Abstract
MicroRNAs (miRNAs) have been widely demonstrated to play fundamental roles in gene regulation in most eukaryotes. To date, there has been no study describing the miRNA composition in genetically modified organisms (GMOs). In this study, small RNAs from dry seeds of two GM soybean lines and their parental cultivars were investigated using deep sequencing technology and bioinformatic approaches. As a result, several differentially expressed gma-miRNAs were found between the GM and non-GM soybeans. Meanwhile, more differentially expressed gma-miRNAs were identified between distantly relatednon-GM soybeans, indicating that the miRNA components of soybean seeds varied among different soybean lines, including the GM and non-GM soybeans, and the extent of difference might be related to their genetic relationship. Additionally, fourteen novel gma-miRNA candidates were predicted in soybean seeds including a potential bidirectionally transcribed miRNA family with two genomic loci (gma-miR-N1). Our findings firstly provided useful data for miRNA composition in edible GM crops and also provided valuable information for soybean miRNA research.
Introduction
MicroRNAs (miRNAs) are a class of genome-encoded, single-stranded, hairpin precursor-derived, small non-coding RNAs that can regulate gene expression at the transcriptional and post-transcriptional levels [1]. Because of their fundamental roles and common features, miRNAs have attracted considerable attention and a great deal of research has been reported in this field in recent years. To date, 35,828 mature miRNAs and 28,645 stem-loop precursors from 223 species including plants, animals, viruses, and even single-celled organisms have been identified and registered in miRBase (Release 21.0) [2]. In the medical field, circulating extracellular miRNAs are used as clinical biomarkers in disease prevention and diagnosis [3, 4]. Due to their endogeneity and high regulatory specificity, the research of miRNAs has resulted in great progress in many medical fields, such as cancer therapy, small RNA medicine, and the treatment of viral disease [5–7]. Further studies on the biogenesis and functions of miRNAs have revealed that the universality and complexity of miRNAs far exceed our current understanding, and additional studies are needed to refine the roles of small RNA and expand our knowledge of gene expression and regulation [8–10].
The introduction of genetically modified (GM) crops has led to significant increases in agricultural output by enhancing plant tolerance to biotic or environmental stress in the past few decades. According to a statistical analysis, the global area of GM cultivars increased to more than 175 million hectares in 2013 [11]. Soybean (Glycine max) is one of the most important economic crops for protein and oil supplements in the world, and it is a major target for transgenic modification. At present, several GM soybean lines have been developed and approved for commercialization. Insect resistance and herbicide tolerance are the main agronomic traits that have been improved in GM soybean [12]. With the rapid increase in the area used for GM crops, more attention has been focused on security issues related to the impact of genetically modified organisms (GMOs) on the environment and food safety for the public. To date, more than 50 countries and areas have established separate legislation and regulations for labeling foods that contain GM ingredients [13]. Although there are only a few reports claiming that GM crops could have negative consequences [14–16], research assessing the potential risks associated with GMOs should continue.
Cross-kingdom regulation of gene expression by miRNA was first discovered and reported by Zhang et al. in 2012 [17]. Experiments performed in vivo and in vitro indicated that rice miR168a could be found in the human blood circulatory system and could inhibit a mammalian gene expression (low-density lipoprotein receptor adapter protein 1, LDLRAP1). Compared to past work focusing on intracellular and intercellular regulation by miRNAs, the discovery of cross-kingdom regulation by miRNAs provided strong evidence for the co-evolving relationship between humans and edible plants, whereby miRNAs play key roles in information exchange at the molecular level. Furthermore, the discovery of cross-kingdom regulation also raised new concerns regarding miRNA components in GM crops.
As with messenger RNAs (mRNAs) in plants and animals, the expression of some miRNAs is variable between tissues or developmental stages, particularly in organisms under stress[18–21]. Considering that transgenic manipulation involves inserting foreign genes or modifying the activity of existing genes, it is reasonable to propose that certain endogenous miRNAs found in GMOs might be affected and differentially expressed. Taking advantage of high-throughput sequencing technology, it is feasible to accurately evaluate the miRNA composition of individual organisms. On the other hand, RNA stored in dry seeds was shown to be relatively stable, and soybean seeds are directly edible after processing, for example as soybean milk [22], which is of practical significance for further research about potential functions for plant miRNAs in human body. Based on these considerations, the dry seeds of GM plants and their parental cultivars were selected for this study.
The current evaluation system for GM crops is focused on proteins, fats, carbohydrates, toxins, and nutritional ingredients. However, miRNAs have not previously been taken into account. Herein, four small RNA libraries were constructed from two GM soybeans (MON89788 and DP-3Ø5423×GTS 40-3-2) and their acceptor lines. After deep sequencing and bioinformatic analysis, several conserved gma-miRNAs were found to be differentially expressed in dry seeds. Combined with miRcheck prediction and experimental verification, fourteen novel gma-miRNA candidates and their stem-loop precursors were also identified [23]. Interestingly, a large proportion of small RNAs generated from ribosomal RNA genes in chromosome Gm13 in soybean seeds was also discovered. Taken together, these results provided useful information about the miRNA composition in GM crops and this work contributes to an improved understanding of miRNAs in soybean seeds.
Materials and Methods
GM soybean samples and RNA isolation
The transgenic soybean line MON89788 and non-GM soybean A3244 were provided by the Qinhuangdao Entry-Exit Inspection and Quarantine Bureau (Qinhuangdao, China). Stacked GM soybean DP-3Ø5423×GTS 40-3-2 and its non-GM counterpart Jack were provided by DuPont Company (USA). Conventional PCR experiments were performed with event-specific primers for verification of the GM soybeans. For each soybean line, three biological replicates were used. For each replicate sample, three aliquots (~ 50 mg each) were randomly drawn and subjected to total RNA extraction. Nine RNA samples were combined for construction of small RNA libraries.
An improved SDS/guanidine isothiocyanate method was used for total RNA isolation from soybean seeds. The details of the procedure are listed as follows. One milliliter of SDS lysis buffer (100 mM NaAc-HAc pH 5.2; 10 mM EDTA; 200 mMNaCl; 1% SDS; 2% PVPP; 1% 2-mercaptoethanol) was added to 50 mg powder and vortexed vigorously. The samples were incubated at 60°C for 10 min, and a one-third volume of 5 M KAc (pH 4.8) was added. The tube was vortexed, placed in ice water for 10 min and then centrifuged for 15 min at 13,000×g at 4°C. The supernatant was transferred to a new tube, and an equal volume of RNA extraction buffer (38% v/v phenol in DEPC-H2O, 1 M guanidinium isothiocyanate, 1 M ammonium thiocyanate, 0.1 M sodium acetate pH 5.0) was added. The tube was vortexed for 3 min and placed at room temperature for 5 min. Then, the samples were extracted once with chloroform:isoamyl alcohol (24:1) and twice with acid phenol:chloroform:isoamyl alcohol (25:24:1). A one-tenth volume of 3 M NaAc (pH 5.0) and an equal volume of ice-cold isopropanol were added. The tube was held at room temperature for 10 min and centrifuged for 15 min at 12,000×g at 4°C. The pellet was washed twice with 75% ethanol, and the RNA was dissolved in DEPC water. The quantity and quality of total RNA were evaluated using a NanoDrop 1000 Spectrophotometer and 1% (w/v) agarose gel electrophoresis.
Small RNA library construction
The small RNA libraries were constructed using the TruSeq Small RNA Sample Preparation kit according to the manufacturer’s instructions. The cDNA products were checked for their quantity and quality using the Agilent 2100 BioAnalyzer and deep sequenced using Illumina/Solexa HiSeq 2000 platform. The raw data from the small RNA libraries were deposited at the NCBI Sequence Read Archive (SRA) under accession no. SRP051931.
Bioinformatic analysis of high-throughput data
For preprocessing the high-throughput data, the raw reads were first screened according to the sequence quality, and individual reads with more than four Phred scores below 15 were discarded. The high-quality reads were then processed by trimming the adaptor sequence, consolidating the identical reads and recording the abundance. Only reads with a reliable 3’ adaptor tail and no ‘N’ or adaptor contaminants were used to generate the clean reads. The soybean (Glycine max) genome and annotation data were downloaded from the plantGDB database[24]. SOAP (v1.11) was used for genome mapping, and only perfectly matched reads were subjected to further analysis [25]. Customized Perl scripts were designed and written to annotate the small RNA loci and analyze their chromosomal distribution.
Differential expression analysis of conserved gma-miRNAs
Known soybean miRNAs (gma-miRNAs) and their stem-loop precursor sequences (gma-MIRs) were downloaded from miRBase [2]. For differential expression analysis, the abundance of each gma-miRNA was normalized to the number of total clean reads in each small RNA library. The P-value was calculated using the formula given below based on previous reports [26, 27]. N1 and N2 are the total numbers of clean reads in the non-GM and GM soybean libraries, and x and y are the numbers of gma-miRNA reads in the non-GM and GM soybean libraries, respectively. The log2 ratio was calculated as log2(number of gma-miRNA reads in GM soybean/number of gma-miRNA reads in non-GM soybean). The Winflat program was integrated into Perl scripts for statistical calculation of the P-value [26].
Novel miRNA prediction
For novel miRNA prediction, the clean reads in the four libraries were combined and BLAST searched against the Rfam database to remove known non-coding RNAs [28]. The RNAfold and miRCheck programs were utilized for stem-loop structure determination and novel miRNA prediction with the default parameters [23]. The details of this process were described in our previous work [29]. The only difference was that double strand testing step was omitted. psRNATarget online was used for target prediction of novel miRNA candidates [30].
RT-PCR assays
Differential expression of conserved gma-miRNAs was verified by quantitative RT-PCR experiments. One microgram of total RNA was reverse-transcribed and amplified using miRcute miRNA First-Strand cDNA synthesis kit (Tiangen, KR201) and miRcute miRNA qPCR detection kit (Tiangen, FP401). The comparative ΔΔCT method was used for relative quantitation of these gma-miRNAs [31]. The U6 snRNA was selected as the endogenous reference gene for normalization. For validation of novel gma-miRNA candidates, a diluted cDNA template was used for RT-PCR assays with specific forward primers and the universal reverse primer. The amplified products were detected by 2% (w/v) agarose gel electrophoresis and validated by Sanger sequencing. The primers used in this section are listed in S3 Table.
Results
Overview of small RNA libraries
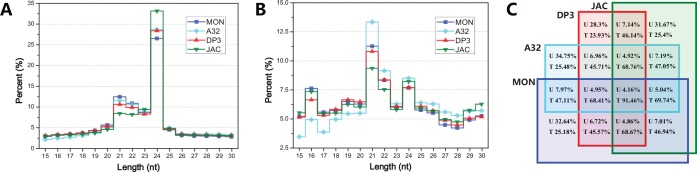
Four small RNA libraries from MON89788, A3244, DP-3Ø5423×GTS 40-3-2 and Jack soybean seeds generated 36,474,511, 36,937,578, 33,667,451 and 40,500,433 raw reads using a deep sequencing platform. After preprocessing, more than 18 million clean reads, corresponding to 1.3~1.6 million unique reads, were obtained from each library. Approximately 17 million clean reads corresponded to at least one perfectly matching locus in the soybean genome, and about 82% of them were located in intergenic regions (Table 1). According to the annotation information, approximately 3 million reads were located in genes or transcripts, and the proportions of exonic and intronic sequences were nearly equal. Intriguingly, a high-density region for small RNA generation was discovered on chromosome Gm13. More than three quarters of the total reads in each library were perfectly matched to a 1 Mb fragment (spanning from 14.7 M to 15.7 M of Gm13) with obvious strand bias (S1 Fig). Further analysis of this genomic locus showed that it’s a super tandem repeat of ribosomal RNA genes, including more than 100 rRNA genes, which was not annotated correctly in Phytozome. A previous report had described the rRNA-derived small RNAs occupying approximately 10% in a soybean small RNA library from germinating cotyledons [32]. However, such a large amount of rRNA-derived small RNAs (~75%) in soybean seeds had never been discovered before. The high proportion of ribosomal related small RNAs as storage ingredient in soybean seeds suggested that they might be functional and play crucial roles during seed germination. As shown in Fig 1A and 1B, 21-nt and 24-nt classes of clean reads represented the majority of the unique and total reads, respectively. There was no obvious difference in the length distribution between the GM and non-GM libraries. In contrast to the 21-nt class, small RNAs in 24-nt class showed more polymorphism. Common read analysis indicated that fewer shared reads were found when more libraries were included. Only 4.16% of the unique reads were shared among the four libraries, accounting for 91.46% of the total reads (Fig 1C).
Table 1. Composition of small RNA libraries from soybean seeds of GM and their parental lines.
| MON | A32 | DP3 | JAC | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Unique | Total | Unique | Total | Unique | Total | Unique | Total | |||
| Raw reads | — | 36,474,511 | — | 36,937,578 | — | 33,667,451 | — | 40,500,433 | ||
| High quality reads | — | 34,851,791 | — | 35,306,825 | — | 32,211,029 | — | 38,797,935 | ||
| Clean reads (15–30 nt) | 1,509,201 | 19,281,725 | 1,607,094 | 19,515,644 | 1,308,765 | 18,327,863 | 1,464,627 | 19,452,871 | ||
| Not matching genome | 791,683 | 2,230,283 | 879,611 | 2,469,019 | 655,168 | 1,858,850 | 723,226 | 2,083,080 | ||
| Matching genome | 717,518 | 17,051,442 | 727,483 | 17,046,625 | 653,597 | 16,469,013 | 741,401 | 17,369,791 | ||
| Intergenic | — | 14,215,639 | — | 14,025,360 | — | 13,571,051 | — | 14,410,600 | ||
| Gene (transcripts) | — | 2,835,803 | — | 3,021,265 | — | 2,897,962 | — | 2,959,191 | ||
| Exon | — | 1,639,965 | — | 1,811,152 | — | 1,713,607 | — | 1,741,190 | ||
| Intron | — | 1,195,838 | — | 1,210,113 | — | 1,184,355 | — | 1,218,001 | ||
| Matching gma-MIRs | 2,655 | 625,399 | 2,649 | 1,103,604 | 2,562 | 742,318 | 2,712 | 613,667 | ||
| Corresponding gma-MIRs | 386 | — | 362 | — | 360 | — | 375 | — | ||
| Mature gma-miRNAs | 144 | 491,705 | 153 | 958,430 | 139 | 621,249 | 138 | 492,651 | ||
| Matching Rfam database | 33,200 | 2,126,950 | 33,153 | 1,964,365 | 32,117 | 2,044,085 | 33,354 | 2,246,844 | ||
| rRNA | 12,381 | 1,783,876 | 12,719 | 1,643,676 | 12,608 | 1,726,000 | 12,665 | 1,895,127 | ||
| tRNA | 2,069 | 199,903 | 2,115 | 194,209 | 1,828 | 191,841 | 2,017 | 221,421 | ||
| snoRNA | 8,365 | 71,627 | 7,247 | 56,350 | 6,477 | 47,812 | 7,691 | 55,875 | ||
| other | 10,385 | 71,544 | 11,072 | 70,130 | 11,204 | 78,432 | 10,981 | 74,421 | ||
| Unknown Reads | 681,663 | 14,299,093 | 691,681 | 13,978,656 | 618,918 | 13,682,610 | 705,335 | 14,509,280 | ||
MON, MON89788; A32, A3244; DP3, DP-3Ø5423×GTS 40-3-2; JAC, Jack.
Fig 1. Length distribution and Venn diagram of small RNA libraries.
A: Length distribution of the unique reads. B: Length distribution of the total reads. C: Venn diagram of four small RNA libraries. All of the small RNA reads from four libraries are combined as whole. Each percentage is calculated by dividing the number of reads in each library. U, unique; T, total; MON, MON89788; A32, A3244; DP3, DP-3Ø5423×GTS 40-3-2; JAC, Jack.
Differential expression analysis of conserved gma-miRNA
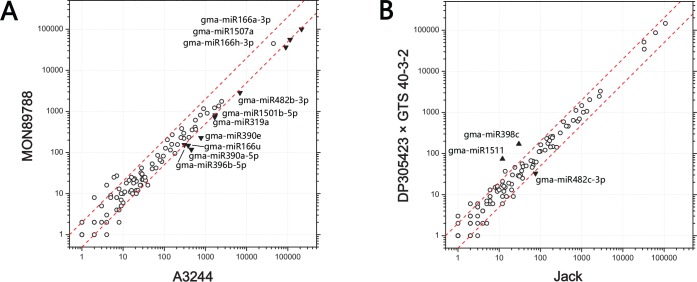
To detect conserved gma-miRNAs, the clean reads were aligned against known gma-miRNAs represented in miRBase. Only identical reads were regarded as mature gma-miRNAs. In total, at least 138 gma-miRNAs were found in each small RNA library. Meanwhile, more than 380 gma-MIR precursors could find perfectly matching reads in each library (S1 Table). For differential profiling expression analysis, the read numbers were normalized to calculate the P-values and log2 ratios. Differences in expression were considered significant at P < 0.001 and a log2 ratio > 1. Totally, three couples were analyzed, including MON89788 and A3244, DP-3Ø5423×GTS 40-3-2 and Jack, A3244 and Jack. As a result, ten gma-miRNAs were down-regulated in MON89788 compared with A3244 soybeans, including mature members of the miR1507, miR1510, miR166, miR319, miR390, miR396 and miR482 families. In DP-3Ø5423×GTS 40-3-2 and Jack soybeans, only miR482c-3p was down-regulated, and two gma-miRNAs, miR398c and miR1511, were up-regulated. Between non-GM soybneas, A3244 and Jack, thirteen varied gma-miRNAs were also identified where ten gma-miRNAs were up-regulated and three gma-miRNAs were down-regulated (Fig 2).
Fig 2. Scatter plot demonstrating the differential expression of gma-miRNAs in GM soybean seeds.
The normalized values were calculated using the following formula: (miRNA reads count/total clean reads) × 107. Red dotted lines indicate the value of |log2| = 1. Down- or up-regulated gma-miRNAs are indicated with black triangles; these gma-miRNAs meet the following three conditions: raw reads of gma-miRNAs > 100, |log2 ratio| > 1, and P < 0.001. A: Results of the comparison between MON89788 and A3244. B: Results of the comparison between DP-3Ø5423×GTS 40-3-2 and Jack.
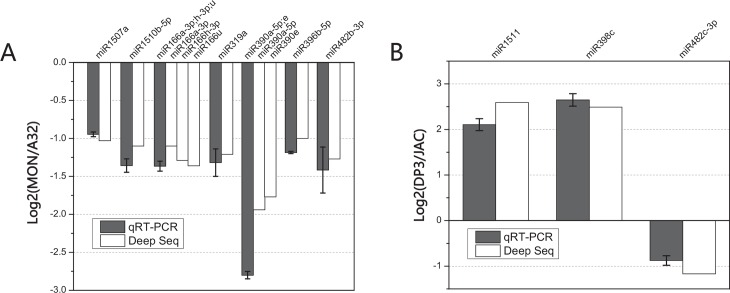
The maximum values of log2 ratio for up- and down-regulation were 2.79 and -1.94 for miR398c and miR390a-5p, respectively (Table 2). With regard to abundance, gma-miR166a, gma-miR166h and gma-miR1507a were the most abundant miRNAs among four small RNA libraries, with 70 ~ 420 thousand clean reads before normalization (S2 Fig). These results indicated that differences of miRNA composition exist between GM soybean seeds and their parental non-GM lines. Meanwhile, differentially expressed miRNAs could also be found between non-GM soybeans, which might be more diverse due to their distant genetic relationship. Although we could not determine the intrinsic factors responsible for these differences, it seemed that differentially expressed miRNAs in soybean seeds are ubiquitous between different cultivars including the GM and non-GM soybeans. Additionally, thirteen differentially expressed gma-miRNAs identified by deep sequencing, two were up-regulated and eleven were down-regulated in GM soybean seeds compared with their parental lines, were confirmed by quantitative real-time PCR (Fig 3).
Table 2. Differential expression of known gma-miRNAs in soybean seeds.
| Sequence (5’-3’) | Mature miRNA | Log2(MON/A32) | Log2(DP3/JAC) | Log2(A32/JAC) | Same to gma-miRNAs | |||
|---|---|---|---|---|---|---|---|---|
| TCTCATTCCATACATCGTCTGA | miR1507a | -1.03 | ↓ | 0.49 | — | 0.88 | — | gma-miR1507a |
| AGGGATAGGTAAAACAACTACT | miR1510b-5p | -1.10 | ↓ | -0.34 | — | 0.98 | — | gma-miR1510b-5p |
| AACCAGGCTCTGATACCATG | miR1511 | 0.06 | — | 2.59 | ↑ | 2.07 | ↑ | gma-miR1511 |
| TTGACAGAAGATAGAGAGCAC | miR156c | -0.44 | — | 0.70 | — | 1.07 | ↑ | gma-miR156c,d,e,i,j,l,m |
| TCGGACCAGGCTTCATTCCCC | miR166a-3p | -1.10 | ↓ | 0.44 | — | 1.00 | ↑ | gma-miR166a-3p,b,c-3p,d,e,f,g,n,o,i-3p |
| TCTCGGACCAGGCTTCATTCC | miR166h-3p | -1.29 | ↓ | 0.66 | — | 1.46 | ↑ | gma-miR166h-3p,k |
| TCTCGGACCAGGCTTCATTC | miR166u | -1.36 | ↓ | 0.41 | — | 0.95 | — | gma-miR166u |
| GGAGATGGGAGGGTCGGTAAAG | miR2118a-5p | -0.38 | — | -0.46 | — | -1.10 | ↓ | gma-miR2118a-5p,b-5p |
| TTGGACTGAAGGGAGCTCCC | miR319a | -1.21 | ↓ | 0.62 | — | 1.21 | ↑ | gma-miR319a,b,e |
| AAGCTCAGGAGGGATAGCGCC | miR390a-5p | -1.94 | ↓ | 0.21 | — | 1.58 | ↑ | gma-miR390a-5p,f,g |
| AGCTCAGGAGGGATAGCGCC | miR390e | -1.77 | ↓ | 0.31 | — | 2.00 | ↑ | gma-miR390e |
| TTCCACAGCTTTCTTGAACTT | miR396b-5p | -1.00 | ↓ | 0.30 | — | 0.88 | — | gma-miR396b-5p,c |
| TGTGTTCTCAGGTCGCCCCTG | miR398c | -0.42 | — | 2.49 | ↑ | 2.79 | ↑ | gma-miR398c |
| TGTTGCGGGTATCTTTGCCTC | miR4412-5p | 0.36 | — | -0.12 | — | -1.57 | ↓ | gma-miR4412-5p |
| TCTTCCCTACACCTCCCATACC | miR482b-3p | -1.27 | ↓ | 0.21 | — | 1.27 | ↑ | gma-miR482b-3p,d-3p |
| TTCCCAATTCCGCCCATTCCT | miR482c-3p | -0.93 | — | -1.17 | ↓ | 1.26 | ↑ | gma-miR482c-3p |
| TGAAGATTTGAAGAATTTGGGA | miR5376 | -0.46 | — | -0.36 | — | -1.19 | ↓ | gma-miR5376 |
MON, MON89788; A32, A3244; DP3, DP-3Ø5423×GTS 40-3-2; JAC, Jack. gma-miRNAs with original abundance more than 100 reads were listed. Log2 ratios of normalized gma-miRNA reads from GM and non-GM soybean seeds were calculated, P < 0.001.
Fig 3. Verification of differentially expressed gma-miRNAs by quantitative RT-PCR.
Identical primers were designed for similar mature miRNAs. The comparative ΔΔCT method was used for the quantitative RT-PCR experiments and the U6 snRNA was selected as the reference gene. A: Results of the comparison between MON89788 and A3244. B: Results of the comparison between DP-3Ø5423×GTS 40-3-2 and Jack.
Novel gma-miRNA prediction
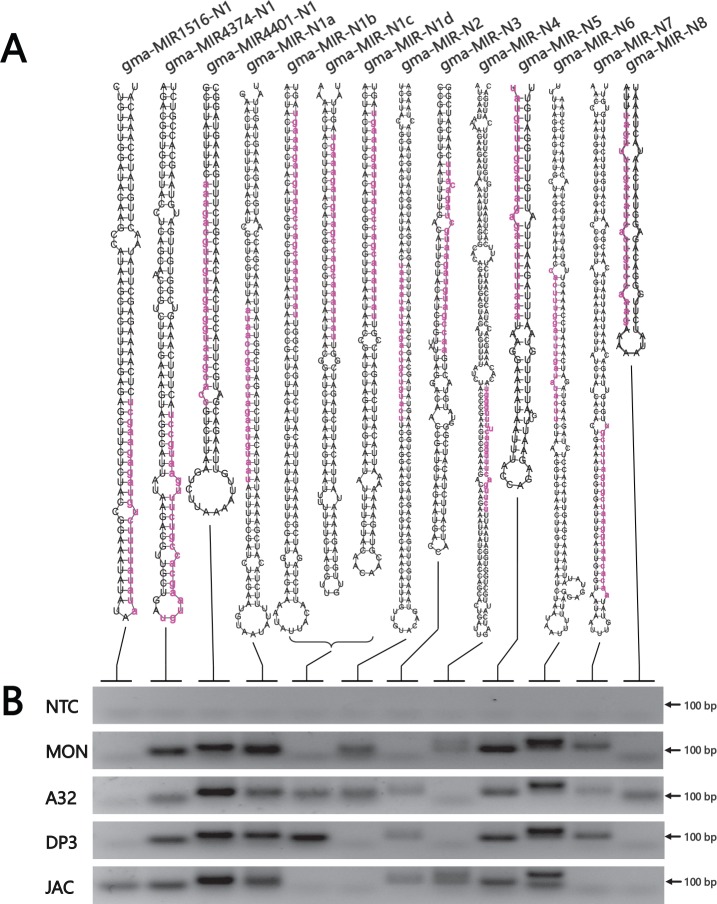
According to the standards of plant miRNA prediction, the miRCheck program was employed to investigate novel miRNAs in soybean. Soybean miRNAs have been extensively surveyed, and 639 mature miRNAs corresponding to 573 precursors have been registered in miRBase. As a result, only fourteen gma-miRNA candidates were finally acquired with relatively low abundance levels based on our optimal pipelines with strict parameters. Among these candidates, 3 stem-loop precursor sequences shared high similarity to the known gma-MIRs, gma-MIR1516a, gma-MIR4374b and gma-MIR4401a (S3 Fig). However, the most abundant reads of these three conserved gma-MIRs were not identical to known gma-miRNAs. The other eleven miRNA precursors were capable to form characteristic hairpin structures without any homology to known gma-MIRs. Notably, gma-miR-N1 corresponded to a potential bidirectionally transcribed miRNA that was first reported in Drosophila [33]. Four members of the gma-miR-N1 family were most likely produced from both the sense and the antisense strands of two genomic loci (S4 Fig). For all of the newly identified gma-miRNAs, the preferred nucleotides at the 5’ end were uracil and adenine, and most of these miRNAs were 24 nt in length. All novel gma-miRNA candidates were subjected to experimental verification by RT-PCR and Sanger sequencing, and they all were positively identified in at least one library (Fig 4, Table 3 and S1 File). Target prediction was performed using psRNATarget online for the novel gma-miRNA candidates [30]. Based on the available annotation information, the target genes of gma-miR-N4 and gma-miR-N5 are an integral membrane protein and the BZIP transcription factor. The results are listed in S2 Table.
Fig 4. Novel gma-miRNA candidates found in this study.
A: Hairpin structures of novel gma-miRNA precursors predicted in GM soybean seeds. Mature miRNAs are indicated with red lowercase letters. B: Expression analysis of novel gma-miRNA candidates by RT-PCR. NTC, no template control; MON, MON89788; A32, A3244; DP3, DP-3Ø5423×GTS 40-3-2; JAC, Jack.
Table 3. Results of novel gma-miRNA prediction in soybean seeds.
| No. | Mature miRNAs | Sequence(5’-3’) | Length | Clean reads in each library | Precursor location | Precursor Conservativeness | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | MON | A32 | DP3 | JAC | ||||||
| 1 | gma-miR1516-N1 | AUAUAUUUUCUGUAGAGAAGCU | 22 nt | 5 | 0 | 0 | 2 | 3 | Gm03_-_47035754_47035862 | gma-MIR1516a |
| 2 | gma-miR4374-N1 | UGUAAGCACCGUCUUUGAAUGCCU | 24 nt | 42 | 18 | 8 | 9 | 7 | Gm01_-_23428261_23428375 | gma-MIR4374b |
| 3 | gma-miR4401-N1 | AAAGACGUUGUUGAGGUAAGCACC | 24 nt | 6 | 3 | 2 | 0 | 1 | Gm05_-_5369728_5369821 | gma-MIR4401a |
| 4 | gma-miR-N1a | AUAACCGAUCUAGAAUGUAAU | 21 nt | 10 | 3 | 5 | 0 | 2 | Gm02_+_39925524_39925657 | None |
| 5 | gma-miR-N1b | UAUUAAACGACCGAUGUAGAAAGU | 24 nt | 13 | 1 | 1 | 10 | 1 | Gm02_-_39925526_39925654 | None |
| 6 | gma-miR-N1c | UAUUAAACGACCGAUGUAGAAAGU | 24 nt | 13 | 1 | 1 | 10 | 1 | Gm03_+_11919586_11919706 | None |
| 7 | gma-miR-N1d | UAUUAAACGACCGAUGUAGAAAGU | 24 nt | 13 | 1 | 1 | 10 | 1 | Gm03_-_11919589_11919703 | None |
| 8 | gma-miR-N2 | UAAAAUAUUAGACUGCUGUACU | 22 nt | 68 | 37 | 24 | 4 | 3 | Gm06_-_46035186_46035339 | None |
| 9 | gma-miR-N3 | AACCGAUGUAGAAUGCUAGACAUU | 24 nt | 25 | 4 | 2 | 5 | 14 | Gm07_-_19042117_19042232 | None |
| 10 | gma-miR-N4 | UCUUGACUUUGGACUUUUGGGU | 22 nt | 20 | 4 | 7 | 1 | 8 | Gm02_-_48422489_48422679 | None |
| 11 | gma-miR-N5 | UAUGUUUGGAUAGAGAAUUUUAAA | 24 nt | 11 | 3 | 5 | 0 | 3 | Gm07_+_44428050_44428136 | None |
| 12 | gma-miR-N6 | CACUUUGGAUUUGAUAUACUU | 21 nt | 6 | 1 | 4 | 0 | 1 | Gm05_+_40112942_40113095 | None |
| 13 | gma-miR-N7 | AACACAAUGGAAUCGUGAUUUCGU | 24 nt | 5 | 1 | 2 | 0 | 2 | Gm07_+_1788665_1788817 | None |
| 14 | gma-miR-N8 | UAGUUUUGAUAUCACUGUCCAAAG | 24 nt | 5 | 1 | 3 | 0 | 1 | Gm08_-_4271026_4271087 | None |
MON, MON89788; A32, A3244; DP3, DP-3Ø5423×GTS 40-3-2; JAC, Jack.
Discussion
MicroRNAs have been a topic of intense interest in current molecular biology, and their biogenesis and functions have been thoroughly studied over the past decade. Nevertheless, no studies have been reported comparing the miRNA composition between GM crops and their near-isogenic lines prior to the discovery of phenomenon of cross-kingdom regulation. Evidence that rice miR168 could enter the human bloodstream and regulate mammalian gene expression has expanded our understanding of miRNA [17]. Although many subsequent reports have questioned this controversial result [34–37], the findings offer a novel perspective for miRNA research. The purpose of this study was to identify the differentially expressed miRNAs in GM soybean seeds using deep sequencing technology and to provide data that will be useful for the future research.
Soybean is one of the most important natural sources for dietary oil and proteins worldwide. The choice to use the dry seeds of GM soybeans for this study was mainly based on the consideration that soybean can be directly consumed as a food, for example as soybean milk, which makes it practical for future research on cross-kingdom regulation of plant miRNA. The MON89788 GM soybean is a genetically engineered variant of the near-isogenic conventional soybean variety A3244. A cp4 EPSPS gene with stronger constitutive viral promoters was introduced via Agrobacterium-mediated transformation to improve the glyphosate tolerance of the plant [38]. The stacked GM soybean DP-3Ø5423×GTS 40-3-2 has the same genetic background as the non-GM Jack soybean except that it contains insertions of the endogenous genes, gm-fad2-1, gm-hra and cp4, which were introduced through crossbreeding with the high oleic acid soybean DP-3Ø5423 and the Roundup Ready soybean 40-3-2. Subchronic feeding studies in rats have been reported using these GM soybeans, and no obvious quantitative differences were found in the proteins, amino acids, nutritional factors or systemic effects [39–41]. Although there were no obvious differences in the contents of biological macromolecules, it’s reasonable to believe that the sustained expression of exogenous genes in vivo might trigger unexpected interactions and alter compositional properties at the RNA level. So far, reports on the RNAs present in dormant soybean seeds are scarce, however the storage proteins, globulins, 7S β-conglycinin and 11S glycinin, have been heavily studied to characterize their patterns of accumulation and associated regulatory factors prior to dormancy [22, 42, 43]. Recent research on miRNAs in plant seeds has addressed their potential regulatory roles in miRNA-mediated gene expression during seed development and germination [44, 45]. However, no study has been performed to investigate differences in the composition of miRNAs in GM soybeans.
In this study, we identified a largely repeated rRNA gene region for small RNA production at the approximately 15 Mb locus on chromosome Gm 13. There is an obvious strand bias and the mapping ratio between sense and antisense strands is about 1:100. Unbalanced mapping depth along the rRNA gene indicates that they are authentic small RNAs instead of rRNA degradation products. Although these rRNA-derived small RNAs had been reported to be involved in various signaling pathways and have potential biological functions, the underlying mechanism is still largely unknown [46]. Similarly to storage proteins, these specific small RNAs might be integral components of soybean seeds that function as molecular regulators to control gene expression during seed germination like miRNA [47]. Further studies on this abundant class of rRNA-derived small RNAs in soybean seeds will provide new insights into seed biology.
For novel miRNA prediction, an optimal pipeline based on plant miRNA criteria and the miRCheck program described in our previous work was used [29, 48]. Considering that miRNAs are known to be tissue-, condition- and developmental stage-specific in many eukaryotes, the current set of registered miRNAs in miRBase might be only part of the complete set of authentic miRNAs in soybean. Although the gma-miRNAs are well annotated in domestic soybean, fourteen novel miRNA candidates were discovered in this study, including three members of known conserved gma-miRNA families. All the precursors of these novel miRNAs could form perfect hairpin structures, indicating that our bioinformatic pipeline is reliable and sensitive. A potential family of bidirectionally transcribed miRNAs was also identified with low abundance, suggesting the complexity and diversity of gma-miRNAs.
In this work, a deep sequencing platform was utilized for comparative profiling of miRNA expression in soybean seeds of GM plants and their near-isogenic acceptor lines. As expected, several conserved gma-miRNAs were found to be differentially expressed more than 2-fold change, and three conserved gma-miRNAs were legume-specific. Compared to A3244, all ten of the differentially expressed gma-miRNAs in MON89788 were down-regulated, including the members of miR1507, miR1510, miR166, miR319, miR390, miR396 and miR482 families. Two of the most abundant down-regulated miRNAs were gma-miR166a-3p and gma-miR166h-3p. miR166 has been reported to regulate the homeodomain leucine zipper (HD-ZIP) gene, which might play significant roles in seedling development [49]. Between DP-3Ø5423×GTS 40-3-2 and Jack soybeans, three varied gma-miRNAs were identified with low or moderate abundance where miR482c-3p was down-regulated, and miR398c and miR1511 were up-regulated. Notably, thirteen differentially expressed gma-miRNAs were also discovered when comparing non-GM soybeans (A3244 and Jack). These results implied that the miRNA components of soybean seeds varied among different soybean lines, and the extent of difference might be related to their genetic relationships. For a better understanding of these differentially expressed miRNAs, further research is required to investigate their functions and whether they could influence human gene expression due to uptake into the bloodstream upon consumption.
Supporting Information
(PDF)
(PDF)
(PDF)
(PDF)
(DOCX)
(XLSX)
(DOCX)
(DOCX)
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31000356), the Presidential Foundation of the Tianjin Academy of Agricultural Sciences (No. 13002) and Tianjin Research Program of Application Foundation and Advanced Technology for Youths (14JCQNJC14800).
Data Availability
The raw data from the small RNA libraries were deposited at the NCBI Sequence Read Archive (SRA) under accession no. SRP051931.
Funding Statement
This work was supported by the National Natural Science Foundation of China (No. 31000356), the Presidential Foundation of the Tianjin Academy of Agricultural Sciences (No. 13002) and Tianjin Research Program of Application Foundation and Advanced Technology for Youths (14JCQNJC14800).
References
- 1.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153(3):516–9. 10.1016/j.cell.2013.04.003 [DOI] [PubMed] [Google Scholar]
- 2.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic acids research. 2014;42(Database issue):D68–73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell research. 2008;18(10):997–1006. 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- 4.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences. 2008;105(30):10513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature Reviews Cancer. 2006;6(11):857–66. [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nature reviews Cancer. 2006;6(4):259–69. [DOI] [PubMed] [Google Scholar]
- 7.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457(7228):426–33. 10.1038/nature07758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138(4):738–49. 10.1016/j.cell.2009.06.014 [DOI] [PubMed] [Google Scholar]
- 9.Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138(4):750–9. 10.1016/j.cell.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu H, Hu F, Wang R, Zhou X, Sze SH, Liou LW, et al. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011;145(2):242–56. 10.1016/j.cell.2011.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.James C. Global Status of Commercialized Biotech/GM Crops: 2013. ISAAA Brief 2013;(No. 46.).
- 12.Barampuram S, Zhang ZJ. Recent advances in plant transformation. Methods Mol Biol. 2011;701:1–35. 10.1007/978-1-61737-957-4_1 [DOI] [PubMed] [Google Scholar]
- 13.Gruère GP, Rao S. A review of international labeling policies of genetically modified food to evaluate India’s proposed rule. AgBioForum. 2007;10(1):51–64. [Google Scholar]
- 14.Ewen SW, Pusztai A. Effect of diets containing genetically modified potatoes expressing Galanthus nivalis lectin on rat small intestine. The Lancet. 1999;354(9187):1353–4. [DOI] [PubMed] [Google Scholar]
- 15.Losey JE, Rayor LS, Carter ME. Transgenic pollen harms monarch larvae. Nature. 1999;399(6733):214. [DOI] [PubMed] [Google Scholar]
- 16.Quist D, Chapela IH. Transgenic DNA introgressed into traditional maize landraces in Oaxaca, Mexico. Nature. 2001;414(6863):541–3. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell research. 2012;22(1):107–26. 10.1038/cr.2011.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Current Biology. 2002;12(9):735–9. [DOI] [PubMed] [Google Scholar]
- 19.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome biology. 2004;5(3):R13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chellappan P, Vanitharani R, Fauquet CM. MicroRNA-binding viral protein interferes with Arabidopsis development. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(29):10381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sunkar R, Chinnusamy V, Zhu J, Zhu J-K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends in plant science. 2007;12(7):301–9. [DOI] [PubMed] [Google Scholar]
- 22.Mori T, Wakabayashi Y, Takagi S. Occurrence of mRNA for storage protein in dry soybean seeds. J Biochem. 1978;84(5):1103–11. [DOI] [PubMed] [Google Scholar]
- 23.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular cell. 2004;14(6):787–99. [DOI] [PubMed] [Google Scholar]
- 24.Duvick J, Fu A, Muppirala U, Sabharwal M, Wilkerson MD, Lawrence CJ, et al. PlantGDB: a resource for comparative plant genomics. Nucleic acids research. 2008;36(Database issue):D959–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24(5):713–4. 10.1093/bioinformatics/btn025 [DOI] [PubMed] [Google Scholar]
- 26.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome research. 1997;7(10):986–95. [DOI] [PubMed] [Google Scholar]
- 27.Man MZ, Wang X, Wang Y. POWER_SAGE: comparing statistical tests for SAGE experiments. Bioinformatics. 2000;16(11):953–9. [DOI] [PubMed] [Google Scholar]
- 28.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, et al. Rfam 11.0: 10 years of RNA families. Nucleic acids research. 2013;41(Database issue):D226–32. 10.1093/nar/gks1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen R, Jiang N, Jiang Q, Sun X, Wang Y, Zhang H, et al. Exploring microRNA-like small RNAs in the filamentous fungus Fusarium oxysporum. PloS one. 2014;9(8):e104956 10.1371/journal.pone.0104956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic acids research. 2011;39(Web Server issue):W155–9. 10.1093/nar/gkr319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3(6):1101–8. [DOI] [PubMed] [Google Scholar]
- 32.Zabala G, Campos E, Varala KK, Bloomfield S, Jones SI, Win H, et al. Divergent patterns of endogenous small RNA populations from seed and vegetative tissues of Glycine max. BMC plant biology. 2012;12:177 10.1186/1471-2229-12-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, Hannon GJ, et al. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes & development. 2008;22(1):26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang M, Sang X, Hong Z. Beyond nutrients: food-derived microRNAs provide cross-kingdom regulation. BioEssays: news and reviews in molecular, cellular and developmental biology. 2012;34(4):280–4. [DOI] [PubMed] [Google Scholar]
- 35.Witwer KW, McAlexander MA, Queen SE, Adams RJ. Real-time quantitative PCR and droplet digital PCR for plant miRNAs in mammalian blood provide little evidence for general uptake of dietary miRNAs: limited evidence for general uptake of dietary plant xenomiRs. RNA biology. 2013;10(7):1080–6. 10.4161/rna.25246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K, Li H, Yuan Y, Etheridge A, Zhou Y, Huang D, et al. The complex exogenous RNA spectra in human plasma: an interface with human gut biota? PloS one. 2012;7(12):e51009 10.1371/journal.pone.0051009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Wiggins BE, Lawrence C, Petrick J, Ivashuta S, Heck G. Analysis of plant-derived miRNAs in animal small RNA datasets. BMC genomics. 2012;13:381 10.1186/1471-2164-13-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watrud LS, Lee EH, Fairbrother A, Burdick C, Reichman JR, Bollman M, et al. Evidence for landscape-level, pollen-mediated gene flow from genetically modified creeping bentgrass with CP4 EPSPS as a marker. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(40):14533–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tutel'ian VA, Gapparov MG, Avren'eva LI, Guseva GV, Zhminchenko VM, Kravchenko LV, et al. Medical and biological safety assessment of genetically modified soybean event MON 89788. Report 1. Toxicologo-hygienic examinations. Voprosy pitaniia. 2010;79(3):4–12. [PubMed] [Google Scholar]
- 40.Tyshko NV, Britsina MV, Gmoshinskii IV, Zakharova NS, Zorin SN, Mazo VK, et al. Medical and biological safety assessment of genetically modified soybean event MON 89788. Report 2. Genotoxicologic, immunologic and allergologic examinations. Voprosy pitaniia. 2010;79(3):13–7. [PubMed] [Google Scholar]
- 41.Qi X, He X, Luo Y, Li S, Zou S, Cao S, et al. Subchronic feeding study of stacked trait genetically-modified soybean (3Ø5423× 40-3-2) in Sprague–Dawley rats. Food and Chemical Toxicology. 2012;50(9):3256–63. 10.1016/j.fct.2012.06.052 [DOI] [PubMed] [Google Scholar]
- 42.Nielsen N. Soybean seed composition. Soybean: genetics, molecular biology and biotechnology. 1996:127–63. [Google Scholar]
- 43.Kim WS, Jez JM, Krishnan HB. Effects of proteome rebalancing and sulfur nutrition on the accumulation of methionine rich delta-zein in transgenic soybeans. Frontiers in plant science. 2014;5:633 10.3389/fpls.2014.00633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin RC, Liu P-P, Nonogaki H. Simple purification of small RNAs from seeds and efficient detection of multiple microRNAs expressed in Arabidopsis thaliana and tomato (Lycopersicon esculentum) seeds. Seed Science Research. 2005;15(04):319–28. [Google Scholar]
- 45.Li D, Wang L, Liu X, Cui D, Chen T, Zhang H, et al. Deep sequencing of maize small RNAs reveals a diverse set of microRNA in dry and imbibed seeds. PloS one. 2013;8(1):e55107 10.1371/journal.pone.0055107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei H, Zhou B, Zhang F, Tu Y, Hu Y, Zhang B, et al. Profiling and identification of small rDNA-derived RNAs and their potential biological functions. PloS one. 2013;8(2):e56842 10.1371/journal.pone.0056842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nonogaki H. Repression of transcription factors by microRNA during seed germination and postgerminaiton: Another level of molecular repression in seeds. Plant signaling & behavior. 2008;3(1):65–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen R, Hu Z, Zhang H. Identification of microRNAs in wild soybean (Glycine soja). Journal of integrative plant biology. 2009;51(12):1071–9. 10.1111/j.1744-7909.2009.00887.x [DOI] [PubMed] [Google Scholar]
- 49.Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428(6978):84–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(DOCX)
(XLSX)
(DOCX)
(DOCX)
Data Availability Statement
The raw data from the small RNA libraries were deposited at the NCBI Sequence Read Archive (SRA) under accession no. SRP051931.