Abstract
Cotton leaf curl disease (CLCuD) is the major biotic constraint to cotton production on the Indian subcontinent, and is caused by monopartite begomoviruses accompanied by a specific DNA satellite, Cotton leaf curl Multan betasatellite (CLCuMB). Since the breakdown of resistance against CLCuD in 2001/2002, only one virus, the “Burewala” strain of Cotton leaf curl Kokhran virus (CLCuKoV-Bur), and a recombinant form of CLCuMB have consistently been identified in cotton across the major cotton growing areas of Pakistan. Unusually a bipartite isolate of the begomovirus Tomato leaf curl virus was identified in CLCuD-affected cotton recently. In the study described here we isolated the bipartite begomovirus Tomato leaf curl New Delhi virus (ToLCNDV) from CLCuD-affected cotton. To assess the frequency and geographic occurrence of ToLCNDV in cotton, CLCuD-symptomatic cotton plants were collected from across the Punjab and Sindh provinces between 2013 and 2015. Analysis of the plants by diagnostic PCR showed the presence of CLCuKoV-Bur in all 31 plants examined and ToLCNDV in 20 of the samples. Additionally, a quantitative real-time PCR analysis of the levels of the two viruses in co-infected plants suggests that coinfection of ToLCNDV with the CLCuKoV-Bur/CLCuMB complex leads to an increase in the levels of CLCuMB, which encodes the major pathogenicity (symptom) determinant of the complex. The significance of these results are discussed.
Introduction
Cotton is an important commodity and the export of cotton products is crucial for the economies of India and, especially, Pakistan. The cultivation of cotton across Pakistan and northwestern India is severely affected by cotton leaf curl disease (CLCuD) [1, 2]. The disease first came to prominence in the late 1980s near the city of Multan, Pakistan, and rapidly spread to almost all cotton growing areas of the country and into northwestern India. In the late 1990s cotton varieties obtained by conventional breeding and selection were introduced and rapidly restored production of cotton in Pakistan to the levels before the CLCuD epidemic. Unfortunately the disease appeared on all previously resistant varieties from 2001/2002 onwards. It was first observed near the town of Burewala Pakistan, indicating that the resistance had been broken [3]. This led to a second epidemic which rapidly spread to most areas of Pakistan and northwestern India.
Viruses of the genus Begomovirus are whitefly (Bemisia tabaci)-transmitted single-stranded (ss)DNA viruses that belong to family Geminiviridae. Begomoviruses occur in all the warmer parts of the World and infect only dicotyledonous plants [4]. In the Old World (OW) a small number of begomoviruses have been identified with genomes consisting of two components, known as DNA-A and DNA B. The majority of begomoviruses in the OW have a genome consisting of only a single component, homologous to the DNA-A component of bipartite viruses. The opposite is true in the New World where only one native monopartite begomovirus has been identified so far [5, 6]. The genomes of monopartite and DNA-A components of bipartite begomoviruses originating from the OW encode the coat protein (CP) and (A)V2 protein in the virion-sense orientation and the replication-associated protein (Rep; a rolling circle replication initiator protein), the replication enhancer protein (REn), the transcriptional activator protein (TrAP) and the C4 protein in the complementary-sense orientation [7]. DNA-B components encode the nuclear shuttle protein (NSP) and movement protein (MP) in the virion- and complementary-sense, respectively. The reading frames in the virion- and complementary-sense of begomovirus genomes/genomic components are separated by a non-coding (intergenic) region which contains cis-acting regulatory elements for gene expression, a predicted hairpin structure containing the conserved (among most geminiviruses) nonanucleotide sequence TAATATTAC as part of the loop and small repeated sequences, known as “iterons”, which are sequence specific binding sites for Rep. Together the iterons and hairpin form the origin of replication (ori) for virion-sense viral DNA replication. The sequence specific interaction between Rep and cognate iterons also ensures that the Rep of one virus will not initiate replication of the genome of a second virus. The DNA-A and DNA-B components of bipartite begomoviruses share a sequence, known as the common region (CR) that usually spans most of the intergenic region. The CR acts to maintain the integrity of the split genome ensuring that the DNA-A-encoded Rep can initiate replication of the virion strands for both components [8].
The majority of monopartite begomoviruses are associated with additional small ssDNA molecules known as betasatellites and alphasatellites [9]. Betasatellites (previously known as DNA-β) have so far only been identified in the OW. They are half the size of begomovirus components (∼1350 nt) and encode a single gene on the complementary-sense strand that codes for an ∼118 amino acid protein known as βC1. Betasatellites mayincrease the accumulation of their helper begomoviruses, as well as enhance symptoms in some host plants [10, 11]. This is likely due to βC1 having suppressor of RNA interference activity [12, 13].
The alphasatellites (previouslyknownasDNA-1; [14]) are not strict satellites, since they are capable of autonomous-replication in permissive plant cells. They are dependent on their helper begomoviruses for movement within plants and insect transmission between plants [15, 16]. Although widespread in the OW, alphasatellites have also been identified in the NW in association with bipartite begomoviruses, in the absence of betasatellites [17, 18]. Recently an alphasatellite and a betasatellite were shown in association with a mastrevirus (genus Mastrevirus, family Geminiviridae) [19].
CLCuD in Pakistan and northwestern India during the 1990s was shown to be associated with at least four monopartite begomoviruses including Cotton leaf curl Multan virus (CLCuMuV), Cotton leaf curl Alabad virus (CLCuAV), Cotton leaf curl Kokhran virus (CLCuKoV) and Papaya leaf curl virus (PaLCuV) [20, 21]. Of these only CLCuMuV, CLCuKoV and PaLCuV have been shown experimentally to cause CLCuD in cotton in the presence of a distinct betasatellite—Cotton leaf curl Multan betasatellite (CLCuMB)[10, 21]. After the breakdown of resistance in 2001–2002, CLCuD across the Punjab province of Pakistan was shown to be associated with a single monopartite begomovirus; the “Burewala” strain of CLCuKoV (CLCuKoV-Bu; previously called Cotton leaf curl Burewala virus). CLCuKoV-Bu is a recombinant virus with some sequences derived from CLCuMuV [22]. Unusually CLCuKoV-Bu associated with resistance breaking lacked one of the usual complement of genes [22, 23] and was associated with a recombinant form of CLCuMB (CLCuMBBur) with some sequence derived from another betasatellite [24].
Tomato leaf curl New Delhi virus (ToLCNDV) is an unusual begomovirus. It is one of very few bipartite begomoviruses in the OW and has been reported from a large number of different plants, including members of the Solanaceae, Cucurbitaceae and Malvaceae [24–28]. The virus is also unusual in sharing its DNA-B component with a number of other bipartite begomoviruses, including Pepper leaf curl Bangladesh virus[29], Tomato leaf curl Palampur virus[30], Bhendi yellow vein mosaic virus [28] and with Tomato leaf curl virus, a begomovirus for which some isolates are monopartite [31].
Recently we have identified an isolate of ToLCV with a DNA-B in CLCuD-affected cotton plants originating from the Punjab; the first time a bipartite begomovirus was identified in cotton on the Indian subcontinent [32]. The study here reports the occurrence of the bipartite ToLCNDV in CLCuD-symptomatic cotton for the first time. Additionally the widespread presence of ToLCNDV in CLCuD-affected cotton over a wide area of the cotton growing regions of the Punjab and Sindh provinces of Pakistan is shown. The effects of the presence of ToLCNDV on the levels of DNA-B, virus and satellites were investigated by quantitative PCR. The implications of these findings are discussed.
Materials and Methods
Origins of plant materials and DNA extraction
Leaf samples from cotton plants with symptoms typical of CLCuD, consisting of leaf curling, vein thickening, vein yellowing, enations and stunted growth (Fig 1), and from apparently healthy plants,were collected from areas of Punjab and Sindh provinces of Pakistan between2013 and 2015 (Table 1). DNA was extracted from samples using a cetyltrimethyl ammonium bromide (CTAB) method [33]. DNA was quantified using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA USA).
Fig 1. Cotton (Gossypium hirsutum) plant exhibiting typical symptoms of cotton leaf curl disease.
Table 1. Origins offield collected cotton samples and presence of viruses.
| Location | Isolate | GPS coordinates | Virus* | |||
|---|---|---|---|---|---|---|
| Province | District | North | East | ToLCNDV | CLCuKoV | |
| Punjab | Bahawalpur | C140 | 29.59636 | 72.83348 | - | ✓ |
| Punjab | Bahawalpur | C150 | 29.56129 | 72.74316 | - | ✓ |
| Punjab | Bahawalpur | K201 | 29.10163 | 71.7427 | ✓ | ✓ |
| Punjab | Bahawalpur | K202 | 29.10163 | 71.7427 | ✓ | ✓ |
| Punjab | Bahawalpur | K203 | 29.20806 | 71.75604 | ✓ | ✓ |
| Punjab | Bahawalpur | K204 | 29.20806 | 71.75604 | ✓ | ✓ |
| Punjab | Bahawalpur | K206 | 29.17652 | 71.86785 | - | ✓ |
| Punjab | Bahawalpur | K207 | 29.17652 | 71.86785 | ✓ | ✓ |
| Punjab | Bhakkar | D11 | 31.43663 | 71.52154 | - | ✓ |
| Punjab | Bhakkar | D26 | 31.48838 | 71.26929 | - | ✓ |
| Punjab | Faisalabad | C31 | 31.35432 | 73.30094 | - | ✓ |
| Punjab | Khanewal | H38 | 30.40681 | 72.12185 | - | ✓ |
| Punjab | Khanewal | H41 | 30.35511 | 72.08303 | - | ✓ |
| Punjab | Khanewal | H42 | 30.35511 | 72.08303 | - | ✓ |
| Punjab | Toba Tek Singh | H1 | 31.31402 | 72.79922 | - | ✓ |
| Punjab | Toba Tek Singh | H11 | 31.05338 | 72.58824 | - | ✓ |
| Punjab | Toba Tek Singh | H20 | 30.87548 | 72.53648 | ✓ | ✓ |
| Punjab | Toba Tek Singh | H9 | 31.05338 | 72.58824 | ✓ | ✓ |
| Punjab | Faisalabad | NIAB1 | 31.3982 | 73.0339 | ✓ | ✓ |
| Punjab | Faisalabad | NIAB2 | 31.3982 | 73.0339 | ✓ | ✓ |
| Sindh | Ghotki | K123 | 28.10269 | 69.71552 | ✓ | ✓ |
| Sindh | Nawabshah | SS23 | 26.1255 | 68.2804 | ✓ | ✓ |
| Sindh | Nawabshah | SS32 | 26.1255 | 68.2804 | ✓ | ✓ |
| Sindh | Nawabshah | SS33 | 26.1255 | 68.2804 | ✓ | ✓ |
| Sindh | Nawabshah | SS34 | 26.1255 | 68.2804 | ✓ | ✓ |
| Sindh | Nawabshah | SS35 | 26.1255 | 68.2804 | ✓ | ✓ |
| Sindh | Tando Allahyar | SS1 | 25.4799 | 68.7900 | ✓ | ✓ |
| Sindh | Mirpur Khas | SS10 | 25.5101 | 68.9301 | ✓ | ✓ |
| Sindh | Mirpur Khas | SS11 | 25.5101 | 68.9301 | ✓ | ✓ |
| Sindh | Hayderabad | SS16 | 25.4997 | 68.4196 | ✓ | ✓ |
| Sindh | Matiari | SS20 | 25.9299 | 68.3800 | ✓ | ✓ |
*The presence of either Tomato leaf curl New Delhi virus (ToLCNDV) or Cotton leaf curl Kokhran virus (CLCuKoV) was determined by diagnostic PCR with ToLCNDV DNA-B specific primers or with the CLCuKoV specific primers described by Shuja et al.[38].
Ethics statement
The National Institute for Biotechnology and Genetic Engineering (NIBGE) is a public sector institute, the employees of which areauthorized to visit farmer’s fields and collect plant samples. However, before going to any private field, verbal permission was sought from the owner of the field. The field studies did not involve endangered or protected species.
Virus Amplification, Cloning and Sequencing
Rolling circle amplification (RCA) using phi 29 DNA polymerase (ThermoFisher Scientific, Waltham, MA USA) was used to amplify all circular DNA molecules in 31cotton samples [34]. The DNA-A and DNA-B components of ToLCNDV were amplified by polymerase chain reaction (PCR)with RCA-enriched nucleic acid samples as the template and using the specific primer pairsToLCNDV_A1_Forward (GATATCATCATTTCAACGCCCGCATCGAA)/ToLCNDV_A2_Reverse (GATATCTGCTGGTCGCTTCGCCATAGTTC), ToLCNDV_A3_Forward (GAGCTCGTGCAGTTGTCCCCATTGCCCGCGTCAC)/ToLCNDV_A4_Reverse(GAGCTCCATAGGGGCTGTCGAAGTTG) for DNA-A and ToLCNDV_B1_Forward (AAGCTTCTGCTCGAACATGGACGGAAATGAC)/ToLCNDV_B2_Reverse (AAGCTTAGCCAGTTGAGGAATAGATGCATG), ToLCNDV_B3_Forward (GGTACCCGTAACGATCTTGAACTATGTCCC)/ToLCNDV_B4_Reverse (GGTACCCTATCTGGCTATAGGTCCGAACG) for DNA-B. The degenerate primers BegomoF and BegomoR were used to PCR amplify the genomes of monopartite and/or DNA-A components of bipartite begomoviruses [35]. Betasatellites and alphasatellites were PCR-amplified using universal primers [36, 37]. The primers described by Shuja et al. [38] were used to specifically detect CLCuKoV.
PCR reactions for cloning used RCA product as the template. Amplification products of ~2.8 kb, for virus, DNA-A or DNA-B, and ~1.4 kb for alphasatellite and betasatellite, were cloned into a T/A cloning vector (pTZ57R/T; ThermoFisher Scientific, Waltham, MA USA). From each isolate 3–5 clones were sequenced. Sequences were determined by dideoxy nucleotide chain termination sequencing on an Applied Biosystems 3730XL DNA sequencer and were assembled and manipulated using the Lasergene package of sequence analysis software (DNAStar Inc., Madison, WI, USA). The MUSCLE option of the Sequence Demarcation Tool [39] was used to analyze the sequenced clones for the identification of distinct geminivirus species according to revised taxonomy of begomoviruses based on pairwise sequence comparisons [40]. Sequences were aligned using CLUSTAL W [41] implemented in MEGA6 [42]. Phylogenetic analyses were conducted on aligned sequences using the neighbor-joining and bootstrap options of CLUSTAL X and visualized in TreeView [43].
Quantitative real-time PCR
Real-time PCR reactions consisted of a total volume of 25 μL with 12.5 μL of SYBER Green Super Mix (Thermo Fisher Scientific, Waltham, MA USA), 0.25 μL of each primer (0.1 μM each), 2.5 μL of DNA (25 ng), and 9.5 μL water. The cycling conditions were an initial 94°C for 10 min, followed by 40 cycles of 30 seconds (s) at 94°C, 30 s at 57°C, 30 s at 72°C, followed by melt curve analyses. Reactions were performed in a 96 well microtitre plate format using an iQ5 thermal cycler (Bio-Rad, Hercules, CA USA). The 18S ribosomal RNA gene was used as a reference gene to normalize DNA levels in samples. Each sample was run in triplicate.
Standard curves for absolute quantification were obtained from five sets of tenfold serial dilutions (starting from 20 ng/μL) of a plasmid containing the cloned full-length CLCuMB (AJ298903; [10]) ToLCNDV DNA-A (U150150) and DNA-B (U15017; [44]) were dissolved in 20 ng/μL of genomic DNA extracted independently from healthy cotton plants, to obtain a range from 20 ng/μL to 0.002 ng/μL of each component. The primers used in the quantitative PCR analyses were DNA_A_qPCR_Forward (CCTTTAATCATGACTGGCTT)/DNA_A_qPCR_Reverse (CATTTCCATCCGAACATTC) for begomovirus genome or DNA-A component, DNA_B_qPCR_Forward (GCCCATGATTCGTTCGGAC)/DNA_B_qPCR_Reverse (CACGTGGTACTGGAATATCGCA) for DNA-B and Betasatellite_qPCR_Forward (GATTTGACTTATATTGGGCCAATTTAAT)/Betasatellite_qPCR_Reverse (GATACTATCCACAAAGTCACCATCGCTAAT) for betasatellites.
Results
Identification of ToLCNDV in CLCuD affected cotton
Clones of ~ 2.8 kb and ~1.4kb were obtained from 10 cotton plants with CLCuD symptoms (Tables 2, 3 and 4). The virus and satellite sequences obtained were analyzed for the presence of potential genes. The analysis showed that the arrangement of genes for 7 clones was typical of either monopartite begomoviruses or the DNA-A component of bipartite begomoviruses (Table 2).
Table 2. Origins and features of monopartitebegomovirus and bipartite begomovirus DNA-A component clones obtained.
| Clone | Virus/virus component | GPS coordinates | Location (province/district) | Accession no. | Size (nt) | Coding sequences [coordinates/no. of amino acids] | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CP | V2 | Rep | TrAP | REn | C4 | ||||||
| SAZ21* | CLCuKoV-Bur | 29.17652N/71.86785E | Punjab/Bahawalpur | LN845931 | 2758 | 291-1061/256 | 131-487/118 | 1504-2595/363 | 1500-1607/35 | 1058-1462/134 | 2241-2681/146 |
| SAZ22* | CLCuKoV-Bur | 29.17652N/71.86785E | Punjab/Bahawalpur | LN845932 | 2758 | 291-1061/256 | 131-487/118 | 1504-2595/363 | 1500-1607/35 | 1058-1462/134 | 2241-2681/146 |
| SAZ26* | CLCuKoV-Bur | 29.17652N/71.86785E | Punjab/Bahawalpur | LN713267 | 2758 | 291-1061/256 | 131-487/118 | 1504-2595/363 | 1500-1607/35 | 1058-1462/134 | 2241-2681/146 |
| SAZ27* | CLCuKoV-Bur | 29.17652N/71.86785E | Punjab/Bahawalpur | LN713268 | 2758 | 291-1061/256 | 131-487/118 | 1504-2595/363 | 1500-1607/35 | 1058-1462/134 | 2241-2681/146 |
| SAZ37* | CLCuKoV-Bur | 29.17652N/71.86785E | Punjab/Bahawalpur | LN713271 | 2759 | 292-1062/256 | 132-488/118 | 1505-2596/363 | 1501-1608/35 | 1059-1463/134 | 2242-2682/146 |
| SAZ33* | CLCuKoV-Bur | 29.10163N/71.7427E | Punjab/Bahawalpur | LN845933 | 2759 | 292-1062/256 | 132-488/118 | 1505-2596/363 | 1501-1608/35 | 1059-1463/134 | 2242-2682/146 |
| SAZ34@ | ToLCNDV DNA-A | 29.17652N/1.86785E | Punjab/Bahawalpur | LN845962 | 2738 | 280-1050/256 | 120-458/112 | 1499-2584/361 | 1177-1596/139 | 1047-1457/136 | 2251-2427/58 |
* Clones produced using primers BegomoF and BegomoR
@ Clone produced using primers ToLCNDV_A3 and ToLCNDV_A4
Table 3. Origins and features of bipartite begomovirus DNA-B component clones obtained.
| Clone | Virus component | Location (province/district) | Accession no. | Size | Coding sequence [coordinates/no. of amino acids] | |
|---|---|---|---|---|---|---|
| MP | NSP | |||||
| SAZ 28 | ToLCNDV DNA-B | Punjab/Bahawalpur | LN713269 | 2695 | 1305-2150/281 | 440-1126/228 |
| SAZ 30 | ToLCNDV DNA-B | Punjab/Bahawalpur | LN845955 | 2693 | 1305-2150/281 | 440-1246/268 |
| SAZ 31 | ToLCNDV DNA-B | Punjab/Bahawalpur | LN713270 | 2692 | 1304-2149/281 | 441-1247/268 |
| SAZ 35 | ToLCNDV DNA-B | Punjab/Bahawalpur | LN845956 | 2692 | 1304-2149/281 | 441-1247/268 |
| SAZ 68 | ToLCNDV DNA-B | Punjab/Bahawalpur | HG983285 | 2688 | 1301-2146/281 | 438-1244/268 |
| SAZ 181 | ToLCNDV DNA-B | Punjab/Bahawalpur | LN845934 | 2687 | 1301-2146/281 | 438-1244/268 |
| SAZ 235 | ToLCNDV DNA-B | Punjab/Bahawalpur | LN854628 | 2694 | 1304-2149/281 | 440-1126/228 |
Table 4. Origins and features of alphasatellite and betasatellite clones obtained.
| Clone | Satellite | Location (province/district) | Accession no. | Size(nt) | Coding sequence [coordinates/no. of amino acids] | |
|---|---|---|---|---|---|---|
| βC1 | Rep | |||||
| SAZ 4 | CLCuMB | Punjab/Bahawalpur | LN845926 | 1370 | 195-551/118 | - |
| SAZ 6 | CLCuMB | Punjab/Bahawalpur | HG934394 | 1369 | 195-551/118 | - |
| SAZ 13 | CLCuMB | Punjab/Khanewal | LN845927 | 1350 | 195-551/118 | - |
| SAZ 14 | CLCuMB | Punjab/Khanewal | LN845928 | 1350 | 195-551/118 | - |
| SAZ 15 | CLCuMB | Punjab/Khanewal | HG934395 | 1350 | 195-551/118 | - |
| SAZ 24 | CLCuMB | Punjab/Khanewal | LN845930 | 1350 | 195-551/118 | - |
| SAZ 25 | CLCuMB | Punjab/Khanewal | HG934396 | 1350 | 195-551/118 | - |
| SAZ 251 | CLCuMB | Punjab/Faisalabad | LN867444 | 1357 | 195-551/118 | - |
| SAZ 253 | CLCuMB | Punjab/Faisalabad | LN867445 | 1370 | 195-551/118 | - |
| SAZ 254 | CLCuMB | Punjab/Faisalabad | LN867446 | 1370 | 195-551/118 | - |
| SAZ 255 | CLCuMB | Punjab/Faisalabad | LN867447 | 1370 | 195-551/118 | - |
| SAZ 257 | CLCuMB | Punjab/Faisalabad | LN867448 | 1351 | 195-551/118 | - |
| SAZ 261 | CLCuMB | Punjab/Faisalabad | LN867449 | 1352 | 195-551/118 | - |
| SAZ 265 | CLCuMB | Punjab/Faisalabad | LN867450 | 1370 | 195-551/118 | - |
| SAZ 1 | CLCuMA | Punjab/Khanewal | LN845921 | 1365 | - | 77-1024/315 |
| SAZ 2 | CLCuMA | Punjab/Khanewal | HG934390 | 1366 | - | 77-1024/315 |
| SAZ 3 | CLCuMA | Punjab/Khanewal | LN845925 | 1366 | - | 77-1024/315 |
| SAZ 10 | CLCuMA | Punjab/Bahawalpur | LN845922 | 1377 | - | 77-1024/315 |
| SAZ 11 | CLCuMA | Punjab/Bahawalpur | LN845923 | 1377 | - | 77-1024/315 |
| SAZ 12 | CLCuMA | Punjab/Bahawalpur | HG934391 | 1377 | - | 77-1024/315 |
| SAZ 17 | CLCuMA | Punjab/Bahawalpur | HG934392 | 1365 | - | 77-1024/315 |
| SAZ 19 | CLCuMA | Punjab/Bahawalpur | LN845924 | 1366 | - | 77-1024/315 |
| SAZ 20 | CLCuMA | Punjab/Bahawalpur | HG934393 | 1365 | - | 77-1024/315 |
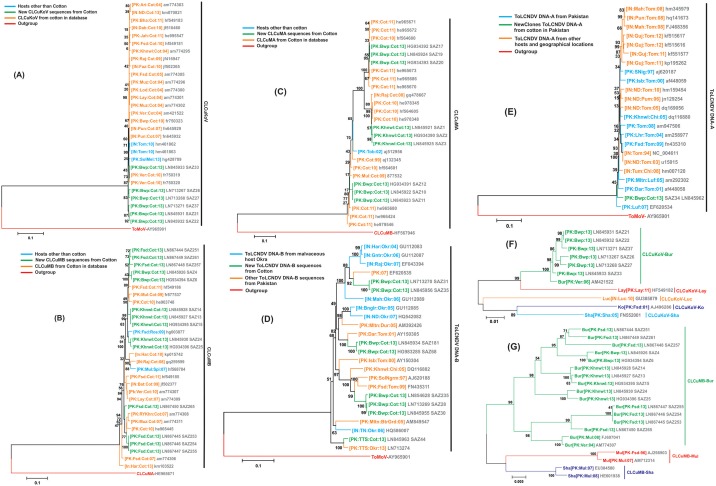
A closer analysis indicated that 6 of these clones have a truncated TrAP gene, with a potential coding capacity of 35 amino acids, whereas one clone (SAZ34) encoded a putatively full-length TrAP predicted to be of 136 amino acids (Table 2). Mutation of the TrAP gene is typical of CLCuKoV-Bur isolates associated with resistance breaking [22]. The six clones obtained here show 98–100% nucleotide sequence identity with CLCuKoV isolates available in the databases. In phylogenetic analyses the sequences obtained here show low branch lengths to previously characterized CLCuKoV isolates (Fig 2, panel A) and segregate with CLCuKoV-Bur isolates (Fig 2, panel F). This confirms that the sequences obtained here are isolates of CLCuKoV-Bur.
Fig 2. Phylogenetic analysis of the sequences of begomoviruses, and satellites obtained in the study.
Trees were constructed from alignments of the sequences of (A and F) Cotton leaf curl Kokhran virus (CLCuKoV), (B and G) Cotton leaf curl Multan betasatellite (CLCuMB), (C) Cotton leaf curl Multan alphasatellite (CLCuMA), (D) Tomato leaf curl New Delhi virus (ToLCNDV) DNA-B and (E) ToLCNDV DNA-A using the Neighbor-Joining method. Isolates in green were obtained in the study described here. The numbers at nodes represent percentage bootstrap scores (1000 replicates). The strain descriptors (in square brackets) in each case give country, location, host and year of sampling. For each isolate the database accession number is given. The trees were arbitrarily rooted on the sequence of Tomato mottle virus (ToMoV) (A, D and E), CLCuMA (B) and CLCuMB (C) as outgroup. The strains of CLCuKoV given in panel F are Burewala (Bur), Layyah (Lay), Lucknow (Luc), Kokhran (Kok) and Shadadpur (Sha). The strains of CLCuMB given in panel G are given as Burewala (Bur), Multan (Mul) and Shadadpur (Sha).
Analysis of the sequence of clone SAZ34 showed it to have 94–96% sequence identity with the sequences of ToLCNDV component DNA-A available in the databases. A phylogenetic analysis showed the sequence to have low branch lengths to the DNA-A components of previously characterized ToLCNDV isolates (Fig 2, panel E). This confirmed that ToLCNDV was present in the cotton plant analysed.
The remaining seven ~2.8kb clones had an arrangement of genes typical of the DNA-B component of bipartite begomoviruses, consisting of one gene encoded in each orientation (Table 3). These sequences showed 82–92% nucleotide sequence identity to the sequences of the DNA-B components of ToLCNDV available in the databases. In phylogenetic analysis the six sequences obtained here had short branch lengths to the sequences of the DNA-B components of ToLCNDV obtained from the databases (Fig 2, panel D). This confirms that the clones are isolates of the DNA-B component of ToLCNDV.
An alphasatellite is associated with cotton leaf curl disease
A total of 23 ~1.4kb clones were obtained from eight CLCuD-affected cotton plants. Analysis of the sequences showed them to encode either one large (~950bp) gene in the virion-sense, typical of alphasatellites, or a small (~350bp) gene in the complementary-sense, typical of betasatellites (Table 4). The 9 presumed alphasatellite clones showed 89–99% nucleotide sequence identity to isolates of Cotton leaf curl Multan alphasatellite (CLCuMA) available in the databases. A phylogenetic analysis also showed the 9 sequences to group with low branch lengths with the sequences of CLCuMA available in the databases (Fig 2, panel C). This confirms that the alphasatellites isolated from cotton here are isolates of CLCuMA.
The 14 presumed betasatellite clones showed 91–99% nucleotide sequence identity to Cotton leaf curl Multan betasatellite (CLCuMB) sequences available in the databases. In a phylogenetic analysis the new sequences group with short branch lengths with the sequences of earlier reported CLCuMB isolates (Fig 2, panel B). Additionally the CLCuMB sequences obtained here segregated with isolates of the “Burewala” strain of CLCuMB (CLCuMBBur) rather than the “Multan” or “Shadadpur” strains (Fig 2, panel G). This shows that the cotton plants examined were infected with CLCuMBBur, which is associated with resistance breaking [45].
Geographic incidence of co-infection of cotton with ToLCNDV and CLCuKoV-Bur
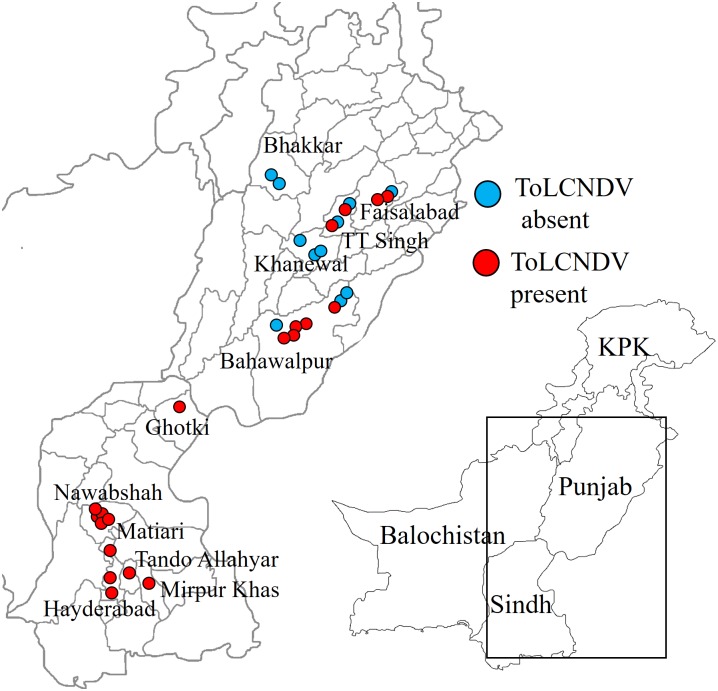
The incidence and area over which coinfection of cotton with ToLCNDV and the CLCuKoV-Bur/CLCuMB complex was investigated by diagnostic PCR with primers specific for ToLCNDVDNA-B and CLCuKoV-Bur on DNA samples extracted from CLCuD symptomatic cotton originating from across the Punjab and northern Sindh provinces of Pakistan (Fig 3; Table 1.). Of the 31 samples examined, 20showed the presence of both viruses. The plants harbouring both viruses originated from geographically widespread areas across the Punjab and Sindh. These results indicate that ToLCNDV is widespread in cotton in Pakistan.
Fig 3. Distribution of samples shown to harbour coinfection of CLCuKoV-Bur and ToLCNDV across Pakistan.
The map shows the origin of collected cotton plant samples with CLCuD symptoms. Plants shown by diagnostic PCR to contain both CLCuKoV-Bur and ToLCNDV are shown as red dots, whereas plants shown only to contain CLCuKoV-Bur are shown as blue dots. The map depicting putative boundaries is drawn by the author and provided here for illustrative purpose only.
Quantitative PCR analysis of CLCuKoV-Bur/CLCuMB and ToLCNDV coinfected cotton plants
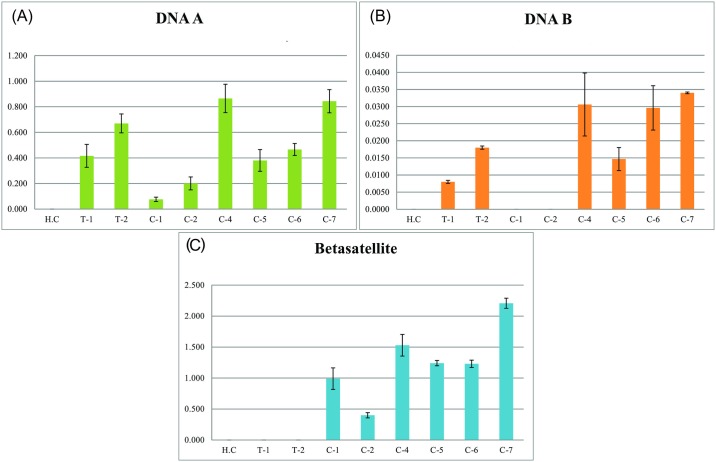
The results of a quantitative PCR analysis of the levels of virus (the primers used amplify both ToLCNDVDNA-A and CLCuKoV-Bur), ToLCNDVDNA-B and CLCuMB in CLCuD symptomatic cotton plants infected with only the CLCuKoV-Bur/CLCuMB complex or co-infected with CLCuKoV-Bur/CLCuMB and ToLCNDV are shown in Fig 4. The analysis indicates that in coinfected cotton plants the levels of CLCuMB are significantly higher than in cotton plants infected with only CLCuKoV-Bur/CLCuMB.
Fig 4. Quantitative real-time PCR analysis of coinfected cotton plants.
Graphs show the determined titers of (A) virus (Tomato leaf curl New Delhi virus [ToLCNDV] DNA-A and Cotton leaf curl Kokhran virus-Burewala CLCuKoV-Bur), (B) ToLCNDV DNA-B and (C) Cotton leaf curl Multan betasatellite (CLCuMB). The DNA samples used in the PCR reactions were extracted from a healthy cotton plant (H.C), tomato plants infected with ToLCNDV (T1 and T2), field collected cotton plants with severe cotton leaf curl disease symptoms either infected with only CLCuKoV-Bur/CLCuMB (C-1 and C-2) or coinfected with CLCuKoV-Bur/CLCuMB and ToLCNDV (C-4 to C-7). The titer of each component is given in ng/μg of genomic DNA on the y-axis and is the mean of three replications. The error bars are the divergence from mean quantified value.
Discussion
Begomovirus disease complexes are evolving rapidly by recombination, component capture and mutation to expand their host range and overcome sources of resistance. The resistance breaking begomovirus-betasatellite complex causing CLCuD evolved by recombination and mutation[22, 23, 45]. The susceptibility of previously resistant/tolerant cotton lines to the disease prompted aninvestigation into possiblechanges in the disease complex since resistance breaking. The results obtained here are consistent with the present belief that CLCuD in resistant cotton varieties across Pakistan and northwestern India is caused by CLCuKoV-Bur and CLCuMBBur. Recently CLCuKoV-Bur and CLCuMBMul have been shown experimentally to be able to cause CLCuD in cotton [46]. However, this study did not investigate whether this combination of virus and betasatellite could break resistance in cotton.
In many of the cotton plants examined here identified the alphasatellite CLCuMA was identified. This indicates that, as was the case before resistance breaking, the virus causing CLCuD is associated with an alphasatellite. The study of Amrao et al. [22] which first identified CLCuKoV-Bur, reported that there was no evidence for the presence of an alphasatellite. The study here is thus the first to report an alphasatellite with the resistance breaking complex. The precise functions of alphasatellites remain unclear, although evidence has been provided to show that alphasatellites may encode a suppressor of gene silencing which overcome host resistance based on small RNAs [47].
The most surprising finding of the study presented here was the presence of the bipartite begomovirus ToLCNDV in cotton affected by CLCuD. A number of other geminiviruses have been identified in cotton including the mastrevirus Chickpea chlorotic dwarf virus [48], ToLCV [32] and Okra enation leaf curl virus [49]. However, these viruses were only identified across a limited area and in a few plants. ToLCNDV, in contrast, was identified in cotton across a wide area of Pakistan, suggesting that it is more than just a fleeting infection.
The quantitative PCR analysis suggests that in cotton there is a synergistic interaction between CLCuKoV-Bur/CLCuMBBur complex and ToLCNDV which leads to an increase in the amount of CLCuMBBur present in coinfected plants. Betasatellites encode a dominant symptom determinant [50, 51] and the βC1 gene of CLCuMB alone has been shown to induce symptoms typical of CLCuD in tobacco [52]. Any increase in betasatellite levels with a concomitant increase in βC1 gene is thus undesirable.
The nature of a possible synergistic interaction between the CLCuKoV-Bur/CLCuMBBur complex and ToLCNDV is unclear. The DNA-A component of ToLCNDV, in the absence of the DNA-B, has been shown to be able to support the replication of CLCuMB in cotton and, at least transiently, induce typical CLCuD symptoms [53]. A study of the interaction of ToLCNDV with CLCuMB in tomato and Nicotiana benthamiana showed the presence of CLCuMB to enhance the viral DNA levels but the presence of DNA-B depressed CLCuMB levels [54]. Nevertheless, the increase in betasatellite and possibly virus levels in coinfected cotton may be due to the movement functions encoded by the DNA-B component of ToLCNDV allowing the infection to spread to tissue which it normally does not reach [55].
CLCuD is a major constraint to cotton production in Pakistan and India. At this time there are no commercially available cotton varieties with resistance to the disease. The appearance of a form of the virus-complex causing the disease with potentially enhanced pathogenicity is thus not good news. Further studies will be needed to monitor the situation and see whether the coinfection persists and precisely what the effects are on the yield of cotton. Additionally, any efforts towards developing resistance to the disease, either by conventional or non-conventional means, would be wise to take into account the possibility of a more complex situation becoming important in cotton in the future.
Acknowledgments
We are thankful to Dr. Zafar Iqbal for useful discussions during the course of these studies.
Data Availability
All 36 files are available from the NCBI nucleotide database (accession numbers LN845931, LN845932, LN713267, LN713268, LN845933, LN713271, LN845962, LN845926, HG934394, LN845927, LN845928, HG934395, LN845930, HG934396, LN867444, LN867445, LN867446, LN867447, LN867448, LN867449, LN867450, LN845921, HG934390, LN845925, LN845922, LN845923, HG934391, HG934392, LN845924, HG934393, LN713269, LN845955, LN713270, LN845956, HG983285, LN845934, LN854628).
Funding Statement
SSZ is supported by Ph.D scholarship from Higher Education Commission (HEC), Govt. of Pakistan under “Indigenous 5000 Ph.D Fellowship Program”. This research was funded in part through the ‘‘Pak-US cotton productivity enhancement program’’ of the International Center for Agricultural Research in the Dry Areas (ICARDA) funded by United States Department of Agriculture (USDA), Agricultural Research Service (ARS), under Agreement No. 58-6402-0-178F. Any opinions, findings, conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the USDA or ICARDA.
References
- 1.Briddon RW, Markham PG. Cotton leaf curl virus disease. Virus Res. 2000;71:151–9. [DOI] [PubMed] [Google Scholar]
- 2.Sattar MN, Kvarnheden A, Saeed M, Briddon RW. Cotton leaf curl disease—an emerging threat to cotton production worldwide. J. Gen. Virol. 2013;94:695–710. 10.1099/vir.0.049627-0 [DOI] [PubMed] [Google Scholar]
- 3.Mansoor S, Amin I, Iram S, Hussain M, Zafar Y, Malik KA, et al. Breakdown of resistance in cotton to cotton leaf curl disease in Pakistan. Plant Pathol. 2003;52:784. [Google Scholar]
- 4.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Virus taxonomy—Ninth Report of the International Committee on Taxonomy of Viruses. London: Elsevire; 2011.
- 5.Sánchez-Campos S, Martínez-Ayala A, Márquez-Martín B, Aragón-Caballero L, Navas-Castillo J, Moriones E. Fulfilling Koch's postulates confirms the monopartite nature of tomato leaf deformation virus: A begomovirus native to the New World. Virus Res. 2013;173:286–93. 10.1016/j.virusres.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 6.Melgarejo TA, Kon T, Rojas MR, Paz-Carrasco L, Zerbini FM, Gilbertson RL. Characterization of a New World monopartite begomovirus causing leaf curl disease of tomato in Ecuador and Peru reveals a new direction in geminivirus evolution. J. Virol. 2013;87:5397–413. 10.1128/JVI.00234-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013;11:777–88. 10.1038/nrmicro3117 [DOI] [PubMed] [Google Scholar]
- 8.Rojas MR, Hagen C, Lucas WJ, Gilbertson RL. Exploiting chinks in the plant's armor: Evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 2005;43:361–94. [DOI] [PubMed] [Google Scholar]
- 9.Briddon RW, Stanley J. Sub-viral agents associated with plant single-stranded DNA viruses. Virology. 2006;344:198–210. [DOI] [PubMed] [Google Scholar]
- 10.Briddon RW, Mansoor S, Bedford ID, Pinner MS, Saunders K, Stanley J, et al. Identification of DNA components required for induction of cotton leaf curl disease. Virology. 2001;285:234–43. [DOI] [PubMed] [Google Scholar]
- 11.Zhou X. Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 2013;51:357–81. 10.1146/annurev-phyto-082712-102234 [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Xie Y, Raja P, Li S, Wolf JN, Shen Q, et al. Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 2011;7:e1002329 10.1371/journal.ppat.1002329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui X, Li G, Wang D, Hu D, Zhou X. A begomovirus DNAβ-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 2005;79:10764–75. 16051868 [Google Scholar]
- 14.Briddon RW, Bull SE, Amin I, Mansoor S, Bedford ID, Rishi N, et al. Diversity of DNA 1; a satellite-like molecule associated with monopartite begomovirus-DNA β complexes. Virology. 2004;324:462–74. [DOI] [PubMed] [Google Scholar]
- 15.Mansoor S, Khan SH, Bashir A, Saeed M, Zafar Y, Malik KA, et al. Identification of a novel circular single-stranded DNA associated with cotton leaf curl disease in Pakistan. Virology. 1999;259:190–9. [DOI] [PubMed] [Google Scholar]
- 16.Saunders K, Stanley J. A nanovirus-like component associated with yellow vein disease of Ageratum conyzoides: evidence for interfamilial recombination between plant DNA viruses. Virology. 1999;264:142–52. [DOI] [PubMed] [Google Scholar]
- 17.Romay G, Chirinos D, Geraud-Pouey F, Desbiez C. Association of an atypical alphasatellite with a bipartite New World begomovirus. Arch. Virol. 2010;155:1843–7. 10.1007/s00705-010-0760-7 [DOI] [PubMed] [Google Scholar]
- 18.Paprotka T, Metzler V, Jeske H. The first DNA 1-like a satellites in association with New World begomoviruses in natural infections. Virology. 2010;404:148–57. 10.1016/j.virol.2010.05.003 [DOI] [PubMed] [Google Scholar]
- 19.Kumar J, Kumar J, Singh SP, Tuli R. Association of satellites with a mastrevirus in natural infection: complexity of wheat dwarf India virus disease. J. Virol. 2014; 88: 7093–104. 10.1128/JVI.02911-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou X, Liu Y, Robinson DJ, Harrison BD. Four DNA-A variants among Pakistani isolates of cotton leaf curl virus and their affinities to DNA-A of geminivirus isolates from okra. J. Gen. Virol. 1998;79:915–23. [DOI] [PubMed] [Google Scholar]
- 21.Mansoor S, Briddon RW, Bull SE, Bedford ID, Bashir A, Hussain M, et al. Cotton leaf curl disease is associated with multiple monopartite begomoviruses supported by single DNA β. Arch. Virol. 2003;148:1969–86. [DOI] [PubMed] [Google Scholar]
- 22.Amrao L, Amin I, Shahid S, Briddon RW, Mansoor S. Cotton leaf curl disease in resistant cotton is associated with a single begomovirus that lacks an intact transcriptional activator protein. Virus Res. 2010;152:153–63. 10.1016/j.virusres.2010.06.019 [DOI] [PubMed] [Google Scholar]
- 23.Briddon RW, Akbar F, Iqbal Z, Amrao L, Amin I, Saeed M, et al. Effects of genetic changes to the begomovirus/betasatellite complex causing cotton leaf curl disease in South Asia post-resistance breaking. Virus Res. 2014;186:114–9. 10.1016/j.virusres.2013.12.008 [DOI] [PubMed] [Google Scholar]
- 24.Haider MS, Tahir M, Latif S, Briddon RW. First report of Tomato leaf curl New Delhi virus infecting Eclipta prostrata in Pakistan. Plant Pathol. 2006;53:285. [Google Scholar]
- 25.Hussain M, Mansoor S, Amin I, Iram S, Zafar Y, Malik KA, et al. First report of cotton leaf curl disease affecting chili peppers. Plant Pathol. 2003;56:809. [Google Scholar]
- 26.Tahir M, Haider MS. First report of Tomato leaf curl New Delhi virus infecting bitter gourd in Pakistan. Plant Pathol. 2005;54:807. [Google Scholar]
- 27.Phaneendra C, Rao K, Jain R, Mandal B. Tomato leaf curl New Delhi virus is associated with pumpkin leaf curl: A new disease in northern India. Indian J. Virol. 2012;23:42–5. 10.1007/s13337-011-0054-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkataravanappa V, Lakshminarayana Reddy C, Jalali S, Krishna Reddy M. Molecular characterization of distinct bipartite begomovirus infecting bhendi (Abelmoschus esculentus L.) in India. Virus Genes. 2012;44:522–35. 10.1007/s11262-012-0732-y [DOI] [PubMed] [Google Scholar]
- 29.Shafiq M, Asad S, Zafar Y, Briddon RW, Mansoor S. Pepper leaf curl Lahore virus requires the DNA B component of Tomato leaf curl New Delhi virus to cause leaf curl symptoms. Virol J. 2010;7:367 10.1186/1743-422X-7-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malik A, Briddon R, Mansoor S. Infectious clones of Tomato leaf curl Palampur virus with a defective DNA B and their pseudo-recombination with Tomato leaf curl New Delhi virus. Virol. J. 2011;8:173 10.1186/1743-422X-8-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mubin M, Akhtar S, Amin I, Briddon RW, Mansoor S. Xanthium strumarium: a weed host of components of begomovirus–betasatellite complexes affecting crops. Virus Genes. 2012;44:112–9. 10.1007/s11262-011-0662-0 [DOI] [PubMed] [Google Scholar]
- 32.Zaidi SSA, Iqbal Z, Amin I, Mansoor S. First report of Tomato leaf curl Gujarat virus, a bipartite begomovirus on cotton showing leaf curl symptoms in Pakistan. Plant Dis. 2015;99:1655. [Google Scholar]
- 33.Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–5. [Google Scholar]
- 34.Haible D, Kober S, Jeske H. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J. Virol. Methods. 2006;135:9–16. [DOI] [PubMed] [Google Scholar]
- 35.Shahid MS, Mansoor S, Briddon RW. Complete nucleotide sequences of Cotton leaf curl Rajasthan virus and its associated DNA β molecule infecting tomato. Arch. Virol. 2007;152:2131–4. [DOI] [PubMed] [Google Scholar]
- 36.Bull SE, Briddon RW, Markham PG. Universal primers for the PCR-mediated amplification of DNA 1: a satellite-like molecule associated with begomovirus-DNA β complexes. Mol Biotechnol. 2003;23:83–6. [DOI] [PubMed] [Google Scholar]
- 37.Briddon RW, Bull SE, Mansoor S, Amin I, Markham PG. Universal primers for the PCR-mediated amplification of DNA β; a molecule associated with some monopartite begomoviruses. Mol Biotechnol. 2002;20:315–8. [DOI] [PubMed] [Google Scholar]
- 38.Shuja MN, Briddon RW, Tahir M. Identification of a distinct strain of cotton leaf curl Burewala virus. Arch. Virol. 2014; 159:2787:90. 10.1007/s00705-014-2097-0 [DOI] [PubMed] [Google Scholar]
- 39.Muhire BM, Varsani A, Martin DP. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PloS ONE. 2014;9:e108277 10.1371/journal.pone.0108277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown J, Zerbini FM, Navas-Castillo J, Moriones E, Ramos-Sobrinho R, Silva JF, et al. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 2015; 160:1593–619. 10.1007/s00705-015-2398-y [DOI] [PubMed] [Google Scholar]
- 41.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. [DOI] [PubMed] [Google Scholar]
- 42.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol.Biol.Evol. 2013; mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Page RDM. TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–8. [DOI] [PubMed] [Google Scholar]
- 44.Padidam M, Beachy RN, Fauquet CM. Tomato leaf curl geminivirus from India has a bipartite genome and coat protein is not essential for infectivity. J. Gen. Virol. 1995;76:25–35. [DOI] [PubMed] [Google Scholar]
- 45.Amin I, Mansoor S, Amrao L, Hussain M, Irum S, Zafar Y, et al. Mobilisation into cotton and spread of a recombinant cotton leaf curl disease satellite. Arch. Virol. 2006;151:2055–65. [DOI] [PubMed] [Google Scholar]
- 46.Kumar J, Gunapati S, Alok A, Lalit A, Gadre R, Sharma N, et al. Cotton leaf curl Burewala virus with intact or mutant transcriptional activator proteins: complexity of cotton leaf curl disease. Arch. Virol. 2015; 160:1219–28. 10.1007/s00705-015-2384-4 [DOI] [PubMed] [Google Scholar]
- 47.Nawaz-ul-Rehman MS, Nahid N, Mansoor S, Briddon RW, Fauquet CM. Post-transcriptional gene silencing suppressor activity of the alpha-Rep of non-pathogenic alphasatellites associated with begomoviruses. Virology. 2010;405:300–8. 10.1016/j.virol.2010.06.024 [DOI] [PubMed] [Google Scholar]
- 48.Manzoor M, Ilyas M, Shafiq M, Haider M, Shahid A, Briddon RW. A distinct strain of Chickpea chlorotic dwarf virus (genus Mastrevirus, family Geminiviridae) identified in cotton plants affected by leaf curl disease. Arch. Virol. 2014; 159:1217–21. 10.1007/s00705-013-1911-4 [DOI] [PubMed] [Google Scholar]
- 49.Hameed U, Zia-Ur-Rehman M, Herrmann HW, Haider M, Brown JK. First report of Okra enation leaf curl virus and associated cotton leaf curl Multan betasatellite and cotton leaf curl Multan alphasatellite infecting cotton in Pakistan: A new member of the cotton leaf curl disease complex. Plant Dis. 2014; 98:1447. [DOI] [PubMed] [Google Scholar]
- 50.Saunders K, Norman A, Gucciardo S, Stanley J. The DNA β satellite component associated with ageratum yellow vein disease encodes an essential pathogenicity protein (βC1). Virology. 2004;324:37–47. [DOI] [PubMed] [Google Scholar]
- 51.Saeed M, Behjatnia SAA, Mansoor S, Zafar Y, Hasnain S, Rezaian MA. A single complementary-sense transcript of a geminiviral DNA β satellite is determinant of pathogenicity. Mol. Plant-Microbe In. 2005;18:7–14. [DOI] [PubMed] [Google Scholar]
- 52.Qazi J, Amin I, Mansoor S, Iqbal J, Briddon RW. Contribution of the satellite encoded gene βC1 to cotton leaf curl disease symptoms. Virus Res. 2007;128:135–139. [DOI] [PubMed] [Google Scholar]
- 53.Saeed S, Y. Zafar Y, Randles JW, Rezaian MA. A monopartite begomovirus-associated DNA β satellite substitutes for the DNA B of a bipartite begomovirus to permit systemic infection. J. Gen. Virol. 2007;88:2881–2889. [DOI] [PubMed] [Google Scholar]
- 54.Jyothsna P, Haq QMI, Singh P, Sumiya KV, Praveen S, Rawat R, et al. Infection of Tomato leaf curl New Delhi virus (ToLCNDV), a bipartite begomovirus with betasatellites, results in enhanced level of helper virus components and antagonistic interaction between DNA B and betasatellites. Appl. Microbiol. Biot. 2013;97:5457–5471. [DOI] [PubMed] [Google Scholar]
- 55.Sanderfoot AA, Lazarowitz SG. Getting it together in plant virus movement: cooperative interactions between bipartite geminivirus movement proteins. Trends Cell Biol. 1996;6:353–358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All 36 files are available from the NCBI nucleotide database (accession numbers LN845931, LN845932, LN713267, LN713268, LN845933, LN713271, LN845962, LN845926, HG934394, LN845927, LN845928, HG934395, LN845930, HG934396, LN867444, LN867445, LN867446, LN867447, LN867448, LN867449, LN867450, LN845921, HG934390, LN845925, LN845922, LN845923, HG934391, HG934392, LN845924, HG934393, LN713269, LN845955, LN713270, LN845956, HG983285, LN845934, LN854628).