Abstract
Although non-invasive bladder tumours (pTa) are the most common group of bladder tumours at presentation, there has until recently been relatively little information on their molecular biology. Thus it was of great interest when mutations in the FGF receptor 3 (FGFR3) were identified in bladder tumours and it became apparent that these were most common in tumours of low grade and stage. Since the initial description of activating mutations of FGFR3, there have been numerous studies confirming the frequency and spectrum of these mutations in bladder cancers of all grades and stages. Mutation screening techniques have evolved and improved. FGFR3 mutation has been assessed as a predictive biomarker in tumour tissues and as a diagnostic biomarker in urine. Efforts have been made to understand the function of FGFR3 in urothelial and other cells. Although our understanding of FGFR3 function is incomplete, it is already apparent that this may represent an important therapeutic target not only in non-invasive bladder cancer but also in a significant number of invasive tumours. This review summarises the current state of knowledge of this interesting receptor in urothelial carcinoma (UC).
Keywords: Bladder cancer, FGFR3, mutation, expression, biomarker, therapeutic target
Mutations of FGFR3
The FGFR family consists of four members (FGFRs 1-4) of high affinity cell surface-associated receptors that are highly conserved both within the family and throughout evolution [1]. These receptors have a common structure, consisting of an extracellular domain that includes an amino terminal hydrophobic signal peptide followed by three immunoglobulin (Ig)-like domains, a hydrophobic transmembrane domain, and an intracellular tyrosine kinase domain. Fibroblast growth factors (FGFs), the ligands for FGFRs, bind to extracellular Ig-like domains II and III [2] resulting in downstream signalling. Specificity of FGF binding is conferred not only by the receptor family member but also by alternative splicing of FGFRs [3]. For example the C-terminal half of FGFR3 Ig domain III may be encoded by either of two exons, resulting in two isoforms, FGFR3b and FGFR3c [1, 4, 5]. The receptors and their isoforms are expressed in a cell and tissue-specific manner, necessary for their differential roles in different tissues and cell lineages and at different stages in development.
Frequency and spectrum of mutations
Activating mutations of FGFR3 are found in the germline in several autosomal dominant human skeletal dysplasia syndromes including achondroplasia, hypochondroplasia, severe achondroplasia with developmental delay and acanthosis nigricans (SADDAN) and thanatophoric dysplasia types I and II (TDI and TDII) [6]. In the germline, activating mutations have profound effects on the growth of the long bones where premature differentiation of chondrocytes in the growth plates is induced, causing shortening. The same phenotype has been induced in mouse models using knock-in approaches [7] and conversely, targeted disruption of the gene in mouse models causes overgrowth [8]. In light of this negative effect of FGFR3 on bone growth, its involvement as an oncogene in human cancers was unexpected.
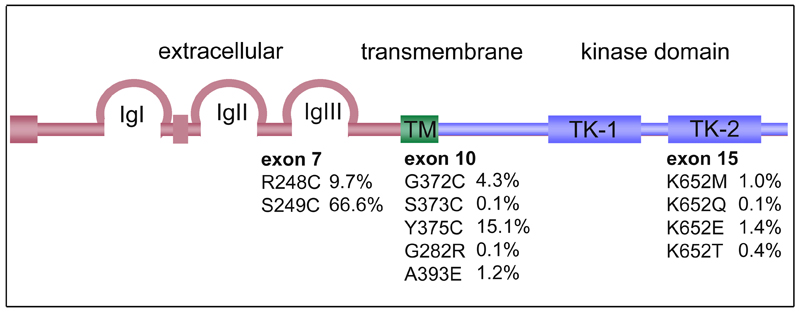
Capellen et al., [9] identified mutations in 9 of 26 (35%) bladder and 3 of 12 (25%) cervical carcinomas that were identical to those found in TDI and TDII (R248C, S249C, G372C and K652E). TDI and TDII are lethal forms of dwarfism, suggesting that these mutations represent highly activated forms of the receptor. Since this initial study there have been several studies that have identified 11 different mutations [9–21]. The frequency and distribution of these is shown in Figure 1.
Figure 1. Relative frequencies and positions of FGFR3 mutations in bladder cancers.
Frequencies are the percentage of all FGFR3 mutations described to date. IgI, IgII, IgIII, immunoglobulin-like domains; TK-1, TK-2, split tyrosine kinase domain.
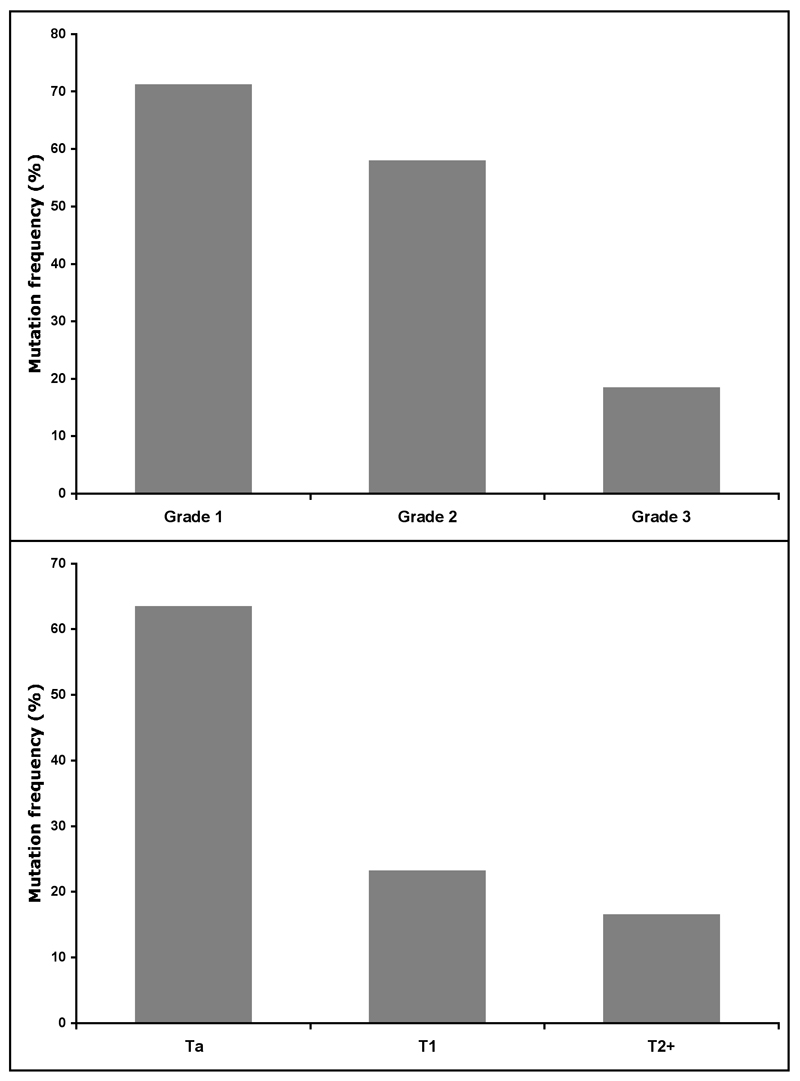
It is now clear that mutation of FGFR3 in bladder cancer is strongly associated with low tumour grade and stage and several studies describe very similar profiles. A compilation of results from 6 studies that contain adequate breakdown of this information is shown in Figure 2. The association of mutation with low tumour grade and stage led to examination of urothelial papilloma, which is a proposed precursor lesion for low-grade papillary UC [18]. Interestingly, 9 of 12 papillomas studied contained mutations. Of 79 pTaG1 tumours examined in the same study, comprising 62 papillary urothelial neoplasms of low malignant potential (PUN-LMP) and 17 low grade papillary urothelial carcinomas (LG-PUC) according to the 2004 WHO classification system [22], 86% contained a mutation. This confirmed the association of mutation with low risk UC and indicated that there is a strong molecular relationship between urothelial papilloma and low-grade UC.
Figure 2. Relationship of FGFR3 mutation to bladder tumour grade and stage.
As different mutations in FGFR3 result in differential activation of the receptor [23], it might be expected that different mutations would be associated with different clinico-pathological status. Several studies have assessed this and to date no evidence for association has been found for the majority of mutations. In the largest analysis of mutations in superficial bladder cancer published to date [24], the rare mutation A393E was found significantly more frequently in tumours classified as LMPM (low malignant potential neoplasms [22]). This study also found an association of the F286L germline polymorphism in FGFR3 with tumours of low grade and stage.
The only other malignancy in which significant involvement of FGFR3 has been reported is multiple myeloma in which 25% of cases have t(4;14)(p16;q32) that translocates FGFR3 on chromosome 4 into the IgH locus on chromosome 14 resulting in overexpression of wildtype FGFR3 from the derivative 14. Mutations in the fusion protein are infrequent but when present, have been associated with tumour progression. Thus, in this cell type both over-expression and mutational activation of the receptor can contribute to tumour development [25].
Although some mutations were initially found in cervical carcinoma, more recent studies of larger sample sets have found only rare cases with mutation [26]. Similarly, there have been several studies of a range of different cancers including prostate, breast, ovarian, lung, skin, brain, renal, oesophageal and stomach and to date no mutations have been found [27–29]. Mutations in 2 colorectal cancer samples were reported, though these did not involve the hotspot codons and it is not clear whether they represent inactivating rather than activating mutations [30]. Mutations in codon 697 (G697C) were reported in 62% of oral squamous cell carcinoma in Japanese patients [31] but this was not confirmed in a recent study from Europe [32]. However, frequent mutation has been detected in seborrheic keratoses, benign skin lesions that are common in older individuals [33, 34] and in epidermal nevi [35]. These findings in skin, together with the preponderance of mutations in benign or non-invasive bladder tumours and the apparent lack of mutations in other malignancies, indicates an association of FGFR3 mutation with low risk cancers and benign epithelial overgrowths.
Mutation detection methods
Initial studies used single strand conformation polymorphism (SSCP) analysis to detect mutations. In several studies, all coding exons were screened and PCR products showing band shifts were sequenced. In these studies, some mutations in codon 375 (Y375C) were detected but it has been shown subsequently that SSCP does not detect this mutation with high efficiency. Direct sequencing and the recently developed SNaPshot assay [36] detect Y375C as up to 25% of all mutations detected compared with ~10% by SSCP analysis. Allele-specific PCR (AS-PCR) has been used to detect the 4 most common mutations (R248C,S249C, G372C and Y375C) [37]. AS-PCR was compared with direct sequencing in 95 bladder tumour samples and matched tumours and urine sediments from 20 patients. The assay detected all mutations in paired tumour-urine samples and found several mutations in tumours that had not been detected by sequencing. It was suggested that these may have been present in only a small fraction of cells and might be detected by this assay but not by sequencing. Currently SNaPshot appears to be the most sensitive method and has been developed to detect the nine most common mutations found in UC in a single assay. The three hot-spot exons (7, 10 and 15) are amplified in a multiplex PCR reaction, followed by annealing and extension of primers specific for each mutation with a labelled dideoxynucleotide. Extended primers, each designed with different lengths of poly(dT) tails are then separated by capillary electrophoresis and all possible mutations are visualised in one electophoretic run. This technique is superior to SSCP analysis in detecting Y275C mutations and is able to detect a mutant FGFR3 heterozygote in a 20-fold excess of wildtype DNA i.e one mutant molecule in the presence of 39 wildtype molecules [36]. This makes the technique suitable for sensitive detection of mutant tumour cells in urine (see below).
Timing of FGFR3 mutation during bladder tumour development
The high frequency of FGFR3 mutation in urothelial papilloma, which is a putative precursor for papillary bladder cancer [18], suggests a role early in disease development. A possible earlier precursor is flat urothelial hyperplasia, which is frequently found in association with UC of both papillary and solid growth patterns. Studies of hyperplastic urothelium and adjacent papillary UC have shown that these lesions often share the same genetic alterations, particularly chromosome 9 deletions [38, 39]. Chromosome 9 loss detected by loss of heterozygosity (LOH) analysis is common in UC of all grades and stages [40]. LOH has not been found in urothelial papilloma, though only a very small number of samples have been analysed [41]. A recent study examined both FGFR3 mutation and chromosome 9 LOH in a series of 30 hyperplasias and simultaneous or consecutive papillary tumours from some of the same patients [42]. FGFR3 mutation was found in 23% of hyperplasias and chromosome 9 LOH in 30%. In two cases, FGFR3 mutations but no chromosome 9 LOH was present and in 6 cases chromosome 9 LOH alone was found. This may indicate that chromosome 9 loss usually occurs earlier than FGFR3 mutation during the development of papillary UC. However, the relationship of flat hyperplasia to papilloma is not yet clear and an alternative explanation for the observed results is that FGFR3 mutations are associated predominantly or only with tumours with a papillary growth pattern (for which papilloma is a precursor) and that chromosome 9 loss, which is known to occur in UC of all grades, stages and growth patterns may occur in some flat hyperplasias which do not subsequently develop a papillary growth pattern and which represent the precursor for non-papillary UC. Certainly, flat dysplasia and carcinoma in situ (CIS), which are predicted precursors for such tumours, commonly show chromosome 9 loss. The availability of robust methods for whole genome amplification suitable for formalin fixed paraffin-embedded samples will allow analysis of larger numbers of samples to clarify these important issues in UC pathogenesis.
Recently we assessed mutation status in different areas that represented different tumour stages within the same tumour block [43]. Laser capture microdissection was used to ensure that tumour DNA extracted from high stage areas was not contaminated with stromal cell DNA. Mutation status was found to be different in different regions of blocks from 8 of 158 patients. In each case mutation was found in the non-invasive and not the invasive component. Where possible we had selected physically separate tumour fragments within each block to facilitate clean dissection of different stages. As all resected tissues were placed in the same container at cystoscopy regardless of the number of separate tumours present, a tumour block could contain fragments of different tumours. Of the eight samples with different mutation status in non-invasive and invasive components, 2 were reported as multifocal, 4 as single tumours and no information was recorded for 2 cases. We assessed whether there was physical continuity between the superficial and invasive components and whether tumour morphology was compatible with a single tumour entity or suggested the possibility of more than one tumour clone within the sample. Six of the eight tumours were predicted to represent single tumours based on number of tumours resected, contiguity of regions of different stage and/or morphological similarity of tumour throughout the sample. This suggests either that the allele containing the FGFR3 mutation was lost in the high stage region or that a small subset of FGFR3 wildtype cells present in the low stage tumour had progressed. This finding of complexity in mutation status within the same patient requires more detailed examination in the future. This should include LOH analysis to determine whether loss of a mutant allele does occur during tumour progression and an assessment of tumour clonality.
Relationship of FGFR3 mutation to other molecular alterations
The strong association of FGFR3 mutation with low tumour grade and stage is reflected in the finding that mutations of FGFR3 and TP53 show an inverse relationship [10, 44, 45]. This almost certainly indicates their involvement in distinct pathogenic pathways that lead to development of low-grade papillary UC and solid invasive UC [46]. Extensive analysis of TP53 inactivation in UC has shown that inactivating mutations and abnormal accumulation of p53 protein is associated with aggressive UC and infrequently found in low grade/stage disease [47]. Recent studies have found that a small number of tumours contain both FGFR3 and TP53 mutations. Van Rhijn et al., [44] and Bakkar et al., [10] reported 5.7% and 3.7% of tumours with both mutations respectively. No association of the presence of mutations in both FGFR3 and TP53 with tumour grade or stage was found in either of these studies. The significance of the presence of both genetic changes is unknown but may identify a subset of tumours with a distinct developmental pathway or may reflect the infrequent progression of low-grade FGFR3 mutant UC. If this were the case, the finding of both mutations in non-invasive UC may represent a marker of high risk. The differential association of FGFR3 and TP53 mutation with tumour grade and stage is also reflected at the protein level. Thus, it has been found that loss of expression of FGFR3 protein and increased p53 protein accumulation is associated with increased tumour stage and grade [48].
The clear relationship of FGFR3 and TP53 mutations with distinct tumour subsets raises questions about those tumours that have molecular and phenotypic characteristics that do not fit well in either subset. These are the T1 tumours that have in the past been termed “superficial” and have been frequently grouped with Ta tumours despite their ability to break through the epithelial basement membrane. These tumours often show molecular alterations in common with muscle invasive tumours [49]. pT1G3 tumours in particular, are considered a high-risk group that present a major clinical management challenge [50]. A series of 119 pT1G3 tumours has been examined for FGFR3 and TP53 mutation status and for the relationship of these events to recurrence and survival [12]. Many TP53 mutations were found (65% of cases) and FGFR3 mutations were found in 16.8% of cases. These frequencies are compatible with findings reported for several smaller groups of pT1 tumours. Mutations of both genes were found in 9% of tumours these genetic alterations were distributed independently and not associated with either recurrence or survival. This indicates that T1G3 tumours do not resemble either of the two major groups of bladder tumours. Possibly they represent a distinct group with a different pathogenesis pathway or possibly some may be pTa tumours that have acquired alterations that now confer an invasive phenotype. This study represents the first large molecular analysis of this interesting group of tumours and highlights the need for further studies to elucidate their pathogenesis and to identify prognostic biomarkers within the group.
One of the predicted effects of FGFR3 mutation is activation of the Ras-MAPK pathway [51]. The relationship between FGFR3 and Ras gene mutations in UC has been examined and these mutations were found to be mutually exclusive [52]. In contrast to the relationship between TP53 and FGFR3 mutations, the absolute mutual exclusivity of FGFR3 and Ras gene mutations is thought to reflect activation of the same pathway by either event. Ras mutations are found in ~13% of UC and in total 85% of low-grade Ta tumours were found to have a Ras or an FGFR3 mutation, suggesting that MAPK pathway activation may be an obligate event in most of these tumours. Interestingly, unlike FGFR3 mutation, no obvious relationship of mutation of a Ras gene with tumour grade of stage has been found. This implies that although both events may fulfil at least one function that precludes selection for both, there is a difference, possibly in the strength and/or duration of signals generated that allows Ras mutation to contribute equally well to the development of both major tumour groups. Studies of much larger series of tumours are now needed, together with comparisons of the downstream effects of Ras and FGFR3 signalling in urothelial cells.
Mutations of the alpha catalytic subunit of PI3 kinase, PIK3CA, have been identified recently in UC [53]. Exons 9 and 20 that contain the mutations hotspot codons of PIK3CA were sequenced and mutations found in 13% of a panel of 87 tumours representing the whole spectrum of disease. An association with low tumour grade and stage was found, though this was much less striking that that found for FGFR3, with mutations identified in approximately 20% of Ta tumours. Inevitably, given this distribution of mutations, an association of PIK3CA mutation with FGFR3 mutation was apparent. Eighteen of 69 (26%) FGFR3 mutant tumours were also PIK3CA mutant compared with 4 of 58 (6.9%) FGFR3-wildtype tumours. The functional relationship between these events, if any, is not known but the known ability of Ras to activate the PI3 kinase pathway will certainly prompt investigation of the relationship to Ras mutations and it will also be of great interest to examine clinical outcome in tumours with both PIK3CA and FGFR3 mutation. LOH and homozygous deletion of PTEN (phosphatase and tensin homologue deleted on chromosome ten) are found in high grade and stage bladder tumours [54, 55]. As PTEN is a key negative regulator of AKT in the PI3 kinase pathway it might be expected that inactivation of PTEN or activation of PIK3CA could fulfil the same role and indeed, in some other tumour types, PTEN involvement and PIK3CA mutation or amplification are mutually exclusive events [56, 57]. However, this is not the case in all tumours, for example in endometrial cancer where co-existent mutations are common and it has been suggested that their combined effect may be required [58]. Thus it will be important to examine this relationship in UC as well as to define relationships to FGFR3 status.
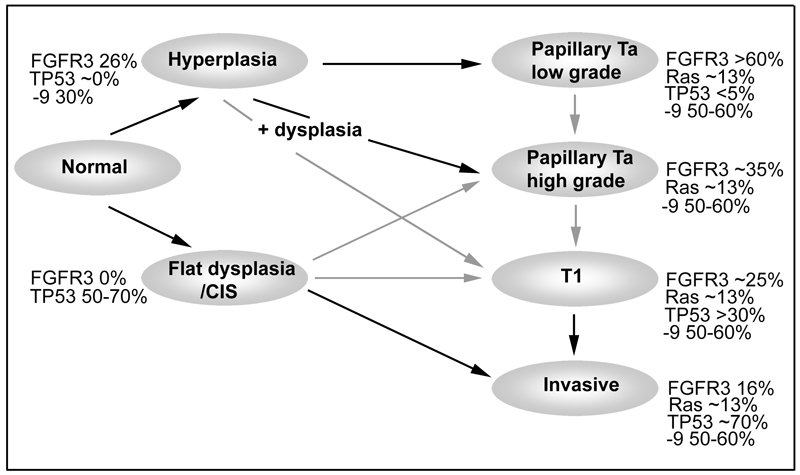
Taken together, these observations on the relationship of FGFR3 mutation to other genetic events contributes significantly to the current view of the molecular pathogenesis of bladder cancer. A representation of the possible timing of some events and their relative frequencies in the two major predicted pathways is shown in Figure 3. The possibility of alternative pathways leading to T1 tumours is also shown.
Figure 3. Potential pathways of urothelial tumorigenesis.
Black arrows indicate likely pathways and grey arrows indicate uncertain relationships. Frequencies are given for mutations in FGFR3, TP53, Ras genes and PIK3CA and for chromosome 9 LOH. PIK3CA data are from a single study [53]. Figures for other genes and chromosome 9 are representative of a much larger literature. Low-grade papillary tumours (top) may arise via simple hyperplasia and minimal dysplasia and these are characterised at the molecular level by deletions of chromosome 9 and activating mutations of FGFR3. Invasive carcinoma (bottom) is believed to arise via the flat high-grade lesion CIS and in this case TP53 mutation occurs early and FGFR3 mutations are infrequent. There is limited data for chromosome 9 LOH in CIS. Muscle invasive tumours are usually genetically unstable and contain many genomic alterations in addition to those shown here. The finding of dysplasia in association with high-grade papillary tumours that lack TP53 mutation but have frequent chromosome 9 losses suggests that an independent route to high-grade papillary tumours may exist (centre). The pathway to development of T1 tumours is uncertain.
Genome-wide analysis of FGFR3 mutant and wildtype bladder tumours
Some genome-wide analyses have been applied to tumours with known FGFR3 mutation status. SNP array analysis was used to assess whether distinct chromosomal changes (LOH and copy number changes) were correlated with mutation status in T1 tumours from 19 patients [59]. FGFR3 mutant and wildtype tumours showed differences in the frequency of alterations of 8p and 17p with few changes in these regions in mutant tumours and more in wildtype tumours. In general, significant copy number gains were found only in FGFR3 wildtype tumours. In this study, genome-wide expression profiling followed by hierarchical cluster analysis divided tumours into groups that contained “benign” (low/medium risk) tumours including most of the FGFR3 mutant tumours and “malignant” (high risk) tumours, which contained most of the wildtype tumours. When a previous CIS-classifier gene signature [60] was applied most wildtype tumours were classified as “CIS” and most mutant tumours as “no CIS”. Interestingly, it was reported that low-risk wildtype “no CIS”-classified tumours showed upregulated expression of FGFR3, unlike the wildtype tumours that had a “CIS” profile. This may indicate a distinct pathogenic pathway for such low-grade Ta FGFR3 wildtype tumours.
In a second study, expression profiling, FGFR3 and TP53 mutation analysis and chromosome 9 LOH analysis was applied to a series of 75 Ta and T1 tumours [61]. Hierarchical cluster analysis divided the tumours into three groups, strongly related to tumour grade, the first of which (cluster I) contained the highest proportion of Ta grade 1 tumours and the third (cluster III), the highest proportion of grade 3 tumours. The distribution of TP53 and FGFR3 mutations with respect to tumour grade and stage was compatible with other studies. Of the 9 TP53 mutations, only 1 was found in a grade 1 tumour whereas of 46 FGFR3 mutations, all but 1were found in grade 1 or 2 tumours. When FGFR3 mutation status was correlated with gene expression profiles, there was a marked polarisation towards clusters I (14/15 tumours mutant) and II (18/27 tumours mutant) with no mutant tumours found in cluster III. Cluster I tumours showed high expression of protein synthesis genes and low expression of cell cycle gene and cluster III showed high expression of cell cycle genes. These studies provide further confirmation of the association of FGFR3 mutation with bladder tumours of low grade and stage and provide gene lists for this group that may provide insights into specific processes and cellular phenotypes that are regulated by mutation of this receptor.
FGFR3 expression in the urothelium
As antibodies are not available to detect all members of the FGFR family by immunohistochemistry, real-time RT-PCR has been used to compare the relative expression of FGFR3 with other FGFRs in the urothelium. In normal urothelial cells freshly isolated from human ureter, FGFR3 was the major FGFR expressed, with very low levels of FGFRs 1, 2 and 4 [62]. Once the cells were cultured, levels of FGFR2 rose and some FGFR4 was detected but FGFR3 remained the predominant FGFR. Interestingly, FGFR3 levels were regulated relative to cell proliferation with lower levels in the exponential phase of population growth at each passage, rising at confluence and reaching highest levels in mortal normal cells at senescence. In cultured cells, protein levels detected western blotting, closely reflect RNA levels [62]. A good antibody for FGFR3 is available and it has been shown by immunohistochemistry that the normal ureteric and bladder urothelium expresses low levels of the protein [43].
Distinct FGFR isoforms, have been described in normal and tumour cells. Two full-length FGFR3 isoforms have been described with differential splicing that alters the C-terminal part of the third Ig domain. FGFR3b, a form that contains exon 8 but not 9 was initially identified in mouse skin [2, 5] and later in human epithelial cells [63]. This isoform shows high affinity for FGF1 and also binds FGFs 9 and 18 but not other FGFs studied to date. Mesenchymal cells including chondrocytes express FGFR3c that contains exon 9 rather than 8 and shows high affinity for a much wider range of FGFs including FGF2 [64]. In addition to these full-length isoforms, several shorter forms have been described in tumour cell lines [65–68].
PCR analysis of cDNA isolated from normal urothelial cells has detected several RNA forms [62]. The only full-length isoform identified was FGFR3b. A major PCR product was found to encode a small isoform lacking exons 8-10 which encode the third Ig domain and the transmembrane domain. This form (FGFR3 Δ8-10) was shown to be expressed, glycosylated and secreted. Most interestingly, when purified, this soluble form of FGFR3 was shown to act as a dominant negative that could inhibit FGF-stimulated proliferation [62]. The mechanism for this dominant negative effect is not yet clear but could be via binding to and sequestering FGFs or by dimerising with full length FGFR3. The biological significance of this form is unclear but as the ratio of FGFR3 Δ8-10:FGFR3b was found to increase in confluent cells, it is possible that it plays a role in downregulating FGFR3 signalling in quiescence.
In many bladder tumour cell lines, an isoform switch from FGFR3b to FGFR3c, the mesenchymal isoform, has been found [62]. In some cases, all transcripts detected were FGFR3c. As this isoform binds a wide range of FGFs including FGF2, this presents the possibility that autocrine or paracrine stimulation may activate FGFR3 signalling in some tumours. The urine of bladder cancer patients has been reported to contain FGFs including FGF2 [69–71] though expression of FGF2 is low in bladder tumour tissues [72]. It has been suggested that the high levels of urinary FGF2 found in cancer patients may be derived from basement membrane, which shows high levels of expression, that this is liberated during tumour invasion and stimulates angiogenesis at the invasion margin [72].
FGFR3 protein expression has been examined in several studies and increased expression found in some tumours. Matsumoto et al., [73] reported expression of moderate or high levels of protein in 49% of tumours but found no relationship to tumour grade or stage. Two other studies found a relationship between higher expression and lower tumour grade and stage [48, 74]. In these studies the mutation status of the tumours was not reported. However, semi-quantitative PCR has been used to examine RNA levels in a panel of tumours of known mutation status [75] and this demonstrated an association of higher levels of RNA expression and mutation status.
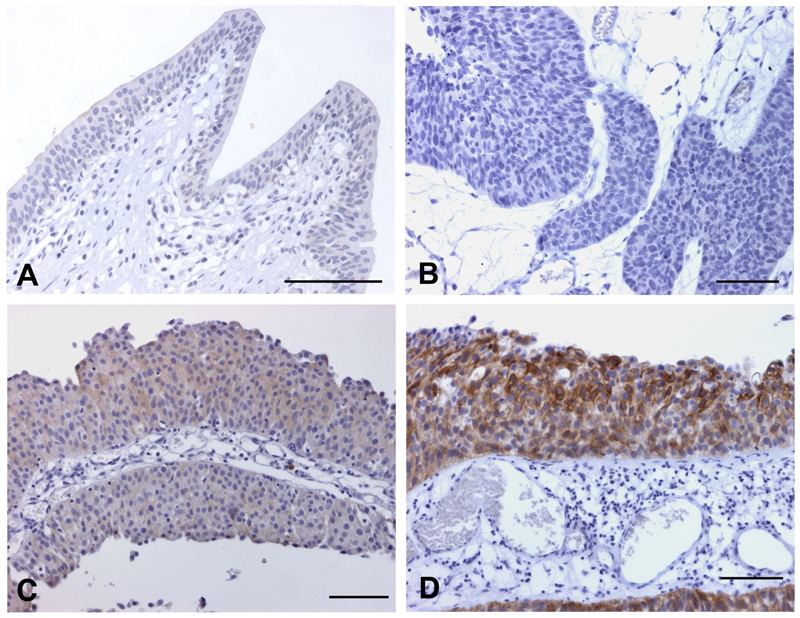
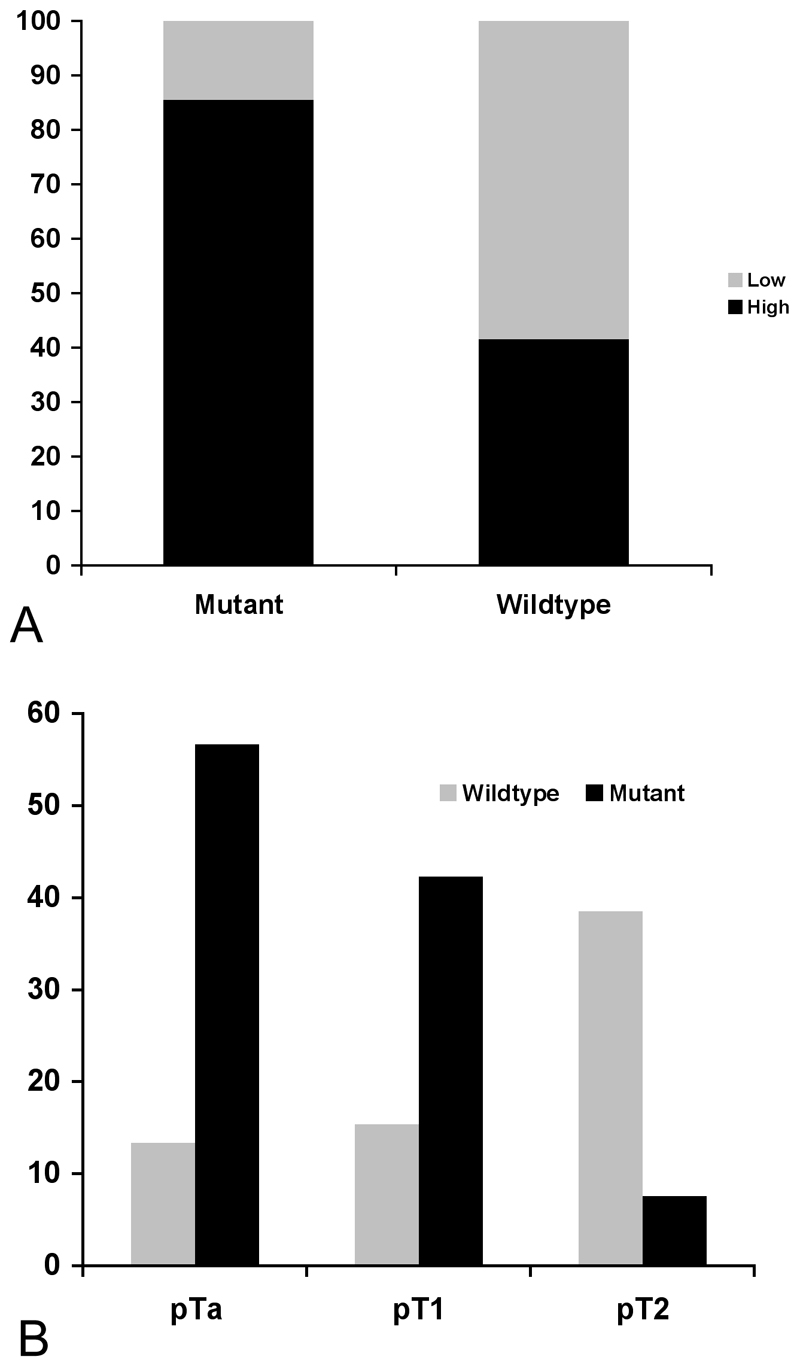
Recently, we assessed FGFR3 protein expression in a population-based series of tumours of known mutation status [43]. Four categories of staining were defined (Figure 4). Over-expression of FGFR3 protein was found in many tumours compared to normal bladder and ureteric controls. Increased expression was associated with mutation (85% of mutant tumours showed high-level expression)(Figure 5A). Overall, as expected from the known distribution of FGFR3 mutation with grade and stage, high FGFR3 expression, which in turn was related to mutation status, was significantly more frequent in non-invasive (pTa) compared to invasive (pT2) tumours and in low grade (grades 1 and 2) compared with high grade (grade 3) tumours. Interestingly, 42% of tumours including 44% of muscle invasive tumours with no detectable mutation showed over-expression (Figure 5B). This may represent a non-mutant subset of tumours in which FGFR3 signalling contributes to the transformed phenotype. An increasingly greater proportion of tumours were negative (score 0) with advancing stage (9% of pTa, 19% of pT1 and 28% of pT2 tumours). This contrasted with the situation for normal (score 1) staining, which was more evenly spread (21% of pTa, 23% of pT1 and 26% of pT2 tumours), suggesting that complete loss of expression has biological significance.
Figure 4. Patterns of FGFR3 staining in normal urothelium and bladder tumours.
Normal ureteric urothelium (A) and bladder tumour samples (B-D). A, staining pattern 1; B, staining pattern 0; C, staining pattern 2; D, staining pattern 3; bars = 100μm.
Reproduced with permission from: DC Tomlinson, O Baldo, P Harnden and MA Knowles. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. Journal of Pathology volume 213, 2007, Copyright Pathological Society of Great Britain and Ireland. Permission is granted by John Wiley & Sons Ltd on behalf of PathSoc.
Figure 5. FGFR3 protein expression in relation to mutation and tumour stage.
A. Distribution of high and low expression of FGFR3 protein in bladder tumours with and without mutation. B. Relationship of high-level expression of FGFR3 to tumour stage and mutation status. Data from [43].
Taken together, data on FGFR3 expression and splicing indicate that these are highly regulated in the urothelium. Further work on the mechanisms and biological significance of the observed alterations are now needed. Clearly there are several possible mechanisms by which FGFR signalling could be increased in bladder tumours, including the involvement of autocrine or paracrine signalling by FGFs. Currently, little is known about expression of FGFs by the normal urothelium, the submucosa or in bladder tumours and this information will be required to gain full understanding of the role of FGFRs in bladder tumour development.
FGFR3 as a biomarker for risk of recurrence and progression
A major issue in the management of non-invasive bladder tumours is prediction of recurrence and progression. Thus the potential application of any molecular alterations found in these tumours as prognostic or predictive biomarkers is of great interest. Several studies have examined FGFR3 mutation as a potential predictive marker for recurrence. Although an initial analysis suggested that there may be an association with low recurrence rate within a group of 57 patients with superficial tumours (pTa, pT1)[17], other studies have not confirmed this [24, 59]. Indeed, a large study (n>700) of Spanish UC, found that mutation was significantly associated with higher recurrence rate in Ta grade 1 tumours [24]. In TaG2 tumours there was also a trend towards higher recurrence rate, though this was not significant. This suggests that when carefully stratified groups are studied, mutant FGFR3 is an oncogene with clinically significant effect. However, no effect on progression has been found. In the latter study, it was found that although within the entire group of tumours, comprising both pTa and pT1 cases, there was an association of mutation with lower risk of progression, when stratification for grade and stage were applied, no significant difference was found. Thus, the accumulated findings to date suggest that within the low risk group of Ta bladder tumours, mutation of FGFR3 confers an increased risk of recurrence but the presence or absence of such a mutation confers no risk of progression in any UC group.
Grade and stage of UC provide a major indication of clinical outcome. Many molecular alterations have been assessed as biomarkers for prediction of prognosis, some of which show potentially useful associations. p53 has been extensively investigated [76] as have several markers related to cell proliferation including Rb, cyclin D1, Ki67/MIB-1, p21 and p27. However, none of these as single markers provide information that is applicable to the management of individual patients [77]. Although FGFR3 mutation as a single marker is not predictive of outcome, a multicentre study of tumours from 286 patients found that mutation status in combination with immunohistochemistry for Ki67 allowed definition of three “molecular grades” of tumour with distinct prognoses in which FGFR3 mutation and low Ki67 index (molecular grade 1) was associated with significantly better disease specific survival, wildtype FGFR3 and high Ki67 index (molecular grade 3) was associated with poor survival, and wildtype FGFR3 and low Ki67 index (molecular grade 3) showed intermediate status [20]. These molecular biomarkers provide objective and reproducible measures that may be superior to assessment of grade, which is well known to be a highly subjective measurement [78, 79].
Abnormal staining pattern for cytokeratin 20 (CK20) has been described in pTa bladder tumours that show low risk of recurrence [80]. In the normal urothelium and in low risk tumours, staining is confined to the mature superficial cells and staining throughout the urothelium represents an abnormal pattern. In a recent study, it was found that 89% of tumours with a normal CK20 pattern of expression showed mutation of FGFR3 whereas only 37% of cases with abnormal CK20 expression showed mutation. Both markers were strong predictors of disease-specific survival in univariate analysis but did not show independence from grade and stage in multivariate analyses [81]. This provides additional evidence for an association with FGFR3 mutation and a relatively benign disease course.
FGFR3 as a urine biomarker
The high recurrence rate (60-80%) of non-invasive UC necessitates long-term patient monitoring. Currently the standard of care in Europe and the USA consists of cystoscopic surveillance with urine cytology every 3-6 months for the first 2 years, followed by less frequent observation in subsequent years. Such invasive monitoring is costly, uncomfortable and carries a relatively high risk of infection. The prevalence of bladder cancer is high and the large numbers of patients requiring monitoring place a great burden on health service providers. This requirement for multiple hospital visits and invasive procedures makes this the most expensive of all cancers to treat [82, 83]. Thus, there is a pressing need for urine-based tests to identify tumour recurrence and reduce the need for cystoscopic surveillance. Urine cytology, though universally used as the gold standard test, fails to detect the majority of low-grade, non-invasive tumours [84]. Many tests have been described during the past decade and several have been approved by the FDA (reviewed in [85]). However, none have yet achieved widespread use in the clinic and these remain adjuncts to cystoscopy [86]. This is in most cases due to lower sensitivity and/or specificity in patients with a history of tumours of low grade and stage. One possible way to improve performance in such patients is to develop assays that are specifically designed to detect molecular alterations that are either very common in this group of patients or have been shown to be present in the primary tumour of an individual patient. Microsatellite alterations are common in most cancers and denote the occurrence of LOH or a form of genomic instability that affects short repeat sequences. In UC, many common regions of LOH have been identified, with chromosome 9 LOH the most common in tumours of low grade and stage [46]. Microsatellite analysis has been successfully applied for non-invasive detection of bladder cancer [87–89] and this provides higher sensitivity than cytology, which detects only 13-50% of tumours. The high frequency of FGFR3 mutation in low grade/stage UC may provide an additional marker that in combination with microsatellite analysis or other assay(s) could raise sensitivity to a clinically acceptable level. This was tested on a prospective series of 59 UC tissues and matched urine samples [16]. Twenty-three microsatellite markers and mutation analysis of FGFR3 exons 7, 10 and 15 were carried out and together these assays showed a sensitivity of 89% and specificity of 93%. In this study that included tumours of all grades and stages, 44% of cases had FGFR3 mutation. The use of combined analysis of FGFR3 with cytology has also been examined on urine sediments and matched tumour samples obtained at transurethral resection (TURBT)(n=72) or cystectomy (n=120) [14]. 67% of patients in the TURBT group had an FGFR3 mutation and 28% in the cystectomy group. It was found that FGFR3 analysis outperformed cytology in the TURBT group whereas cytology outperformed FGFR3 analysis in the cystectomy group. Combined analysis identified tumour presence in 86% of patients.
In both of these studies, assays were carried out on urine collected from patients with a known concurrent tumour. The ability of such assays to detect recurrence must now be tested in the prospective setting. The development of the SNaPshot assay should increase the ability to detect mutant DNA molecules in an excess of normal DNA, which is a common situation in urine sediments where leukocytes and normal cells derived from the urinary tract may predominate.
FGFR3 as a therapeutic target
Activated receptor tyrosine kinases are druggable targets for small molecule-based and antibody-based approaches to therapy. Thus, there is great interest in FGFR3 as a potential therapeutic target in bladder cancer. Several approaches to targeting FGFR3 have been examined in multiple myeloma and promising responses have been obtained with small molecule inhibitors and antibodies [90–94]. In these cancers, it has been demonstrated that several of the agents tested, directly affect the function of both over-expressed wildtype and mutant FGFR3. Recent results on bladder tumour cells provide evidence that FGFR3 is a valid therapeutic target and that these approaches to receptor inhibition are applicable to bladder tumours.
To validate FGFR3 as a therapeutic target, we used RNAi to examine the effects of downregulation of receptor in bladder tumour cells that express high levels of mutant (S249C) receptor [95]. The S249C mutation is predicted to induce disulphide bond formation by the introduction of an additional cysteine in the extracellular domain of FGFR3b. This was confirmed in cells expressing S249C FGFR3 protein which formed stable homodimers and was constitutively phosphorylated [95]. Stable expression of an FGFR3 shRNA was used to knock down S249C FGFR3b in tumour cells and this resulted not only in inhibition of proliferation but also of anchorage independent growth and clonogenicity at low density, both of which are phenotypic markers of transformation. Knockdown also caused profound cell flattening. In contrast to the effect of mutant FGFR3b knockdown in tumour cell lines, knockdown in telomerase-immortalised normal human urothelial cells (TERT-NHUC) caused no significant phenotypic effects. This implies a dependence of the tumour cells on continued FGFR3b signalling as described for oncogenic stimuli in other tumour cell types, so-called “oncogene addiction” [96]. Significantly this also shows that although TERT-NHUC express FGFR3b, there is no dependence on this for either survival or proliferation. This differential sensitivity may allow non-toxic targeted therapies to be developed.
Both antibodies and small molecules have been used to assess the effects of inhibition of FGFR3 in bladder tumour cells. The tumour cell line MGH-U3 expresses Y375C mutant FGFR3, which like the S249C mutant receptor shows constitutive phosphorylation. When treated with the small molecule FGFR inhibitor SU5402 [97] or siRNAs, these cells showed reduced receptor phosphorylation, reduced proliferation and reduced colony formation in soft agarose [75].
Human single chain Fv antibody fragments that recognise the extracellular domain of FGFR3c have been isolated and characterised [98]. These specifically bound both wildtype and mutant FGFR3 but not FGFR1 and were able to block proliferation of the bladder tumour cell line RT112, which expresses high levels of FGFR3. In another study that focussed on multiple myeloma, a human anti-FGFR3-neutralising Fab was shown to inhibit FGFR3 autophosphorylation and downstream signalling and was cytotoxic to multiple myeloma cells that over-express wildtype FGFR3 [94]. This antibody inhibited ligand-binding by wildtype receptor and significantly, it inhibited the growth of xenografts of tumour cells expressing FGFR3-S249C, the most common FGFR3 mutant form found in bladder cancer.
Conclusions
It is clear that FGFR3 plays a major role in the development of low-grade non-invasive bladder tumours. Although the exact phenotypic consequences of FGFR3 mutation are currently unknown, as are details of the downstream signalling consequences of receptor activation in the urothelium, this receptor is predicted to have important applications both as a biomarker and a therapeutic target. Future work must address the precise consequences of FGFR3 signalling in both the normal cell context and in situations where signalling is constitutively activated by over-expression or mutation. At present there is very little information on the cellular consequences of such aberrant signalling and how this may contribute at an early stage in the development of bladder cancer and this must be explored in relevant in vitro and in vivo models. Currently there is very little information on the expression of FGFs either in urothelial cells or in the bladder stroma and it will be important to assess the potential role of paracrine or autocrine signalling via FGF receptors in the urothelium. However, whilst these aspects of the function and biological consequences of FGFR3 signalling require further detailed study, rapid application of FGFR3 as a clinical biomarker and therapeutic target is likely. The ease with which mutations can be detected provides an objective biomarker that may have application in urine-based disease monitoring. If ongoing studies confirm initial observations, this assay could be implemented rapidly for disease monitoring and/or screening and may have major impact on patient care. Exploitation of this receptor as a therapeutic target also holds much promise in the treatment of bladder cancer, particularly low-grade non-invasive tumours. Recent evidence indicates that FGFR3-targeted therapies may also be applicable in the significant numbers of patients with invasive tumours in which high-level expression of non-mutant receptor can be detected. The availability of some small molecule inhibitors of FGFRs will allow further assessment of the likely efficacy of this approach to therapy in the preclinical setting. This, combined with the rapidity of screening for small molecule inhibitors of receptor tyrosine kinases should ensure that targeted agents suitable for clinical testing become available in the near future.
Acknowledgements
I am indebted to all members of my laboratory who have contributed to our work on FGF receptors, particularly to Darren Tomlinson, Wendy Kennedy, Carolyn Hurst, Erica di Martino and Fiona Lamont and to Patricia Harnden for her expert support in histopathology. Work on FGFRs in this laboratory is funded by Cancer Research UK and the Association for International Cancer Research.
References
- 1.Johnson DE, Williams LT. Structural and functional diversity in the FGF receptor multigene family. Adv Cancer Res. 1993;60:1–41. doi: 10.1016/s0065-230x(08)60821-0. [DOI] [PubMed] [Google Scholar]
- 2.Avivi A, Yayon A, Givol D. A novel form of FGF receptor-3 using an alternative exon in the immunoglobulin domain III. FEBS Lett. 1993;330:249–52. doi: 10.1016/0014-5793(93)80882-u. [DOI] [PubMed] [Google Scholar]
- 3.Ornitz DM, Xu J, Colvin JS, et al. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–7. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 4.Werner S, Duan DS, de Vries C, Peters KG, Johnson DE, Williams LT. Differential splicing in the extracellular region of fibroblast growth factor receptor 1 generates receptor variants with different ligand- binding specificities. Mol Cell Biol. 1992;12:82–8. doi: 10.1128/mcb.12.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chellaiah AT, McEwen DG, Werner S, Xu J, Ornitz DM. Fibroblast growth factor receptor (FGFR) 3. Alternative splicing in immunoglobulin-like domain III creates a receptor highly specific for acidic FGF/FGF-1. J Biol Chem. 1994;269:11620–7. [PubMed] [Google Scholar]
- 6.Passos-Bueno MR, Wilcox WR, Jabs EW, Sertie AL, Alonso LG, Kitoh H. Clinical spectrum of fibroblast growth factor receptor mutations. Hum Mutat. 1999;14:115–25. doi: 10.1002/(SICI)1098-1004(1999)14:2<115::AID-HUMU3>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Li C, Qiao W, Xu X, Deng C. A Ser(365)-->Cys mutation of fibroblast growth factor receptor 3 in mouse downregulates Ihh/PTHrP signals and causes severe achondroplasia. Hum Mol Genet. 2001;10:457–65. doi: 10.1093/hmg/10.5.457. [DOI] [PubMed] [Google Scholar]
- 8.Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–7. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 9.Cappellen D, De Oliveira C, Ricol D, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 10.Bakkar AA, Wallerand H, Radvanyi F, et al. FGFR3 and TP53 gene mutations define two distinct pathways in urothelial cell carcinoma of the bladder. Cancer Res. 2003;63:8108–12. [PubMed] [Google Scholar]
- 11.Billerey C, Chopin D, Aubriot-Lorton MH, et al. Frequent FGFR3 mutations in papillary non-invasive bladder (pTa) tumors. Am J Pathol. 2001;158:1955–9. doi: 10.1016/S0002-9440(10)64665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez S, Lopez-Knowles E, Lloreta J, et al. FGFR3 and Tp53 mutations in T1G3 transitional bladder carcinomas: independent distribution and lack of association with prognosis. Clin Cancer Res. 2005;11:5444. doi: 10.1158/1078-0432.CCR-05-0122. [DOI] [PubMed] [Google Scholar]
- 13.Kimura T, Suzuki H, Ohashi T, Asano K, Kiyota H, Eto Y. The incidence of thanatophoric dysplasia mutations in FGFR3 gene is higher in low-grade or superficial bladder carcinomas. Cancer. 2001;92:2555–61. doi: 10.1002/1097-0142(20011115)92:10<2555::aid-cncr1607>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 14.Rieger-Christ KM, Mourtzinos A, Lee PJ, et al. Identification of fibroblast growth factor receptor 3 mutations in urine sediment DNA samples complements cytology in bladder tumor detection. Cancer. 2003;98:737–44. doi: 10.1002/cncr.11536. [DOI] [PubMed] [Google Scholar]
- 15.Sibley K, Cuthbert-Heavens D, Knowles MA. Loss of heterozygosity at 4p16.3 and mutation of FGFR3 in transitional cell carcinoma. Oncogene. 2001;20:686–91. doi: 10.1038/sj.onc.1204110. [DOI] [PubMed] [Google Scholar]
- 16.van Rhijn BW, Lurkin I, Chopin DK, et al. Combined microsatellite and FGFR3 mutation analysis enables a highly sensitive detection of urothelial cell carcinoma in voided urine. Clin Cancer Res. 2003;9:257–63. [PubMed] [Google Scholar]
- 17.van Rhijn BW, Lurkin I, Radvanyi F, Kirkels WJ, van der Kwast TH, Zwarthoff EC. The fibroblast growth factor receptor 3 (FGFR3) mutation is a strong indicator of superficial bladder cancer with low recurrence rate. Cancer Res. 2001;61:1265–8. [PubMed] [Google Scholar]
- 18.van Rhijn BW, Montironi R, Zwarthoff EC, Jobsis AC, van der Kwast TH. Frequent FGFR3 mutations in urothelial papilloma. J Pathol. 2002;198:245–51. doi: 10.1002/path.1202. [DOI] [PubMed] [Google Scholar]
- 19.van Rhijn BW, van Tilborg AA, Lurkin I, et al. Novel fibroblast growth factor receptor 3 (FGFR3) mutations in bladder cancer previously identified in non-lethal skeletal disorders. Eur J Hum Genet. 2002;10:819–24. doi: 10.1038/sj.ejhg.5200883. [DOI] [PubMed] [Google Scholar]
- 20.van Rhijn BW, Vis AN, van der Kwast TH, et al. Molecular grading of urothelial cell carcinoma with fibroblast growth factor receptor 3 and MIB-1 is superior to pathologic grade for the prediction of clinical outcome. J Clin Oncol. 2003;21:1912–21. doi: 10.1200/JCO.2003.05.073. [DOI] [PubMed] [Google Scholar]
- 21.Wallerand H, Bakkar AA, de Medina SG, et al. Mutations in TP53, but not FGFR3, in urothelial cell carcinoma of the bladder are influenced by smoking: contribution of exogenous versus endogenous carcinogens. Carcinogenesis. 2005;26:177. doi: 10.1093/carcin/bgh275. [DOI] [PubMed] [Google Scholar]
- 22.WHO. WHO Classification Tumours of the Urinary System and Male Genital Organs. Lyon: IARC Press; 2004. [Google Scholar]
- 23.Adar R, Monsonego-Ornan E, David P, Yayon A. Differential activation of cysteine-substitution mutants of fibroblast growth factor receptor 3 is determined by cysteine localization. J Bone Miner Res. 2002;17:860–8. doi: 10.1359/jbmr.2002.17.5.860. [DOI] [PubMed] [Google Scholar]
- 24.Hernandez S, Lopez-Knowles E, Lloreta J, et al. Prospective study of FGFR3 mutations as a prognostic factor in nonmuscle invasive urothelial bladder carcinomas. J Clin Oncol. 2006;24:3664–71. doi: 10.1200/JCO.2005.05.1771. [DOI] [PubMed] [Google Scholar]
- 25.Chesi M, Brents LA, Ely SA, et al. Activated fibroblast growth factor receptor 3 is an oncogene that contributes to tumor progression in multiple myeloma. Blood. 2001;97:729–36. doi: 10.1182/blood.v97.3.729. [DOI] [PubMed] [Google Scholar]
- 26.Wu R, Connolly D, Ngelangel C, Bosch FX, Munoz N, Cho KR. Somatic mutations of fibroblast growth factor receptor 3 (FGFR3) are uncommon in carcinomas of the uterine cervix. Oncogene. 2000;19:5543–6. doi: 10.1038/sj.onc.1203934. [DOI] [PubMed] [Google Scholar]
- 27.Karoui M, Hofmann-Radvanyi H, Zimmermann U, et al. No evidence of somatic FGFR3 mutation in various types of carcinoma. Oncogene. 2001;20:5059–61. doi: 10.1038/sj.onc.1204651. [DOI] [PubMed] [Google Scholar]
- 28.Naimi B, Latil A, Berthon P, Cussenot O. No evidence for fibroblast growth factor receptor 3 (FGFR-3) R248C/S249C mutations in human prostate cancer. Int J Cancer. 2000;87:455–6. [PubMed] [Google Scholar]
- 29.Sibley K, Stern P, Knowles MA. Frequency of fibroblast growth factor receptor 3 mutations in sporadic tumours. Oncogene. 2001;20:4416–8. doi: 10.1038/sj.onc.1204543. [DOI] [PubMed] [Google Scholar]
- 30.Jang JH, Shin KH, Park JG. Mutations in fibroblast growth factor receptor 2 and fibroblast growth factor receptor 3 genes associated with human gastric and colorectal cancers. Cancer Res. 2001;61:3541–3. [PubMed] [Google Scholar]
- 31.Zhang Y, Hiraishi Y, Wang H, et al. Constitutive activating mutation of the FGFR3b in oral squamous cell carcinomas. Int J Cancer. 2005;117:166–8. doi: 10.1002/ijc.21145. [DOI] [PubMed] [Google Scholar]
- 32.Aubertin J, Tourpin S, Janot F, Ahomadegbe JC, Radvanyi F. Analysis of fibroblast growth factor receptor 3 G697C mutation in oral squamous cell carcinomas. Int J Cancer. 2007;120:2058–9. doi: 10.1002/ijc.22285. author reply 60. [DOI] [PubMed] [Google Scholar]
- 33.Hafner C, van Oers JM, Hartmann A, et al. High Frequency of FGFR3 Mutations in Adenoid Seborrheic Keratoses. J Invest Dermatol. 2006;126:2404–7. doi: 10.1038/sj.jid.5700422. [DOI] [PubMed] [Google Scholar]
- 34.Logie A, Dunois-Larde C, Rosty C, et al. Activating mutations of the tyrosine kinase receptor FGFR3 are associated with benign skin tumors in mice and humans. Hum Mol Genet. 2005;14:1153. doi: 10.1093/hmg/ddi127. [DOI] [PubMed] [Google Scholar]
- 35.Hafner C, van Oers JM, Vogt T, et al. Mosaicism of activating FGFR3 mutations in human skin causes epidermal nevi. J Clin Invest. 2006;116:2201–7. doi: 10.1172/JCI28163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Oers JM, Lurkin I, van Exsel AJ, et al. A simple and fast method for the simultaneous detection of nine fibroblast growth factor receptor 3 mutations in bladder cancer and voided urine. Clin Cancer Res. 2005;11:7743–8. doi: 10.1158/1078-0432.CCR-05-1045. [DOI] [PubMed] [Google Scholar]
- 37.Bakkar AA, Quach V, Le Borgne A, et al. Sensitive Allele-Specific PCR Assay Able to Detect FGFR3 Mutations in Tumors and Urine from Patients with Urothelial Cell Carcinoma of the Bladder. Clin Chem. 2005;51:1555. doi: 10.1373/clinchem.2005.049619. [DOI] [PubMed] [Google Scholar]
- 38.Hartmann A, Moser K, Kriegmair M, Hofstetter A, Hofstaedter F, Knuechel R. Frequent genetic alterations in simple urothelial hyperplasias of the bladder in patients with papillary urothelial carcinoma [In Process Citation] Am J Pathol. 1999;154:721–7. doi: 10.1016/S0002-9440(10)65318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obermann EC, Junker K, Stoehr R, et al. Frequent genetic alterations in flat urothelial hyperplasias and concomitant papillary bladder cancer as detected by CGH, LOH, and FISH analyses. J Pathol. 2003;199:50–7. doi: 10.1002/path.1259. [DOI] [PubMed] [Google Scholar]
- 40.Cairns P, Shaw ME, Knowles MA. Initiation of bladder cancer may involve deletion of a tumour-suppressor gene on chromosome 9. Oncogene. 1993;8:1083–5. [PubMed] [Google Scholar]
- 41.Chow NH, Cairns P, Eisenberger CF, et al. Papillary urothelial hyperplasia is a clonal precursor to papillary transitional cell bladder cancer. Int J Cancer. 2000;89:514–8. [PubMed] [Google Scholar]
- 42.van Oers JM, Adam C, Denzinger S, et al. Chromosome 9 deletions are more frequent than FGFR3 mutations in flat urothelial hyperplasias of the bladder. Int J Cancer. 2006;119:1212–5. doi: 10.1002/ijc.21958. [DOI] [PubMed] [Google Scholar]
- 43.Tomlinson D, Baldo O, Knowles MA. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. Journal of Pathology. 2007 doi: 10.1002/path.2207. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Rhijn BW, van der Kwast TH, Vis AN, et al. FGFR3 and P53 characterize alternative genetic pathways in the pathogenesis of urothelial cell carcinoma. Cancer Res. 2004;64:1911–4. doi: 10.1158/0008-5472.can-03-2421. [DOI] [PubMed] [Google Scholar]
- 45.Lamy A, Gobet F, Laurent M, et al. Molecular profiling of bladder tumors based on the detection of FGFR3 and TP53 mutations. J Urol. 2006;176:2686–9. doi: 10.1016/j.juro.2006.07.132. [DOI] [PubMed] [Google Scholar]
- 46.Knowles MA. Molecular subtypes of bladder cancer: Jekyll and Hyde or chalk and cheese? Carcinogenesis. 2006;27:361–73. doi: 10.1093/carcin/bgi310. [DOI] [PubMed] [Google Scholar]
- 47.Esrig D, Spruck CHd, Nichols PW, et al. p53 nuclear protein accumulation correlates with mutations in the p53 gene, tumor grade, and stage in bladder cancer. Am J Pathol. 1993;143:1389–97. [PMC free article] [PubMed] [Google Scholar]
- 48.Mhawech-Fauceglia P, Cheney RT, Fischer G, Beck A, Herrmann FR. FGFR3 and p53 protein expressions in patients with pTa and pT1 urothelial bladder cancer. Eur J Surg Oncol. 2006;32:231–7. doi: 10.1016/j.ejso.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 49.Richter J, Jiang F, Gorog JP, et al. Marked genetic differences between stage pTa and stage pT1 papillary bladder cancer detected by comparative genomic hybridization. Cancer Res. 1997;57:2860–4. [PubMed] [Google Scholar]
- 50.Dalbagni G. The management of superficial bladder cancer. Nat Clin Pract Urol. 2007;4:254–60. doi: 10.1038/ncpuro0784. [DOI] [PubMed] [Google Scholar]
- 51.Hart KC, Robertson SC, Kanemitsu MY, Meyer AN, Tynan JA, Donoghue DJ. Transformation and Stat activation by derivatives of FGFR1, FGFR3, and FGFR4. Oncogene. 2000;19:3309–20. doi: 10.1038/sj.onc.1203650. [DOI] [PubMed] [Google Scholar]
- 52.Jebar AH, Hurst CD, Tomlinson DC, Johnston C, Taylor CF, Knowles MA. FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene. 2005;24:5218–25. doi: 10.1038/sj.onc.1208705. [DOI] [PubMed] [Google Scholar]
- 53.Lopez-Knowles E, Hernandez S, Malats N, et al. PIK3CA mutations are an early genetic alteration associated with FGFR3 mutations in superficial papillary bladder tumors. Cancer Res. 2006;66:7401–4. doi: 10.1158/0008-5472.CAN-06-1182. [DOI] [PubMed] [Google Scholar]
- 54.Cairns P, Evron E, Okami K, et al. Point mutation and homozygous deletion of PTEN/MMAC1 in primary bladder cancers. Oncogene. 1998;16:3215–8. doi: 10.1038/sj.onc.1201855. [DOI] [PubMed] [Google Scholar]
- 55.Aveyard JS, Skilleter A, Habuchi T, Knowles MA. Somatic mutation of PTEN in bladder carcinoma. Br J Cancer. 1999;80:904–8. doi: 10.1038/sj.bjc.6690439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saal LH, Holm K, Maurer M, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–9. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 57.Byun DS, Cho K, Ryu BK, et al. Frequent monoallelic deletion of PTEN and its reciprocal associatioin with PIK3CA amplification in gastric carcinoma. Int J Cancer. 2003;104:318–27. doi: 10.1002/ijc.10962. [DOI] [PubMed] [Google Scholar]
- 58.Oda K, Stokoe D, Taketani Y, McCormick F. High frequency of coexistent mutations of PIK3CA and PTEN genes in endometrial carcinoma. Cancer Res. 2005;65:10669–73. doi: 10.1158/0008-5472.CAN-05-2620. [DOI] [PubMed] [Google Scholar]
- 59.Zieger K, Dyrskjot L, Wiuf C, et al. Role of activating fibroblast growth factor receptor 3 mutations in the development of bladder tumors. Clin Cancer Res. 2005;11:7709–19. doi: 10.1158/1078-0432.CCR-05-1130. [DOI] [PubMed] [Google Scholar]
- 60.Dyrskjot L, Kruhoffer M, Thykjaer T, et al. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–8. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 61.Lindgren D, Liedberg F, Andersson A, et al. Molecular characterization of early-stage bladder carcinomas by expression profiles, FGFR3 mutation status, and loss of 9q. Oncogene. 2006;25:2685–96. doi: 10.1038/sj.onc.1209249. [DOI] [PubMed] [Google Scholar]
- 62.Tomlinson DC, L'Hote CG, Kennedy W, Pitt E, Knowles MA. Alternative splicing of fibroblast growth factor receptor 3 produces a secreted isoform that inhibits fibroblast growth factor-induced proliferation and is repressed in urothelial carcinoma cell lines. Cancer Res. 2005;65:10441–9. doi: 10.1158/0008-5472.CAN-05-1718. [DOI] [PubMed] [Google Scholar]
- 63.Murgue B, Tsunekawa S, Rosenberg I, deBeaumont M, Podolsky DK. Identification of a novel variant form of fibroblast growth factor receptor 3 (FGFR3 IIIb) in human colonic epithelium. Cancer Res. 1994;54:5206–11. [PubMed] [Google Scholar]
- 64.Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jang JH. Identification and characterization of soluble isoform of fibroblast growth factor receptor 3 in human SaOS-2 osteosarcoma cells. Biochem Biophys Res Commun. 2002;292:378–82. doi: 10.1006/bbrc.2002.6668. [DOI] [PubMed] [Google Scholar]
- 66.Johnston CL, Cox HC, Gomm JJ, Coombes RC. Fibroblast growth factor receptors (FGFRs) localize in different cellular compartments. A splice variant of FGFR-3 localizes to the nucleus. J Biol Chem. 1995;270:30643–50. doi: 10.1074/jbc.270.51.30643. [DOI] [PubMed] [Google Scholar]
- 67.Sturla LM, Merrick AE, Burchill SA. FGFR3IIIS: a novel soluble FGFR3 spliced variant that modulates growth is frequently expressed in tumour cells. Br J Cancer. 2003;89:1276–84. doi: 10.1038/sj.bjc.6601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Terada M, Shimizu A, Sato N, Miyakaze SI, Katayama H, Kurokawa-Seo M. Fibroblast growth factor receptor 3 lacking the Ig IIIb and transmembrane domains secreted from human squamous cell carcinoma DJM-1 binds to FGFs. Mol Cell Biol Res Commun. 2001;4:365–73. doi: 10.1006/mcbr.2001.0306. [DOI] [PubMed] [Google Scholar]
- 69.Chodak GW, Hospelhorn V, Judge SM, Mayforth R, Koeppen H, Sasse J. Increased levels of fibroblast growth factor-like activity in urine from patients with bladder or kidney cancer. Cancer Res. 1988;48:2083–8. [PubMed] [Google Scholar]
- 70.Chopin DK, Caruelle JP, Colombel M, et al. Increased immunodetection of acidic fibroblast growth factor in bladder cancer, detectable in urine. J Urol. 1993;150:1126–30. doi: 10.1016/s0022-5347(17)35705-1. [DOI] [PubMed] [Google Scholar]
- 71.Gravas S, Bosinakou I, Kehayas P, Giannopoulos A. Urinary basic fibroblast growth factor in bladder cancer patients. Histopathological correlation and clinical potential. Urol Int. 2004;73:173–7. doi: 10.1159/000079700. [DOI] [PubMed] [Google Scholar]
- 72.O'Brien T, Cranston D, Fuggle S, Bicknell R, Harris AL. Two mechanisms of basic fibroblast growth factor-induced angiogenesis in bladder cancer. Cancer Res. 1997;57:136–40. [PubMed] [Google Scholar]
- 73.Matsumoto M, Ohtsuki Y, Ochii K, et al. Fibroblast growth factor receptor 3 protein expression in urothelial carcinoma of the urinary bladder, exhibiting no association with low-grade and/or non-invasive lesions. Oncol Rep. 2004;12:967. [PubMed] [Google Scholar]
- 74.Gomez-Roman JJ, Saenz P, Molina M, et al. Fibroblast growth factor receptor 3 is overexpressed in urinary tract carcinomas and modulates the neoplastic cell growth. Clin Cancer Res. 2005;11:459. [PubMed] [Google Scholar]
- 75.Bernard-Pierrot I, Brams A, Dunois-Larde C, et al. Oncogenic properties of the mutated forms of fibroblast growth factor receptor 3b. Carcinogenesis. 2006;27:740–7. doi: 10.1093/carcin/bgi290. [DOI] [PubMed] [Google Scholar]
- 76.Malats N, Bustos A, Nascimento CM, et al. P53 as a prognostic marker for bladder cancer: a meta-analysis and review. Lancet Oncol. 2005;6:678–86. doi: 10.1016/S1470-2045(05)70315-6. [DOI] [PubMed] [Google Scholar]
- 77.Buscarini M, Quek ML, Gill P, Xia G, Quinn DI, Stein JP. Molecular prognostic factors in bladder cancer. BJU Int. 2005;95:739–42. doi: 10.1111/j.1464-410X.2005.05393.x. [DOI] [PubMed] [Google Scholar]
- 78.Ooms EC, Anderson WA, Alons CL, Boon ME, Veldhuizen RW. Analysis of the performance of pathologists in the grading of bladder tumors. Hum Pathol. 1983;14:140–3. doi: 10.1016/s0046-8177(83)80242-1. [DOI] [PubMed] [Google Scholar]
- 79.Tosoni I, Wagner U, Sauter G, et al. Clinical significance of interobserver differences in the staging and grading of superficial bladder cancer. BJU Int. 2000;85:48–53. doi: 10.1046/j.1464-410x.2000.00356.x. [DOI] [PubMed] [Google Scholar]
- 80.Harnden P, Mahmood N, Southgate J. Expression of cytokeratin 20 redefines urothelial papillomas of the bladder. Lancet. 1999;353:974–7. doi: 10.1016/S0140-6736(98)05383-5. [DOI] [PubMed] [Google Scholar]
- 81.van Oers JM, Wild PJ, Burger M, et al. FGFR3 Mutations and a Normal CK20 Staining Pattern Define Low-Grade Noninvasive Urothelial Bladder Tumours. Eur Urol. 2007 doi: 10.1016/j.eururo.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 82.Botteman MF, Pashos CL, Redaelli A, Laskin B, Hauser R. The health economics of bladder cancer: a comprehensive review of the published literature. Pharmacoeconomics. 2003;21:1315–30. doi: 10.1007/BF03262330. [DOI] [PubMed] [Google Scholar]
- 83.Sangar VK, Ragavan N, Matanhelia SS, Watson MW, Blades RA. The economic consequences of prostate and bladder cancer in the UK. BJU Int. 2005;95:59–63. doi: 10.1111/j.1464-410X.2005.05249.x. [DOI] [PubMed] [Google Scholar]
- 84.Bastacky S, Ibrahim S, Wilczynski SP, Murphy WM. The accuracy of urinary cytology in daily practice. Cancer. 1999;87:118–28. doi: 10.1002/(sici)1097-0142(19990625)87:3<118::aid-cncr4>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 85.van Rhijn BW, van der Poel HG, van der Kwast TH. Urine markers for bladder cancer surveillance: a systematic review. Eur Urol. 2005;47:736–48. doi: 10.1016/j.eururo.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 86.Lokeshwar VB, Soloway MS. Current bladder tumor tests: does their projected utility fulfill clinical necessity? J Urol. 2001;165:1067–77. [PubMed] [Google Scholar]
- 87.Steiner G, Schoenberg MP, Linn JF, Mao L, Sidransky D. Detection of bladder cancer recurrence by microsatellite analysis of urine. Nat Med. 1997;3:621–4. doi: 10.1038/nm0697-621. [DOI] [PubMed] [Google Scholar]
- 88.Hoque MO, Lee J, Begum S, et al. High-throughput molecular analysis of urine sediment for the detection of bladder cancer by high-density single-nucleotide polymorphism array. Cancer Res. 2003;63:5723–6. [PubMed] [Google Scholar]
- 89.Schneider A, Borgnat S, Lang H, et al. Evaluation of microsatellite analysis in urine sediment for diagnosis of bladder cancer. Cancer Res. 2000;60:4617–22. [PubMed] [Google Scholar]
- 90.Chen J, Williams IR, Lee BH, et al. Constitutively activated FGFR3 mutants signal through PLC{gamma}-dependent and -independent pathways for hematopoietic transformation. Blood. 2005;106:328–37. doi: 10.1182/blood-2004-09-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grand EK, Chase AJ, Heath C, Rahemtulla A, Cross NC. Targeting FGFR3 in multiple myeloma: inhibition of t(4;14)-positive cells by SU5402 and PD173074. Leukemia. 2004;18:962–6. doi: 10.1038/sj.leu.2403347. [DOI] [PubMed] [Google Scholar]
- 92.Trudel S, Ely S, Farooqi Y, et al. Inhibition of fibroblast growth factor receptor 3 induces differentiation and apoptosis in t(4;14) myeloma. Blood. 2004;103:3521–8. doi: 10.1182/blood-2003-10-3650. [DOI] [PubMed] [Google Scholar]
- 93.Trudel S, Li ZH, Wei E, et al. CHIR-258, a novel, multitargeted tyrosine kinase inhibitor for the potential treatment of t(4;14) multiple myeloma. Blood. 2005;105:2941. doi: 10.1182/blood-2004-10-3913. [DOI] [PubMed] [Google Scholar]
- 94.Trudel S, Stewart AK, Rom E, et al. The inhibitory anti-FGFR3 antibody, PRO-001, is cytotoxic to t(4;14) multiple myeloma cells. Blood. 2006;107:4039–46. doi: 10.1182/blood-2005-10-4179. [DOI] [PubMed] [Google Scholar]
- 95.Tomlinson DC, Hurst CD, Knowles MA. Knockdown by shRNA identifies S249C mutant FGFR3 as a potential therapeutic target in bladder cancer. Oncogene. 2007 doi: 10.1038/sj.onc.1210399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–4. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 97.Mohammadi M, McMahon G, Sun L, et al. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–60. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- 98.Martinez-Torrecuadrada J, Cifuentes G, Lopez-Serra P, Saenz P, Martinez A, Casal JI. Targeting the extracellular domain of fibroblast growth factor receptor 3 with human single-chain Fv antibodies inhibits bladder carcinoma cell line proliferation. Clin Cancer Res. 2005;11:6280–90. doi: 10.1158/1078-0432.CCR-05-0282. [DOI] [PubMed] [Google Scholar]