Abstract
We present comprehensive single amino acid resolution maps of the residues stabilising the human Gαi1 subunit in nucleotide- and receptor-bound states. We generated these maps by measuring the effects of alanine mutations on the stability of Gαi1 and of the rhodopsin-Gαi1 complex. We identified stabilization clusters in the GTPase and helical domains responsible for structural integrity and the conformational changes associated with activation. In activation cluster I, helices α1 and α5 pack against strands β1-3 to stabilize the nucleotide-bound states. In the receptor-bound state, these interactions are replaced by interactions between α5 and strands β4-6. Key residues in this cluster are Y320, crucial for the stabilization of the receptor-bound state, and F336, which stabilizes nucleotide-bound states. Destabilization of helix α1, caused by rearrangement of this activation cluster, leads to the weakening of the inter-domain interface and release of GDP.
Introduction
G protein coupled receptors (GPCRs) turn extracellular signals into intracellular responses by activating heterotrimeric G proteins 1–3. Upon binding an activating ligand, receptors catalyse the release of GDP bound to the Gα subunit. Subsequent binding of GTP causes dissociation of the Gα and Gβγ subunits from the receptor. A large number of mutagenesis studies proposed the C-terminal helix α5 of Gα as a key interaction site for receptor binding and as a conduit for signal transduction 4–9. These data, in combination with crystal structures of individual G protein subunits and of trimetric G proteins provided a broad understanding of the G protein activation mechanism 2,10–15. More recently, the crystal structure of the β2 adrenergic receptor-Gs complex (β2AR-Gs) 16 confirmed that the main site of interaction between the receptor and the G protein is the C-terminus of helix α5, and revealed additional contacts between intracellular loop 2 (ICL2) of the receptor and helix αN of the Gαs. The largest conformational change in the GTPase domain was a rotation of helix α5 and its displacement towards the receptor, accompanied by rearrangements of the α5-β6 interface, the phosphate binding β1-α1 loop (P-loop) and helix α1. This structure also showed the dissociation between the GTPase and helical domains of the G protein, consistent with previous BRET, DEER and single particle electron microscopy data 17–19. Furthermore, analysis of hydrogen–deuterium exchange mass spectrometry data 20 has led to suggest that G protein activation is also associated with an increased disorder around the β1 strand and the nucleotide binding pocket, especially the P-loop and the adjacent N-terminal part of helix α5, while the C-terminus of Gα was protected upon binding the receptor.
A recent modelling study 21 has suggested that G protein activation is associated with the rearrangement of the interfaces between helices α1 and α5, and between α5 and the loop α5-β6. Subsequent experimental mutagenesis studies 22 pinpointed residue F336 in helix α5 of Gαi1 as a particularly important for G protein activation, as its mutation increases the rate of spontaneous GDP release. The proposed mechanism involves F336 acting as a relay, transmitting conformational changes via strands β2, β3 and helix α1 to the phosphate binding loop.
These combined data suggest a mechanism that involves binding of the C terminus of Gα to the receptor accompanied by the formation of additional interactions between the helix αN and the receptor, and transmission of the allosteric signal via the strand β1 or via β2, β3 and helix α1 to destabilize the nucleotide binding site. However, the exact details of the molecular mechanism of the activation remain unclear.
Here, we set out to establish a detailed and comprehensive understanding of the G protein activation mechanism at the residue level that consolidates and extends the existing knowledge. To do this, we characterized the influence of each amino acid of Gαi1 on the stability of the GDP- and GTP-bound states of Gαi1 alone, and of the signaling complex between heterotrimeric Gi (Gαi1β1γ1) and rhodopsin (Rho), a prototypical GPCR. The aggregated analysis of these data allowed us to draw a complete functional map of the Gαi1 subunit stability at different stages of its activation cycle that allowed us to propose an activation mechanism at single amino acid resolution.
Results
We have recently showed that the complex between the heterotrimeric Gi (Gαi1β1γ1) and rhodopsin (Rho) is more stable than the native Rho-Gt complex and is suitable for biophysical studies23. In this work we mutated each amino acid of Gαi1 to alanine or glycine and quantified 1) the thermal stability of each mutant in the inactive GDP-bound and the active GTP(GTPγS)-bound states (Fig. 1 & 2, Supplementary Fig. S1, Supplementary Table 1, methods); and 2) the efficiency of formation (relative abundance) and relative stability of the reconstituted Rho-Gi protein complex (Fig. 1, Supplementary Fig. S2, S3, S4, Supplementary Table 1, methods).
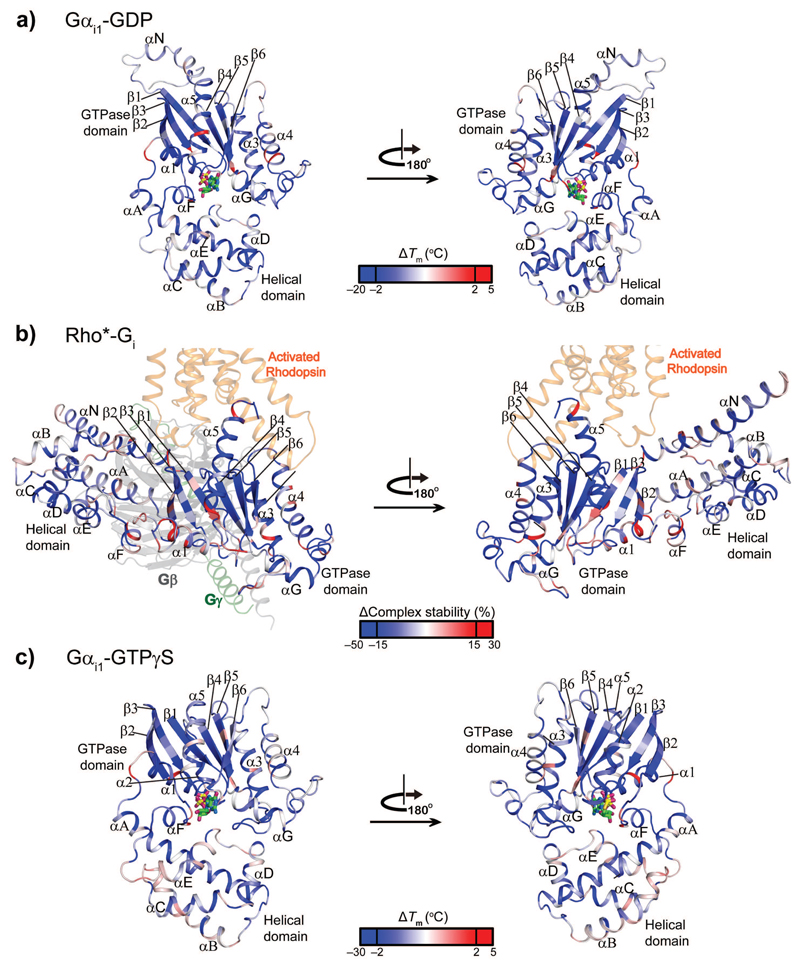
Figure 1. Stability effects of Gαi1 alanine mutants on the nucleotide-bound and receptor-bound states.
a-c, Effects of alanine substitutions in Gαi1 on stability of the (a) GDP-, (b) receptor- and (c) GTPγS–bound states. In the GDP- (a)and GTPγS-bound (b) states, the ∆Tm values for each single alanine mutant are mapped onto the crystal structure of GDP-bound Gαi1 (PDB 1GDD11) and GTPγS-bound Gαi1 (PDB 1GIA10), as a spectrum ranging from blue over white to red. In the receptor-bound state (b), the change in the complex stability (∆complex stability) is mapped onto the homology model of Rho*-Gi complex (see supplementary methods) as spectrum ranging from blue over white to red. Rhodopsin is shown in orange. β and γ subunits are displayed in grey and forest green, respectively. GDP and GTPγS are shown as sticks. The view of GTPase domain of Gαi1 in the complex-bound state is the same as in the GDP- and GTP-bound state, while the helical domain is significantly displaced relative to the GTPase domain in the receptor-bound state.
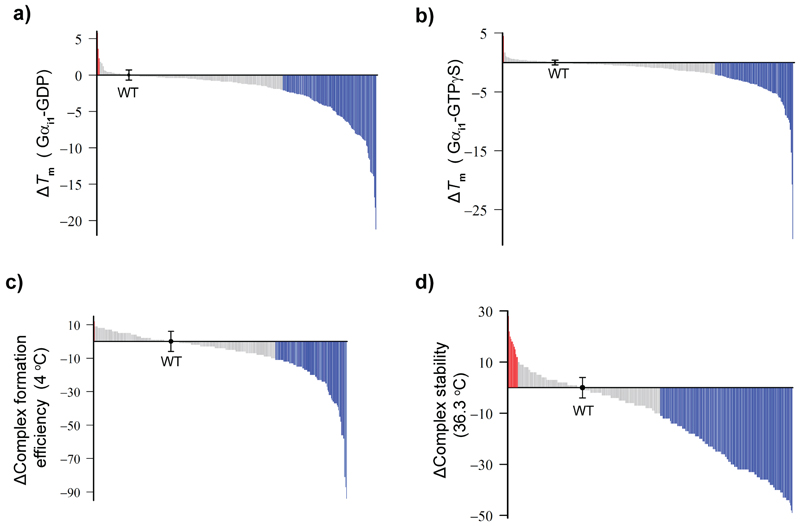
Figure 2. Distribution of effects of Gαi1 alanine mutants on the nucleotide-bound and receptor-bound states.
a, b, Changes in stability (∆Tm) of Gαi1(Ala)-GDP (w/ 1mM GDP) (a) and ∆Tm of Gαi1(Ala)-GTPγS (w/ 0.1mM GTPγS) (b). Gray: -2°C < ∆Tm < 2°C; blue: ∆Tm < -2°C; red: 2°C < ∆Tm. The ∆Tm of Gαi1(WT)-GDP or -GTPγS was shown as the black dot. c, Distribution of ∆ complex formation efficiency of Rho*-Gi(Ala). Blue: ∆ complex formation efficiency is less than -10%; gray: between -10% and 10%; red: more than 10%. d, Distribution of ∆complex stability of Rho*-Gi(Ala). Blue: ∆complex stability is less than -10%; gray: between -10% and 10%; red: more than 10%. The definition of ∆Tm, ∆complex formation efficiency, and ∆complex stability are described in the methods section. All data are presented in Supplementary Table 1. As for ∆Tm of Gαi1 (WT)-GDP and Gαi1(Ala)-GTPγS, data points represent mean ± s.d. from 25 and 24 individual experiments, respectively. As for ∆complex formation efficiency and ∆complex stability of Rho*-Gi(WT), data points represent mean ± s.d. of 33 and 38 individual experiments, respectively.
Interpretation of the changes in stability upon mutation
Mutation of an amino acid to alanine (or, to glycine, if the original amino acid is alanine) results in the elimination of the side chain, which leads to an alteration of the local structure that changes the stability of a protein or complex. If this mutation has a large effect on protein stability, this suggests that the side chain was involved in many local interactions, indicating a structured environment. Conversely, a small change in stability implies that the side chain is not involved in many local interactions. Thus, if point mutations along a stretch of residues do not affect protein stability, this region is likely to be unstructured.
Importantly, changes in protein stability are sensitive to conformational rearrangements. This is the basis of phi-value analysis, a technique developed by Alan Fersht and colleagues to study the energetic and structural details of protein folding intermediates 24. Here, we have adapted this method to study conformational changes of Gi in the GTP-bound and Rho*-Gi (where Rho* denotes light-activated state of rhodopsin) complex relative to Gi in the GDP-bound state, which we used as a reference state.
Comparison of the effects of mutations on stability for several conformational states of the protein substantially increases the “interrogating” power of the alanine scanning technique. Importantly, this technique requires a wide coverage of the protein sequence, ideally approaching 100% of mutated residues. Through the integration of this exhaustive data set, we generated a detailed interpretation of the conformational changes during protein activation, which allowed us to expand, test, or rule out existing hypothesis on the activation mechanism of G proteins.
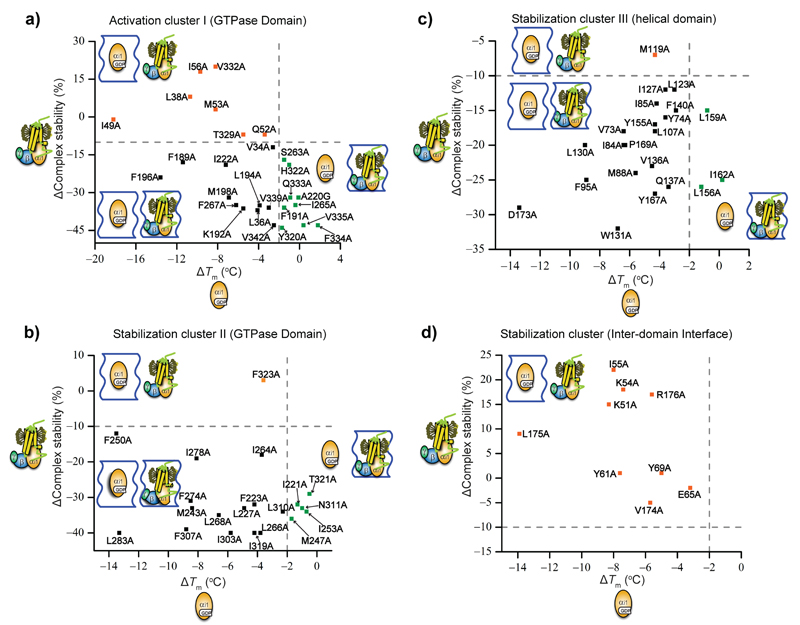
These simple considerations formed the basis for interpreting the measured stability changes in structural terms. For instance, we found that many mutations (30-50%) destabilize both GDP-bound Gαi1 and the Rho*-Gi complex (Fig. 1 & 2, Supplementary Table 1). These residues are located in regions with the same local environment (i.e. conformation) in both states, and are thus important for the stability and integrity of the protein. However, mutations at several positions have different effects on GDP-bound Gαi1 and the Rho*-Gi complex, indicating that they are in regions that undergo conformational changes upon formation of the complex. This way we identified positions that contributed specifically to the stability of Gαi1 in each conformation (Fig. 3).
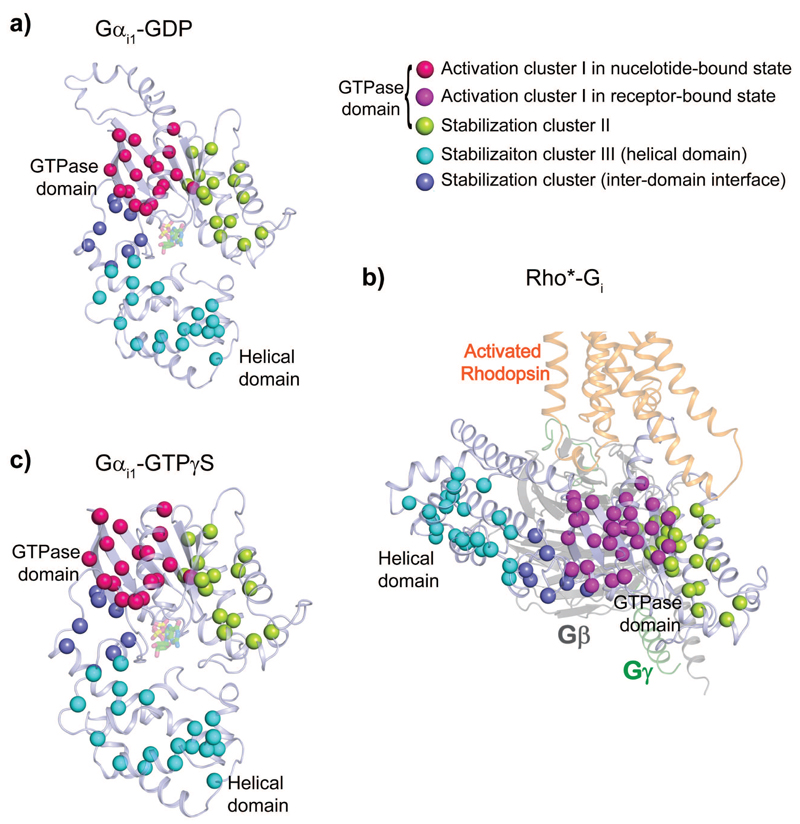
Figure 3. Stabilization clusters in the GTPase, helical domain and the inter-domain interface in the nucleotide- and receptor-bound states.
a-c, Identified stabilization clusters derived from stability effects of Gαi1 alanine mutants on GDP- (a), receptor- (b) and GTPγS-bound (c) states. The identified activation cluster I and stabilization cluster II in the GTPase domain, the stabilization cluster III in the helical domain, and the stabilization cluster in the inter-domain interface are shown as spheres and mapped to GDP-bound Gαi1 state (PDB 1GDD 11) (a), homology model of Rho*-Gi complex state (b) and GTPγS-bound Gαi1 state (PDB 1GIA 10) (c). The stabilization cluster II, III and the stabilization cluster in the inter-domain interface are coloured in lemon, cyan and slate blue spheres in all three states, respectively. The activation cluster I is displayed as hot pink spheres in nucleotide-bound state (a, c), and as magenta spheres in the receptor-bound state (b) to indicate its conformational change upon coupling to the receptor. The relative orientation of the GTPase domain is identical in all states, while the helical domain is displaced in the receptor-bound state.
In order to compare and extrapolate our findings to other Gα proteins, throughout this paper we used the common G protein numbering system (CGN) proposed in Flock et al.25 In this system, the superscript next to the residue number denotes: i) either the GTPase (G) or helical (H) domain, ii) the secondary structure element within each domain (e.g. HN for helix N or S1 for beta sheet β1), and iii) its position within this structural element (e.g. 1), according to a sequence alignment of 973 G protein sequences. For example, L353G.H5.25 corresponds to the L353 in Gai1, GTPase domain, helix 5, and position 25 of the helix 5 in the universal alignment.
N- and C-termini become ordered in the Rho*-Gi complex
The N-terminus provided an excellent benchmark to test the capabilities of our method. Mutations at positions 1G.HN.8 to 32G.HNS1.3 had little impact on the stability of Gαi1 alone, suggesting that this region was unstructured in the absence of the Gβγ subunit. However, mutations of the residues that form the interface with Gβγ in the G protein trimer (e.g., L5AG.HN.12, S16AG.HN.40, I19AG.HN.43, D20AG.HN.44 and L23AG.HN.47) had a severe impact on the stability of the Rho*-Gi complex, while mutation of the residues in this region facing the solvent did not have such effect (Fig 1, Supplementary Table 1).
Mutation of R32G.HNS1.3 at the base of the N-terminus stabilized the Rho*-Gi complex, most likely by improving its interactions with intracellular loop 2 (ICL2) of the receptor. This stabilization effect is probably receptor specific and thus may contribute to receptor-G protein specificity. It is also possible that this position is interacting with the helical domain in its most “open” conformation as shown in Fig 1b.
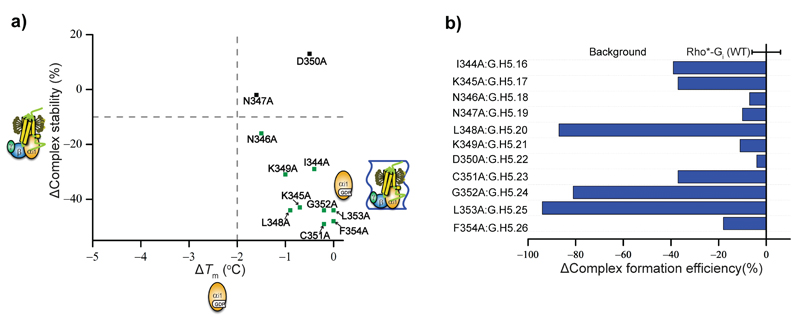
Numerous studies have shown that the last eleven residues in the C-terminus of Gα play a critical role in receptor binding8,9,26,27. Accordingly, most alanine mutations at positions 344G.H5.16-354G.H5.26 considerably affected the formation of the Rho*-Gi complex (Fig. 1 & 4). Particularly, substitution of the conserved L348G.H5.20 and L353G.H5.25 and the less conserved G352G.H5.24 at the end of the C-terminus severely impaired coupling with the receptor without affecting the stability of the nucleotide-bound states. These data agreed with NMR and crystallography studies on a Gt C-terminal peptide bound to rhodopsin, and with the crystal structure of the β2AR-Gs complex, which showed that the C-terminus of Gαs becomes helical and penetrates into a crevice formed in the cytoplasmic side of the transmembrane bundle upon receptor activation 16,20,28,29. Interestingly, N347AG.H5.19 did not affect the formation and stability of the Rho*-Gi complex, and D350AG.H5.22 even slightly stabilized it (Fig. 4a), showing that not all amino acids in the C-terminus have the same effect on receptor binding. Finally, the absence of a destabilizing effect upon mutating positions 344G.H5.16-354G.H5.26 in the nucleotide bound states strongly suggested that this region was unstructured in the absence of the receptor.
Figure 4. Effect on the nucleotide-bound and receptor-bound states of alanine mutation of the last 11 amino acids of Gαi1.
a, Effect on the thermal stability of the GDP-bound and receptor-bound states of alanine mutation of the last 11 resides of the C-terminus of Gαi1. The CGN of the labelled residues is listed in Supplementary Table 1. b, Effect on Rho*-Gi complex formation of alanine mutation of the last 11 resides of the C-terminus of Gαi1. The increase in ∆complex formation efficiency is coloured in red and the decrease is coloured in blue. The definition of ∆Tm, ∆ complex formation efficiency, and ∆complex stability are provided in the supplementary methods and the derived numbers are shown in Supplementary Table 1. Data points represent mean ± s.d. of 33 individual experiments.
Rearrangement of activation cluster I upon complex formation
The movement of helix α5 in the GTPase domain upon formation of the complex 4,16,21 results in significant conformational changes around its base, which is packed against the β sheet consisting of strands β1-β6, and helix α1. Our data are in agreement with such rearrangements, as shown by the different effect of mutations on the nucleotide-bound state and on the complex. Importantly, our analysis allowed us to focus on the individual contribution to the stability of the different Gαi1 states of each amino acid of this entire region. We detected a number of residues with a concerted role, which we termed as activation cluster I (Fig 3 and 5a), formed by several highly conserved hydrophobic residues from β1-3 strands, helix α1 and inward-facing residues of helix α5. Alanine substitutions of these residues considerably destabilized the GDP-bound conformation (3-18 ºC) and moderately affected the GTPγS-bound state (1-5 ºC) (Fig. 5a & 6a, Supplementary Fig. S5a, Supplementary Table 1). Importantly, mutation of F336G.H5.8 (universally conserved in Gα subfamilies; see Flock et al25) in helix α5 is the only substitution that resulted in a complete impairment of Gαi1 stability and of its ability to bind nucleotides. F336AG.H5.8 also caused protein aggregation and a severe impairment in reconstitution of Gαi1 with Gβγ to form the Gαβγ heterotrimer (Supplementary Fig. S6). Interestingly, this mutant still formed a relatively stable complex with the receptor. In the structure of the β2AR-Gs complex, the corresponding phenylalanine moved from the buried hydrophobic core of Gsα to contact ICL2 of the receptor16. This suggests that F336G.H5.8 plays a critical role in stabilizing the Gαi1 subunit in the nucleotide-bound conformation, consistent with the observation that its mutation increases the rate of spontaneous nucleotide release22. We hypothesized that relocation of F336G.H5.8 concomitant with the upward movement and twist of α5 triggers the reorganization of the cluster I into the receptor-bound state.
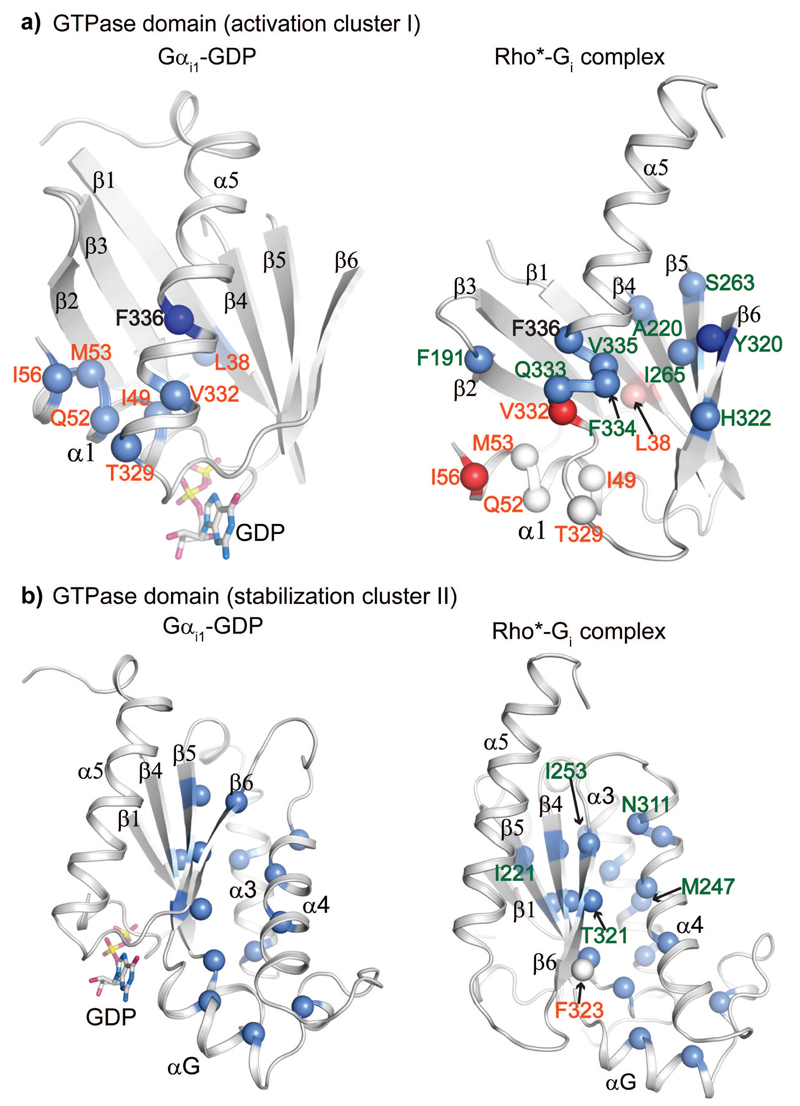
Figure 5. Close-up view of activation cluster I and stabilization cluster II.
a-b, Residues involved in the activation cluster I (a) and the stabilization cluster II (b) of the GTPase domain in GDP-bound and receptor-bound states. The involved residues are shown as spheres in both the GDP-bound and receptor-bound states. Light blue: destabilizing effect by mutation to alanine; white: stability comparable to WT after mutation to alanine; light red: stabilizing effect due to mutation to alanine. Residues labelled in orange: alanine mutations dramatically destabilize the GDP-bound state but not the receptor-bound state; residues labelled in forest green: alanine mutations do not affect the GDP-bound state, but significantly destabilize the receptor-bound state; residues without lableing: alanine mutation destablizie both GDP- and receptor-bound state. I56AG.H1.11 and V332AG.H5.4 (shown in red) significantly stabilize the complex. The alanine mutation of F336G.H5.8 completely impairs the stability of the GDP-bound state (shown in deep blue). The alanine mutation of Y320G.S6.2 severely impairs the complex formation (shown in deep blue). The CGN of the labelled residues is listed in Supplementary Table 1.
Figure 6. Stability effect of alanine mutation of the residues involved in the activation and stabilization clusters.
a-d, Effect on stability of mutation of the residues involved in the activation cluster I (a), stabilization clusters II (b) and III (c), and the inter-domain interface (d) on the GDP- and receptor-bound states. Orange: mutations that dramatically destabilize the GDP-bound state, but do not affect the stability of receptor-bound state; forest green: mutations that do not destabilize the GDP-bound state, but significantly destabilize the receptor-bound state; black: mutation that destabilize both nucleotide- and receptor-bound states. The alanine mutants represented by the orange and the forest green box correspond to the colour of residue number shown in Fig. 5a-b and Fig. 7a-b. The blue wavy box cartoons represent the nucleotide- or receptor-bound states affected by mutations.
Upon binding the receptor, a structural reorganization of cluster I disrupts the interactions that stabilize helix α1 (Fig. 5a). This is suggested by the fact that mutation of residues I49G.H1.4, M53G.H1.8, and I56G.H1.11 in α1, L38G.S1.6 of β1, T329G.H5.1 and V332G.H5.4 of α5, which tether helix α1 in the Gαi1-GDP state, severely impaired its stability, but did not affect the stability of Rho*-Gi complex (Fig. 6a, Supplementary Table 1). Moreover, mutation of the conserved N331G.H5.3 and V332G.H5.4 in helix α5, which stabilizes the nucleotide-bound state by making connections to the helix α1, increased the stability of the complex by 30% and 20%, respectively. Movement of these residues disrupted contacts between the base of helix α5 and helix α1, which would lead to the loss of helicity at the base of helix α5 observed in the β2AR-Gs complex 16. This order-to-disorder transition potentially increased the flexibility of the loop β6-α5, which contains the guanine-ring-binding TCAT motif, thus perturbing its interaction with GDP.
The loss of local structural stability associated with an increased disorder in the C-terminal part of helix α1 and the N-terminal part of helix α5 is compensated by the strengthening of their interactions with the β4, β5 and β6 strands and the relocated helix α5. This is suggested by the fact that mutation of A220G.S4.1 of β4, S263G.S5.1 and I265G.S5.3of β5, Y320G.S6.2 and H322G.S6.4 of β6, Q333G.H5.5, F334G.H5.6, V335G.H5.7, and V342G.H5.14 of α5 dramatically destabilized the Rho*-Gi complex (20-50%) without affecting the stability of both nucleotide-bound states. Additionally, many of these mutants showed competent heterotrimer reconstitution, while the efficiency in forming the Rho*-Gi complex was reduced by 20-80% (Supplementary Fig. S6). A sequence alignment of human G proteins showed that these residues are highly conserved in the Gα subfamily25. Taken together, this indicates that these residues are not only important for stabilizing the G protein conformation in the receptor-bound state, but also crucial for allosteric regulation of receptor-mediated G protein activation.
Y320 in activation cluster I as a signal transduction hub
Mutation of Y320G.S6.2 in the β6 strand, which is a conserved tyrosine or phenylalanine in the Gα subfamily, severely impaired the Rho*-Gi complex formation (Fig. 5a & 6a) while having only a very moderate effect on the nucleotide bound states. Remarkably, Y320AG.S6.2, L348AG.H5.20, G352AG.H5.24 and L353AG.H5.25 had a similarly strong impact on the formation of the complex, but Y320G.S6.2 is the only position that does not interact directly with the receptor. Also, Y320AG.S6.2 showed a well-preserved ability to bind nucleotides and form the heterotrimer (Supplementary Fig. S6). We hypothesized that mutation of Y320CGN prevented the formation of an allosteric activation pathway that propagates the signal for GDP release transmitted from the receptor, making Y320G.S6.2 a key signal transduction hub in the mechanism of receptor-mediated G protein activation.
Cluster II is the structural scaffold of GTPase domain
We identified a second cluster of residues with a common role in the GTPase domain formed by residues in helices α3, α4 and αG packed against residues in strands β4, β5 and β6 (Fig 3 & 5b). While cluster II partially overlapped with cluster I, most mutations here destabilized both receptor- and nucleotide-bound states of Gαi1. Most mutations destabilized the GDP-bound state by 3-13 ºC, the receptor-bound state by 30-40%, and the GTPγS-bound state by 1-5 ºC (Fig. 5b & 6b). Residues in cluster II are highly conserved among G proteins, and likely form the structural scaffold of the Gα subunit25. It should be noted that mutation of residues I221G.S4.2 of β4, T321G.S6.3, M247G.H3.6 and I253G.H3.12, I264G.S5.2, N311G.h4s6.2 and I319G.S6.1 dramatically destabilized R*-Gi complex without affecting the stability of the GDP-bound state (Fig. 6b, Supplementary Table 1). Mutation of K248G.H3.7 and D251G.H3.10, which are located in solvent-exposed surface, also showed similar effect. We hypothesized that these residues may form additional stabilizing contacts in the receptor-bound conformation, or are involved in direct interactions with the receptor.
Helical domain behaves as a rigid body
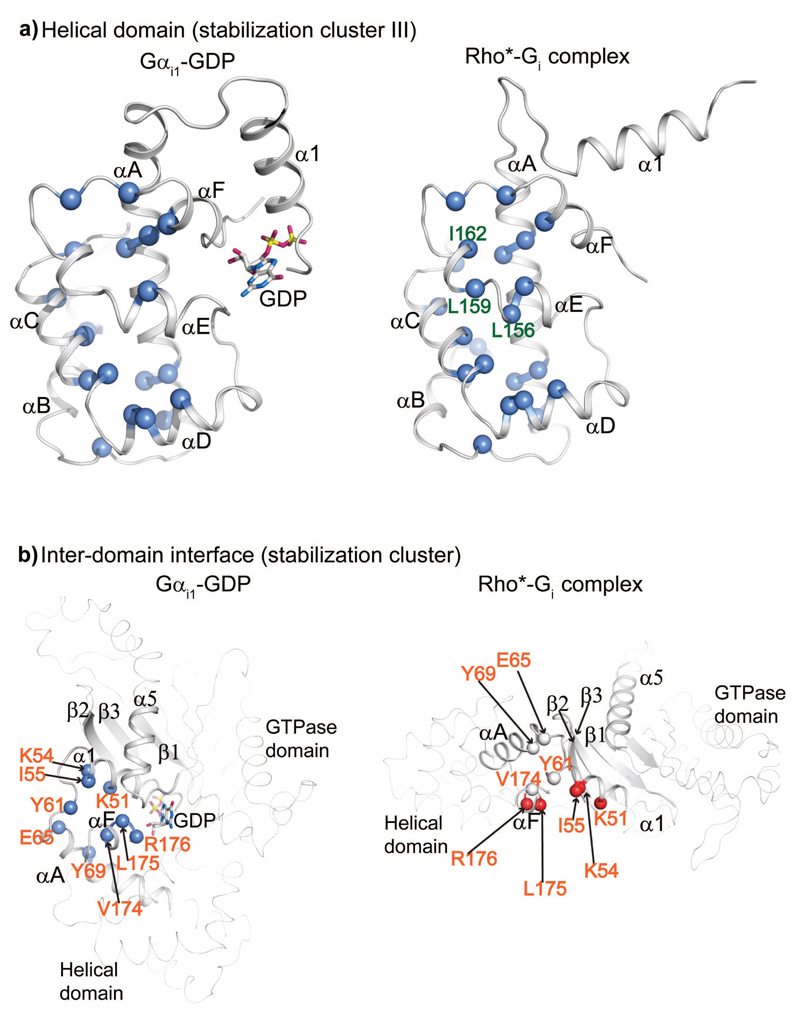
A hallmark of G protein activation by the receptor is the release of GDP accompanied by the separation of the GTPase and helical domains. The helical domain consequently displays dynamic equilibrium between multiple orientations relative to the GTPase domain 16,18,19,30. We showed that a cluster of mostly hydrophobic residues (63H.HA.1-176H.HF.6; stabilization cluster III) of Gαi1 stabilizes the helical domain (Fig. 3 & 7a, Table. S1). In contrast to the activation and stabilization clusters in the GTPase domain, where most mutants affected both the stability and formation of the Rho*-Gi complex, mutations in the stabilization cluster III did not affect the formation of the Rho*-Gi complex (Fig. 6c & 7a). A sequence alignment shows that hydrophobic residues are preferred at these positions in all Gα subtypes (Supplementary File F1). This is consistent with the observation that the helical domain can be expressed independently from the GTPase domain while retaining its ability to activate cGMP phosphodiesterase 31.
Figure 7. Close-up view of stabilization cluster III and stabilization cluster in the inter-domain interface.
a-b, Reisudes invloved in stabilization clusters in the helical domain (a) and inter-domain interface (b) in GDP-bound and receptor-bound states. The involved residues are shown as spheres in both the GDP-bound and the receptor-bound states. Light blue: destabilising effect by mutation to alanine; white: stability comparable to WT after mutation to alanine; light red: stabilization after mutation to alanine. Residues labelled in orange: alanine mutations dramatically destabilize the GDP-bound state but not the receptor-bound state; residues labelled in forest green: alanine mutations do not affect the GDP-bound state, but significantly destabilize the receptor-bound state; residues without lableing: alanine mutation destabilize both GDP- and receptor-bound state.
However, there were some exceptions. Mutation of A138H.HD.5, L156H.HE.6, L159H.HE.9, R161H.HE.11and I162H.HE.12 destabilized the Rho*-Gi complex without affecting the stability of GDP- and GTP-bound states (Fig. 6c, 7a, S6, Supplementary Table 1). Also, I78AH.HA.16 reduced complex formation by 20% and L175AH.HF.5 destabilized GDP-bound state and reduced complex formation. This suggest that subtle internal rearrangements of the AH domain are required to keep its integrity in the Rho*-Gi complex.
Weakening of the inter-domain interface promotes activation
The inter-domain interface in Gαi1 is composed by the N-terminal part of helices αA, αF, α1 and loop of αF/α1 (Fig. 3). Mutation of the residues in this interface dramatically destabilized the GDP-bound state (5-14 ºC), but did not destabilize the Rho*-Gi complex. In fact, mutations K51AG.H1.6, K54AG.H1.9 and I55AG.H1.10 increased the relative stability of Rho*-Gi complex by 15-20%. We also observed a similar effect for L175AH.HF.5 and R176AH.HF.6, which increased complex stability by 9% and 17%, respectively (Fig. 6d, 7b, Table. S1). A sequence alignment showed that the residues located in the inter-domain interface of Gαi1 are highly conserved in all Gα subfamilies (Supplementary File F1, Flock et al25). Our data suggest that subtle conformational perturbations in the inter-domain interface of GDP-bound state can facilitate the domain separation and the release of GDP, in agreement with previous observations that the helical domain dissociates from the GTPase domain upon binding to the receptor 16–18. The importance of weakening the inter-domain interface for G protein activation is further supported by the structure of the Gα subunit from Arabidopsis thaliana (AtGPA1) 32,33. This protein has a structure that is very similar to Gαi1 (RMSD of 1.8 Å between backbone atoms). However, due to the absence of classical GPCRs in plants, AtGPA1 exchanges nucleotides by a self-activation mechanism attributed to the marginally stable helical domain, which shows a tendency to dissociate from the GTPases domain and unfold. Comparison between the Gαi1-GDP and AtGPA1 structures shows that they contain similar residues at the inter-domain interface, whereas the cross-interface hydrogen bonds in AtGPA1 are weaker compared to those in Gαi1-GDP (Supplementary Fig. S7).
Differences between GDP and GTP states
The GTPγS-bound state of Gαi1 was more stable than the GDP-bound state. The apparent melting temperatures were, respectively, 70 °C and 63 °C at saturating concentrations of the corresponding nucleotides. In addition, GTPγS had a much higher affinity for Gαi1 compared to GDP, as judged by a steeper concentration dependence of the stabilizing effect (Supplementary Fig. S1). We observed that most mutations destabilized both GDP and GTPγS states, consistent with the relatively minor differences between the GDP- and GTPγS-bound crystallographic structures of Gαi1. 34. One interesting observation is that the GTPγS bound state was on average two-fold less sensitive to mutations (Supplementary Fig. S8), again suggesting that this is a more stable state. Also, several mutations concentrated around the third phosphate group and at the Gβγ interface had a disproportionally large effect on the GTPγS-bound state. This is precisely the area that undergoes conformational changes associated with the activation of the Gα, causing the dissociation of the α and βγ subunits.
Discussion and Conclusions
The exhaustive coverage (the entire sequence of the Gαi1 subunit) and single amino acid resolution of our mutagenesis analysis, combined with the stability measurements obtained for Gαi1 in its GDP-, receptor- and GTPγS-bound states, allowed us to obtain an extremely detailed data set to understand the molecular mechanisms of G protein activation. Our results showed with that the interactions involved in the stabilization of the receptor-bound conformation of Gαi1 are broader and more complex that were previously suggested.
First, we identified two clusters of residues that confer stability to the GTPase domain. The activation–cluster I consists of residues in helices α1 and α5 packed against residues in strands β1-3 in the nucleotide-bound states. In the receptor-bound state, the interactions between α5/α1 and β1-3 are weakened and compensated by a new set of interactions between α5 and strands β4-6. The most prominent examples of residues involved in this rearrangement are Y320G.S6.2 and H322G.S6.4, which are crucial for the stabilization of the receptor-bound state but have no effect on the nucleotide bound state. Conversely, F336G.H5.8 is important for the stability of the GDP- and GTP-bound states, but plays little role in the stabilization of the Rho*-Gi complex. Helix α1 is likely to become mostly unstructured in the Rho*-Gi complex, as judged by the absence of significant effect of mutations on complex stability. However, some mutations towards its C-terminal part result in stabilization of the complex. The helix α1 is a recipient of the conformational changes that lead to the destabilization of the inter-domain interface and to nucleotide release, a key step in G protein activation. Mutations in the inter-domain interface between the GTPase and the helical domains, consisting of αA, αF, α1 and loop of αF/α1, dramatically destabilize nucleotide-bound states but do not affect, and some even stabilize, the complex. The above-mentioned residues are just some of the most noticeable examples, but we found a network of residues that contribute to the stabilization of these distinct conformational states (Fig. 3, 5 & 7).
In addition to being able to change its conformation, the G protein also has to maintain its structural integrity and identity during the signaling cycle. Stabilization cluster II includes residues in helices α3, α4 and αG packed against residues in strands β4, β5 and β6. The majority of mutations in this cluster affected similarly both states, and we concluded that this cluster provides a steady structural scaffold to the GTPase domain. As this cluster partially overlaps with the activation cluster I, there were several mutations in strands β4-6, such as I319G.S6.1, which preferentially affected the receptor-bound state. A third cluster of residues maintains the structural integrity of the helical domain. Most mutations in this domain resulted in similar effects on the stability of the nucleotide-bound states or the Rho*-Gi complex. Overall, these mutations were less detrimental to the stability than mutations in the GTPase domain. Several mutations, mostly located in helix αH, destabilized the complex without affecting either the GDP- or the GTP-bound state, suggesting that this region undergoes some conformational changes upon receptor binding.
Recently, long-scale molecular dynamics simulations by Dror et al35 suggested that the key events in G protein activation are structural rearrangements in the nucleotide binding site, especially the repositioning of the β6-α5 loop, caused by the movement of helix α5 away from the nucleotide binding site, and a concomitant weakening of the inter-domain interface. They also found that the GDP could only dissociate if the helical domain is in the open conformation. Interestingly, the helical domain remains mostly rigid in the simulations. These findings are very complimentary to our results.
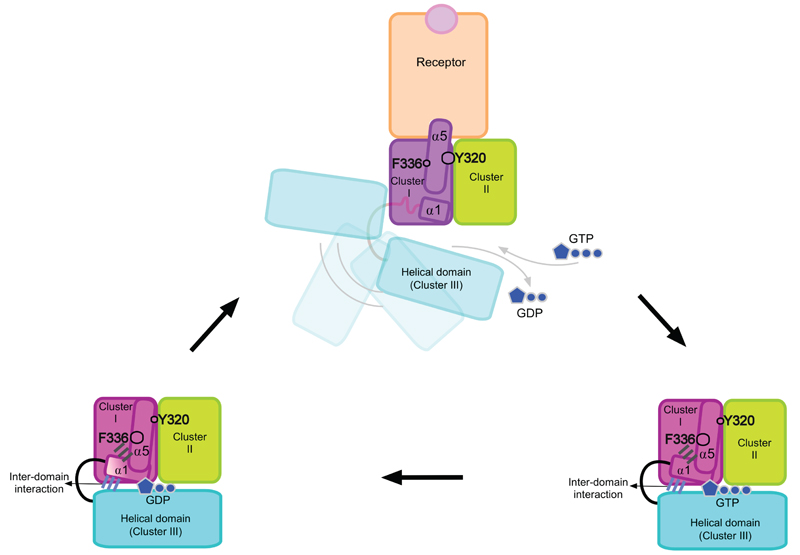
Overall, our data suggest that the most significant event in activation of Gαi1 is the destabilization of helix α1 caused by a rearrangement on the activation cluster I. This leads to a perturbation and weakening of the inter-domain interface, dissociation of the helical domain from the GTPase domain in a rigid body movement, and release of the GDP (Fig. 8). How does the subsequent binding of GTP trigger dissociation of the complex? The answer may be found in the relative stability of the GDP-, GTP- and receptor bound states. The GTP-bound state of Gαi1 is thermodynamically the most stable state of the protein, as reflected by the considerably higher thermal stability of the GTPγS-bound state. Due to GTP hydrolysis, the G protein is kinetically trapped in a less stable GDP-bound state. This is a meta-stable state because the nucleotide exchange rate is very low in the absence of the receptor, consistent with proposed role of the helical domain to protect the GDP from exchange with GTP13 readily available in the cytoplasm. As the complex is formed and the nucleotide binding site becomes accessible, the additional stabilization by GTP overcomes the stability of the complex, leads to the stabilization of the helix α1, the inter-domain interface and reverts the helix α5 to its conformation in the nucleotide bound state.
Figure 8. Nucleotide exchange in the Gαi1 subunit mediated by the activation and stabilization clusters.
Cluster I is coloured in hot pink in both the GDP- and GTP-bound states, and in magenta in the receptor-bound state. Cluster II and III are coloured in lemon and cyan in the three states, respectively. The inter-domain interaction and the interactions between helices α1 and α5 are coloured in slate blue and grey, respectively. GDP and GTP are shown in dark blue. Cluster I consists of helices α1 and α5 packed against stands β1-3 in the nucleotide-bound states. In the receptor-bound state, these interactions are weakened and compensated by new interactions between helix α5 and stands β4-6. The most prominent examples of the residues involved in this rearrangement are Y320G.S6.2, which is crucial for the stabilization of the receptor bound state, and F336G.H5.8, important for the stability of the GDP- and GTP-bound states. Destabilization of helix α1 results in weakening of the inter-domain interface, separation of the helical domain from the GTPase domain and release of GDP.
It is still an open question to what extent our findings can be generalized to other GPCR-G protein combinations, and which aspects are specific to the Gαi1 or the Rho*-Gi complex. However, in Flock et al.25 we show that many of the residues identified here may play similar roles in all G proteins.
Online Materials and Methods
1). Alanine scanning mutagenesis
The alanine scanning expression library of Gαi1 was prepared as we have reported before 36. The wild-type (WT) plasmid was constructed by inserting human G protein alpha-subunit (Gαi1) into pJ411 vector (DNA 2.0) which incorporated a N-terminal 10-histidine tag followed by a TEV cleavage site. The alanine mutants were produced based on the WT plasmid by high-throughput (HTP) alanine mutagenesis as we have reported previously 36. All 354 amino acid residues in Gαi1 were mutated. All non-alanine residues were replaced to alanine and alanine residues were substituted to glycine. The exact protein sequence of the construct is provided in the supplementary note.
2). Preparation of native βγ subunit (Gβγ)
Gβγ was separated from endogenous transducin (Gt) as previously described 23. Briefly, dark-adapted bovine retinas (W L Lawson, USA) were exposed to room light at 4 ºC overnight. The rod outer segment (ROS) membranes were collected by centrifugation in a 25-30% (w/w) sucrose gradient. After isotonic and hypotonic washes, Gt was dissociated from ROS membrane by adding GTP (Sigma-Aldrich). The collected Gt was filtered through 0.22 µm membrane (Millipore Corp) and dialyzed against the dialysis buffer (10mM Tris-HCl, pH 7.4, 2 mM MgCl2, 1 mM DTT,) containing 50% glycerol. Gβγ was further separated from the Gαt on a Blue-Sepharose column (GE Healthcare) by a linear salt gradient (0-500 mM NaCl) in the dialysis buffer supplemented with 30% glycerol. The Gβγ was concentrated to 1-5 mg/ml and stored at -80 °C.
3). Preparation of bovine rhodopsin
Bovine rhodopsin was extracted from dark-adapted ROS membranes which were prepared according to Okada’s method 37. The dark-adapted ROS membranes were solubilized in solubilization buffer (50 mM sodium acetate, pH 6, 1 mM EDTA, 2 mM 2-mercaptoethanol, 3 mM CaCl2, 3 mM MgCl2, 3 mM MnCl2, 100 mM NaCl) supplemented with 80 mM (4.1%) β-dodecyl-D-n-maltoside (DDM) at 4 ºC overnight. After centrifugation at 30,000 rpm in a Ti70 rotor, the supernatant was diluted with solubilization buffer to a concentration of DDM as 0.4%. The diluted sample was loaded to a column packed with ConA Sepharose resin (GE Healthcare) which was equilibrated with washing buffer (solubilization buffer supplemented with 0.02% DDM). After extensive washing, bovine rhodopsin was eluted with solubilization buffer supplemented with 0.02% DDM and 0.2 M α-D-methylmannoside. The eluted bovine rhodopsin was concentrated to 1-4 mg/ml and stored at -80 °C.
4). High throughput (HTP) culturing and purification of Gαi1 alanine mutants
The recombinant Gαi1 alanine mutants were expressed in BL21 (DE3) competent cells. The cultures were grown at 37 ºC in TB media (GERBU Biotechnik GmbH) by using 24 well plates (mutant/well) (Whatman UniFilter Microplates, GE Healthcare). The culture volume was 5 ml/well. When the OD600 reached 0.6, cells were induced with 0.5 mM IPTG and continued to grow for 20 hours at 20 ºC. The cell were harvested by centrifugation resuspended in the binding buffer (25 mM Tris-HCl, pH 7.4, 500 mM NaCl, 10% glycerol, 50 mM imidazole, 5 mM 2-mercaptoethanol) and transferred to a 96 deep-well plate (Thermo Scientific). The re-suspended cells were disrupted by sonication for 1 min using a SONICS VCX-600 sonicator equipped with an 8-pin probe. After clarifying cell lysates by centrifugation, the supernatants were loaded to a 96 deep-well filter plate (one mutant per well) pre-loaded with 500 µl cobalt chelating resin (GE Healthcare) and equilibrated with binding buffer. After extensive washing with binding buffer, the recombinant Gαi1 alanine mutants were eluted with elution buffer (25 mM Tris-HCl, pH 7.4, 500 mM NaCl, 10% glycerol, 500 mM imidazole, 5 mM 2-mercaptoethanol). The eluted proteins were dialyzed against 25 mM Hepes, pH 7.4, 100 mM NaCl and 2 mM DTT using Slide-A-Lyzer MINI Dialysis Device (Thermo Scientific). Among of 354 alanine mutants, the purified R142AH.HD.9, Y230AG.s4h3.4, K270AG.s5hg.1 and D272AG.HG.2 were severely aggregated and could not be used in the further assays. The flowchart of HTP purification is shown in Supplementary Fig. S1.
5). Characterization of the effect of Gαi1 alanine mutants on the receptor-bound state by a HTP assay
In each round, WT Gαi1 was always prepared in parallel with the Gαi1 alanine mutants [Gαi1(Ala)] to form rhodopsin-Gi protein complex [Rho*-Gi(WT)] as the reference control. The recombinant Gαi1 alanine mutants (12.5 µM) from HTP purification and the native Gβγt (10 µM) were reconstituted to form heterotrimer (Gi) by incubation in a 96-well PCR plate (one mutant per well) (Eppendorf) on ice for 2 h. Under the dim-red light in the dark room, purified rhodopsin (18 µM) was added and mixed with Gi in ice-cold assay buffer (25 mM Hepes, pH 7.4, 100 mM NaCl, 2mM DTT, 0.02% DDM, 1 mM MgCl2, 0.16 unit/ml apyrase). After irradiation with orange light (>495 nm) on ice for 10 min, the tetramer complex Rho*-Gi(Ala) was formed by coupling the activated rhodopsin with Gi and the formed Rho*-Gi(Ala) complex was further incubated in the dark at 4 ºC overnight. The reaction volume was 50 µl for each alanine mutant. 20 µl of each Rho*-Gi(Ala) complex was transferred to another 96-well PCR plate and heated for 30 min in a PCR machine (Eppendorf Mastercycler Gradient) at 36.3 ºC. After centrifugation at 3000 rpm for 10 min at 4 ºC, 14 µl of formed Rho*-Gi(Ala) complex (4 ºC) and 14 µl of heated Rho*-Gi(Ala) complex (36.3 ºC) were mixed with NativePAGE Sample Buffer (4×) (Invitrogen) and NativePAGE 5% G-250 Sample Additive (Invitrogen), respectively. The mixtures were loaded onto 4-16% NativePAGE Bis-Tris-HCl Gels (Invitrogen) and gel electrophoresis was performed in a 4 ºC cold room according to the manufacturer’s protocol (Invitrogen). Protein markers were used with NativeMark Unstained Protein Standard (Invitrogen). The gel bands of Rho*-Gi complex were integrated and quantified using the ImageJ software. The complex formation efficiency (CF) (%) was obtained from the normalization of integrated density of Rho*-Gi complex band [IDC(Ala or WT), 4 ºC] with integrated density of Rho*-Gi(WT) complex band [IDC(WT), 4 ºC]. The complex stability (CS) (%) was defined as the normalization of integrated density of Rho*-Gi complex band [IDC(Ala or WT), 36.3 ºC] with integrated density of Rho*-Gi(WT) complex band [IDC(Ala or WT), 4 ºC].
The ∆CF (%) and ∆CS (%) were defined as:
The distribution and summary of ∆CF efficiency and ∆CS of each Gαi1 alanine mutant are listed in Fig. 2 and Supplementary Table 1. The flowchart diagram of HTP assay is shown in Supplementary Fig. S2.
6). HTP measurements of thermal stability Gαi1 alanine mutants by differential scanning fluorimetry (DSF)
The thermostability of each Gαi1 alanine mutant in the nucleotide-bound states was measured by HTP differential scanning fluorimetry (DSF). The samples were prepared on ice. 10 µl of recombinant Gαi1 alanine mutant stocks (0.7 µg/µl) were dispensed into a 96-well PCR plate (one mutant per well) (Eppendorf) and mixed with 100 µl ice-cold assay buffer (25mM Hepes, pH 7.4, 100mM NaCl, 2mM DTT) containing 5× SYPRO-orange (Invitrogen) and nucleotides (1 mM GDP or 100 µM GTPγS). After mixing, 110 µl reaction mixture of each alanine mutant was divided into 0.2 ml PCR tubes (Qiagen) as three samples of 35 µl. The DSF experiments were performed with Rotor GeneQ (Qiagen) by ramping from 25 °C to 95 °C at a rate of 3 °C/min. The melting temperature (Tm) was defined as the inflection point of the melting curve as analyzed by the Rotor Gene Q Series Software. The Tm value of each Gαi1 alanine mutant [Tm(Ala)] upon addition of the nucleotides was averaged from three individual experiments.
The ∆Tm value was defined as:
In each round, WT Gαi1 was always prepared in parallel with Gαi1 alanine mutants as a reference control.
In addition, the thermal shift of WT Gαi1 in titration with GDP and GTPγS were also performed with HTP DSF.
7). Analysis of heterotrimer formation by fluorescence assisted size exclusion chromatography (FSEC)
The recombinant Gαi1 alanine mutants (6 µM) and Gβγt (2 µM) were reconstituted to form heterotrimer (Gi) in 100 µl running buffer (25 mM Hepes, pH 7.4, 100 mM NaCl) overnight on ice. 80 µl of reconstituted Gi was injected to Superdex 200 packed in a Tricorn 10/200 column (GE Healthcare) equilibrated with the running buffer. The elution profile was monitored by protein-intrinsic fluorescence with λex: 280 nm and λem: 340 nm at a flow rate of 1 ml/min. The retention time of the reconstituted Gi was integrated with UNICORN 5.2 software (GE Healthcare).
8). Modelling of the rhodopsin/Gi complex
Homology modelling of Gαi1
The sequences of Gαi1 and Gαs were aligned using Clustal Omega 38. This initial alignment was manually refined using Chimera 39 to adjust some of the gaps in the loop regions. Using this alignment, Gαi1 was modelled with Modeller 40 using the structure of Gs bound to the beta 2 adrenergic receptor 16 as a template. Residues missing in the template were refined using the loop optimization method in Modeller. All models were subjected to 300 iterations of variable target function method optimization and thorough molecular dynamics and simulated annealing optimization and scored using the discrete optimized protein energy potential. The 20 best-scoring models were analyzed visually, and a suitable model (in terms of low score and structure of the loops) was selected.
Homology modelling of active rhodopsin
The sequences of bovine rhodopsin and the human beta 2 adrenergic receptor were aligned using Clustal Omega 38. This initial alignment was manually refined using Chimera 39 to adjust some of the gaps in the loop regions. Using this alignment, rhodopsin was modelled with Modeller 40 using the structure of the beta 2 adrenergic receptor bound to Gs 16 as a template. Residues missing in the template were refined using the loop optimization method in Modeller. All models were subjected to 300 iterations of variable target function method optimization and thorough molecular dynamics and simulated annealing optimization and scored using the discrete optimized protein energy potential. The 20 best-scoring models were analyzed visually, and a suitable model (in terms of low score and structure of the loops) was selected.
Modelling of the rhodopsin - Gi complex
The models of Gαi1 and rhodopsin were superimposed to the structures of Gαs and the beta 2 adrenergic receptor 16, keeping the G beta and gamma subunits. In addition to the crystallographic waters resolved in the structure of rhodopsin, we added additional ordered water molecules, as observed in the high-resolution structure of the adenosine A2A receptor 41. Cysteines 322 and 323 were palmitoylated. Glu, Asp, Arg and Lys residues were set as charged, except Glu122(3.37) and Asp83(2.50) 42. Topology and parameter definitions for palmitoyl-cysteine and retinal bound via protonated Schiff-base link to lysine 43,44 were obtained from the parameter/topology repository of NAMD 45. The complex was embedded in a solvated and pre-equilibrated lipid bilayer consisting of 360 molecules of 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphatidylcholine (POPC) and approx. 50.000 water molecules. Sodium and chloride ions were added to a concentration of 0.15M NaCl, and then additional ions were added to achieve charge neutrality. The system measured roughly 120 × 120 × 160 Å^3, with a total of approximately 215'000 atoms. This system was equilibrated as follows: first a short (0.5 ns) simulation was performed in which only the lipid tails were allowed to move, in order to induce the appropriate disorder of a fluid-like bilayer. Then, the geometry of the entire system was optimized by 1000 steps of energy minimization, followed by two equilibration steps with the protein constrained (0.5 ns) and without constraints (0.5 ns). In order to equilibrate the complex, the system was subjected to 20 ns of unrestrained molecular dynamics. Simulations were carried out using NAMD 2.8 45 with the CHARMM27 all-hydrogen force field 46 at constant pressure (1 atm), and using a time step of 2 fs.
Supplementary Material
Acknowledgements
We thank F. Heydenreich, V. Korkhov, R. Kammerer and C. Piscitelli for the critical reading of the manuscript, and V. Panneels and R. Jaussi for technical support. S. Maeda was supported by the Roche postdoctoral fellowship RPF113. T.F. is Boehringer Ingelheim Fonds PhD fellow. D. M. and S. Mendieta were supported by the Paul Scherrer Institute internship program. This work was supported by Swiss National Science Foundation grants Sinergia 141898 (D.B.V., G.F.X.S.), 133810 31-135754 (D.B.V.), 31-153145 (G.F.X.S.), 31003A-146520 (X.D.), National Centre of Competence in Research (NCCR) Structural Biology, NCCR Molecular Systems Engineering, European Cooperation in Science and Technology Action CM1207 GLISTEN: GPCR-Ligand Interactions, Structures, and Transmembrane Signalling: a European Research Network (X.D. and G.F.X.S.), the Marie Curie Initial Training Network NanoMem (D.B.V., G.F.X.S.), the Medical Research Council (MC_U105185859; M.M.B., T.F.) and the. M.M.B. is a Lister Institute Research Prize Fellow. Molecular dynamics simulations were run at the Swiss National Supercomputing Centre (CSCS).
Footnotes
Author Contributions
DS collected and performed all data analysis. TF helped with analysis and interpretation. SMaeda, SMendieta and DM helped with experiments. XD built the molecular models, and MM helped with the interpretation of the structural effects of mutations. RD, GFXS and MMB contributed to the discussion and writing the manuscript. DS, XD and DBV wrote the manuscript. All authors read and provided their comments on the draft. DS and DBV conceived research and DBV supervised the project.
References
- 1.Gilman AG. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–49. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 2.Wall MA, et al. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995;83:1047–58. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 3.Sprang SR. G proteins, effectors and GAPs: structure and mechanism. Curr Opin Struct Biol. 1997;7:849–56. doi: 10.1016/s0959-440x(97)80157-1. [DOI] [PubMed] [Google Scholar]
- 4.Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nature structural & molecular biology. 2006;13:772–7. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- 5.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2007 doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 6.Marin EP, Krishna AG, Sakmar TP. Rapid activation of transducin by mutations distant from the nucleotide-binding site: evidence for a mechanistic model of receptor-catalyzed nucleotide exchange by G proteins. The Journal of biological chemistry. 2001;276:27400–5. doi: 10.1074/jbc.C100198200. [DOI] [PubMed] [Google Scholar]
- 7.Marin EP, Krishna AG, Sakmar TP. Disruption of the alpha5 helix of transducin impairs rhodopsin-catalyzed nucleotide exchange. Biochemistry. 2002;41:6988–94. doi: 10.1021/bi025514k. [DOI] [PubMed] [Google Scholar]
- 8.Garcia PD, Onrust R, Bell SM, Sakmar TP, Bourne HR. Transducin-alpha C-terminal mutations prevent activation by rhodopsin: a new assay using recombinant proteins expressed in cultured cells. The EMBO journal. 1995;14:4460–9. doi: 10.1002/j.1460-2075.1995.tb00125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onrust R, et al. Receptor and betagamma binding sites in the alpha subunit of the retinal G protein transducin. Science. 1997;275:381–4. doi: 10.1126/science.275.5298.381. [DOI] [PubMed] [Google Scholar]
- 10.Coleman DE, et al. Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis. Science. 1994;265:1405–12. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 11.Mixon MB, et al. Tertiary and quaternary structural changes in Gi alpha 1 induced by GTP hydrolysis. Science. 1995;270:954–60. doi: 10.1126/science.270.5238.954. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, Singer WD, Danesh SM, Sternweis PC, Sprang SR. Recognition of the activated states of Galpha13 by the rgRGS domain of PDZRhoGEF. Structure. 2008;16:1532–43. doi: 10.1016/j.str.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noel JP, Hamm HE, Sigler PB. The 2.2 A crystal structure of transducin-alpha complexed with GTP gamma S. Nature. 1993;366:654–63. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- 14.Lambright DG, Noel JP, Hamm HE, Sigler PB. Structural determinants for activation of the alpha-subunit of a heterotrimeric G protein. Nature. 1994;369:621–8. doi: 10.1038/369621a0. [DOI] [PubMed] [Google Scholar]
- 15.Lambright DG, et al. The 2.0 A crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–9. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 16.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–55. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Eps N, et al. Interaction of a G protein with an activated receptor opens the interdomain interface in the alpha subunit. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1105810108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Westfield GH, et al. Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proc Natl Acad Sci U S A. 2011;108:16086–91. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gales C, et al. Probing the activation-promoted structural rearrangements in preassembled receptor-G protein complexes. Nature structural & molecular biology. 2006;13:778–86. doi: 10.1038/nsmb1134. [DOI] [PubMed] [Google Scholar]
- 20.Chung KY, et al. Conformational changes in the G protein Gs induced by the beta2 adrenergic receptor. Nature. 2011;477:611–5. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexander NS, et al. Energetic analysis of the rhodopsin-G-protein complex links the alpha5 helix to GDP release. Nature structural & molecular biology. 2014;21:56–63. doi: 10.1038/nsmb.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaya AI, et al. A conserved phenylalanine as a relay between the alpha5 helix and the GDP binding region of heterotrimeric Gi protein alpha subunit. The Journal of biological chemistry. 2014;289:24475–87. doi: 10.1074/jbc.M114.572875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda S, et al. Crystallization scale preparation of a stable GPCR signaling complex between constitutively active rhodopsin and G-protein. PLoS ONE. 2014;9:e98714. doi: 10.1371/journal.pone.0098714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matouschek A, Kellis JT, Jr, Serrano L, Fersht AR. Mapping the transition state and pathway of protein folding by protein engineering. Nature. 1989;340:122–6. doi: 10.1038/340122a0. [DOI] [PubMed] [Google Scholar]
- 25.Flock T, et al. Universal allosteric mechanism for Galpha activation by GPCRs. Nature. 2015 doi: 10.1038/nature14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamm HE, et al. Site of G protein binding to rhodopsin mapped with synthetic peptides from the alpha subunit. Science. 1988;241:832–5. doi: 10.1126/science.3136547. [DOI] [PubMed] [Google Scholar]
- 27.Osawa S, Weiss ER. The effect of carboxyl-terminal mutagenesis of Gt alpha on rhodopsin and guanine nucleotide binding. J Biol Chem. 1995;270:31052–8. doi: 10.1074/jbc.270.52.31052. [DOI] [PubMed] [Google Scholar]
- 28.Standfuss J, et al. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–60. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dratz EA, et al. NMR structure of a receptor-bound G-protein peptide. Nature. 1993;363:276–81. doi: 10.1038/363276a0. [DOI] [PubMed] [Google Scholar]
- 30.Van Eps N, et al. Interaction of a G protein with an activated receptor opens the interdomain interface in the alpha subunit. Proc Natl Acad Sci U S A. 2011;108:9420–4. doi: 10.1073/pnas.1105810108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu W, Northup JK. The helical domain of a G protein alpha subunit is a regulator of its effector. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12878–83. doi: 10.1073/pnas.95.22.12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones JC, et al. The crystal structure of a self-activating G protein alpha subunit reveals its distinct mechanism of signal initiation. Sci Signal. 2011;4:ra8. doi: 10.1126/scisignal.2001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urano D, Chen JG, Botella JR, Jones AM. Heterotrimeric G protein signalling in the plant kingdom. Open Biol. 2013;3:120186. doi: 10.1098/rsob.120186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raw AS, Coleman DE, Gilman AG, Sprang SR. Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42 --> Val mutant of Gialpha1. Biochemistry. 1997;36:15660–9. doi: 10.1021/bi971912p. [DOI] [PubMed] [Google Scholar]
- 35.Dror RO, et al. SIGNAL TRANSDUCTION. Structural basis for nucleotide exchange in heterotrimeric G proteins. Science. 2015;348:1361–5. doi: 10.1126/science.aaa5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun D, et al. AAscan, PCRdesign and MutantChecker: a suite of programs for primer design and sequence analysis for high-throughput scanning mutagenesis. PLoS ONE. 2013;8:e78878. doi: 10.1371/journal.pone.0078878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwata S. Methods and results in crystallization of membrane proteins. International University Line; La Jolla, CA: 2003. [Google Scholar]
- 38.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 40.Webb B, Sali A. Current Protocols in Bioinformatics. John Wiley & Sons, Inc.; 2002. Comparative Protein Structure Modeling Using MODELLER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu W, et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–6. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahalingam M, Martinez-Mayorga K, Brown MF, Vogel R. Two protonation switches control rhodopsin activation in membranes. Proc Natl Acad Sci U S A. 2008;105:17795–800. doi: 10.1073/pnas.0804541105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tajkhorshid E, Baudry J, Schulten K, Suhai S. Molecular dynamics study of the nature and origin of retinal's twisted structure in bacteriorhodopsin. Biophys J. 2000;78:683–93. doi: 10.1016/S0006-3495(00)76626-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nina M, Roux B, Smith JC. Functional interactions in bacteriorhodopsin: a theoretical analysis of retinal hydrogen bonding with water. Biophys J. 1995;68:25–39. doi: 10.1016/S0006-3495(95)80184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.