Abstract
Frost tolerance is critical for wheat survival during cold winters. Natural variation for this trait is mainly associated with allelic differences at the VERNALIZATION 1 (VRN1) and FROST RESISTANCE 2 (FR2) loci. VRN1 regulates the transition between vegetative and reproductive stages and FR2, a locus including several tandemly duplicated C-REPEAT BINDING FACTOR (CBF) transcription factors, regulates the expression of Cold regulated genes. We identified sequence and copy number variation at these two loci among winter and spring wheat varieties and characterized their association with frost tolerance. We identified two FR-A2 haplotypes – ‘FR-A2-S’ and ‘FR-A2-T’ – distinguished by two insertion/deletions and ten single nucleotide polymorphisms within the CBF-A12 and CBF-A15 genes. Increased copy number of CBF-A14 was frequently associated with the FR-A2-T haplotypes and with higher CBF14 transcript levels in response to cold. Factorial ANOVAs revealed significant interactions between VRN1 and FR-A2 for frost tolerance in both winter and spring panels suggesting a crosstalk between vernalization and cold acclimation pathways. The model including these two loci and their interaction explained 32.0 and 20.7% of the variation in frost tolerance in the winter and spring panels, respectively. The interaction was validated in a winter wheat F4:5 population segregating for both genes. Increased VRN-A1 copy number was associated with improved frost tolerance among varieties carrying the FR-A2-T allele but not among those carrying the FR-A2-S allele. These results suggest that selection of varieties carrying the FR-A2-T allele and three copies of the recessive vrn-A1 allele would be a good strategy to improve frost tolerance in wheat.
Keywords: VRN-A1, CBF, haplotype, copy number variation, frost tolerance, Triticum aestivum
Introduction
Temperate cereals such as wheat (Triticum spp.) and barley (Hordeum vulgare L.) can be classified into spring or winter types based on their growth habit. Winter varieties are sown in autumn and must be able to withstand freezing temperatures for their reproductive success. These varieties require long exposures to low non-freezing temperatures (vernalization) to accelerate flowering whereas spring varieties, generally sown after the risk of exposure to freezing temperatures has passed, do not have this requirement. Cold acclimation is the process by which gradual exposure to low, non-freezing temperatures leads to an increase in frost tolerance. Two loci, FROST RESISTANCE 1 (FR1) and FROST RESISTANCE 2 (FR2), both located on the long arm of homoeologous group 5 chromosomes, are key components of the cold acclimation regulatory network in wheat and barley (Vágújfalvi et al. 2003; Francia et al. 2004; reviewed in Galiba et al. 2009). An additional smaller QTL for frost tolerance, distinct from FR2, has recently been reported on chromosome 5B (Zhao et al. 2013). This QTL, identified in wheat germplasm adapted to central Europe, was not part of this study.
The FR1 locus was isolated during a study to identify loci contributing to differential cold acclimation in winter vs. spring wheat (Galiba et al. 1995; Sutka et al. 1999). FR1 is most likely a pleiotropic effect of the VERNALIZATION1 (VRN1) gene (Limin and Fowler 2006; Stockinger et al. 2007; Dhillon et al. 2010) and, therefore, will be referred to hereafter as VRN1 (with an additional letter indicating the specific wheat genome e.g. VRN-A1, VRN-B1, and VRN-D1). VRN1 transcript levels increase during exposure to low temperatures and trigger the transition from the vegetative to reproductive growth stage. This transition is associated with suppressed induction of C-REPEAT BINDING FACTOR (CBF) genes in response to cold, resulting in reduced frost tolerance (Kobayashi et al. 2005; Limin and Fowler 2006; Stockinger et al. 2007; Dhillon et al. 2010).
The presence of dominant Vrn-A1a or Vrn-A1b alleles eliminates the need for vernalization prior to flowering, resulting in a spring growth habit. Plants carrying dominant Vrn-B1 and Vrn-D1 alleles also exhibit a spring growth habit, although these alleles are weaker than the Vrn-A1 alleles and plants maintain some residual response to vernalization (Santra et al. 2009). Spring wheat varieties carrying the dominant Vrn-A1a or Vrn-A1b alleles are more sensitive to frost damage than those carrying the Vrn-B1 or Vrn-D1 alleles and both are more sensitive than varieties with a winter growth habit (Koemel et al. 2004; Reddy et al. 2006). Different recessive winter vrn-A1 alleles have been identified using various nomenclature; including ‘V’ and ‘W’ which are distinguished by a C/T single nucleotide polymorphism (SNP) in the fourth exon of this gene (Table 1). These alleles have been shown to be associated with differences in vegetative growth patterns (Chen et al. 2009, 2010) and it has been reported that they may also be associated with differences in frost tolerance (Eagles et al. 2011). Wheat genotypes carrying the ‘V’ allele had a lower vrn-A1 copy number and reduced vernalization requirement to induce flowering, than those carrying the ‘W’ allele, which were characterized by higher vrn-A1 copy number and a greater vernalization requirement (Table 1) (Díaz et al. 2012).
Table 1.
Variable nomenclature and characteristics of two vrn-A1 alleles in different studies. Nucleotide and amino acid residue numbers are relative to ‘Jagger’ VRN-A1 coding sequence and VRN-A1 protein
|
vrn- A1 Allele |
Chen et al. 2009 | Eagles et al. 2011 | Exon 4 SNP, Nucleotide 349 |
Amino acid 117 |
Vernalization requirement |
Copy number in the winter panel |
|---|---|---|---|---|---|---|
| V |
vrn- A1a |
Jagger (J) | C | L | Shorter | Seven out of eight varieties < 3 copies |
| W |
vrn- A1b |
Wichita (K) |
C/T | L/F | Longer | 53 out of 58 varieties = 3 copies |
The second frost tolerance locus, FR2, was mapped ~30 cM proximal to VRN1 on homoeologous group 5 chromosomes of both wheat and barley and is associated with frost tolerance and expression of the COLD REGULATED (COR) gene, Cor14b (Tóth et al. 2003; Vágújfalvi et al. 2003; McIntosh et al. 2004; Francia et al. 2004; Båga et al. 2007; Motomura et al. 2013). In hexaploid wheat, the differential CBF expression detected between frost-sensitive and frost-tolerant varieties was tightly associated with the FR-A2 locus (Vágújfalvi et al. 2005). QTL for frost tolerance have been identified in several independent studies at this locus (Båga et al. 2007; Motomura et al. 2013). The FR-Am2 locus from diploid wheat Triticcum monococcum (Am genome related to the A genome of polyploid wheat) was later found to comprise 11 tandemly duplicated CBF genes clustered closely together on chromosome 5Am (Miller et al. 2006).
CBF proteins are AP2/ERF transcription factors which contain five conserved amino acid motifs (AP2, CMIII-1, CMIII-2, CMIII-3 and CMIII-4) that distinguish them from other members of the ERF family (Nakano et al. 2006). Among these five motifs, the AP2 domain is a critical functional domain required for the CBF proteins to bind to the C-repeat/dehydration-responsive elements (CRT/DRE) located in the promoter regions of their target COR genes (Stockinger et al. 1997; Liu et al. 1998). The CBF genes have been best characterized in Arabidopsis, where three tandemly duplicated CBF genes are present in a cluster on chromosome 4 (Stockinger et al. 1997; Gilmour et al. 1998; Medina et al. 1999). The CBF gene family in the temperate grasses has undergone an expansion dated to the Eocene-Oligocene transition during a period of global cooling ~ 33 MYA (Sandve and Fjellheim 2010). Interestingly, deletions and duplications of several CBF genes have been detected at the FR2 locus in wheat and barley (Skinner et al. 2005; Francia et al. 2007; Fricano et al. 2009; Knox et al. 2010; Pearce et al. 2013), potentially allowing for a more flexible response to cold stress and suggesting that this gene cluster is subject to dynamic expansions and contractions.
Three adjacent genes, CBF12, 14 and 15, located at the center of the eleven-gene cluster at the Fr-Am2 locus were previously shown to be completely linked to differences in frost survival and accumulation of Cor14b and DEHYDRIN 5 (DHN5) transcripts at 12–15 °C in diploid wheat (Knox et al. 2008). Their critical role in frost tolerance was also supported by several expression studies. In hexaploid wheat recombinant substitution lines, transcript levels of CBF14 and 15 were more than fourfold higher when the FR-A2 allele from the frost-tolerant wheat cultivar ‘Cheyenne’ was present as compared to the frost-sensitive allele from T. spelta (CBF12 was not included in this study) (Vágújfalvi et al. 2005). Expression of CBF12 and CBF14 after long-term cold acclimation differed between frost-tolerant and frost-sensitive mutant lines derived from the wheat cultivar ‘Winoka’ (Sutton et al. 2009). In barley, HvCBF12 and 14 were more highly expressed in the frost-tolerant cultivar ‘Nure’ than in the frost-sensitive cultivar ‘Tremois’ (Stockinger et al. 2007). Studies of CBF nucleotide sequence variation, insertion/deletions (indels), and copy number variation (CNV) also support an important role of these genes in frost tolerance (Knox et al. 2008, 2010). In barley, allelic variation at HvCBF14 was associated with differences in frost tolerance (Fricano et al. 2009). In wheat, the copy number of CBF14 was found to be higher in winter wheat than spring wheat (Knox et al. 2010; Dhillon and Stockinger 2013). More recently, a large multi-gene deletion at the Fr-B2 locus, which included CBF-B12, CBF-B14 and CBF-B15, was shown to be associated with reduced frost tolerance in both tetraploid durum (Triticum durum L.) and hexaploid bread wheat (Pearce et al. 2013). Taken together, the expression experiments and the genetic data suggest that CBF12, CBF14 and CBF15 play a critical role in frost tolerance in the temperate cereals.
In the current study, we analyzed the sequences of CBF12, CBF14 and CBF15 homoeologs from 146 hexaploid wheat varieties and identified SNP and indel allelic variation in VRN-A1 and the CBF genes located at the central cluster of FR-A2. We determine frost survival for these varieties and for a bi-parental population segregating for the allelic variation at the VRN-A1 and FR-A2 loci. We also identify CNV at VRN-A1 and FR-A2 and determine that interactions between these two loci are associated with differential frost survival in both winter and spring wheat.
Materials and methods
Plant materials
Two wheat diversity panels and one bi-parental F4:5 segregating population were analyzed during the current study:
Winter panel: A diversity panel of 65 hexaploid winter wheat varieties was selected to represent varieties grown in diverse regions of the world, including accessions originating from the USA (36), Russia (8), Romania (6), Canada (4), Ukraine (2), Finland (2), Denmark (1), France (1), Germany (1), Serbia (1), Switzerland (1) and Poland (1) and one accession of undetermined geographic origin (Supplemental Material Table S1).
Spring panel: A second diversity panel of 81 hexaploid spring wheat varieties from North America (34), Australia (22), South America (8), Europe (8), Africa (5) and Asia (4) was selected to represent spring wheat varieties from different regions of the world (Supplemental Material Table S2).
The growth habit of varieties in both panels was determined by genotyping the vernalization loci VRN-A1, VRN-B1 and VRN-D1 (see “Molecular markers” below) and confirmed with information provided by the Germplasm Resources Information Network (GRIN) database of the National Plant Germplasm System (http://www.ars-grin.gov/npgs/).
Bi-parental F4:5 segregating population (ORFW/Eltan): The unreleased, soft white wheat genotype ‘Oregon Feed Wheat No. 5’ (ORFW) carries a major genetic factor conditioning freezing sensitivity (Skinner and Campbell 2008). A heterozygous F2:4 family derived from a cross between the frost-tolerant winter wheat cultivar ‘Eltan’ (PI 536994) (Peterson et al. 1991) and ORFW was self-pollinated and the progeny were genotyped for sequence variation at the FR-A2 and VRN-A1 loci. Seventy individuals homozygous for each of the two FR-A2 haplotypes (S and T) and each of the two vrn-A1 alleles segregating in this population (V = 2 copies and W = 3 copies) were selected to generate a segregating F4:5 population. No recombination event was detected between CBF-A12, CBF-A14 and CBF-A15, so the effects of the three central CBF genes at the FR-A2 locus could not be separated in this population.
In all frost tolerance assays for winter wheat accessions, the soft white wheat cultivars ‘Eltan’ and ‘Stephens’ (PI 658243) (Kronstad et al. 1978) developed in the Pacific Northwest (PNW) of the US were used as frost-tolerant and frost-susceptible controls, respectively. In the frost-tolerant assay for the 81 spring wheat cultivars, the cultivars ‘Alpowa’ (PI 566596) and ‘Louise’ (PI 634865) developed in the PNW (Kidwell et al. 2006) were included as frost tolerant and sensitive controls, respectively.
To screen for diversity within ancestors of modern wheat, we selected 25 Triticum turgidum spp. dicoccoides accessions from the University of Haifa germplasm collection (Supplemental Material Table S5, (Nevo and Beiles 1989; Peleg et al. 2005) and two accessions of Triticum urartu (‘ICWT 500612’ and ‘G1812’).
DNA extraction and gene sequencing
Leaf tissue was harvested from 3-week-old seedlings and genomic DNA extracted using the BioSprint 96 DNA Plant Kit (Qiagen, Valencia, CA, USA).
Genome-specific primers were developed for each of the three homoeologous copies of CBF12, CBF14 and CBF15 (Supplemental Material Table S3). All PCRs were carried out in 20 µl volumes containing 5 % DMSO, 10 mM Tris-HCl, 50 mM KCl, 3 mM MgCl2, 0.2 mM dNTPs, 0.375 µM of each primer, 50–100 ng genomic DNA and 0.75 U Taq DNA polymerase. Amplifications were performed using a standard touchdown PCR protocol with the appropriate annealing temperature (Supplemental Material Table S3).
All PCR products were purified using ExoSAP-IT® (Affymetrix, Santa Clara, CA, USA) and sequencing templates were prepared with BigDye® Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). All samples were sequenced at the USDA-ARS Western Regional Small Grains Genotyping Laboratory, Pullman WA, USA.
Molecular markers
Previously developed PCR markers were employed to genotype the Vrn-A1 (Vrn-A1a, Vrn-A1b alleles, Yan et al. 2004), Vrn-B1 and Vrn-D1 alleles (Fu et al. 2005). The two allelic variants of the recessive vrn-A1 allele in the winter wheat varieties were classified into ‘V’ and ‘W’ alleles based on the markers described previously [(forward: 5’-CAACTTGTTTGGGACTAAAGGC-3’; reverse: 5’-CTGCAACTCCTTGAGATTCAAAG-3’ (Chen et al. 2009)] and confirmed by analyzing the sequencing traces of the 375-bp amplified product.
Two cleaved amplified polymorphic sequences (CAPS) markers were developed to distinguish the ‘FR-A2-S’ and ‘FR-A2-T’ haplotypes, based on the SNPs that were discovered through sequencing CBF-A12 and CBF-A15 (details in Results below). The PCR primers for CBF-A12 and CBF-A15 were those used for sequencing and are listed in Supplemental Material Table S3. For the CAPS-A12 marker, ZraI was used to digest the CBF-A12 PCR product into fragments of 706/476 bp (FR-A2-S) or 400/304/476 bp (FR-A2-T). For the CAPS-A15 marker, SalI was used to digest the CBF-A15 PCR product into fragments of 403/605 bp (FR-A2-S) or an undigested band of 1,017 bp (FR-A2-T) (Supplemental Material Figure S1).
Taqman® assays to determine VRN-A1 and CBF copy number
VRN-A1 copy number was determined using the Taqman® assay described by Díaz et al. (2012) with the exception that FAM-BHQ1 and CY5-BHQ3 were used as the 5’fluorophore and 3’ quencher in target (VRN-A1) and control CONSTANS2 (CO2) probes, respectively (Supplemental Material Table S4). Homoeologue-specific Taqman® assays were developed for CBF-A12, CBF-A14 and CBF-A15 genes. For each assay, primers to amplify the template were redundant for all homoeologs, while probes were designed to be homoeolog-specific (Supplemental Material Table S4). Probe specificity was confirmed by testing each assay for amplification in T. aestivum nullisomic-tetrasomic lines missing each of the homoeologous group 5 chromosomes. For example, primers were considered A-genome specific, when no fluorescence from the Taqman® assay was detected in the line missing chromosome 5A. Each 20 µl reaction consisted of 0.2 µM of both forward and reverse primers of target and control genes, 0.1 µM of both probes, 1x Taqman® Fast Universal PCR master mix (Applied Biosystems, Foster City, CA, USA) and 20 ng of sample DNA. Samples were run as multiplex reactions, including the target gene and control assays in each well. In all cases, the single-copy wheat gene CONSTANS 2 (CO2) was used as the control. Reactions were run in an ABI 7500 Fast qRT-PCR cycler (Applied Biosystems, Foster City, CA, USA). Copy number of each target genes was estimated from the average fold-change ratio between target gene relative to CO2 based on four biological replicates. Comparisons between control and target gene assays are relative because they use different fluorophores (FAM and CY5), so the ratio between a single copy target and a single copy control could be different from one.
Quantitative real- time PCR (qRT-PCR)
Six biological replicates of individuals from the Eltan/ORFW F4:5 segregating population were grown under greenhouse conditions for 4 weeks and leaf tissue from the most recently deployed leaf was harvested at 4 p.m. on day one of the experiment. At 8 a.m. on day two, plants were transferred to 4 °C and leaf tissue harvested again at four p.m. Harvested tissue was immediately frozen in liquid nitrogen and then ground into a fine powder. Total RNA was extracted with the Spectrum™ Plant Total RNA Kit (Sigma-Aldrich, St. Louis, MO, USA) and 1 µg of the RNA equivalent of cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit with RNAse Inhibitor (Applied Biosystems, Foster City, CA, USA). Each 20 µl qRT-PCR reaction consisted of 1x USB® VeriQuest™ SYBR® Green qPCR Master Mix (Affymetrix, Santa Clara, CA, USA) 0.5 µM of F and R primers and 10 ng of cDNA. qRT-PCR was carried out using an Applied Biosystems 7500 Fast Real-Time PCR System machine (Applied Biosystems, Foster City, CA, USA). ACTIN was used as an endogenous control. All qRT-PCR primers used in this study are listed in Supplemental Material Table S6. Primer efficiency and specificity were determined by analyzing amplification in a fourfold dilution series and checking dissociation curves for a single amplified product. Transcript levels are reported as linearized fold-ACTIN levels calculated by the formula 2(ACTIN CT – TARGET CT) ± SE. The reported value represents the ratio between the initial number of molecules of the target gene and the number of molecules of ACTIN and therefore, the Y-axes are comparable across genes.
Frost tolerance test
For each genotype tested, 20 seeds were planted into a single cell of a six-cell pack with randomization, in soil-less potting mix (Sunshine Mix #1/LC1, SunGro Horticulture, Seba Beach, CA, USA). Eight of these cell packs were evaluated in each run of the programmable temperature cabinet, so that 48 genotypes were evaluated in each run. Prior to freezing, the seeds were germinated under a long-day regime (16 h light, 8 h dark) under cool-white fluorescent lights (200 µmol/m2/s) at 22/15 °C day/night temperatures, for 7 days in a Conviron PGR-15 growth chamber (Conviron, Manitoba, Canada) at the WSU Plant Growth Facility, and plants were then cold acclimated at a constant temperature of 4 °C for 5 weeks under a long-day regime. At the end of this acclimation period, the leaves of each plant were clipped 2.5 cm above the crown. The planting substrate was saturated with water containing 10 mg/L Snowmax® (a commercial product that results in uniform ice nucleation at about −3 °C) and a layer of crushed ice was placed on the soil surface. Then, the plants were placed into a LU113 programmable temperature cabinet (Espec Na, Hudsonville MI, USA) in darkness and a temperature probe (Sensatronics, Austin TX, USA) was placed into each cell of 20 plants, with a capacity of 48 cells per run. Because all of our panels were larger than 48, each panel was split into 2 blocks and the experiment for each panel was designed as a partially balanced incomplete block with 2–6 six replications. Appropriate checks for each panel were included in each block. The progress of each freeze test was monitored at 2-min intervals using the temperature probes which were attached to a Senturion Environmental Monitor. The temperature of the programmable chamber was lowered from 4 to −3 °C over 1 h and held at −3 °C for 16 hours to allow the heat produced during ice formation to dissipate. The temperature was then decreased to a target temperature at a rate of −4 °C/h, held at the target temperature for one hour and then increased to 4 °C at a rate of 4 °C per hour. Target temperatures were determined based on preliminary assays to consistently discern differences between susceptible and resistant check genotypes within each set of genotypes evaluated. After these steps, the trays were transferred into a growth chamber with a constant temperature of 4 °C under a long-day regime for the first 24 h, before being moved into a greenhouse at ~22 °C. Survival rate (survival/emergence) in each cell was scored 5 weeks after the freezing.
For the winter panel, two replications were assayed at a target temperature of −13 °C, and two replications assayed at a target temperature of −14 °C. For the spring panel, four replications were assayed at a target temperature of −6 °C. For the ‘Eltan/ORFW’ F4:5 segregating population, six replications were assayed at a target temperature of −10 °C.
Statistical analysis
The survival data were analyzed within each panel using the GLM procedure of ‘SAS 9.3’ (SAS Institute, Cary, NC, USA) with the model Yijk= X +Gi(Bj) + Rk + eijk; where Yijk represents survival for genotype i in block j and replication k, X represents the mean for survival within a panel, Gi represents the effect of genotype i in block (B)j, Rk represents the replication effect and eijk is the experimental error. In these models, genotypes were considered to be fixed effects, and replications were random effects. Least squares (LS) means for survival, adjusted for the incomplete blocks, were obtained for each genotype within each panel and these were used in subsequent analyses of FR-A2 and VRN-A1 allelic effects. Negative LS means were corrected to 0.
The statistical model used within each panel to determine FR-A2 and VRN-A1 allelic effects was Yij= X +Fi+ Vj + FVij + eij ; where Yijrepresents the LS mean for percent survival of genotypes with FR-A2 alleles indexed by i, and VRN-A1 alleles indexed by j, X represents the average survival within a panel as above, Fi represents the effect of allele i at FR-A2; Vj represents the effect of allele j at VRN-A1; FVij represents their interaction and eij is the experimental error. For the winter panel, the alleles tested at the FR-A2 locus were designated FR-A2-S and FR-A2-T while the alleles for VRN-A1 were either V or W (VRN-A1 SNP) or the CNV at VRN-A1 (VRN-A1 CNV) which we scored as 1, 2, or 3 based on the relative VRN-A1:CO2 ratio obtained from the Taqman® assay. For the ORFW/Eltan population, the FR-A2 locus alleles were scored as above and the VRN-A1 CNV (2 or 3) was used to score the alleles at the VRN-A1 locus because complete linkage was observed between the VRN-A1 SNP and VRN-A1 CNV in that panel. For the spring panel, the alleles at the FR-A2 locus were scored as above but those at the VRN-A1 locus were scored as the presence of either Vrn-A1a or Vrn-A1b conditioning spring growth habit (group A) or the absence of those two alleles (group B).
For all models, homogeneity of variances was tested using Levene’s test and normality of residuals using the Shapiro-Wilk tests as implemented in SAS 9.3. For the three simple effect ANOVAS, in which Normality of the residuals could not be restored by transformation, we used non-parametric Kruskal-Wallis ANOVAs (Using the NPAR1WAY procedure in SAS 9.3) to test the significance of the differences. The results of the three non-parametric tests were consistent with the P values obtained in the corresponding parametric ANOVAs.
Accession numbers of the CBF genes sequenced in the current study
Twelve distinct sequences of the A, B and D genome homeologs of CBF12, CBF14 and CBF15 were submitted to the European Nucleotide Archive of European Molecular Biology Laboratory (EMBL) (http://www.ebi.ac.uk/ena/). Accession numbers: CBF-A12 (ORFW), HG530924; CBF-A12 (Eltan), HG530925; CBF-B12, HG530926; CBF-D12, HG530927; CBF-A14 (Eltan), HG530928; CBF-A14 (Bussard), pending; CBF-B14, HG530929; CBF-D14, HG530930; CBF-A15 (ORFW), HG530931; CBF-A15 (Eltan), HG530932; CBF-B15, HG530933; CBF-D15, HG530934.
Results
Copy number and sequence variation at the VRN-A1 locus in winter wheat
The 65 varieties in the winter panel were originally selected to represent several global environments with severe winters. Eight of these varieties carried the V allele and 57 the W allele at Vrn-A1 (Table 2). We also sequenced the amplified fragment from this marker, which revealed that those lines carrying the V allele had only ‘C’ at the SNP position, while overlapping ‘C’ and ‘T’ peaks were detected in lines carrying the W allele, indicative of multiple copies of vrn-A1 (see Table 1 for allele nomenclature). There were clear differences in VRN-A1 copy number based on the ratio of fluorescent signal from the VRN-A1/CO2 Taqman® assays. The 65 winter varieties were divided into three groups according to their VRN-A1 copy number: the first group included only the variety ‘Jagger’ with a VRN-A1:CO2 ratio of 0.4, corresponding to a haploid copy number of one. Eleven varieties had a ratio of 0.7–0.9 (~ 2 haploid copies), while the remaining 54 varieties had a ratio of 1.1–1.3 (~ 3 haploid copies) (Table 2).
Table 2.
Five alleles of recessive vrn-A1 conditioning differential survival in freezing trials were identified among the winter panel varieties.
| Type ‘V’ | Type ‘W’ | |||
|---|---|---|---|---|
| Copy number estimate |
Variety Names | Mean Survival (% ± SE) |
Variety Names | Mean Survival (% ± SE) |
| 1 copy (0.4–0.6)a |
Jagger | 5.9±14.9 | ||
| 2 copies (0.7–0.9) |
Chernova, Fundulea 174–71, Munstertaler, Olympia, ORFW, Sava |
16.8±6.4 | Alma, Ilichevka, Mira, Simon, Stephens |
8.4±5.4 |
| 3 copies (1.1–1.3) |
Vakka | 47.4±14.9 | AGS2000, Alabaskaja, Albatros Odesskii, Arthur, Bavaria, Besostaja 1, Boundary, Bussard, CDC Kestrel , CDC Clair, Centurk 78, Chukar, Crimson, Daws, Eltan, Endurance, Expedition, F26-67, Frankenmuth, Freedom, Fundulea 133, Fundulea 174, Fundulea 63–70, Geneva, Harding, Hopewell, IDO621, Inna, Jerry, Karl, Kavkaz, Malakov, Millenium, Minhardi, Mironovskaja, Moldova, Moro, Norstar, Odesskaja, OH552, Purkov, Redwin, SD97250, Seward, Tiber, Trego, Viking, Volgogradskaja 84, Wahoo, Wanser, Wesley, Xerpha |
44.8±2.9 |
Copy number based on average fold-change ratio between target gene relative to CONSTANS2 based on four biological replicates.
VRN-A1 copy number was associated with the presence of the V or W allele in this winter panel. The V allele was associated with a lower VRN-A1 copy number and the W allele with a higher copy number (P < 0.0001). In total, 53 of the 58 lines carrying the W allele had an estimated haploid copy number of 3, whereas 6 of the 8 lines carrying the V allele had an estimated copy number of 2. Of the other two varieties with the V allele, one carried a single copy of VRN-A1 with the ‘C’ SNP (Jagger) and the other one three copies of VRN-A1, all with the ‘C’ SNP (Vakka) (Table 2).
Two FR-A2 haplotypes in the central CBF cluster
We characterized variation in the three central CBF genes of the FR2 locus by sequencing the coding region and a short region of 5’ and 3’ UTR from the CBF12, CBF14 and CBF15 A, B and D homoeologs for all 65 accessions in the winter panel (see “Materials and methods” for the accession numbers of all the sequences). We found no sequence diversity in either the B or D homoeologs of any of these genes. On the A genome, we found a rare SNP polymorphism (A/G) at nucleotide 576 in CBF-A14 (coordinates based on start codon at position ‘1’ of the CBF-A14 gene as a reference). Among the 65 accessions sequenced, only one - ‘Bussard’ - had an ‘A’ at the SNP point (EMBL accession number pending), whereas the other 64 accessions all had a ‘G’ (EMBL accession number HG530928). However, at CBF-A12 and CBF-A15, we identified four SNPs plus one indel polymorphism in CBF-A12 and six SNPs plus one indel polymorphism in CBF-A15. These SNPs and indels at CBF-A12 and CBF-A15 were linked and grouped the winter panel into just two haplotypes (Fig. 1). Seven winter accessions ‘Alma’, ‘Bussard’, ‘Malakov’, ‘Mira’, ‘ORFW’, ‘Sava’ and ‘Viking’ carried the same haplotype at FR-A2, designated ‘FR-A2-S’ (EMBL accession numbers: CBF-A12, HG530924; CBF-A15, HG530931) (Supplemental Material Table S1). The second haplotype was shared by the remaining 58 winter accessions and was designated ‘FR-A2-T’ (EMBL accession numbers: CBF-A12, HG530925; CBF-A15, HG530932).
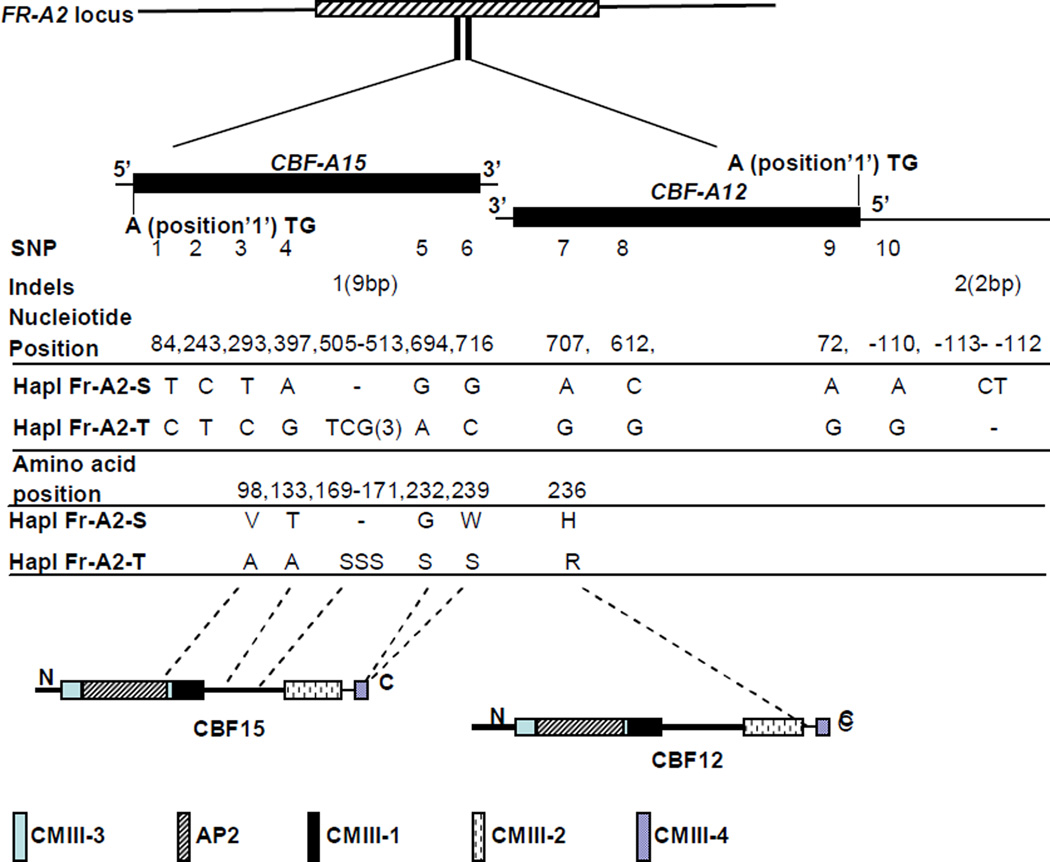
Fig. 1.
Nucleotide and amino acid sequence variation distinguishing FR-A2 haplotypes Nucleotide positions of the two indel and ten SNP polymorphisms of CBF-A12 and CBF-A15 are listed. Six of the 12 sequence polymorphisms lead to amino acid changes and the positions and detail of each amino acid substitution in CBF-12 and CBF-15 are displayed. AP2, CMIII-1, CMIII-2, CMIII-3 and CMIII-4 are the five conserved amino acid motifs which distinguish CBF genes from the other members of ERF family. These five domains are indicated by different colors and filling types in the diagrammatic sketch of CBF12 and CBF15 proteins
In CBF-A12, one indel polymorphism and one SNP were located in the promoter region resulting in a change from ‘CTAA’ (‘FR-A2-S’) to ‘AG’ (‘FR-A2-T’) at nucleotides -110 to -113 (coordinates based on start codon at position ‘1’ of the CBF-A12 gene using the ‘FR-A2-S’ haplotype as a reference). There were two synonymous SNP mutations located at nucleotide positions 72 and 612 and one SNP that resulted in an H/R amino acid polymorphism at position 236 between the CBF conserved motif CMIII-2 and CMIII-4 (Fig. 1). This and other amino acid changes are designated with a two-letter and one number code (e.g. ‘H236R’), where the first and last letters represent the amino acids at the ‘FR-A2-S’ and ‘FR-A2-T’ haplotypes, respectively, and the number the position from the initial methionine in the ‘FR-A2-S’ haplotype.
In CBF-A15, we identified two synonymous mutations at nucleotide positions 84 and 243. A three amino acid deletion (SSS) was detected in the ‘FR-A2-S’ haplotype after position 170 between the CMIII-1 and CMIII-2 motifs. There were also four SNPs which resulted in conservative amino acid changes; ‘V98A’ in the AP2 DNA binding domain (BLOSUM 62= 0), ‘T133A’ between the CMIII-1 and CMIII-2 motifs (BLOSUM 62= 0), ‘G229S’ (BLOSUM 62= 0) and ‘W236S’ (BLOSUM 62= −3) both located in the CMIII-4 motif (Fig. 1). To summarize, we identified twelve polymorphisms distinguishing ‘FR-A2-S’ from ‘FR-A2-T’, six of which result in amino acid substitutions (one in CBF-A12 and five in CBF-A15).
To determine if sequence diversity at the FR-A2 locus also existed in the ancestors of modern wheat, we sequenced these three CBF genes in 25 accessions of T. dicoccoides [tetraploid wild emmer wheat (genomes AABB)] and two accessions of T. urartu, the diploid A-genome progenitor of tetraploid and hexaploid wheat. In contrast to modern hexaploid varieties, we identified some sequence variation in the CBF-A14 gene. Three different versions of the CBF-A14 gene were identified within the T. dicoccoides population, but, with the exception of the variety ‘Bussard’, which carries a G – A SNP at position 576, none of these polymorphisms were found within the hexaploid wheat varieties in our study (EMBL accession numbers CBF-A14 (Bussard), HG939430; CBF-A14 (Nesher), HG939431; CBF-A14 (Tabigha), HG939432). At the CBF-A12 and CBF-A15 genes we identified the same FR-A2 haplotypes as described in hexaploid wheat. T. dicoccoides accessions can be split into two geographically distinct sub-populations, one northern, centered on Iraq and Turkey, and one southern, centered on Israel and Syria (Luo et al. 2007). In both populations, we identified accessions with the ‘FR-A2-S’ and ‘FR-A2-T’ haplotypes (Supplemental Material Table S5). Within the northern sub-population, 8 out of 12 accessions carried the ‘FR-A2-T’ haplotype, whereas in the southern sub-population, the ‘FR-A2-S’ haplotype was more common (8 out of 13 accessions). We also detected both ‘FR-A2’ haplotypes in T. urartu with the accession ‘G1812’ (originating in Lebanon) carrying the ‘FR-A2-S’ haplotype and the accession ‘ICWT 500612’ (originating in Turkey) carrying the ‘FR-A2-T’ haplotype. These results suggest that these haplotypes at the FR-A2 locus are ancient and pre-date the origin of tetraploid wheat. Based on the 10 SNPs that differentiate the ‘FR-A2-S’ and ‘FR-A2-T’ haplotypes (8 transitions and 2 transversions in 1,810 aligned bases), we calculated a minimum divergence time of 661,000 ± 209,000 years using Kimura’s two-parameter model (Kimura 1980) and a conservative divergence rate of 4.2 × 10−9 substitutions/nt/year (Ramakrishna et al. 2002). This estimated divergence time places the origin of the ‘FR-A2-S’ and ‘FR-A2-T’ haplotypes before the origin of tetraploid wheat [<500,000 years (Huang 2002)]. We developed two CAPS markers to distinguish the two FR-A2 haplotypes (Supplemental Material Fig. S1) and used them to screen the 81 varieties in the spring wheat panel. The frequency of the FR-A2 alleles differed significantly between the winter and spring panels (χ2 test P < 0.0001). The ‘FR-A2-T’ haplotype was more frequent among winter wheat varieties (89.3%) and the ‘FR-A2-S’ haplotype was more frequent among spring wheat varieties (66.7%) (Supplemental Material Tables S1, S2).
CBF CNV is correlated with FR-A2 haplotype
Previous studies have shown that CNV exists at the FR-A2 locus in temperate cereals, so we analyzed the CBF-A12, CBF-A14 and CBF-A15 gene copy number and found evidence of CNV for all three genes in both winter and spring panels. In the winter panel, significant correlations between CNV and frost tolerance were discovered for both CBF-A12 (R = 0.36, P = 0.0035) and CBF-A14 (R = 0.35, P = 0.0040). In the spring panel, correlations between CBF CNV and frost tolerance were not significant. In both spring and winter panels, we observed a good correlation between CBF-A12 and CBF-A15 CNV within both ‘FR-A2-S’ and ‘FR-A2-T’ haplotype groups (Table 3). Both these genes were also highly correlated individually with CBF-A14 CNV in the ‘FR-A2-T’ haplotype, but strikingly, this correlation was greatly reduced in the ‘FR-A2-S’ haplotype (Table 3). These patterns of correlation suggest that a key component of the FR-A2 haplotypes is a change in CBF-A14 copy number relative to the other genes at this locus.
Table 3.
Pearson correlations among the CNVs of three CBF genes within each FR-A2 haplotype in the winter and spring panels
| Winter | Spring | |||||
|---|---|---|---|---|---|---|
| CBF genes |
FR-A2-S n=7 |
FR-A2-T n=58 |
All n=65 |
FR-A2-S n=53 |
FR-A2-T n=28 |
All n=81 |
|
A12 vs A15 |
0.55 (P=0.19) |
0.48 (P=0.0002) |
0.33 (P=0.0008) |
0.81 (P=0.0001) |
0.87 (P=0.0001) |
0.81 (P=0.0001) |
|
A12 vs A14 |
-0.04 (P=0.19) |
0.50 (P=0.0001) |
0.56 (P=0.0001) |
0.33 (P=0.0184) |
0.72 (P=0.0001) |
0.35 (P=0.0015) |
|
A14 vs A15 |
0.02 (P=0.27) |
0.59 (P=0.0001) |
0.20 (P=0.10) |
0.09 (P=0.4995) |
0.83 (P=0.0001) |
0.14 (P=0.2244) |
The average copy number ratio of all three CBF genes was significantly different between FR-A2 haplotypes in the winter panel, with the strongest association observed for CBF-A14 (P = 0.0002) (Table 4). The ‘FR-A2-T’ haplotype was associated with increased CBF-A12 and CBF-A14 copy number, and reduced CBF-A15 copy number relative to the ‘FR-A2-S’ haplotype. Within the spring panel, differences in copy number between the two haplotypes were also significant for CBF-A14 and CBF-A15, but there was no significant difference in CBF-A12 copy number (P = 0.38) (Table 4).
Table 4.
Least squares means of CBF copy number within each FR-A2 haplotype in the winter and spring panels
| Winter |
Spring |
|||||
|---|---|---|---|---|---|---|
| CBF gene | FR-A2-S | FR-A2-T | Probability of a significant difference |
FR-A2-S | FR-A2-T | Probability of a significant difference |
| CBF-A12 | 0.77 | 1.02 | 0.005 | 0.59 | 0.55 | 0.38 |
| CBF-A14 | 1.01 | 1.98 | 0.0002 | 0.78 | 1.19 | 0.0001 |
| CBF-A15 | 3.10 | 2.48 | 0.03 | 1.94 | 1.48 | 0.0085 |
In summary, this data show that the ‘FR-A2-S’ and ‘FR-A2-T’ haplotypes are associated with differences in copy number of all three CBF genes in the central region of the FR-A2 locus, and that the ‘FR-A2-T’ haplotype can be defined as having an increased CBF-A14 copy number and a decreased CBF-A15 copy number relative to the ‘FR-A2-S’ haplotype.
Interactions between the VRN-A1 and FR-A2 loci and their association with frost tolerance
Winter panel
The survival rates of the 65 lines in the winter panel after exposure to freezing temperatures were associated with different alleles at the VRN-A1 and FR-A2 loci. The 3×2 factorial ANOVA using VRN-A1 CNV (1, 2, 3) and FR-A2 haplotype (S, T) as classification variables (Table 5) explained 32.0% of the variation in frost tolerance in the winter panel. We observed significant differences in frost tolerance between the FR-A2 alleles (P = 0.022) but not between the VRN-A1 copy number classes (P = 0.424). The interaction between these two factors was significant (P = 0.025) (Table 5) reflecting the contrasting effects of the FR-A2 alleles on frost tolerance in the presence of the different VRN-A1 alleles.
Table 5.
Significance of the effects of the VRN-A1 and FR-A2 loci and their interactions on percent survival in freezing tests. ANOVAs were based on factorial models within each of the three populations.
| Population | VRN-A1 | FR-A2 | Factorial ANOVA |
R2 (%) |
P (VRN-A1 X FR-A2) |
P (VRN-A1) |
P (FR-A2) |
|---|---|---|---|---|---|---|---|
| Winter panel | 1, 2 or 3 copies | S vs T | 3×2 | 31.97 | 0.0246 | 0.4243 | 0.0223 |
| Eltan/ ORFW | V=2 copies vs W=3 copies |
S vs T | 2×2 | 45.26 | 0.0026 | 0.1485 | <0.0001 |
| Spring panel | Group A: Vrn- A1 vs Group B: VrnB1+D1 |
S vs T | 2×2 | 20.72 | 0.0247 | <0.0001 | 0.0562 |
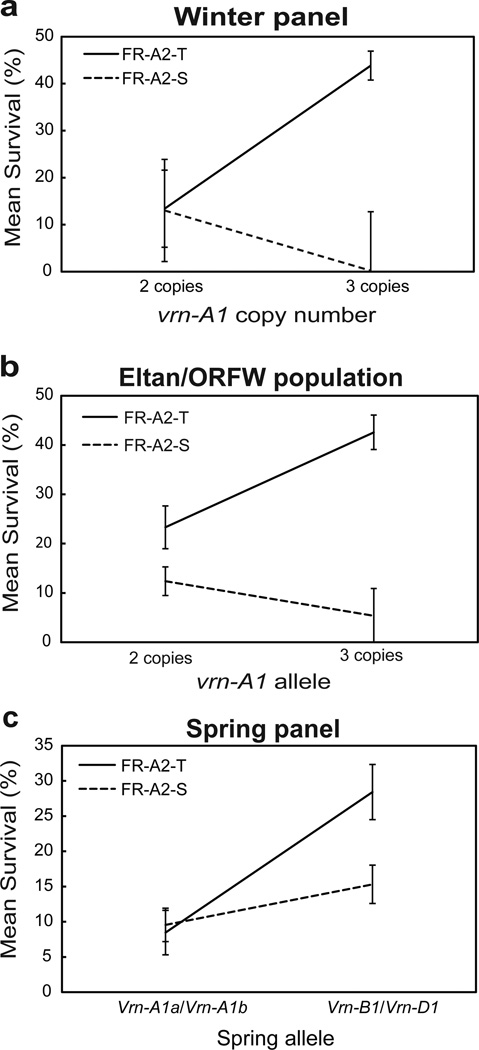
To analyze the simple effects, we ran four one-way ANOVAs within each FR-A2 haplotype and within VRN-A1 CNV groups 2 and 3. The effect of FR-A2 within the VRN-A1 class with one copy could not be tested statistically because only one variety was found in this class (Jagger). Within the ‘FR-A2-S’ group, lines that carried two copies of vrn-A1 had greater survival after freezing (13.03 ± 10.86%) than lines that carried three copies (0.20% ± 12.54%) but the differences were not significant (P = 0.152) (Fig. 2a). In contrast, within the ‘FR-A2-T’ group, lines that carried three copies of vrn-A1 (43.83 ± 3.07%) had about threefold greater survival than lines that carried two copies of vrn-A1 (13.40 ± 8.21%) and the differences were highly significant (P = 0.004). In the reciprocal analysis, differences in frost survival between the ‘FR-A2-S’ and ‘FR-A2-T’alleles were only significant within the VRN-A1 ‘three copy’ group (P = 0.005) and not within the ‘two copy’ group (P = 0.864, Fig. 2a).
Fig. 2.
Interaction between VRN-A1 and FR-A2 within the winter panel (a), Eltan/ORFW F4:5 population (b) and the spring panel (c). For each class and allele combination, average LS means survival rates ± SE are presented. All the interactions were highly significant (P<0.05, based on ANOVA including VRN-A1 and FR-A2 for each population, Table 5).
We then performed an additional factorial ANOVA using FR-A2 and the VRN-A1 SNP allele (V/W) rather than VRN-A1 CNV as a classification variable, to investigate which of the two factors was more predictive of the differences in frost tolerance. When VRN-A1 CNV was used as a classification variable, the model explained 32.0% of the variation in frost tolerance, whereas when the two VRN-A1 SNP classes were used, the model explained just 21.6% of the variation. In addition, the ANOVA using VRN-A1 CNV classes showed a more significant interaction with FR-A2 (P = 0.0246) than the ANOVA using VRN-A1 SNP as classification criteria (P = 0.123). These results suggest that the differences in frost tolerance were more closely associated with the differences in copy number at VRN-A1 than with the SNP differences.
This result was further validated by two additional one-way ANOVAs within the ‘FR-A2-T’ haplotype. First, we compared the effects of VRN-A1 CNV among varieties fixed for the VRN-A1 W allele. The 49 varieties carrying three copies of vrn-A1 (and W/FR-A2-T alleles) exhibited average frost survival rates (41.68 ± 3.68%) fourfold higher than the five varieties carrying two vrn-A1 copies (and W/FR-A2-T alleles) (9.63 ± 2.65%, P = 0.0116). In contrast, no significant differences in frost tolerance were detected between V and W alleles among the varieties carrying two copies of vrn-A1 and the ‘FR-A2-T’ haplotype (P = 0.5111). Taken together the above analysis consistently suggests that it is the CNV at VRN-A1 rather than the SNP variation that is responsible for the observed differences in frost tolerance. However, we cannot rule out the possibility of an effect from other closely linked genes.
Bi-parental ‘Eltan/ORFW’ F4:5 population
To validate the interaction between VRN-A1 and FR-A2 observed in the winter panel, we analyzed the data from the ‘Eltan/ORFW’ F4:5 population using a 2×2 factorial ANOVA. In this population, VRN-A1 CNV co-segregated with the VRN-A1 SNP which distinguished V and W alleles. The ANOVA using VRN-A1 (V= 2 vs. W= 3 in this panel) and FR-A2 haplotype (S, T) as classification variables and their respective interaction (Table 5) explained 45.3% of the variation in frost tolerance. As in the winter panel, significant differences were detected for FR-A2 (P < 0.0001) and no significant differences were detected between the two VRN-A1 classes (P = 0.1485) in the bi-parental population (Table 5). This ANOVA showed a highly significant interaction (P = 0.0026, Fig. 2b) due to the differential responses of the VRN-A1 alleles within the different FR-A2 classes.
The analysis of the simple effects, showed that lines within the ‘FR-A2-S’ group that carried the VRN-A1 V allele (=2 copies) had ~ twofold greater survival (12.39 ± 2.91 %) than the lines that carried the VRN-A1 W allele (=3 copies, 5.38 ± 5.53%) but the differences were not significant (P = 0.131). In contrast, within the ‘FR-A2-T’ group (Fig. 2b) lines that carried the VRN-A1 W allele (42.57 ± 3.50%) had twofold greater frost survival than lines that carried two copies of VRN-A1 (23.31 ± 4.34%) and the differences were highly significant (P = 0.0123, Fig. 2b). In the reciprocal analyses, the FR-A2 haplotypes showed significant differences in frost survival within both VRN1 classes, but the differences were larger within the W allele class (P < 0.0001) than within the V allele class (P = 0.0387).
Spring panel
In spring wheat varieties, different dominant VRN1 alleles have significantly different effects on frost tolerance and survival. Therefore, we divided the varieties in the spring panel into two groups. The first group (group A, 47 accessions) included accessions that carried the strongest dominant Vrn-A1a or Vrn-A1b alleles, which completely eliminate the vernalization requirement. The second group (group B, 34 accessions) included the remaining accessions carrying the dominant Vrn-B1 or Vrn-D1 spring alleles, which exhibit a residual vernalization requirement and a slightly later flowering time than the Vrn-A1 alleles. The interaction between these two loci was analyzed using a 2×2 factorial ANOVA with VRN-A1 groups (A, B) and FR-A2 haplotype (S, T) as classification variables (Table 5). This model explained 20.7% of the variation in frost tolerance. In this analysis, VRN-A1 (P < 0.0001) and the interaction between FR-A2 and VRN-A1 (P = 0.032) both showed significant differences in frost tolerance, but FR-A2 alone did not (P = 0.056) (Table 5). Taken together, our results show that there is a significant interaction between the VRN-A1 and FR-A2 loci in both winter and spring wheat and that, therefore, models considering both loci and their interaction are necessary to correctly classify wheat genotypes for frost tolerance.
Within the ‘FR-A2-S’ group, lines that carried Vrn-B1 or Vrn-D1 (Group B) showed twofold higher survival (15.31 ± 2.71%) than lines that carried Vrn-A1a or Vrn-A1b alleles (group A) (9.54 ± 2.38%), and the difference was significant (P = 0.05). Within the ‘FR-A2-T’ group, lines that carried the Vrn-B1 or Vrn-D1 alleles (group B) showed 3.4-fold greater frost survival (28.41 ± 3.92%) than the lines which carried the Vrn-A1a or Vrn-A1b alleles (group A - 8.45 ± 3.16%, Fig. 2c) and the differences were highly significant (P = 0.0005). Differences between the FR-A2 alleles were not significant in the presence of Vrn-A1a or Vrn-A1b alleles (P = 0.3976) but were highly significant in the presence of the Vrn-B1 or Vrn-D1 alleles (P = 0.0266).
FR-A2 haplotype and CBF transcript levels
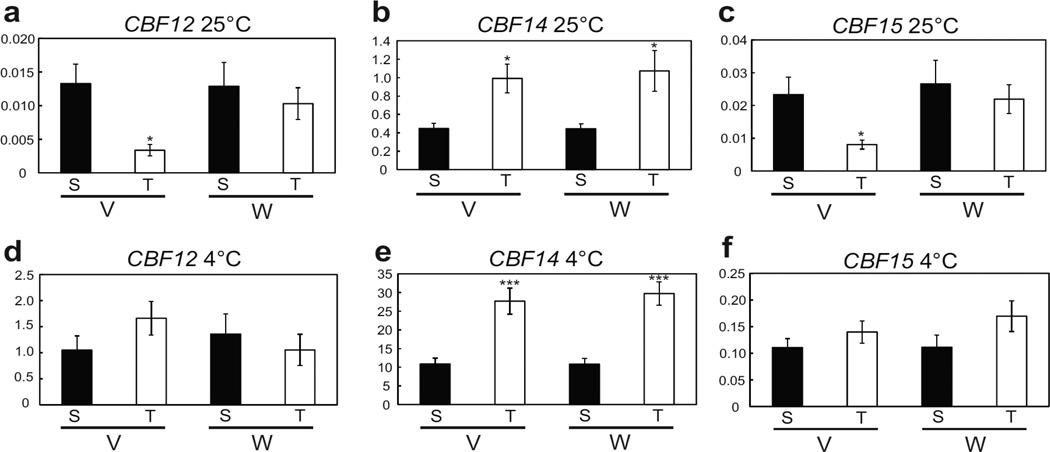
To determine if the detected differences in CBF copy number associated with FR-A2 haplotypes result in differential expression of these genes, we compared CBF transcript levels in ‘FR-A2-S’ and ‘FR-A2-T’ genotypes within the segregating Eltan/ORFW F4:5 population after exposure to a short period of cold (8 h). Transcript levels of CBF14 were more than 10-fold higher than those of CBF12 or CBF15 and all three CBF genes were significantly induced in response to cold treatment in all genotypic combinations (Fig. 3). At 4 °C, we found no significant differences in CBF12 or CBF15 expression levels between the ‘FR-A2-S’ or ‘FR-A2-T’ genotypes (Fig. 3d, f), despite the association of ‘FR-A2-S’ with a higher copy number of CBF-A15. However, CBF14 expression was significantly higher (P ≤0.001) in plants carrying the FR-A2-T allele than those carrying the FR-A2-S allele, both in genotypes carrying the V or the W allele at VRN-A1 and both at 25 °C (Fig. 3c) and 4 °C (Fig. 3e). These results show that the increased CBF-A14 genomic copy number associated with the ‘FR-A2-T’ genotype is associated with an increase in CBF14 transcript levels in response to cold treatment.
Fig. 3.
Expression levels of CBF12, CBF14 and CBF15 at 25 °C (a – c) and after 8 h exposure to 4 °C (d – f). Y-axis values represent the ratio between the initial number of molecules of the target gene and the number of molecules of ACTIN and are the average of six biological replicates ± SE. V/W = Vrn-A1 allele, S = ‘FR-A2-S’, T = ‘FR-A2-T’. * = P ≤ 0.05, *** = P ≤ 0.001, significance of differences between ‘FR-A2-S’ and ‘FR-A2-T’.
Discussion
VRN-A1 allelic diversity and copy number
Previous studies describing variation at the VRN-A1 locus among winter varieties identified different vrn-A1 alleles, which are distinguished by differences in both sequence and copy number (Table 1). In addition to the three alleles described in previous studies (Díaz et al. 2012), we identified six ‘V’ individuals carrying two copies and one variety with three copies of the ‘C’ form of vrn-A1. We demonstrate here that CNV at the VRN-A1 locus is closely associated with the V and W allele classes. By taking advantage of a small number of historic recombination events that have separated these two sources of variation, and carrying out separate ANOVAs within each allele and copy number class, we show that VRN-A1 CNV is more likely responsible than the V and W alleles for the differences in frost tolerance associated with this locus. It is therefore possible that the previously observed association between vrn-A1 V and W alleles and flowering time (Chen et al. 2009) is also an indirect effect of the close association between these alleles and VRN-A1 CNV.
Copy number and sequence variation at the FR-A2 locus
We also identified sequence and CNV at the central cluster of the FR-A2 locus. Previous studies have documented CBF-A14 CNV in wheat, but the association between this variation and frost tolerance was not determined (Knox et al. 2010; Dhillon and Stockinger 2013). In addition to differences in CBF-A14 copy number, we also identified CNV in the two other genes in the central FR-A2 cluster, CBF-A12 and CFB-A15. Despite sequencing many varieties with multiple copies of these two genes, we did not identify double peaks in the sequencing traces at the SNP or indel positions which define the FR-A2 S/T alleles. This indicates that the events resulting in differences in CBF gene copy number between the FR-A2 haplotype classes occurred after the divergence of the ‘FR-A2-S’ and ‘FR-A2-T’ haplotypes.
Furthermore, the large number of linked polymorphisms between ‘FR-A2-S’ and ‘FR-A2-T’ suggests that these are relatively old haplotypes. In support of this hypothesis, we identified both FR-A2 haplotypes within T. dicoccoides accessions originating from both northern and southern sub-populations and within different T. urartu accessions, demonstrating that this sequence diversity at the FR-A2 locus already existed within the wild donor of the A genome in polyploid wheat before the polyploidization event that originated tetraploid wheat < 500,000 years ago (Huang et al. 2002). This agrees with the estimation of the divergence times between the ‘FR-A2–S’ and ‘FR-A2-T’ haplotypes at least 600,000 years ago. The presence of ‘FR-A2-S’ and ‘FR-A2-T’ haplotypes in both diploid and tetraploid wheat suggests either multiple origins of tetraploid wheat involving different T. urartu accessions or gene flow between the sympatric populations of T. urartu and tetraploid wheat. Interestingly, we did not identify any polymorphisms in any of the B or D homoeologous copies of these genes within the modern wheat panels (we did find some instances of sequence diversity within the CBF-A14 gene among T. dicoccoides accessions). CBF-A14 CNV was closely associated with ‘FR-A2-S’ and ‘FR-A2-T’ haplotypes in both winter and spring panels, but CBF-A12 CNV was associated with the FR-A2 haplotypes only in the winter panel. The tight association between CBF-A14 CNV and FR-A2 haplotypes within the varieties tested in the current study limited our ability to determine if the observed differences in frost tolerance were associated with CBF CNV or with the FR-A2 S and T haplotypes.
We compared CBF expression levels between ‘FR-A2-S’ and ‘FR-A2-T’ genotypes within the segregating Eltan/ORFW F4:5 population and found that the ‘FR-A2-T’ genotype exhibited significantly higher CBF14 expression levels in response to cold treatment than the ‘FR-A2-S’ genotype. Despite the association of ‘FR-A2-S’ genotypes with a higher CBF-A15 copy number, this did not result in higher transcript levels of this gene under the conditions tested and neither did we detect differences in CBF-A12 transcript levels. These findings suggest that the increased frost tolerance associated with the ‘FR-A2-T’ haplotype may be explained in part by an increase in CBF14 expression levels in response to cold treatment. However, we cannot rule out a potential role of the observed amino acid changes in CBF12 and CBF15 as alternative causes for the differences in cold tolerance associated with these three tightly linked genes.
To study the potential effect of allelic differences on CBF12 and CBF15 gene function, we analyzed those changes resulting in amino acid substitutions within the conserved domains of these proteins. Mutations with negative BLOSUM 62 scores (negative values indicate different amino acid properties) are generally more likely to disrupt the protein function. However, amino acid substitutions in critical conserved positions may have a disruptive effect even if they have a neutral BLOSUM 62 score. For example, an A37V (BLOSUM 62 = 0) substitution in a critical amino acid of the conserved AP2 domain of DREBIII-4 in Brassica napus abolished the protein’s function by preventing it from binding either the CRT/DRE element or the GCC box (Liu et al. 2006). In our study, the ‘FR-A2-S’ haplotype carries a ‘V’ at position 98, which corresponds to the same position in the AP2 domain as in the substitution described by Liu et al. (2006), whereas the ‘FR-A2-T’ haplotype carries an ‘A’.. The amino acid ‘A’ is conserved in CBF2, CBF3, CBF6, CBF9, CBF13 and CBF14 in common wheat and is also largely conserved in CBF proteins from Arabidopsis, rice, oat, barley, perennial ryegrass and rye (data not shown). The CBF15 ‘FR-A2-S’ haplotype also has a unique T133A (BLOSUM62 = 0) in a position that is conserved in barley, rye and other diploid and polyploid wheat CBF15 proteins. It would be interesting to investigate if the CBF12 and CBF15 proteins encoded by the ‘FR-A2-S’ haplotype are functional.
In Arabidopsis, the last 98 amino acids of the CBF1 C-terminal region have been shown to be important for the trans-activation function of this protein (Wang et al. 2005). In our study, two amino acid changes in the conserved CMIII-4 motif (S229G, BLOSUM 62 = 0 and S236W, BLOSUM 62 = −3) were detected in the C-terminal regions of the CBF15 ‘FR-A2-T’ haplotype (Fig. 1), compared to the CBF15 protein sequences of diploid and hexaploid wheat and barley. This result suggests that the amino acid changes in the ‘FR-A2-T’ haplotype are derived. Since this allele is associated with increased frost tolerance in hexaploid wheat varieties with three VRN-A1 copies it is likely that selection for frost tolerance resulted in an increase of the frequency of these allele combination in winter wheat.
The C-terminal mutation between motif CMIII-2 and CMIII-4 predicted in the CBF12 protein resulted in the change from a histidine to an arginine at position 236 (H236R). The ‘H’ allele is also present in barley suggesting that this amino acid represents the ancestral state. Interestingly, the H236R polymorphism in CBF12 is also present in T. monococcum with the frost susceptible accession DV92 allele carrying the residue ‘H’ and the frost-tolerant G3116 accession carrying the ‘R’ (Knox et al. 2008). This result suggests that this polymorphism might precede the divergence of the A and Am genomes ~ 1 million years ago. Because of the complete linkage between the CBF-A12 and CBF-A15 polymorphisms in the varieties analyzed in this study, it is not possible to separate the effect of the detected mutations in the individual genes on frost survival.
Interaction between VRN-A1 and FR-A2 in hexaploid wheat
Within all three analyzed populations, we identified a significant interaction between the effects of VRN-A1 and FR-A2 loci on frost tolerance. Previous studies of this interaction compared the effect of spring and winter VRN1 alleles on the FR2 alleles. For example, the expression of CBF2 and COR/LEA protein levels was lower in isogenic lines with dominant Vrn-1 alleles (spring growth habit) than in those with recessive vrn-1 alleles (winter growth habit) both in wheat (Kobayashi et al. 2005) and barley (Stockinger et al. 2007). Similarly, Limin and Fowler (2006) found that NILs carrying the recessive vrn-A1 allele showed greater frost tolerance than lines carrying the dominant Vrn-A1 allele. Dhillon et al. (2010) showed that the expression of VRN1 was necessary but not sufficient to induce the down-regulation of the CBF and COR genes. It was only after the initiation of the reproductive development that the cold acclimation process was interrupted.
Our study expands upon previous interaction experiments by characterizing FR-A2 haplotypes and by studying their effects within different dominant Vrn-1 alleles and different recessive vrn-A1 alleles. Within the spring panel, results were consistent with previous analyses in that the strongest spring alleles have a larger epistatic effect on FR-A2, eliminating or greatly reducing the effect of the FR-A2 alleles on frost survival. This could be related to the earlier and higher levels of expression of Vrn-A1 relative to Vrn-B1 and Vrn-D1, which results in an earlier transition to the reproductive stage in the plants carrying the Vrn-A1 allele (Loukoianov et al. 2005).
Among the winter varieties, CNV in VRN-A1 showed opposite effects on frost survival depending on the FR-A2 haplotype. Increased VRN-A1 copy number was associated with increased frost tolerance in varieties carrying the ‘FR-A2-T’ haplotype, but with decreased frost tolerance in varieties carrying the ‘FR-A2-S’ haplotype. This interaction, validated using a bi-parental segregating population, highlights the significant crosstalk that exists between these two loci and provides opportunities for future research to investigate the molecular mechanisms responsible for these interactions.
The interactions between VRN-A1 and FR-A2 on frost tolerance also have practical implications. For example, our results suggest that combining the vrn-A1 haplotype with three copies with the ‘FR-A2-T’ haplotype may be a good combination for winter wheat breeders selecting for high frost tolerance. An important conclusion from this study is that VRN-A1 and FR-A2 must be considered together in the design of strategies to increase frost tolerance in winter wheat.
In contrast, since increased expression of CBF genes is associated with negative pleiotropic effects, including slower growth, delayed flowering and lower grain yields (Morran et al. 2011), in regions where cold does not pose a major threat for survival, varieties carrying the ‘FR-A2-S’ haplotype in a spring background may have a selective advantage. This could potentially explain the higher observed frequency of the ‘FR-A2-S’ in spring as opposed to winter wheat, since spring-sown plants are not exposed to severe low temperatures.
The models including VRN-A1, FR-A2 and their interactions explain a considerable proportion of frost tolerance variation in each of our experiments suggesting that selection for the correct allele combination of these two genes can have a significant impact on frost tolerance improvement in winter wheat breeding programs while these alleles are segregating. The genetic markers for the FR-A2 haplotypes developed in this study together with previous markers for VRN1 (Yan et al. 2004; Fu et al. 2005; Chen et al. 2009) will aid breeder’s efforts to select the best allele combinations for their particular environment.
Supplementary Material
Key message.
The interaction between VRN-A1 and FR-A2 largely affect the frost tolerance of hexaploid wheat
Acknowledgments
This project was supported by the National Research Initiative Competitive Grants WNR-2008-01010 and 2011-68002-30029 (Triticeae-CAP) from the USDA National Institute of Food and Agriculture, the Washington Grains Commission Project no. 5345, and USDA, ARS (in house) CWU: 5348-21220-003-00D. JD also acknowledges support from the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation.
Footnotes
Accession numbers: CBF-A12 (ORFW), HG530924; CBF-A12 (Eltan), HG530925; CBF-B12, HG530926; CBF-D12, HG530927; CBF-A14 (Eltan), HG530928; CBF-A14 (Bussard), pending; CBF-B14, HG530929; CBF-D14, HG530930; CBF-A15 (ORFW), HG530931; CBF-A15 (Eltan), HG530932; CBF-B15, HG530933; CBF-D15, HG530934.
Declaration
The experiments in this manuscript comply with the current laws of the United States. The authors declare that they have no conflict of interest.
Author contribution statement
JZ and SP carried out the experimental part of the work, wrote manuscript drafts and reviewed later versions. AB contributed to the frost tolerance tests. DRS offered the support and facilities for sequencing. DZS produced the Bi-parental ‘Eltan/ORFW’ population. KGC and JD designed the project, wrote the grants to fund it, supervised, contributed to the statistical analyses, and extensively revised the manuscript.
Table S1 List of the 65 winter wheat accessions in the winter panel
Table S2 List of the 81 spring wheat accessions in the spring panel
Table S3 Genome specific primers and PCR conditions used to amplify and sequence the CBF12CBF14 and CBF15 homoeologs from the A, B and D genomes of hexaploid wheat
Table S4 Primers and probes used in the Taqman® assays for the CNV analysis at VRN-A1 and the central cluster of the FR-A2 locus. VRN-A1 and CO2 assays were adapted from Díaz et al. (2012).
Table S5: T. dicoccoides accessions used in this study and their FR-A2 haplotype
Table S6: qRT-PCR primers and their amplification efficiencies
Fig. S1 CAPS markers used to distinguish FR-A2 haplotypes
Lane M: DNA marker. Lane 1: CBF-A12 band type for ‘FR-A2-S’. Lane 2: CBF-A12 band type for ‘FR-A2-T’ (CBF-A12 was digested with ZraI). Lane 3: CBF-A15 band type for ‘FR-A2-S’. Lane 5: CBF-A15 band type for ‘FR-A2-T’ (CBF-A15 was digested with SalI).
References
- Båga M, Chodaparambil SV, Limin AE, Pecar M, Fowler DB, Chibbar RN. Identification of quantitative trait loci and associated candidate genes for low-temperature tolerance in cold-hardy winter wheat. Funct Integr Genomics. 2007;7:53–68. doi: 10.1007/s10142-006-0030-7. [DOI] [PubMed] [Google Scholar]
- Chen Y, Carver BF, Wang S, Zhang F, Yan L. Genetic loci associated with stem elongation and winter dormancy release in wheat. Theor Appl Genet. 2009;118:881–889. doi: 10.1007/s00122-008-0946-5. [DOI] [PubMed] [Google Scholar]
- Chen Y, Carver BF, Wang S, Cao S, Yan L. Genetic regulation of developmental phases in winter wheat. Mol Breed. 2010;26:573–582. [Google Scholar]
- Dhillon T, Stockinger EJ. Cbf14 copy number variation in the A, B and D genomes of diploid and polyploid wheat. Theor Appl Genet. 2013;126:2777–2789. doi: 10.1007/s00122-013-2171-0. [DOI] [PubMed] [Google Scholar]
- Dhillon T, Pearce SP, Stockinger EJ, Distelfeld A, Li C, Knox AK, Vashegyi I, Vágújfalvi A, Galiba G, Dubcovsky J. Regulation of freezing tolerance and flowering in temperate cereals: the VRN-1 connection. Plant Physiol. 2010;153:1846–1858. doi: 10.1104/pp.110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz A, Zikhali M, Turner AS, Isaac P, Laurie DA. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum) PLoS One. 2012;7:e33234. doi: 10.1371/journal.pone.0033234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagles HA, Cane K, Trevaskis B. Veery wheats carry an allele of Vrn-A1 that has implications for freezing tolerance in winter wheats. Plant Breed. 2011;130:413–418. [Google Scholar]
- Francia E, Rizza F, Cattivelli L, Staca AM, Galiba G, Tóth B, Hayes PM, Skinner JS, Pecchioni N. Two loci on chromosome 5H determine low-temperature tolerance in a ‘Nure’ (winter) x ‘Tremois’ (spring) barley map. Theor Appl Genet. 2004;108:670–680. doi: 10.1007/s00122-003-1468-9. [DOI] [PubMed] [Google Scholar]
- Francia E, Barabaschi D, Tondelli A, Laidò G, Rizza F, Stanca AM, Busconi M, Fogher C, Stockinger EJ, Pecchioni N. Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor Appl Genet. 2007;115:1083–1091. doi: 10.1007/s00122-007-0634-x. [DOI] [PubMed] [Google Scholar]
- Fricano A, Rizza F, Faccioli P, Pagani D, Pavan P, Stella A, Rossini L, Piffanelli P, Cattiveli L. Genetic variants of HvCbf14 are statistically associated with frost tolerance in a European germplasm collection of Hordeum vulgare . Theor Appl Genet. 2009;119:1335–1348. doi: 10.1007/s00122-009-1138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Szűcs P, Yan L, Helguera M, Skinner J, Zitzewitz JV, Hayes PM, Dubcovsky J. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Gen Genomics. 2005;273:54–65. doi: 10.1007/s00438-004-1095-4. [DOI] [PubMed] [Google Scholar]
- Galiba G, Quarrie SA, Sutka J, Morgounov A, Snape JW. RFLP mapping of the vernalization (Vrn1) and frost resistance (Fr1) genes on chromosome 5A of wheat. Theor Appl Genet. 1995;90:1174–1179. doi: 10.1007/BF00222940. [DOI] [PubMed] [Google Scholar]
- Galiba G, Vágújfalvi A, Li C, Soltész A, Dubcovsky J. Regulatory genes involved in the determination of frost tolerance in temperate cereals. Plant Sci. 2009;176:12–19. [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- Huang S, Sirikhachornkit A, Su XJ, Faris J, Gill B, Haselkorn R, Gornicki P. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Nat Acad Sci USA. 2002;99:8133–8138. doi: 10.1073/pnas.072223799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell KK, Shelton GB, Demacon VL, Burns JW, Carter BP, Chen XM, Morris CF, Pérez NAB. Registration of ‘Louise’ wheat. Crop Sci. 2006;46:1384–1385. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Knox AK, Li C, Vágújfalvi A, Galiba G, Stockinger EJ, Dubcovsky J. Identification of candidate CBF genes for the frost tolerance locus Fr-Am2 in Triticum monococcum . Plant Mol Biol. 2008;67:257–270. doi: 10.1007/s11103-008-9316-6. [DOI] [PubMed] [Google Scholar]
- Knox AK, Dhillon T, Cheng H, Tondelli A, Pecchioni N, Stockinger EJ. CBF gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor Appl Genet. 2010;121:21–35. doi: 10.1007/s00122-010-1288-7. [DOI] [PubMed] [Google Scholar]
- Kobayashi F, Takumi S, Kume S, Ishibashi M, Ohno R, Murai K, Nakamura C. Regulation by Vrn-1/FR-1 chromosomal intervals of CBF-mediated Cor/Lea gene expression and freezing tolerance in common wheat. J Exp Bot. 2005;56:887–895. doi: 10.1093/jxb/eri081. [DOI] [PubMed] [Google Scholar]
- Koemel JE, Guenzi AC, Anderson JA, Smith EL. Cold hardiness of wheat near-isogenic lines differing in vernalization alleles. Theor Appl Genet. 2004;109:839–846. doi: 10.1007/s00122-004-1686-9. [DOI] [PubMed] [Google Scholar]
- Kronstad WE, Rohde CR, Kolding MF, Metzger RJ. Registration of ‘Stephens’ wheat. Crop Sci. 1978;18:1097. [Google Scholar]
- Limin AE, Fowler DB. Low-temperature tolerance and genetic potential in wheat (Triticum aestivum L.): response to photoperiod, vernalization, and plant development. Planta. 2006;224:360–366. doi: 10.1007/s00425-006-0219-y. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis . Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhao TJ, Liu JM, Liu WQ, Liu Q, Yan YB, Zhou HM. The conserved Ala37 in the ERF/AP2 domain is essential for binding with the DRE element and the GCC box. FEBS Lett. 2006;580:1303–1308. doi: 10.1016/j.febslet.2006.01.048. [DOI] [PubMed] [Google Scholar]
- Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J. Regulation of VRN-1 vernalization genes in normal an transgenic polyploid wheat. Plant Physiol. 2005;138:2364–2373. doi: 10.1104/pp.105.064287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MC, Yang ZL, You FM, Kawahara T, Waines JG, Dvorak J. The structure of wild and domesticated emmer wheat populations, gene flow between them, and the site of emmer domestication. Theor Appl Genet. 2007;114:947–959. doi: 10.1007/s00122-006-0474-0. [DOI] [PubMed] [Google Scholar]
- McIntosh RA, Devos KM, Dubcovsky J, Rogers WJ. Catalogue of genes symbols for wheat: 2004 supplement. Wheat Information Service. 2004 http://wheat.pw.usda.gov/ggpages/wgc/2004upd.html.
- Medina J, Bargues M, Terol J, Perez-Alonso M, Salinas J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 1999;119:463–469. doi: 10.1104/pp.119.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AK, Galiba G, Dubcovsky J. A cluster of 11 CBF transcription factors is located at the frost tolerance locus Fr-Am2 in Triticum monococcum . Mol Gen Genomics. 2006;275:193–203. doi: 10.1007/s00438-005-0076-6. [DOI] [PubMed] [Google Scholar]
- Morran S, Eini O, Pyvovarenko T, Parent B, Singh R, Ismagul A, Eliby S, Shirley N, Langridge P, Lopato S. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol J. 2011;9:230–249. doi: 10.1111/j.1467-7652.2010.00547.x. [DOI] [PubMed] [Google Scholar]
- Motomura Y, Kobayashi F, Iehisa JCM, Takumi S. A major quantitative trait locus for cold-responsive gene expression is linked to frost-resistance gene FR-A2 in common wheat. Breed Sci. 2013;63:58–67. doi: 10.1270/jsbbs.63.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo E, Beiles A. Genetic diversity of wild emmer wheat in Israel and Turkey - Structure, evolution and applications in breeding. Theor Appl Genet. 1989;77:421–455. doi: 10.1007/BF00305839. [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce S, Zhu J, Boldizsár Á, Vágújfalvi A, Burke A, Campbell KG, Galiba G, Dubcovsky J. Large deletions in the CBF gene cluster at the Fr-B2 locus are associated with reduced frost tolerance in wheat. Theor Appl Genet. 2013;126:2683–2697. doi: 10.1007/s00122-013-2165-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Z, Fahima T, Abbo S, Krugman T, Nevo E, Yakir D, Saranga Y. Genetic diversity for drought resistance in wild emmer wheat and its ecogeographical associations. Plant Cell Environ. 1991;28:176–191. [Google Scholar]
- Peterson CJJ, Allan RE, Rubenthaler GL, Line RF. Registration of ‘Eltan’ wheat. Crop Sci. 1991;31:1704. [Google Scholar]
- Ramakrishna W, Dubcovsky J, Park YJ, Busso CS, Emberton J, SanMiguel P, Bennetzen JL. Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics. 2002;162:1389–1400. doi: 10.1093/genetics/162.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy L, Allan RE, Campbell KAG. Evaluation of cold hardiness in two sets of near-isogenic lines of wheat (Triticum aestivum) with polymorphic vernalization alleles. Plant Breed. 2006;125:448–456. [Google Scholar]
- Sandve SR, Fjellheim S. Did gene family expansions during the Eocene-Oligocene boundary climate cooling play a role in Pooideae adaptation to cool climates? Mol Ecol. 2010;19:2075–2088. doi: 10.1111/j.1365-294X.2010.04629.x. [DOI] [PubMed] [Google Scholar]
- Santra DK, Santra M, Allen RE, Campbell KG, Kidwell KK. Genetic and molecular characterization of vernalization genes Vrn-A1, Vrn-B1, and Vrn-D1 in spring wheat germplasm from the Pacific Northwest region of the USA. Plant Breed. 2009;128:576–584. [Google Scholar]
- Skinner DZ, Campbell KAG. Evidence of a major genetic factor conditioning freezing sensitivity in winter wheat. Plant Breed. 2008;127:228–234. [Google Scholar]
- Skinner JS, von Zitzewitz J, Szucs P, Maruez-Cedillo L, Filichkin T, Amundsen K, Stockinger EJ, Thomashow MF, Chen TH, Hayes PM. Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol Biol. 2005;59:533–551. doi: 10.1007/s11103-005-2498-2. [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Skinner JS, Gardner KG, Francia E, Pecchioni N. Expression levels of barley Cbf genes at the Frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2 . The Plant J. 2007;51:308–321. doi: 10.1111/j.1365-313X.2007.0141.x. [DOI] [PubMed] [Google Scholar]
- Sutka J, Galiba G, Vágújfalvi A, Gill BS, Snape JW. Physical mapping of the Vrn-A1 and Fr1 genes on chromosome 5A of wheat using deletion lines. Theor Appl Genet. 1999;99:199–202. [Google Scholar]
- Sutton F, Chen DG, Ge X, Kenefick D. CBF genes of the FR-A2 allele are differentially regulated between long-term cold acclimated crown tissue of freeze-resistant and susceptible, winter wheat mutant lines. BMC Plant Biol. 2009;9:34–42. doi: 10.1186/1471-2229-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth B, Galiba G, Fehér E, Sutka J, Snape JW. Mapping genes affecting flowering time and frost resistance on chromosome 5B of wheat. Theor Appl Genet. 2003;107:509–514. doi: 10.1007/s00122-003-1275-3. [DOI] [PubMed] [Google Scholar]
- Vágújfalvi A, Galiba G, Cattivelli L, Dubcovsky J. The cold-regulated transcriptional activator Cbf3 is linked to the frost-tolerance locus FR-A2 on wheat chroosome 5A. Mol Gen Genomics. 2003;269:60–67. doi: 10.1007/s00438-003-0806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vágújfalvi A, Aprile A, Miller A, Dubcovsky J, Delugu G, Galiba G, Cattivelli L. The expression of several Cbf genes at the FR-A2 locus is linked to frost resistance in wheat. Mol Gen Genomics. 2005;274:506–514. doi: 10.1007/s00438-005-0047-y. [DOI] [PubMed] [Google Scholar]
- Wang Z, Triezenberg SJ, Thomashow MF, Stockinger EJ. Multiple hydrophobic motifs in Arabidopsis CBF1 COOH-terminus provide functinal redundancy in trans-activation. Plant Mol Biol. 2005;58:543–559. doi: 10.1007/s11103-005-6760-4. [DOI] [PubMed] [Google Scholar]
- Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J. Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Genet. 2004;109:1677–1686. doi: 10.1007/s00122-004-1796-4. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Gowda M, Würechum T, Longin CF, Korzun V, Kollers S, Schachschneider R, Zeng J, Fernando R, Dubcovsky J, Reif JC. Dissecting the genetic architecture of frost tolerance in central European winter wheat. J Exp Bot. 2013;64:4453–4460. doi: 10.1093/jxb/ert259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.