Health, GE Healthcare, or Janssen Research & Development. The UCSF study was funded by the U.S. National Institutes of Health grants K23-AG031861, R01-AG027859, R01-AG032306, R01-AG038791, P01-AG1972403 and P50-AG023501; the State of California Department of Health Services; the Alzheimer's Association, and the John Douglas French Alzheimer's Foundation. We also thank William Jagust, MD, for PiB-PET imagery. Finally, we wish to express our appreciation to all study participants of the AIBL and UCSF studies.
Footnotes
Conflict of Interest: The authors have no conflicts of interest to disclose.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.dadm.2014.11.005.
Supplementary data
References
- 1.Blennow K., Hampel H., Weiner M., Zetterberg H. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat Rev Neurol. 2010;6:131–144. doi: 10.1038/nrneurol.2010.4. [DOI] [PubMed] [Google Scholar]
- 2.Villemagne V.L., Pike K.E., Chetelat G., Ellis K.A., Mulligan R.S., Bourgeat P. Longitudinal assessment of Aβ and cognition in aging and Alzheimer disease. Ann Neurol. 2011;69:181–192. doi: 10.1002/ana.22248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Villemagne V.L., Burnham S., Bourgeat P., Brown B., Ellis K.A., Salvado O. Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer's disease: A prospective cohort study. Lancet Neurol. 2013;12:357–367. doi: 10.1016/S1474-4422(13)70044-9. [DOI] [PubMed] [Google Scholar]
- 4.Corbett A., Smith J., Ballard C. New and emerging treatments for Alzheimer's disease. Expert Rev Neurother. 2012;12:535–543. doi: 10.1586/ern.12.43. [DOI] [PubMed] [Google Scholar]
- 5.Aisen P.S., Vellas B., Hampel H. Moving towards early clinical trials for amyloid-targeted therapy in Alzheimer's disease. Nat Rev Drug Discov. 2013;12:324. doi: 10.1038/nrd3842-c1. [DOI] [PubMed] [Google Scholar]
- 6.Rowe C.C., Ng S., Ackermann U., Gong S.J., Pike K., Savage G. Imaging beta-amyloid burden in aging and dementia. Neurology. 2007;68:1718–1725. doi: 10.1212/01.wnl.0000261919.22630.ea. [DOI] [PubMed] [Google Scholar]
- 7.Ikonomovic M.D., Klunk W.E., Abrahamson E.E., Mathis C.A., Price J.C., Tsopelas N.D. Post-mortem correlates of in vivo PiB-PET amyloid imaging in a typical case of Alzheimer's disease. Brain. 2008;131:1630–1645. doi: 10.1093/brain/awn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelissen N., Van Laere K., Thurfjell L., Owenius R., Vandenbulcke M., Koole M. Phase 1 study of the Pittsburgh compound B derivative 18F-flutemetamol in healthy volunteers and patients with probable Alzheimer disease. J Nucl Med. 2009;50:1251–1259. doi: 10.2967/jnumed.109.063305. [DOI] [PubMed] [Google Scholar]
- 9.Johnson K.A., Sperling R.A., Gidicsin C.M., Carmasin J.S., Maye J.E., Coleman R.E. Florbetapir (F18-AV-45) PET to assess amyloid burden in Alzheimer's disease dementia, mild cognitive impairment, and normal aging. Alzheimers Dement. 2013;9(5 Suppl):S72–S83. doi: 10.1016/j.jalz.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordberg A., Rinne J.O., Kadir A., Langstrom B. The use of PET in Alzheimer disease. Nat Rev Neurol. 2010;6:78–87. doi: 10.1038/nrneurol.2009.217. [DOI] [PubMed] [Google Scholar]
- 11.Klunk W.E., Engler H., Nordberg A., Wang Y., Blomqvist G., Holt D.P. Imaging brain amyloid in Alzheimer's disease with Pittsburgh compound-B. Ann Neurol. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 12.Ng S., Villemagne V.L., Berlangieri S., Lee S.T., Cherk M., Gong S.J. Visual assessment versus quantitative assessment of 11C-PIB PET and 18F-FDG PET for detection of Alzheimer's disease. J Nucl Med. 2007;48:547–552. doi: 10.2967/jnumed.106.037762. [DOI] [PubMed] [Google Scholar]
- 13.Rabinovici G.D., Rosen H.J., Alkalay A., Kornak J., Furst A.J., Agarwal N. Amyloid vs FDG-PET in the differential diagnosis of AD and FTLD. Neurology. 2011;77:2034–2042. doi: 10.1212/WNL.0b013e31823b9c5e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagust W.J., Landau S.M., Shaw L.M., Trojanowski J.Q., Koeppe R.A., Reiman E.M. Relationships between biomarkers in aging and dementia. Neurology. 2009;73:1193–1199. doi: 10.1212/WNL.0b013e3181bc010c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheinin N.M., Aalto S., Koikkalainen J., Lotjonen J., Karrasch M., Kemppainen N. Follow-up of [11C]PIB uptake and brain volume in patients with Alzheimer disease and controls. Neurology. 2009;73:1186–1192. doi: 10.1212/WNL.0b013e3181bacf1b. [DOI] [PubMed] [Google Scholar]
- 16.Villain N., Chetelat G., Grassiot B., Bourgeat P., Jones G., Ellis K.A. Regional dynamics of amyloid-β deposition in healthy elderly, mild cognitive impairment and Alzheimer's disease: A voxelwise PiB-PET longitudinal study. Brain. 2012;135:2126–2139. doi: 10.1093/brain/aws125. [DOI] [PubMed] [Google Scholar]
- 17.Blennow K., Hampel H., Zetterberg H. Biomarkers in amyloid-β immunotherapy trials in Alzheimer's disease. Neuropsychopharmacology. 2014;39:189–201. doi: 10.1038/npp.2013.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salloway S., Sperling R., Fox N.C., Blennow K., Klunk W., Raskind M. Two phase 3 trials of bapineuzumab in mild-to-moderate Alzheimer's disease. N Engl J Med. 2014;370:322–333. doi: 10.1056/NEJMoa1304839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koyama A., Okereke O.I., Yang T., Blacker D., Selkoe D.J., Grodstein F. Plasma amyloid-β as a predictor of dementia and cognitive decline: A systematic review and meta-analysis. Arch Neurol. 2012;69:824–831. doi: 10.1001/archneurol.2011.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiddle S.J., Sattlecker M., Proitsi P., Simmons A., Westman E., Bazenet C. Candidate blood proteome markers of Alzheimer's disease onset and progression: A systematic review and replication study. J Alzheimers Dis. 2014;38:515–531. doi: 10.3233/JAD-130380. [DOI] [PubMed] [Google Scholar]
- 21.Lista S., Faltraco F., Prvulovic D., Hampel H. Blood and plasma-based proteomic biomarker research in Alzheimer's disease. Prog Neurobiol. 2013;101-102:1–17. doi: 10.1016/j.pneurobio.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Zurbig P., Jahn H. Use of proteomic methods in the analysis of human body fluids in Alzheimer research. Electrophoresis. 2012;33:3617–3630. doi: 10.1002/elps.201200360. [DOI] [PubMed] [Google Scholar]
- 23.Guntert A., Campbell J., Saleem M., O'Brien D.P., Thompson A.J., Beyers H.L. Plasma gelsolin is decreased and correlates with rate of decline in Alzheimer's disease. J Alzheimers Dis. 2010;21:585–596. doi: 10.3233/JAD-2010-100279. [DOI] [PubMed] [Google Scholar]
- 24.Sattlecker M., Kiddle S.J., Newhouse S., Proitsi P., Nelson S., Williams S. Alzheimer's disease biomarker discovery using SOMAscan multiplexed protein technology. Alzheimers Dement. 2014;10:724–734. doi: 10.1016/j.jalz.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 25.Song F., Poljak A., Crawford J., Kochan N.A., Wen W., Cameron B. Plasma apolipoprotein levels are associated with cognitive status and decline in a community cohort of older individuals. PLoS One. 2012;7:e34078. doi: 10.1371/journal.pone.0034078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thambisetty M., Simmons A., Hye A., Campbell J., Westman E., Zhang Y. Plasma biomarkers of brain atrophy in Alzheimer's disease. PLoS One. 2011;6:e28527. doi: 10.1371/journal.pone.0028527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hye A., Riddoch-Contreras J., Baird A.L., Ashton N.J., Bazenet C., Leung R. Plasma proteins predict conversion to dementia from prodromal disease. Alzheimers Dement. 2014;10:799–807. doi: 10.1016/j.jalz.2014.05.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thambisetty M., Hye A., Foy C., Daly E., Glover A., Cooper A. Proteome-based identification of plasma proteins associated with hippocampal metabolism in early Alzheimer's disease. J Neurol. 2008;255:1712–1720. doi: 10.1007/s00415-008-0006-8. [DOI] [PubMed] [Google Scholar]
- 29.Thambisetty M., Tripaldi R., Riddoch-Contreras J., Hye A., An Y., Campbell J. Proteome-based plasma markers of brain amyloid-β deposition in non-demented older individuals. J Alzheimers Dis. 2010;22:1099–1109. doi: 10.3233/JAD-2010-101350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiddle S.J., Thambisetty M., Simmons A., Riddoch-Contreras J., Hye A., Westman R. Plasma based markers of [11C] PiB-PET brain amyloid burden. PLoS One. 2012;7:e44260. doi: 10.1371/journal.pone.0044260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burnham S.C., Faux N.G., Wilson W., Laws S.M., Ames D., Bedo J. A blood-based predictor for neocortical Aβ burden in Alzheimer's disease: Results from the AIBL study. Mol Psychiatry. 2014;19:519–526. doi: 10.1038/mp.2013.40. [DOI] [PubMed] [Google Scholar]
- 32.Ellis K.A., Bush A.I., Darby D., De Fazio D., Foster J., Hudson P. The Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging: Methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer's disease. Int Psychogeriatr. 2009;21:672–687. doi: 10.1017/S1041610209009405. [DOI] [PubMed] [Google Scholar]
- 33.Rowe C.C., Ellis K.A., Rimajova M., Bourgeat P., Pike K.E., Jones G. Amyloid imaging results from the Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging. Neurobiol Aging. 2010;31:1275–1283. doi: 10.1016/j.neurobiolaging.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Dayon L., Hainard A., Licker V., Turck N., Kuhn K., Hochstrasser D.F. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem. 2008;80:2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 35.Carrillo B., Yanofsky C., Laboissiere S., Nadon R., Kearney R.E. Methods for combining peptide intensities to estimate relative protein abundance. Bioinformatics. 2010;26:98–103. doi: 10.1093/bioinformatics/btp610. [DOI] [PubMed] [Google Scholar]
- 36.Bettcher B.M., Watson C.L., Walsh C.M., Lobach I.V., Neuhaus J., Miller J.W. Interleukin-6, age, and corpus callosum integrity. PLoS One. 2014;9:e106521. doi: 10.1371/journal.pone.0106521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 38.Petersen R.C., Smith G.E., Waring S.C., Ivnik R.J., Tangalos E.G., Kokmen E. Mild cognitive impairment: Clinical characterization and outcome. Arch Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 39.Neary D., Snowden J.S., Gustafson L., Passant U., Stuss D., Black S. Frontotemporal lobar degeneration: A consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- 40.Lehmann M., Ghosh P.M., Madison C., Laforce R., Jr., Corbetta-Rastelli C., Weiner M.W. Diverging patterns of amyloid deposition and hypometabolism in clinical variants of probable Alzheimer's disease. Brain. 2013;136:844–858. doi: 10.1093/brain/aws327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blacker D., Haines J.L., Rodes L., Terwedow H., Go R.C., Harrell L.E. ApoE-4 and age at onset of Alzheimer's disease: The NIMH genetics initiative. Neurology. 1997;48:139–147. doi: 10.1212/wnl.48.1.139. [DOI] [PubMed] [Google Scholar]
- 42.Thambisetty M., An Y., Nalls M., Sojkova J., Swaminathan S., Zhou Y. Effect of complement CR1 on brain amyloid burden during aging and its modification by APOE genotype. Biol Psychiatry. 2013;73:422–428. doi: 10.1016/j.biopsych.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Oijen M., Witteman J.C., Hofman A., Koudstaal P.J., Breteler M.M. Fibrinogen is associated with an increased risk of Alzheimer disease and vascular dementia. Stroke. 2005;36:2637–2641. doi: 10.1161/01.STR.0000189721.31432.26. [DOI] [PubMed] [Google Scholar]
- 44.Xu G., Zhang H., Zhang S., Fan X., Liu X. Plasma fibrinogen is associated with cognitive decline and risk for dementia in patients with mild cognitive impairment. Int J Clin Pract. 2008;62:1070–1075. doi: 10.1111/j.1742-1241.2007.01268.x. [DOI] [PubMed] [Google Scholar]
- 45.Cortes-Canteli M., Paul J., Norris E.H., Bronstein R., Ahn H.J., Zamolodchikov D. Fibrinogen and beta-amyloid association alters thrombosis and fibrinolysis: A possible contributing factor to Alzheimer's disease. Neuron. 2010;66:695–709. doi: 10.1016/j.neuron.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryu J.K., McLarnon J.G. A leaky blood-brain barrier, fibrinogen infiltration and microglial reactivity in inflamed Alzheimer's disease brain. J Cell Mol Med. 2009;13:2911–2925. doi: 10.1111/j.1582-4934.2008.00434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paul J., Strickland S., Melchor J.P. Fibrin deposition accelerates neurovascular damage and neuroinflammation in mouse models of Alzheimer's disease. J Exp Med. 2007;204:1999–2008. doi: 10.1084/jem.20070304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marques F., Sousa J.C., Sousa N., Palha J.A. Blood–brain-barriers in aging and in Alzheimer's disease. Mol Neurodegener. 2013;8:38. doi: 10.1186/1750-1326-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sagare A., Deane R., Bell R.D., Johnson B., Hamm K., Pendu R. Clearance of amyloid-beta by circulating lipoprotein receptors. Nat Med. 2007;13:1029–1031. doi: 10.1038/nm1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klohs J., Baltes C., Princz-Kranz F., Ratering D., Nitsch R.M., Kneusel I. Contrast-enhanced magnetic resonance microangiography reveals remodeling of the cerebral microvasculature in transgenic ArcAβ mice. J Neurosci. 2012;32:1705–1713. doi: 10.1523/JNEUROSCI.5626-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahn H.J., Zamolodchikov D., Cortes-Canteli M., Norris E.H., Glickman J.F., Strickland S. Alzheimer's disease peptide beta-amyloid interacts with fibrinogen and induces its oligomerization. Proc Natl Acad Sci U S A. 2010;107:21812–21817. doi: 10.1073/pnas.1010373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jedynak B.M., Lang A., Liu B., Katz E., Zhang Y., Wyman B.T. A computational neurodegenerative disease progression score: Method and results with the Alzheimer's Disease Neuroimaging Initiative cohort. Neuroimage. 2012;63:1478–1486. doi: 10.1016/j.neuroimage.2012.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.