Abstract
Objective
The aim of this study was to identify early proteomic biomarkers of spontaneous preterm delivery (PTD) in mid-trimester amniotic fluid from asymptomatic women.
Methods
This is a case-cohort study. Amniotic fluid from mid-trimester genetic amniocentesis (14–19 weeks of gestation) was collected from 2008 to 2011. The analysis was conducted in 24 healthy women with subsequent spontaneous PTD (cases) and 40 randomly selected healthy women delivering at term (controls). An exploratory phase with proteomics analysis of pooled samples was followed by a verification phase with ELISA of individual case and control samples.
Results
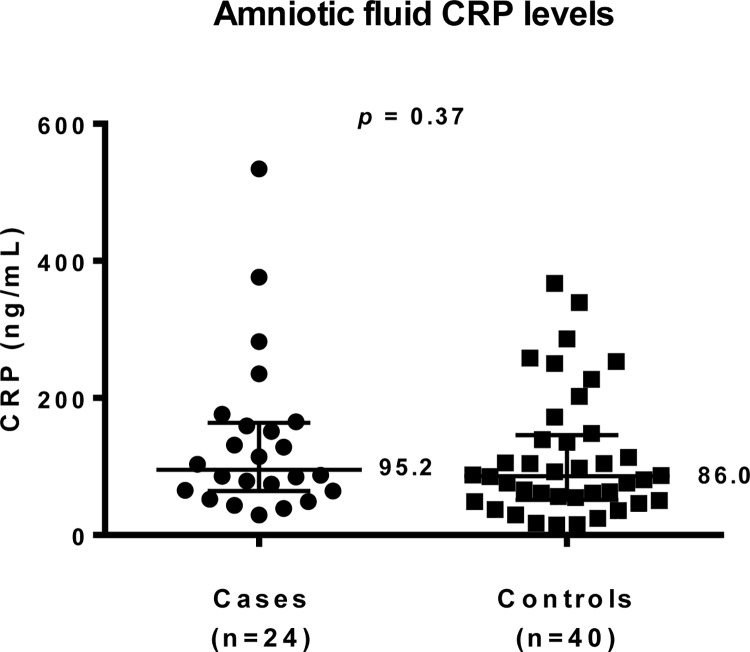
The median (interquartile range (IQR: 25th; 75th percentiles) gestational age at delivery was 35+5 (33+6–36+6) weeks in women with spontaneous PTD and 40+0 (39+1–40+5) weeks in women who delivered at term. In the exploratory phase, the most pronounced differences were found in C-reactive protein (CRP) levels, that were approximately two-fold higher in the pooled case samples than in the pooled control samples. However, we could not verify these differences with ELISA. The median (25th; 75th IQR) CRP level was 95.2 ng/mL (64.3; 163.5) in women with spontaneous PTD and 86.0 ng/mL (51.2; 145.8) in women delivering at term (p = 0.37; t-test).
Conclusions
Proteomic analysis with mass spectrometry of mid-trimester amniotic fluid suggests CRP as a potential marker of spontaneous preterm delivery, but this prognostic potential was not verified with ELISA.
Introduction
Preterm delivery (PTD) is a current global concern in obstetric and neonatal care [1, 2]. It is related to short- and long-term morbidity in neonates [3] and is a leading cause of child death worldwide. Approximately two-thirds of PTDs are spontaneous [4]. The etiology behind spontaneous PTD is complex, and understanding of the sequence and timing of events preceding the condition is incomplete [5].
Proteomics, one of the most promising platforms for biomarker detection, provides insight into the basic biological mechanisms involved in a condition. It constitutes an alternative unbiased approach, compared to the widely used hypothesis-based biomarker discovery approach [6, 7]. Previous studies have explored the maternal, fetal [8–13] and amniotic fluid proteomes and their associations with intra-amniotic inflammation, intra-amniotic infection and PTD in women with preterm labor or preterm prelabor rupture of membranes [8, 10, 11, 14–21]. However, there are only a few published studies exploring the potential of proteomics early in pregnancy, before onset of symptoms. To the best of our knowledge, Fotopoulou et al. [15] are the only researchers who have investigated the protein composition in mid-trimester amniotic fluid in relation to spontaneous PTD, using mass spectrometry profiling. However, their results have not been verified, as is considered mandatory for proteomic analysis [22], representing a considerable limitation.
Due to the complexity and incompletely identified pathophysiological pathways of spontaneous PTD, clinical applications of proteomic technology are still in the early stages [5]. Moreover, the translation of proteomic knowledge into the clinical setting requires verification with an easier, rapid, cost-effective method.
Identification of early prognostic or diagnostic biomarkers, before onset of clinical symptoms, is important. The main aim of this study was therefore to explore potential early biomarkers for spontaneous PTD during the mid-trimester of pregnancy in an exploratory proteomics phase with a pooled sample strategy, utilizing liquid chromatography-tandem mass spectrometry (LC-MS/MS). The second aim was to verify the difference in candidate protein levels found between cases and controls, using ELISA in individual samples from the same cohort.
Materials and Methods
Study design
This study was a case-cohort study of women, aged over 18, who underwent a mid-trimester transabdominal genetic amniocentesis at 14–19 weeks of gestation in a viable singleton pregnancy. Amniocentesis indications were advanced maternal age, anxiety, abnormal first-trimester combined screening or family history of chromosomal abnormalities or genetic diseases. Age under 18 years, multiple pregnancy, positive HIV or hepatitis B test and known or suspected fetal malformation were exclusion criteria. Women who could not understand the written and oral information in Swedish, who declined participation or from whom insufficient amniotic fluid was retrieved at amniocentesis were also excluded.
After medical review, women with chronic diseases (e.g. severe rheumatism, hypo- or hyperthyroidism, severe asthma, diabetes mellitus, hypertension, multiple sclerosis, hereditary chromosomal defects, vitamin D deficiency and severe neurological disorders) were excluded from analysis.
Women with a subsequent spontaneous PTD were compared with women who delivered at term. For the sake of homogeneity, the term delivery group was limited to women giving birth at gestational weeks 38+0 to 41+6, among whom a random selection was made in order to constitute the control group. Medical records were scrutinized at inclusion and after delivery.
Sample collection
An additional 3 mL of amniotic fluid was collected during mid-trimester genetic transabdominal amniocentesis and immediately stored at 4-8°C. The samples were centrifuged for 20 minutes at 12°000 g, at 4°C. The supernatant was separated from the pellet and divided into aliquots that were frozen and stored at -80°C. All samples were handled according to a standardized protocol at the same laboratory.
Exploratory proteomics phase: sample preparation and LC-MS/MS analysis
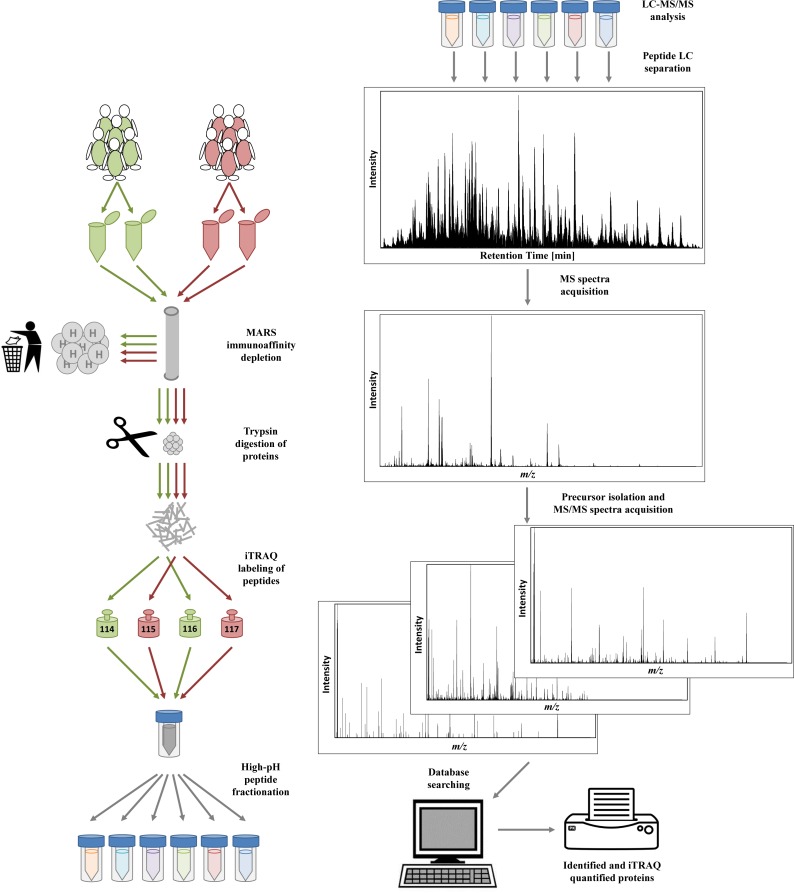
For the exploratory proteomic analysis, pooled amniotic fluid supernatant samples from cases and controls were prepared according to the approach reported by Tambor et al. [23]. Briefly, the samples were supplemented with protease inhibitors and filtered. An equal amount of protein was taken from each sample to create a pooled representative control sample and a representative case sample; both were created in duplicate. The most abundant proteins in the samples were removed using a MARS Hu-14 immunoaffinity column (Agilent, Palo Alto, CA). The proteins were then reduced with tris (2-carboxyethyl) phosphine hydrochloride and the thiol groups were blocked with methyl methane thiosulfonate. Proteins were digested with trypsin at 37°C overnight. The peptides in the representative control samples were labeled with isobaric tags for relative and absolute quantification using iTRAQ 114 and 116 tags, while representative case samples were labeled with iTRAQ 115 and 117 tags (AB Sciex, Foster City, CA). High-pH reversed-phase chromatography was then employed to fractionate the peptides into six 2-minute fractions. An UltiMate 3000 RSLCnano system hyphenated to a Q-Exactive mass spectrometer (Thermo Scientific, Bremen, Germany) was used for protein identification and quantification, using the full MS/Top10 experimental setup. The recorded HCD fragmentation spectra were evaluated in Proteome Discoverer software (Thermo Scientific), using MASCOT (Matrix Science, London, UK) against the human UniProt database. When it came to quantification, only proteins up to 1% FDR threshold were reported and only proteins quantified with at least three peptides were evaluated. The most significantly regulated proteins were found by global ranking method [24]. For both duplicates, 25 proteins were evaluated from the top and from the bottom of list of proteins ranked based on the relative quantitative change (Fig 1). Further specific details regarding the exploratory proteomics phase can be found in the supporting materials and methods (S1 File).
Fig 1. Graphic presentation of the methodology of sample processing and LC-MS/MS analysis of pooled case and control samples.
Verification of amniotic fluid CRP levels by ELISA immunoassay
Amniotic fluid C-reactive protein (CRP) levels were determined in individual case samples and control samples with commercially available ELISA kits (Quantikine ELISA, R & D Systems, Minneapolis, MN). All samples were initially diluted 1/100, but if the subsequent absorbance was below the detection limit for the assay, a 1/10 dilution was used instead. The detection range of the assay was 0.78–50 ng/mL, but dilutions accommodated a practical detection range of 7.8–5000 ng/mL. All of the samples were run in duplicate. The interassay coefficient of variation (CV) was <15%, and the intraassay CV was <5% on all three plates. If duplicates varied more than 15%, the samples were rerun. The resulting absorbance was read at 450 nm (Multiskan FC Microplate Photometer).
Ethics statement
This study was approved by the Central Ethical Review Board at the University of Gothenburg, Sweden (Dnr. Ö 639–03, T 318–08, T 694–11, T 958–11). Written informed consent was obtained from all participants.
Statistical analyses
Continuous variables were compared using a nonparametric Mann-Whitney U Test and presented as median and interquartile range (IQR: 25th; 75th percentiles). Categorical variables were compared using Fisher’s Exact Test and presented as the number (%). Differences were considered statistically significant at p < 0.05 using a two-sided alternative hypothesis. All statistical analyses were performed with SPSS 20.0 for Windows XP OS (SPSS Inc., USA).
Results
Characteristics of the entire study group
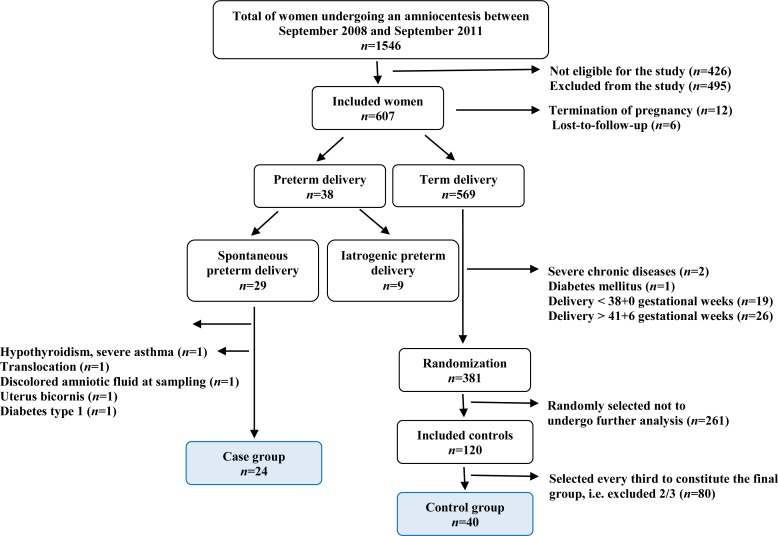
From September 2008 to September 2011, 1546 women underwent mid-trimester amniocentesis at Sahlgrenska University Hospital/Östra, Gothenburg, Sweden. Of this population, 426 women did not fulfill the strict inclusion criteria and 495 were excluded, the majority of whom because they declined participation. An additional 18 women were excluded from analysis; twelve requested a termination of pregnancy due to chromosomal abnormality and six were lost to follow-up.
The remaining women (n = 607) were included in the study. Their medical records were scrutinized at inclusion and after delivery. PTD occurred in 6.3% (38/607), of which 4.8% (29/607) were spontaneous PTD and 1.5% (9/607) were iatrogenic PTD. Of the group with spontaneous PTD, five women were excluded due to hypothyroidism, severe asthma, bicornuate uterus, type 1 diabetes, translocation and/or discolored amniotic fluid at sampling. Therefore, the remaining women with spontaneous PTD (n = 24) constitute the cases in this study.
Of the 569 women who delivered at ≥37 gestational weeks, 381 were healthy and gave birth at gestational week 38+0 to 41+6. Forty were randomly selected and they constituted the final control group in this study (Fig 2).
Fig 2. Flow chart showing selection of study participants.
The respective maternal and neonatal characteristics of the spontaneous PTD and term delivery groups are presented in Table 1. There were no significant differences between the groups, except for the obvious difference in birth weight.
Table 1. Maternal and neonatal characteristics in the group of women with spontaneous preterm delivery and the group of women with term delivery.
| Variable | Spontaneous preterm delivery (n = 24) | Term delivery (n = 40) | p |
|---|---|---|---|
| Gestational age at delivery (weeks+days) | 35+5 (33+6–36+6) | 40+0 (39+1–40+5) | |
| Maternal age at sampling (years) | 37 (36–40) | 36 (35–38) | 0.69 |
| Nulliparous | 9 (37.5%) | 12 (30.0%) | 0.59 |
| IVF | 4 (16.7%) | 1 (2.5%) | 0.06 |
| Maternal BMI at first prenatal visit | 24.6 (23.6–26.9) | 23.5 (21.2–26.3) | 0.23 |
| Smoking at first prenatal visit | 2 (8.3%) | 2 (5.0%) | 0.63 |
| Previous preterm delivery | 3 (12.5%) | 1 (2.5%) | 0.14 |
| Gestational age at sampling (weeks+days) | 15+5 (15+2–16+3) | 15+5 (15+2–16+2) | 0.69 |
| Mode of delivery | |||
| Vaginal delivery | 21 (87.5%) | 28 (70.0%) | 0.14 |
| Caesarean section | 3 (12.5%) | 8 (20.0%) | 0.51 |
| Vacuum extraction | 0 (0%) | 4 (10.0%) | 0.29 |
| Birth weight (grams) | 2505 (2378–2848) | 3530 (3215–3713) | <0.0001 |
| Gender | 0.18 | ||
| Male | 9 (37.5%) | 21 (52.5%) | |
| Female | 15 (62.5%) | 19 (47.5%) | |
| Apgar score < 7 at 5 min | 0 (0%) | 1 (2.6%) | 0.62 |
Continuous variables were compared using a nonparametric Mann-Whitney U Test and presented as the median (interquartile range). Categorical variables were compared using Fisher’s Exact Test and presented as the number (%).
Exploratory proteomics phase: LC-MS/MS analysis
In the exploratory phase, a total of 77411 spectra were searched, and an amino sequence was assigned in 24641 spectra (1% false discovery rate (FDR)), resulting in the identification of 7267 distinct peptides and 1088 protein groups. Of the protein groups, 594 were quantified with at least three peptides. Of these, 17 proteins were reproducibly upregulated (FDR 0.029) and 19 proteins were downregulated (FDR 0.026), as shown in S1 and S2 Tables. Five upregulated and five downregulated proteins were among top 10 dysregulated proteins independently on the direction of the change (Table 2). The most pronounced difference between levels in the pooled control and pooled case samples was found in the case of CRP (P02741, CRP), which was selected for further verification with ELISA.
Table 2. The ten most dysregulated proteins in both duplicates (115/114 and 117/116) from the exploratory proteomics analysis, where 115 and 117 represent the channels for the cases and 114 and 116 represent the channels for the controls.
| Prot. Acc. # | Gene | Description | 115/114 | 117/116 |
|---|---|---|---|---|
| P02741 | CRP | C-reactive protein | 2.27 | 2.30 |
| P60174 | TPI1 | Triosephosphate isomerase | 2.26 | 1.95 |
| A8K7I4 | CLCA1 | Calcium-activated chloride channel regulator 1 | 1.75 | 1.98 |
| P40925 | MDH1 | Malate dehydrogenase, cytoplasmic | 1.86 | 1.63 |
| P01037 | CST1 | Cystatin-SN | 1.46 | 1.51 |
| P02042 | HBD | Hemoglobin subunit delta | 0.56 | 0.52 |
| P69905 | HBA1 | Hemoglobin subunit alpha | 0.60 | 0.51 |
| P68871 | HBB | Hemoglobin subunit beta | 0.61 | 0.52 |
| P09466 | PAEP | Glycodelin | 0.61 | 0.59 |
| P49913 | CAMP | Cathelicidin antimicrobial peptide | 0.65 | 0.71 |
As reported in the table, five upregulated and five downregulated proteins were among top 10 dysregulated proteins independently on the direction of the change. The level of CRP was roughly two-fold higher in the pooled samples from cases (115, 117), compared to the pooled samples from controls (114, 116).
Verification of amniotic fluid CRP level difference from the exploratory phase
In the exploratory proteomics analysis, CRP levels were approximately two-fold higher in cases than in controls in both duplicates. We attempted to verify this difference in the same women, but with an individual ELISA analysis of each case and control sample. The median (IQR: 25th; 75th percentiles) CRP level (ng/mL) in the spontaneous PTD group was 95.2 (64.3; 163.5) compared to 86.0 (51.2; 145.8) in the term delivery group, a difference that was not significant (p = 0.37) (Fig 3).
Fig 3. A two-column scatter graph of median (IQR: 25th; 75th percentiles) amniotic fluid CRP levels in the groups.
Discussion
The exploratory proteomics phase of this study, analyzing pooled samples from mid-trimester amniotic fluid in asymptomatic women, revealed rather few dysregulated proteins. CRP was selected for verification since it had the most pronounced difference in levels between the control and case groups. Furthermore, it was the subject of previous publications and was readily available as a commercial human ELISA test. However, the case-control CRP level difference we initially found with proteomics, and the related prognostic potential, was not verified on subsequent individual analysis with ELISA.
CRP is a very well described marker of systemic inflammation [25] and it is associated with both PTD and preterm prelabor rupture of membranes [26, 27]. Several studies have previously performed targeted analyses of CRP in mid-trimester amniotic fluid [26, 28–31], with contradictory results. While three studies [28, 29, 31] did not find any significant differences between amniotic fluid CRP levels in cases of spontaneous PTD and term deliveries, Ozer et al. [30] and Ghezzi et al. [26] did find significant differences. These conflicting results highlight our effort to analyze this protein using a different but complementary approach.
The mid-trimester amniotic fluid proteome has been explored in relation to spontaneous PTD in twins [32] but there is, to our knowledge, only one published study exploring mid-trimester amniotic fluid as a predictor of spontaneous PTD in singletons. Fotopoulou et al. [15] investigated a small group of women with both spontaneous PTD and term deliveries. Using a combination of mass spectrometry and different solid-phase chromatography protein chips, they identified seven clusters that differed significantly when women with a subsequent spontaneous PTD were compared with women who delivered at term. However, the individual proteins/peptides could not be identified by SELDI TOF analysis.
A major strength of our study is the verification phase, otherwise frequently neglected. The standardized protocol for sample handling and the strict selection criteria for the case and control groups are additional strengths. Furthermore, this study had a relatively large cohort of healthy, asymptomatic women with subsequent spontaneous PTD and a very low lost-to-follow-up rate.
An important limitation of the study was that one single biomarker (CRP) was selected for further verification. We refrained from considering other potential biomarkers that had, in contrast to CRP, not been the objects of previous published mid-trimester studies. A pooling strategy provides an initial proteomic evaluation, albeit not as expensive or time-consuming as proteomic analysis of each and every individual sample. However, the pooled sample strategy inevitably has certain limitations. One such limitation is the absence of estimates of individual variability, required to facilitate significance testing. We hypothesized that the discrepancy between the exploratory phase and the verification stage might have been caused by the fact that some samples may have contained extremely low/high analyte levels (outliers) and may thus have shifted levels in the entire pool. The key prerequisite if samples are pooled is thus results verification in individual samples with an alternative method, which we attempted using ELISA. Finally, another limitation of this study might be that the spontaneous PTD group consisted of women with both preterm labor and preterm prelabor rupture of membranes; the respective phenotypes may differ somewhat.
Conclusions
The exploratory proteomic phase of the experiment revealed six dysregulated proteins, of which CRP levels were approximately two-fold higher among women with spontaneous PTD, compared to controls. However, when individual cases and controls were examined with ELISA, there was no significant difference in CRP levels between the case and control groups. This finding is interesting in light of how extensively this test is used, and confirms the current ambiguity concerning its status. Further proteomics studies are needed, on individual samples and with similar strict criteria for the groups as in this study.
Supporting Information
(DOCX)
The proteins are ranked according to the absolute value of log2 of average of 115/114 and 117/116 ratio.
(DOCX)
The proteins are ranked according to the absolute value of log2 of average of 115/114 and 117/116 ratio.
(DOCX)
Acknowledgments
The authors would like to acknowledge the Antenatal Clinic at the Department of Obstetrics and Gynecology, Sahlgrenska University Hospital/Östra, Gothenburg, Sweden for their help with recruitment and sampling and the staff at the Perinatal Laboratory for their help with recruitment and processing of samples.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The research underlying this study has been funded by grants as detailed below: The Health & Medical Care Committee of the Regional Executive Board, Region Västra Götaland, Sweden, Grant numbers: VGFOUREG-231311, VGFOUREG-308151 and VGFOUREG-368351 (URL: http://www.fou.nu/is/vgregion); Agreement concerning research and education of doctors, Sweden, Grant numbers: ALFGBG-136431, ALFGBG-426411 and ALFGBG-507701 2 (URL: http://www.fou.nu/is/alfgbg/); Sahlgrenska University Hospital Foundations, No specific grant number (URL: https://www.sahlgrenska.se/sv/SU/Forskning/Stiftelser-och-gavor/); Linnéa and Josef Carlsson’s Foundation, Sweden, Grant numbers: 2011:1 nr:21, 2012 nr:15 and 2014 nr:12 (URL: http://www.carlssonsstiftelse.se/); and The Internal Grant Agency of the Ministry of Health, Czech Republic, Grant number: NT/13599 (URL: http://www.cuni.cz/UKEN-65.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bryce J, Boschi-Pinto C, Shibuya K, Black RE. WHO estimates of the causes of death in children. Lancet. 2005;365(9465):1147–52. [DOI] [PubMed] [Google Scholar]
- 2.Goldenberg RL, Andrews WW, Hauth JC. Choriodecidual infection and preterm birth. Nutrition reviews. 2002;60(5 Pt 2):S19–25. [DOI] [PubMed] [Google Scholar]
- 3.Woodward LJ, Moor S, Hood KM, Champion PR, Foster-Cohen S, Inder TE, et al. Very preterm children show impairments across multiple neurodevelopmental domains by age 4 years. Archives of disease in childhood Fetal and neonatal edition. 2009;94(5):F339–44. 10.1136/adc.2008.146282 [DOI] [PubMed] [Google Scholar]
- 4.Morken NH, Kallen K, Hagberg H, Jacobsson B. Preterm birth in Sweden 1973–2001: rate, subgroups, and effect of changing patterns in multiple births, maternal age, and smoking. Acta obstetricia et gynecologica Scandinavica. 2005;84(6):558–65. [DOI] [PubMed] [Google Scholar]
- 5.Kacerovsky M, Lenco J, Musilova I, Tambor V, Lamont R, Torloni MR, et al. Proteomic biomarkers for spontaneous preterm birth: a systematic review of the literature. Reproductive sciences. 2014;21(3):283–95. 10.1177/1933719113503415 [DOI] [PubMed] [Google Scholar]
- 6.Dasilva N, Diez P, Matarraz S, Gonzalez-Gonzalez M, Paradinas S, Orfao A, et al. Biomarker discovery by novel sensors based on nanoproteomics approaches. Sensors. 2012;12(2):2284–308. 10.3390/s120202284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kacerovsky M, Andrys C, Drahosova M, Musilova I, Hornychova H, Lesko D, et al. Soluble Toll-like receptor 1 family members in the amniotic fluid of women with preterm prelabor rupture of the membranes. The journal of maternal-fetal & neonatal medicine: the official journal of the European Association of Perinatal Medicine, the Federation of Asia and Oceania Perinatal Societies, the International Society of Perinatal Obstet. 2012;25(9):1699–704. [DOI] [PubMed] [Google Scholar]
- 8.Buhimschi CS, Bhandari V, Hamar BD, Bahtiyar MO, Zhao G, Sfakianaki AK, et al. Proteomic profiling of the amniotic fluid to detect inflammation, infection, and neonatal sepsis. PLoS medicine. 2007;4(1):e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buhimschi CS, Dulay AT, Abdel-Razeq S, Zhao G, Lee S, Hodgson EJ, et al. Fetal inflammatory response in women with proteomic biomarkers characteristic of intra-amniotic inflammation and preterm birth. BJOG: an international journal of obstetrics and gynaecology. 2009;116(2):257–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buhimschi IA, Christner R, Buhimschi CS. Proteomic biomarker analysis of amniotic fluid for identification of intra-amniotic inflammation. BJOG: an international journal of obstetrics and gynaecology. 2005;112(2):173–81. [DOI] [PubMed] [Google Scholar]
- 11.Romero R, Espinoza J, Gotsch F, Kusanovic JP, Friel LA, Erez O, et al. The use of high-dimensional biology (genomics, transcriptomics, proteomics, and metabolomics) to understand the preterm parturition syndrome. BJOG: an international journal of obstetrics and gynaecology. 2006;113 Suppl 3:118–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romero R, Kusanovic JP, Gotsch F, Erez O, Vaisbuch E, Mazaki-Tovi S, et al. Isobaric labeling and tandem mass spectrometry: a novel approach for profiling and quantifying proteins differentially expressed in amniotic fluid in preterm labor with and without intra-amniotic infection/inflammation. The journal of maternal-fetal & neonatal medicine: the official journal of the European Association of Perinatal Medicine, the Federation of Asia and Oceania Perinatal Societies, the International Society of Perinatal Obstet. 2010;23(4):261–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein J, Buffin-Meyer B, Mullen W, Carty DM, Delles C, Vlahou A, et al. Clinical proteomics in obstetrics and neonatology. Expert review of proteomics. 2014;11(1):75–89. 10.1586/14789450.2014.872564 [DOI] [PubMed] [Google Scholar]
- 14.Buhimschi IA, Zhao G, Rosenberg VA, Abdel-Razeq S, Thung S, Buhimschi CS. Multidimensional proteomics analysis of amniotic fluid to provide insight into the mechanisms of idiopathic preterm birth. PloS one. 2008;3(4):e2049 10.1371/journal.pone.0002049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fotopoulou C, Kyeyamwa S, Linder M, Thieme D, Hartenstein S, Klein O, et al. Proteomic analysis of midtrimester amniotic fluid to identify novel biomarkers for preterm delivery. The journal of maternal-fetal & neonatal medicine: the official journal of the European Association of Perinatal Medicine, the Federation of Asia and Oceania Perinatal Societies, the International Society of Perinatal Obstet. 2012;25(12):2488–93. [DOI] [PubMed] [Google Scholar]
- 16.Mercer BM, Crouse DT, Goldenberg RL, Miodovnik M, Mapp DC, Meis PJ, et al. The antibiotic treatment of PPROM study: systemic maternal and fetal markers and perinatal outcomes. American journal of obstetrics and gynecology. 2012;206(2):145 e1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tambor V, Fucikova A, Lenco J, Kacerovsky M, Rehacek V, Stulik J, et al. Application of proteomics in biomarker discovery: a primer for the clinician. Physiological research / Academia Scientiarum Bohemoslovaca. 2010;59(4):471–97. [DOI] [PubMed] [Google Scholar]
- 18.Tambor V, Kacerovsky M, Lenco J, Bhat G, Menon R. Proteomics and bioinformatics analysis reveal underlying pathways of infection associated histologic chorioamnionitis in pPROM. Placenta. 2013;34(2):155–61. 10.1016/j.placenta.2012.11.028 [DOI] [PubMed] [Google Scholar]
- 19.Vuadens F, Benay C, Crettaz D, Gallot D, Sapin V, Schneider P, et al. Identification of biologic markers of the premature rupture of fetal membranes: proteomic approach. Proteomics. 2003;3(8):1521–5. [DOI] [PubMed] [Google Scholar]
- 20.Gravett MG, Novy MJ, Rosenfeld RG, Reddy AP, Jacob T, Turner M, et al. Diagnosis of intra-amniotic infection by proteomic profiling and identification of novel biomarkers. JAMA: the journal of the American Medical Association. 2004;292(4):462–9. [DOI] [PubMed] [Google Scholar]
- 21.Ruetschi U, Rosen A, Karlsson G, Zetterberg H, Rymo L, Hagberg H, et al. Proteomic analysis using protein chips to detect biomarkers in cervical and amniotic fluid in women with intra-amniotic inflammation. Journal of proteome research. 2005;4(6):2236–42. [DOI] [PubMed] [Google Scholar]
- 22.Bea JW, Wright NC, Thompson P, Hu C, Guerra S, Chen Z. Performance evaluation of a multiplex assay for future use in biomarker discovery efforts to predict body composition. Clinical chemistry and laboratory medicine: CCLM / FESCC. 2011;49(5):817–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tambor V, Kacerovsky M, Andrys C, Musilova I, Hornychova H, Pliskova L, et al. Amniotic fluid cathelicidin in PPROM pregnancies: from proteomic discovery to assessing its potential in inflammatory complications diagnosis. PloS one. 2012;7(7):e41164 10.1371/journal.pone.0041164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Cras-Meneur C, Ohsugi M, Stormo GD, Permutt MA. A global approach to identify differentially expressed genes in cDNA (two-color) microarray experiments. Bioinformatics. 2007;23(16):2073–9. [DOI] [PubMed] [Google Scholar]
- 25.Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. The New England journal of medicine. 1999;340(6):448–54. [DOI] [PubMed] [Google Scholar]
- 26.Ghezzi F, Franchi M, Raio L, Di Naro E, Bossi G, D'Eril GV, et al. Elevated amniotic fluid C-reactive protein at the time of genetic amniocentesis is a marker for preterm delivery. American journal of obstetrics and gynecology. 2002;186(2):268–73. [DOI] [PubMed] [Google Scholar]
- 27.Gomez R, Ghezzi F, Romero R, Munoz H, Tolosa JE, Rojas I. Premature labor and intra-amniotic infection. Clinical aspects and role of the cytokines in diagnosis and pathophysiology. Clinics in perinatology. 1995;22(2):281–342. [PubMed] [Google Scholar]
- 28.Borna S, Mirzaie F, Abdollahi A. Mid-trimester amniotic fluid C-reactive protein, ferritin and lactate dehydrogenase concentrations and subsequent risk of spontaneous preterm labour. Aust N Z J Obstet Gynaecol. 2009;49(4):400–3. 10.1111/j.1479-828X.2009.01019.x [DOI] [PubMed] [Google Scholar]
- 29.Kim A, Lee ES, Shin JC, Kim HY. Identification of biomarkers for preterm delivery in mid-trimester amniotic fluid. Placenta. 2013. [DOI] [PubMed] [Google Scholar]
- 30.Ozer KT, Kavak ZN, Gokaslan H, Elter K, Pekin T. Predictive power of maternal serum and amniotic fluid CRP and PAPP-A concentrations at the time of genetic amniocentesis for the preterm delivery. European journal of obstetrics, gynecology, and reproductive biology. 2005;122(2):187–90. [DOI] [PubMed] [Google Scholar]
- 31.Tarim E, Bagis T, Kilicdag EB, Sezgin N, Yanik F. Are amniotic fluid C-reactive protein and glucose levels, and white blood cell counts at the time of genetic amniocentesis related with preterm delivery? Journal of perinatal medicine. 2005;33(6):524–9. [DOI] [PubMed] [Google Scholar]
- 32.Lee SM, Park JS, Norwitz ER, Oh S, Kim EJ, Kim SM, et al. Mid-trimester amniotic fluid pro-inflammatory biomarkers predict the risk of spontaneous preterm delivery in twins: a retrospective cohort study. Journal of perinatology: official journal of the California Perinatal Association. 2015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
The proteins are ranked according to the absolute value of log2 of average of 115/114 and 117/116 ratio.
(DOCX)
The proteins are ranked according to the absolute value of log2 of average of 115/114 and 117/116 ratio.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.