Abstract
The mechanisms that lead to phenotypic antibacterial tolerance in bacteria remain poorly understood. We investigate whether changes in NaCl concentration toward physiologically higher values affect antibacterial efficacy against Mycobacterium tuberculosis (Mtb), the causal agent of human tuberculosis. Indeed, multiclass phenotypic antibacterial tolerance is observed during Mtb growth in physiologic saline. This includes changes in sensitivity to ethionamide, ethambutol, d-cycloserine, several aminoglycosides, and quinolones. By employing organism-wide metabolomic and lipidomic approaches combined with phenotypic tests, we identified a time-dependent biphasic adaptive response after exposure of Mtb to physiological levels of NaCl. A first rapid, extensive, and reversible phase was associated with changes in core and amino acid metabolism. In a second phase, Mtb responded with a substantial remodelling of plasma membrane and outer lipid membrane composition. We demonstrate that phenotypic tolerance at physiological concentrations of NaCl is the result of changes in plasma and outer membrane lipid remodeling and not changes in core metabolism. Altogether, these results indicate that physiologic saline-induced antibacterial tolerance is kinetically coupled to cell envelope changes and demonstrate that metabolic changes and growth arrest are not the cause of phenotypic tolerance observed in Mtb exposed to physiologic concentrations of NaCl. Importantly, this work uncovers a role for bacterial cell envelope remodeling in antibacterial tolerance, alongside well-documented allterations in respiration, metabolism, and growth rate.
Keywords: Mycobacterium tuberculosis, cell envelope remodeling, osmolarity, phenotypic antibacterial tolerance, lipids
Phenotypic antibacterial tolerance (persistence) is the phenomenon by which a few members of a larger population of sensitive bacteria escape killing by otherwise lethal concentrations of antibacterial agents.1 In contrast to genetic resistance, which often confers resistance to a single antibacterial or antibacterial class, phenotypic tolerance usually protects bacteria against mechanistic and structurally diverse classes of antibacterial agents. Despite the wealth of information on genetic resistance to specific antibacterial agents, much less is known about the cellular determinants of phenotypic tolerance. What is clear in several bacterial species investigated to date is that phenotypic antibacterial tolerance is often associated with decreased flux through the electron transport chain, defects in respiration, and altered core metabolism2−4 or, alternatively, with the action of toxin–antitoxin systems, some of which induce growth arrest5,6
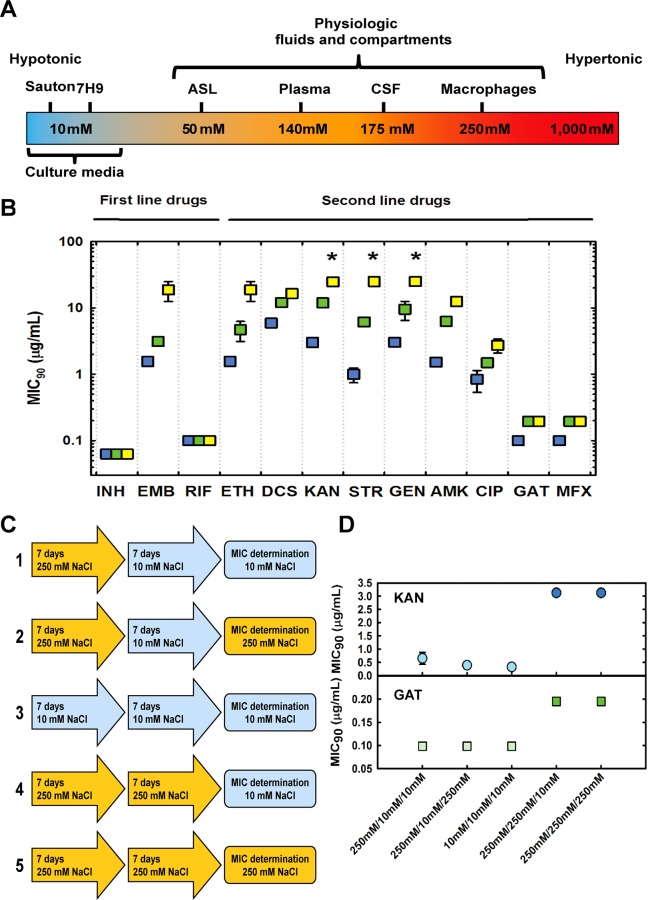
Mycobacterium tuberculosis (Mtb), the cause of human tuberculosis, is the deadliest bacterium affecting mankind, claiming 1.4 million lives yearly.7 Throughout the cycle of infection, Mtb encounters and survives in distinct environments in the human body including nutrient-poor, acidic, oxidative, nitrosative, and hypoxic niches, found intracellularly and extracellularly.8 In addition to these extensively studied physiologic stresses imposed by the host, mycobacteria encounter electrolyte and osmolar stress within the host. Although much less is known about the outcomes of osmolar stress, it could plausibly determine the outcome of infection. For example, only recently, chloride has been demonstrated to be a new cue utilized by Mtb to sense immune-mediated changes in the phagosomal environment.9 In line with this finding, concentrations of NaCl vary within the niches occupied by this pathogen during natural infection, from 50 mM in airway surface liquid to 250 mM in macrophages (Figure 1A),9−11 indicating that Mtb experiences different concentrations of electrolytes during its life in the host. Importantly, these concentrations are significantly higher than the concentrations of NaCl (<10 mM) found in standard microbiologic media used to culture Mtb (Figure 1A). Consistent with the notion that fluctuations in osmolarity are present during Mtb’s infection cycle, a recent study described a serine/threonine kinase-dependent sensing/signaling network involved in osmosensing in Mtb.12 In that study, changes in osmolarity sensing, probed by deletion of PknD substrate Rv0516c, did not affect the sensitivity to isoniazid (INH) and d-cycloserine (DCS), two clinically used antitubercular agents, but weakened the effect of vancomycin.
Figure 1.
Changes in osmolarity induce phenotypic antibacterial tolerance in Mtb. (A) Range of NaCl concentrations experienced by Mtb in humans and culture media. ASL, airway surface liquid; CSF cerebrospinal fluid. (B) MIC90 values for antitubercular drugs obtained in the presence of 14.5 (blue squares), 125 (green squares), and 250 mM (yellow squares) NaCl with Mtb (H37Rv). Asterisks indicate that the values obtained are the lower limit (maximum concentration of drug present in the assay) and not true values. Data are the average of three independent experiments. (C) Experimental settings to probe reversibility of phenotypic antibacterial tolerance at 10 and 250 mM NaCl. (D) MIC90 values for KAN and GAT against Mtb evaluated following an initial period of pre-adaptation (at 10 and 250 mM NaCl) and a second period of pre-adaptation prior to the test (at 10 and 250 mM NaCl), which was also carried out at 10 and 250 mM NaCl. Data are the average of three biological replicates, and results are representative of two independent experiments.
Here we describe a combination of antibacterial pharmacology with time-dependent, organism-wide measurements of polar metabolites and cell envelope lipids to uncover substantial changes in antibacterial sensitivity (tolerance), caused by physiologic concentrations of NaCl. On the basis of the time dependence and reversibility of phenotypes, we identify strong correlations between cell envelope composition and antibacterial tolerance, highlighting osmolality as a physiologic cause of phenotypic tolerance in Mtb, a finding with broad implications for antibacterial drug discovery.
Results and Discussion
Physiologic Concentrations of NaCl Induce Phenotypic Antibacterial Tolerance in Mtb
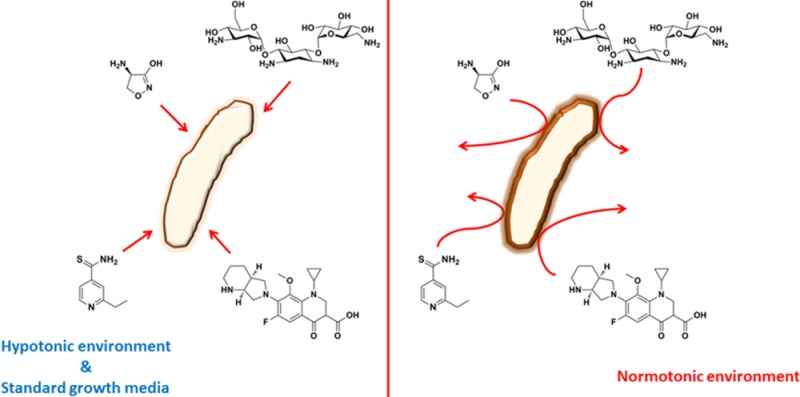
In their recent study, Hatzios and colleagues12 observed a shift in the MIC value for vancomycin when Mtb harboring a genetic disruption on the PknD-substrate Rv0516 was tested in hypotonic media. Inspired by this observation, we investigated the effect of adaptation to physiologic saline concentrations in drug sensitivity in Mtb. We reasoned that without pre-adaptation to physiologic salt concentrations, antibacterial agents might be taken up by Mtb and rapidly inhibit their targets, prior to any delayed effects of salt on cellular metabolism. Consequently, we precultured Mtb H37Rv in medium containing 14.5, 125, and 250 mM NaCl for 7 days. We chose these concentrations because they mimic common hypotonic medium formulations and physiologic concentrations of NaCl encountered in the host niches where Mtb resides. We determined the MIC90 of three first-line drugs (isoniazid (INH), ethambutol (EMB), and rifampicin (RIF)) and nine second-line drugs (d-cycloserine (DCS), ethionamide (ETH), kanamycin (KAN), streptomycin (STR), gentamycin (GEN), amikacin (AMK), ciprofloxacin (CIP), gatifloxacin (GAT), and moxifloxacin (MFX)), in medium with 14.5 mM NaCl (Figure 1B). INH and RIF did not exhibit any change in MIC90, indicating that these changes are not caused by growth rate effects caused by physiologic salt concentrations, as MIC90 values of INH and RIF would have been substantially altered if the growth rate was substantially decreased.13 In contrast, Mtb sensitivity to EMB and ETH exhibited up to a 25-fold increase in MIC90, indicating considerably weaker antibacterial activity at physiologic salt concentrations, suggesting salt-induced drug resistance. Similar results were obtained with second-line drugs. MIC90 values for DCS and quinolones (CIP, GAT, and MFX) displayed a modest increase with increasing concentrations of NaCl, whereas MIC90 values for aminoglycosides (KAN, STR, GEN, and AMK) were drastically increased at physiologic concentrations of NaCl (Figure 1B). These results indicate that physiologic concentrations of NaCl alter the dose at which Mtb is killed by clinically relevant antibacterial agents, diminishing their efficacy. It is worth noting that in the short period (<2 weeks) employed for selection and propagation, genetic resistance is unlikely the cause of the observed phenotypes; hence, these shifts in MIC90 are due to phenotypic tolerance triggered by changes in osmolarity. In line with this conclusion, reversion of sensitivity can be clearly demonstrated (see below).
We next sought to investigate the time dependence of the MIC90 shifts observed in the presence of physiologic concentrations of NaCl. To accomplish this, we designed an experiment in which cells were differentially treated, prior to MIC90 determination with one aminoglycoside and one quinolone (Figure 1C). As can be seen in Figure 1D, carrying out the MIC90 assay at 10 or 250 mM NaCl did not alter the MIC90 for KAN and GAT (condition 1 vs 2 and condition 4 vs 5, respectively). Also, no selection of mutants took place, as a pre-incubation at 250 mM NaCl followed by culture at 10 mM NaCl prior to MIC90 determination did not alter the MIC90 values (condition 1 vs 3). This result indicates that these changes are caused by phenotypic (reversible) tolerance rather than selection of resistant mutants. If we had selected mutants in the first pre-incubation period, MIC90 values from conditions 1, 2, 4, and 5 should be identical and greater than the MIC90 value in column 3, which is not the case. Exposure to high salinity 7 days prior to the assay led to a marked increase in the MIC90 values (condition 1 vs 4 and condition 2 vs 5). Taken together, these results demonstrate that a period of pre-incubation at high salinity immediately preceding MIC90 determination is necessary and sufficient to induce antibacterial tolerance to chemically and mechanistically distinct types of antibacterial drugs.
To investigate whether phenotypic tolerance was a characteristic present only in the laboratory-adapted H37Rv reference strain or also present in clinically derived Mtb, we employed a panel of clinical Mtb strains, with known patterns of genetic drug resistance.14 Five strains (one drug sensitive and four multidrug resistant) were tested for sensitivity to ETH, KAN, STR, GEN, AMK, CIP, GAT, and MFX at 14.5, 125, and 250 mM NaCl. Results indicate that the lower sensitivity against ETH, aminoglycosides (KAN, STR, GEN, and AMK), and quinolones (CIP, GAT, and MFX), seen with H37Rv strains, is also observed in all clinical strains tested (Supporting Information, SI Appendix Figure S1). Importantly, changes in sensitivity to ETH, aminoglycosides, and quinolones observed correlate well with the concentrations of NaCl employed (i.e., increased NaCl concentration always leads to increased MIC). Of note, MIC90 values GAT and MFX were significantly more affected by changes in NaCl levels in the clinical strains tested than in H37Rv. This indicates that quinolone sensitivity in clinical strains might be an issue that is not really well modeled by the laboratory strain. Therefore, phenotypic antibacterial tolerance induced by physiologic concentrations of NaCl is present in both sensitive and multidrug-resistant clinical strains of Mtb.
Physiologic Concentrations of NaCl Reversibly Alter Mtb Growth Characteristics
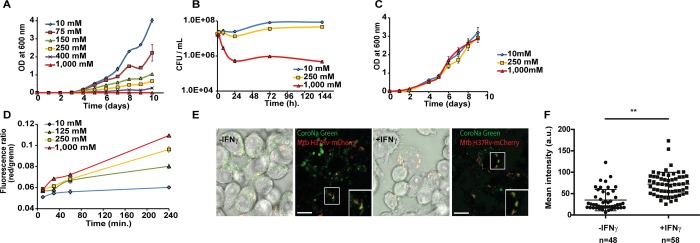
We next sought to identify the mechanisms that impart salt-induced antibacterial tolerance by cultivating Mtb H37Rv at 37 °C in Middlebrook 7H9 medium supplemented over a broad range of NaCl concentrations that span those present in media formulations, cells, and other body fluids, ranging from 10 to 1000 mM. There is a clear inverse correlation between the concentration of NaCl in the culture medium and the growth kinetics of Mtb (Figure 2A). Generation time increases with increasing concentrations of NaCl up to 400 mM, with no detectable growth at 1000 mM NaCl. These findings show that the difference between the saline concentrations routinely used in vitro (<10 mM) and those experienced in vivo (50–250 mM) has important consequences for growth. To evaluate the effect of saline concentration on bacterial viability, we monitored colony-forming units (CFUs) over 7 days of incubation in the presence of various concentrations of NaCl (Figure 2B). Incubation of Mtb with 250 mM NaCl modestly affected viability, indicating that 250 mM NaCl slows growth, with minimal killing. Interestingly, incubation of Mtb at 1000 mM NaCl caused a biphasic response with rapid killing of ∼2 log10 CFUs over 24 h, followed by a static period where no death is observed for days. This behavior suggests that cells adapt in the course of 24 h (approximately one cell division) to become virtually resistant to death due to high salinity, even at concentrations 7-fold higher than physiological concentrations in the blood (140 mM). Similar results were obtained with Mycobacterium bovis BCG and Mycobacterium smegmatis (SI Appendix, Figure S2). To define further the dynamic nature of the response to changes in saline concentration, we tested whether or not the saline-induced growth defect was reversible. Mtb was exposed to medium containing 0, 250, and 1000 mM NaCl for 4 days, after which time extracellular NaCl was removed and inoculum-matched outgrowth was monitored. Growth kinetics of Mtb exposed to 0, 250, and 1000 mM NaCl are indistinguishable once Mtb has been switched to 10 mM NaCl-containing medium, indicating fast and complete reversibility of growth impairment cause by high salinity (Figure 2C), further indicating that the reduced drug effect relates to the bacterial adaptation rather than genetic resistance, which is typically nonreversible. We next characterized the effect of increasing NaCl concentrations on the membrane potential, employing DiOC2.15,16 DiOC2 is a fluorescent dye that exhibits green fluorescence in bacterial cells, including M. tuberculosis, and undergoes a shift to red emission at high concentrations, reached in the cytosol. The ratio of red over green intensity correlates with membrane potential. Changes in saline concentration increased Mtb’s membrane potential, in a time- and concentration-dependent fashion (Figure 2D). In addition, acid-fast staining increases when Mtb is exposed to higher concentrations of saline (SI Appendix, Figure S3), indicating differential dye retention. Increased membrane potential and altered acid-fast staining hint to alteration in the cell envelope, caused by physiologic saline concentrations. To determine whether or not these phenotypes are relevant during infection, we examined the salinity of Mtb-mCherry-containing intracellular compartments using CoroNa Green sodium indicator in infected RAW264.7 macrophages. We observed that Mtb-mCherry co-localized with CoroNa Green, indicating that there are high levels of Na+ in Mtb-containing compartments (Figure 2E). Importantly, treatment with IFN-γ increased the levels of Na+ in Mtb-containing compartments (Figure 2E,F), illustrating intracellular variations on Na+ concentrations around Mtb. Collectively, these results indicate that physiological concentrations of NaCl (e.g., 50–250 mM) impair Mtb growth in a reversible manner and suggest that changes in salinity are present even at the cellular level, during macrophage infection.
Figure 2.
Changes in osmolarity affect Mtb growth. (A) Mtb growth at various NaCl concentrations in Middlebrook 7H9 medium, monitored by OD 600 nm. (B) Effect of various NaCl concentrations on Mtb viability (CFU/mL). (C) Reversible changes in growth observed after exposure to various NaCl concentrations followed by an outgrowth at 10 mM NaCl, monitored by OD 600 nm. (D) Changes in membrane potential over time of Mtb H37Rv cultivated at 10, 125, 250, and 1000 mM NaCl. (E) Association of CoroNa Green (Na+ probe) with Mtb-mCherry phagosomes in resting and activated RAW264.7 macrophages. (F) Quantitative analysis of the levels of Na+ (arbitrary units) in macrophages treated or not with 5 ng/mL IFN-γ. (∗∗) p ≤ 0.01 (Student’s t test). All data shown are representative of at least two independent experiments. Scale bar = 10 μm.
Physiologic Concentrations of NaCl Rapidly and Reversibly Alter Mtb Metabolism
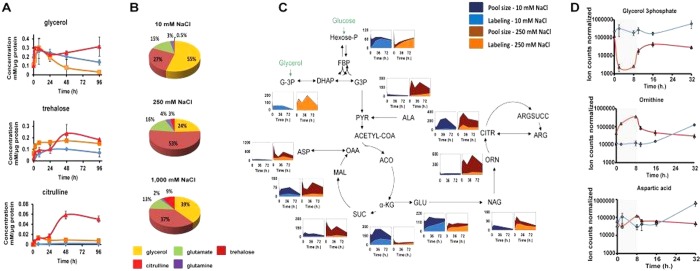
Knowing that osmotic fluctuations can impair protein folding and metabolic activity,17 we first investigated changes in metabolism at different concentrations of NaCl. We focused first on compatible osmolytes, organic biomolecules whose level can be modulated over a broad range without affecting cellular viability (SI Appendix, Figure S4; Figure 3A). NMR metabolomics was chosen as compatible osmolytes should be present in high abundance, and polyols such as trehalose, glucose, and glycerol, amino acids, and their derivatives can be easily monitored with this method. A biphasic response to NaCl is observed with an initial phase that lasts 24 h and a second phase studied for up to 96 h (Figure 3A). Signals for glycerol and trehalose rapidly increase, linking these two osmolytes in early adaptation to high salinity. In addition to these polyols, altered NMR signals corresponding to a nonproteinogenic amino acid, citrulline, increased substantially in the early initial phase. Citrulline is a rare compatible osmolyte reported only in plants to date, and so its presence in Mtb might represent a previously unidentified component of osmotic adaptation in mycobacteria.18,19 In the second phase, NMR signals corresponding to glycerol levels decrease (250 mM NaCl) or increase (1000 mM NaCl), with those for trehalose and citrulline increasing further. No changes were observed in the NMR signals reporting on glutamate, glutamine, glycine betaine, and proline, known compatible osmolytes found in bacteria,17,20 decreasing the likelihood that these metabolites act as osmolytes in Mtb. In contrast to M. smegmatis and other bacteria, Mtb lacks the genes encoding the enzymes that synthesize known compatible osmolytes such as ectoine and N-acetyl-Gln-Gln-amide,21,22 which were also not detected in the metabolome of Mtb. Figure 3B summarizes the full composition of the five most abundant osmolytes in Mtb at 48 h.
Figure 3.
Metabolism is affected by changes in osmolarity. (A) Absolute quantification of glycerol, trehalose, and citrulline, at 10 (blue line), 250 (orange line) and 1000 mM (red line) NaCl. (B) Pie charts illustrating the distribution of the compatible osmolytes from Mtb H37Rv. (C) Metabolic changes during halotolerance: pool size and 13C labeling of selected metabolites during exposure to 10 or 250 mM NaCl. Pool size is expressed as a ratio compared to time 0 h after normalization to the amount of proteins (Y-axis), and X-axis represents the time in hours after exposure to 10 or 250 mM NaCl. All data shown are representative of three independent experiments. G-3P, glycerol 3-phosphate; DHAP, dihydroxyacetone phosphate; G3P, glyceraldehyde 3-phosphate; FBP, fructose 1,6-bisphosphate; PYR, pyruvate; ALA, alanine; ACO, aconitate; α-KG, α-ketoglutarate; SUC, succinate; MAL, malate; OAA, oxaloacetate; ASP, aspartate; GLU, glutamate; NAG, N-acetyl-glutamate; ORN, ornithine; CITR, citrulline; ARGSUCC, l-arginosuccinate; ARG, arginine. (D) Wash-out experiments for NaCl concentration shifts after 8 h of exposure to 1000 mM NaCl for Mtb H37Rv followed by exposure to 10 mM NaCl.
To confirm these results and more specifically characterize changes in labeling and pool sizes of metabolites from intermediary and secondary metabolism, we employed liquid chromatography–mass spectrometry (LC-MS) metabolomics, combined with 13C labeling.23,24 Similar to changes previously observed after substituting carbon sources23 and during exposure to low oxygen,25 abundance and labeling of molecules involved in intermediary metabolism (glycolysis, gluconeogenesis, and the Krebs cycle (SI Appendix, Table S1) were broadly altered at 250 mM NaCl (Figure 3C), including significant changes in pool size and labeling of hexose-phosphate, glycerol-phosphate, alanine, α-ketoglutarate, succinate, and malate. Flux through the Krebs cycle appears to be concentrated in the reductive branch, at physiologic NaCl concentrations. Succinate production and secretion has been shown to be required in Mtb to maintain a viable membrane potential under hypoxia.25,26 In agreement with this notion, we have observed an increase in membrane potential at physiologic salt concentrations (Figure 2D). Importantly, this interpretation is in line with the current view of nonreplication states in Mtb, in which cells remain metabolically active despite an increase in generation time or lack of replication.27
To a greater extent than intermediary metabolism, nonproteinogenic amino acid metabolism is drastically affected at physiologic concentrations of NaCl. The synthesis of N-acetyl-glutamate, ornithine, and citrulline, but not arginine, is significantly up-regulated, as illustrated by an increase in both pool size and 13C labeling (Figure 3C; SI Appendix, extended data), in close agreement with our NMR metabolomics data. Importantly, the metabolic changes observed are carbon-source independent (SI Appendix, Figure S4) and rapidly reversible (Figure 3D). It is noteworthy that our time-dependent metabolic results indicate that Mtb metabolism is substantially perturbed by osmolarity. However, all changes observed are reversible and rapidly revert to basal levels after decreasing salinity. Because these metabolic changes occur and reverse more rapidly than drug effects (Figure 1D), these particular metabolite perturbations are not likely sufficient to cause the drug tolerance observed, although they might represent intermediate events that control or signal downstream events in phenotypic remodeling.
Physiologic Concentrations of NaCl Slowly Change Cell Envelope Composition
Alterations in cell envelope composition have been described during adaptation of other bacterial species to higher salinity.28−34 It is also well documented that growth in high-salinity medium alters the phospholipid head groups and fatty acid composition of bacterial cytoplasmic membranes, by altering the ratio of anionic to zwitterionic lipids.28−32 However, Mtb and other actinobacteria have a unique cell wall composed of separate cytoplasmic and mycolate outer membranes (MOM) that is fundamentally different from the phospholipid bilayers present in other bacteria. To assess lipid remodeling linked to changes in saline concentration, we employed a normal phase HPLC-MS lipidomics approach,35 which measures whole cell lipidomes composed of >10,000 combined ion features in positive and negative ion modes (Figure 4A,B). After generating lipidomes in triplicate and aligning the signals for each lipid present in Mtb grown in low- or high-salt media, comparative lipidomics analysis assesses 12,062 molecular features, which are defined as linked m/z, retention time, and intensity values. Individual lipids are considered changed by salt when they show a 2-fold change in intensity that meets previously validated statistical tests (corrected p < 0.05).35 Of these, 1688 (14%) features were altered when Mtb was cultured in the presence of 1000 mM NaCl for 24 h compared to 10 mM NaCl. Diacylphosphatidylinositol-hexamannoside abundance increased 5-fold in Mtb exposed to high saline (SI Appendix, Figure S6a,b), whereas plasma membrane associated zwitterionic and anionic lipids such as phosphatidylethanolamine and phosphatidylglycerol display 2–5-fold decrease in abundance at 1000 mM NaCl, respectively (Figure 4C; SI Appendix, Figure S6c,d). These results are consistent with previous observations showing that plasma membrane glycerophospholipids are dynamic in Mtb.24 On the basis of the changes observed in glycerophospholipid polar heads, in particular the enrichment with bulkier diacylphosphatidylinositol-hexamannoside, and the sensitivity of mechanosensitive ion channels to plasma membrane lateral pressure, it is likely that under high saline concentrations these channels are going to be closed.
Figure 4.
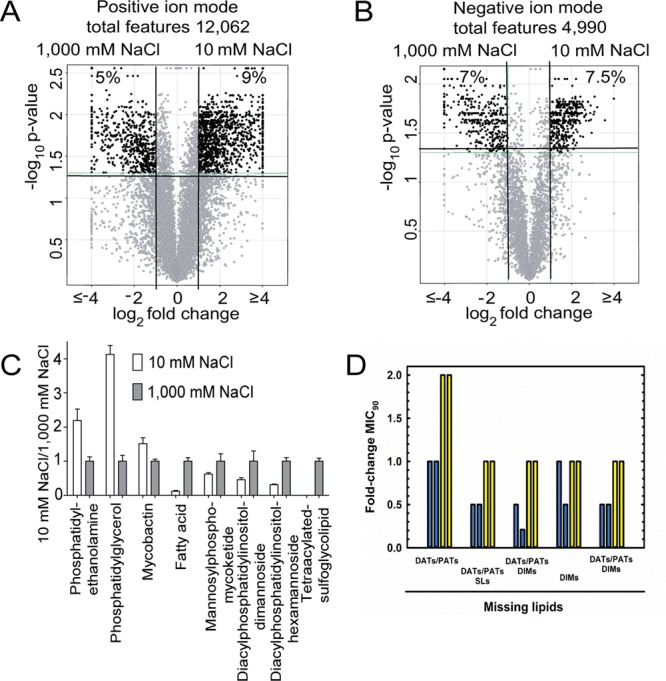
Cell envelope composition is profoundly affected by changes in osmolarity. (A, B) After pairing ions that show equivalent mass and retention time in negative and positive modes, volcano plots illustrate the fold-change in intensity for each paired molecular event. Percent change is calculated as the number of molecular events with intensity fold-change >2 (corrected p < 0.05) divided by the total number of paired events. (C) Selected named lipids of interest were evaluated by comparing their mass and retention times to those of known values in the MycoMass or MycoMap databases and followed by manual reanalysis with ion chromatograms and confirmation of structures by collision induced dissociation mass spectrometry (Figure S5). Data are shown as the mass intensity ratios detected when Mtb is grown at 10 and 1000 mM NaCl. Data are generated from biological triplicate samples and are representative of at least two independent experiments. (D) MIC90 values for GAT against Mtb mutants defective in cell-envelope lipid biosynthesis. Mtb strains were pre-adapted in 10 (blue bars) or 250 (yellow bars) mM NaCl. Each bar indicates data from one independent experiment.
Considering lipids in the MOM, a large increase was observed in the signal for tetra-acylated sulfolipids (5–10-fold) (SI Appendix, Figure S6e), which are abundant surface-exposed polyketides that mediate direct interactions with the host.36 In measuring phosphatidyl-myo-inositol mannosides (PIMs) with two or more fatty acids, we noted a significant increase in signal for structures with higher acylation state (e.g., Ac2PIM2 and Ac2PIM6) when NaCl concentration shifts to 250 and 1000 mM (Figure 4C; SI Appendix, Figure S6h–n). Consistent with this observation, 13C labeling experiments indicate that even at 250 mM, glycerophosphoinositol is turning over 4-fold more rapidly than at 10 mM (33% labeling in 24 h in contrast to 15% labeling in 48 h at 10 mM NaCl) (SI Appendix, Figure S7). Of note, PIMs account for 45% of the total lipids found in the mycobacterial plasma membrane,37 and therefore these changes are likely to have significant effects on permeability and membrane fluidity, which in turn protect the cell against increased salinity and osmotic pressure. Despite reports of the roles of ornithine-containing lipids in Rhizobium tropici,38 their existence in Mtb39,40 and the recently discovered osmo-protective role of ubiquinone in Escherichia coli(41) (with menaquinone as a counterpart in Mtb), no changes in their intensity were observed here (SI Appendix, Figure S6n–q). Taken together, these results indicate a substantial remodeling of Mtb plasma and outer membrane lipids during adaptation to high salinity. Of note, these alterations are substantially slower (Figure 5) than metabolic alterations and therefore kinetically matched with the time frame required for the phenotypic antibacterial tolerance observed. These slower, day-long kinetics of changes in response to salt concentrations are similar to the slow cell envelope alterations that occur in response to Mtb’s entry and exit from bacteriostasis in vitro.42
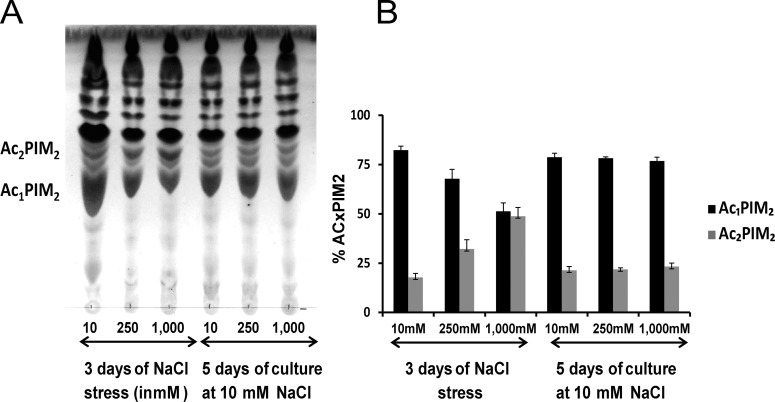
Figure 5.
Cell envelope composition affected by changes in osmolarity is fully reversible. (A) Thin layer chromatography of total lipids from Mtb H37Rv for 3 days of incubation at 10, 250, and 1000 mM NaCl, followed by incubation at 10 mM NaCl for 5 days. The TLC was run in the solvent system 60:25:4 CHCl3/CH3OH/H2O (v/v/v) and developed by phosphomolybdic acid 5% in ethanol followed by heat 10 min at 100 °C. (B) Bar graph showing the percentage of AcxPIM2 during recovery experiment.
To further investigate cell envelope remodeling, we employed Mtb mutants defective in the synthesis of three important types of lipids: acyltrehaloses (DATs/PATs), sulfolipids (SLs), and dimycocerosates (DIMs).43 We evaluate MIC90 values for these strains alongside their parent strains in low and physiologic saline concentrations. If cell envelope remodeling elicited by higher saline concentrations causes phenotypic antibacterial tolerance, any changes that are caused by deletion of single lipids or families of lipids should be minimized in the presence of physiologic concentrations of saline, as a substantial fraction of cell envelope lipids will be altered and likely compensate for individual absences. In contrast, if metabolism or another process dissociated from cell envelope composition triggers phenotypic tolerance, changes should be maintained at both low and high saline. As can be seen in Figure 4D, loss of SLs and DIMs alters sensitivity to GAT at 14.5 mM NaCl but not loss of DATs/PATs, indicating that changes in cell envelope composition might be a cause of altered sensitivity to antibacterial at low salinity. In agreement with our hypothesis, when these strains are pre-incubated at physiologic saline concentrations (250 mM), all MIC values are higher, indicating weaker sensitivity to GAT caused by cell envelope remodeling. In summary, these results are consistent with the notion that the full complement of cell envelope changes (14% of the lipidome) is contributing to phenotypic tolerance observed at physiologic saline.
Conclusion
Currently, antibacterial resistance poses a serious threat to human health, with multidrug and extensive drug resistant tuberculosis emerging as one of the world’s most serious epidemics. Our data characterize Mtb’s response to osmotic stress within the physiologic range of NaCl and demonstrate its effects on Mtb growth, intermediary and secondary metabolism, and cell envelope composition. The changes in plasma membrane and cell wall lipid composition ultimately trigger phenotypic antibacterial tolerance, instead of growth rate and metabolic changes. Importantly, our results also highlight a disconnection between the in vitro conditions most often used to study Mtb and to evaluate antibacterial response and physiological saline conditions, making a case for drug screening at physiologic NaCl concentrations. Finally, these results indicate that an understanding of Mtb’s response to changes in salinity associated with different body fluids and compartments will provide new insights into Mtb’s ability to cause disease and resist antibacterial therapy.
Methods
Materials, methods, and detailed experimental procedures are provided in the SI Appendix. The SI Appendix includes one material and methods document. The SI Appendix also contains MIC90 results obtained with a panel of clinical strains of Mtb (SI Figure 1), CFU measurement of Mtb, M. bovis BCG, and M. smegmatis exposed to different concentrations of NaCl (SI Figure 2), acid-fast staining of Mtb exposed to various NaCl concentrations (SI Figure 3), representative NMR metabolomics data illustrating changes accompanying adaptation to high NaCl concentrations (SI Figure 4), additional metabolomic data demonstrating that the changes observed are carbon source-independent and that the changes in ornithine and citrulline pool sizes are distinct in M. bovis BCG and in M. smegmatis (SI Figure 5), structures, extracted ion currents and collision-induced fragmentation data for the main lipids described in this study; structures, thin-layer chromatography, and MALDI-ToF results for acylated PIMs (SI Figure 6), and 13C labeling and pool size profiles for lipid polar heads (SI Figure 7); mass spectrometry data on metabolites analyzed (SI Table S1); and labeling data for metabolites obtained from Mtb exposed to physiologic and hypotonic conditions (SI extended data).
Acknowledgments
We thank Douglas B. Young, Jonathan P. Stoye, Anthony A. Holder, Alex Gould, and Angelika Gründling for critical reading of the manuscript. We also thank Christophe Guilhot and his laboratory for the Mtb knockout strains used in this work. This work was supported by funds from the Francis Crick Institute, which receives its core funding principally from Wellcome Trust, Cancer Research UK, and the UK Medical Research Council (to L.P.S.C. and M.G.G., MC_UP_A253_1111 and MC_UP_1202/11, respectively), funds from Wellcome Trust (Investigator Award to L.P.S.C. 104785/B/14/Z), start-up funds from Imperial College London (to G.L.M.), funds from FAPESP to F.R.P. (2013/14957-5), funds from FAPESP, CNPq, and CAPES-PDSE to L.B.M. (2011/21232-1, 140079/2013-0, 99999.003125/2014-09), and funds from the NIH to D.B.M. (R01 AI071155 and AI049313). The MRC Biomedical NMR Centre is funded by MRC Grant-in-Aid U117533887.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.5b00148.
Materials, bacterial strains and conditions, acid-fast staining, membrane potential, metabolomics, lipidomics, antibiotics, references, figures, and tables (PDF)
Author Contributions
G.L.-M., L.B.M, A.V.R.M., L.B., T.J.R., D.M.H., and M.G.G. designed and conducted experiments. G.L.-M., L.B.M, A.V.R.M., T.J.R., D.M.H., M.G.G., D.B.M., F.R.P., and L.P.S.C. analyzed and interpreted data. G.L.-M, L.B.M., D.B.M., and L.P.S.C. wrote the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Levin B. R.; Rozen D. E. (2006) Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 4, 556–562. 10.1038/nrmicro1445. [DOI] [PubMed] [Google Scholar]
- Martinez J. L.; Rojo F. (2011) Metabolic regulation of antibiotic resistance. FEMS Microbiol Rev. 35, 768–789. 10.1111/j.1574-6976.2011.00282.x. [DOI] [PubMed] [Google Scholar]
- Proctor R. A.; Kriegeskorte A.; Kahl B. C.; Becker K.; Loffler B.; Peters G. (2014) Staphylococcus aureus small colony variants (SCVs): a road map for the metabolic pathways involved in persistent infections. Front. Cell. Infect. Microbiol. 4, 99. 10.3389/fcimb.2014.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobritz M. A.; Belenky P.; Porter C. B.; Gutierrez A.; Yang J. H.; Schwarz E. G.; Dwyer D. J.; Khalil A. S.; Collins J. J. (2015) Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. U.S.A. 112, 8173–8180. 10.1073/pnas.1509743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I.; Shah D.; Spoering A.; Kaldalu N.; Lewis K. (2004) Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186, 8172–8180. 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyed H. S.; Bertrand K. P. (1983) hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155, 768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye C.; Williams B. G. (2010) The population dynamics and control of tuberculosis. Science 328, 856–861. 10.1126/science.1185449. [DOI] [PubMed] [Google Scholar]
- Ehrt S.; Schnappinger D. (2009) Mycobacterial survival strategies in the phagosome: defence against host stresses. Cell. Microbiol. 11, 1170–1178. 10.1111/j.1462-5822.2009.01335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S.; Sukumar N.; Abramovitch R. B.; Parish T.; Russell D. G. (2013) Mycobacterium tuberculosis responds to chloride and pH as synergistic cues to the immune status of its host cell. PLoS Pathog. 9, e1003282. 10.1371/journal.ppat.1003282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabner J.; Smith J. J.; Karp P. H.; Widdicombe J. H.; Welsh M. J. (1998) Loss of CFTR chloride channels alters salt absorption by cystic fibrosis airway epithelia in vitro. Mol. Cell 2, 397–403. 10.1016/S1097-2765(00)80284-1. [DOI] [PubMed] [Google Scholar]
- Bourque C. W. (2008) Central mechanisms of osmosensation and systemic osmoregulation. Nat. Rev. Neurosci. 9, 519–531. 10.1038/nrn2400. [DOI] [PubMed] [Google Scholar]
- Hatzios S. K.; Baer C. E.; Rustad T. R.; Siegrist M. S.; Pang J. M.; Ortega C.; Alber T.; Grundner C.; Sherman D. R.; Bertozzi C. R. (2013) Osmosensory signaling in Mycobacterium tuberculosis mediated by a eukaryotic-like Ser/Thr protein kinase. Proc. Natl. Acad. Sci. U.S.A. 110, E5069–5077. 10.1073/pnas.1321205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne L. G.; Sramek H. A. (1994) Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 38, 2054–2058. 10.1128/AAC.38.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata M.; Pavan F. R.; Sato D. N.; Marino L. B.; Hirata M. H.; Cardoso R. F.; de Melo F. A.; Zanelli C. F.; Leite C. Q. (2011) Drug resistance in Mycobacterium tuberculosis clinical isolates from Brazil: phenotypic and genotypic methods. Biomed. Pharmacother. 65, 456–459. 10.1016/j.biopha.2011.04.021. [DOI] [PubMed] [Google Scholar]
- Shapiro H. M. (2008) Flow cytometry of bacterial membrane potential and permeability. Methods Mol. Med. 142, 175–186. 10.1007/978-1-59745-246-5_14. [DOI] [PubMed] [Google Scholar]
- de Carvalho L. P.; Darby C. M.; Rhee K. Y.; Nathan C. (2011) Nitazoxanide disrupts membrane potential and intrabacterial pH homeostasis of Mycobacterium tuberculosis. ACS Med. Chem. Lett. 2, 849–854. 10.1021/ml200157f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleator R. D.; Hill C. (2002) Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev. 26, 49–71. 10.1111/j.1574-6976.2002.tb00598.x. [DOI] [PubMed] [Google Scholar]
- Kawasaki S.; Miyake C.; Kohchi T.; Fujii S.; Uchida M.; Yokota A. (2000) Responses of wild watermelon to drought stress: accumulation of an ArgE homologue and citrulline in leaves during water deficits. Plant Cell Physiol. 41, 864–873. 10.1093/pcp/pcd005. [DOI] [PubMed] [Google Scholar]
- Kusvuran S.; Dasgan H. Y.; Abak K. (2013) Citrulline is an important biochemical indicator in tolerance to saline and drought stresses in melon. Sci. World J. 2013, 253414. 10.1155/2013/253414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. M.; Bremer E.; Csonka L. N.; Kraemer R.; Poolman B.; van der Heide T.; Smith L. T. (2001) Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol., Part A: Mol. Integr. Physiol. 130, 437–460. 10.1016/S1095-6433(01)00442-1. [DOI] [PubMed] [Google Scholar]
- Cole S. T.; Brosch R.; Parkhill J.; Garnier T.; Churcher C.; Harris D.; Gordon S. V.; Eiglmeier K.; Gas S.; Barry C. E. 3rd; Tekaia F.; Badcock K.; Basham D.; Brown D.; Chillingworth T.; Connor R.; Davies R.; Devlin K.; Feltwell T.; Gentles S.; Hamlin N.; Holroyd S.; Hornsby T.; Jagels K.; Krogh A.; McLean J.; Moule S.; Murphy L.; Oliver K.; Osborne J.; Quail M. A.; Rajandream M. A.; Rogers J.; Rutter S.; Seeger K.; Skelton J.; Squares R.; Squares S.; Sulston J. E.; Taylor K.; Whitehead S.; Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Sagot B.; Gaysinski M.; Mehiri M.; Guigonis J. M.; Le Rudulier D.; Alloing G. (2010) Osmotically induced synthesis of the dipeptide N-acetylglutaminylglutamine amide is mediated by a new pathway conserved among bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 12652–12657. 10.1073/pnas.1003063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho L. P.; Fischer S. M.; Marrero J.; Nathan C.; Ehrt S.; Rhee K. Y. (2010) Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem. Biol. 17, 1122–1131. 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Larrouy-Maumus G.; Biswas T.; Hunt D. M.; Kelly G.; Tsodikov O. V.; de Carvalho L. P. (2013) Discovery of a glycerol 3-phosphate phosphatase reveals glycerophospholipid polar head recycling in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 110, 11320–11325. 10.1073/pnas.1221597110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eoh H.; Rhee K. Y. (2013) Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 110, 6554–6559. 10.1073/pnas.1219375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S.; Zimmermann M.; Goodwin M. B.; Sauer U.; Barry C. E. 3rd; Boshoff H. I. (2011) Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog. 7, e1002287. 10.1371/journal.ppat.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eoh H.; Rhee K. Y. (2014) Methylcitrate cycle defines the bactericidal essentiality of isocitrate lyase for survival of Mycobacterium tuberculosis on fatty acids. Proc. Natl. Acad. Sci. U.S.A. 111, 4976–4981. 10.1073/pnas.1400390111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romantsov T.; Guan Z.; Wood J. M. (2009) Cardiolipin and the osmotic stress responses of bacteria. Biochim. Biophys. Acta, Biomembr. 1788, 2092–2100. 10.1016/j.bbamem.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajbidor J. (1997) Effect of some environmental factors on the content and composition of microbial membrane lipids. Crit. Rev. Biotechnol. 17, 87–103. 10.3109/07388559709146608. [DOI] [PubMed] [Google Scholar]
- Lopez C. S.; Alice A. F.; Heras H.; Rivas E. A.; Sanchez-Rivas C. (2006) Role of anionic phospholipids in the adaptation of Bacillus subtilis to high salinity. Microbiology 152, 605–616. 10.1099/mic.0.28345-0. [DOI] [PubMed] [Google Scholar]
- Lopez C. S.; Heras H.; Ruzal S. M.; Sanchez-Rivas C.; Rivas E. A. (1998) Variations of the envelope composition of Bacillus subtilis during growth in hyperosmotic medium. Curr. Microbiol. 36, 55–61. 10.1007/s002849900279. [DOI] [PubMed] [Google Scholar]
- Vargas C.; Kallimanis A.; Koukkou A. I.; Calderon M. I.; Canovas D.; Iglesias-Guerra F.; Drainas C.; Ventosa A.; Nieto J. J. (2005) Contribution of chemical changes in membrane lipids to the osmoadaptation of the halophilic bacterium Chromohalobacter salexigens. Syst. Appl. Microbiol. 28, 571–581. 10.1016/j.syapm.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Allakhverdiev S. I.; Kinoshita M.; Inaba M.; Suzuki I.; Murata N. (2001) Unsaturated fatty acids in membrane lipids protect the photosynthetic machinery against salt-induced damage in Synechococcus. Plant Physiol. 125, 1842–1853. 10.1104/pp.125.4.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y. S.; Yamaryo-Botte Y.; Miyanagi K.; Callaghan J. M.; Patterson J. H.; Crellin P. K.; Coppel R. L.; Billman-Jacobe H.; Kinoshita T.; McConville M. J. (2010) Stress-induced synthesis of phosphatidylinositol 3-phosphate in mycobacteria. J. Biol. Chem. 285, 16643–16650. 10.1074/jbc.M110.119263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layre E.; Sweet L.; Hong S.; Madigan C. A.; Desjardins D.; Young D. C.; Cheng T. Y.; Annand J. W.; Kim K.; Shamputa I. C.; McConnell M. J.; Debono C. A.; Behar S. M.; Minnaard A. J.; Murray M.; Barry C. E. 3rd; Matsunaga I.; Moody D. B. (2011) A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chem. Biol. 18, 1537–1549. 10.1016/j.chembiol.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenech P.; Reed M. B.; Dowd C. S.; Manca C.; Kaplan G.; Barry C. E. 3rd (2004) The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J. Biol. Chem. 279, 21257–21265. 10.1074/jbc.M400324200. [DOI] [PubMed] [Google Scholar]
- Bansal-Mutalik R.; Nikaido H. (2014) Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proc. Natl. Acad. Sci. U.S.A. 111, 4958–4963. 10.1073/pnas.1403078111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vences-Guzman M. A.; Guan Z.; Ormeno-Orrillo E.; Gonzalez-Silva N.; Lopez-Lara I. M.; Martinez-Romero E.; Geiger O.; Sohlenkamp C. (2011) Hydroxylated ornithine lipids increase stress tolerance in Rhizobium tropici CIAT899. Mol. Microbiol. 79, 1496–1514. 10.1111/j.1365-2958.2011.07535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laneelle M. A.; Laneelle G.; Asselineau J. (1963) [On the presence of ornithine in bacterial lipids]. Biochim. Biophys. Acta 70, 99–101. 10.1016/0006-3002(63)90727-3. [DOI] [PubMed] [Google Scholar]
- Laneelle M. A.; Prome D.; Laneelle G.; Prome J. C. (1990) Ornithine lipid of Mycobacterium tuberculosis: its distribution in some slow- and fast-growing mycobacteria. J. Gen. Microbiol. 136, 773–778. 10.1099/00221287-136-4-773. [DOI] [PubMed] [Google Scholar]
- Sevin D. C.; Sauer U. (2014) Ubiquinone accumulation improves osmotic-stress tolerance in Escherichia coli. Nat. Chem. Biol. 10, 266–272. 10.1038/nchembio.1437. [DOI] [PubMed] [Google Scholar]
- Galagan J. E.; Minch K.; Peterson M.; Lyubetskaya A.; Azizi E.; Sweet L.; Gomes A.; Rustad T.; Dolganov G.; Glotova I.; Abeel T.; Mahwinney C.; Kennedy A. D.; Allard R.; Brabant W.; Krueger A.; Jaini S.; Honda B.; Yu W. H.; Hickey M. J.; Zucker J.; Garay C.; Weiner B.; Sisk P.; Stolte C.; Winkler J. K.; Van de Peer Y.; Iazzetti P.; Camacho D.; Dreyfuss J.; Liu Y.; Dorhoi A.; Mollenkopf H. J.; Drogaris P.; Lamontagne J.; Zhou Y.; Piquenot J.; Park S. T.; Raman S.; Kaufmann S. H.; Mohney R. P.; Chelsky D.; Moody D. B.; Sherman D. R.; Schoolnik G. K. (2013) The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499, 178–183. 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passemar C.; Arbues A.; Malaga W.; Mercier I.; Moreau F.; Lepourry L.; Neyrolles O.; Guilhot C.; Astarie-Dequeker C. (2014) Multiple deletions in the polyketide synthase gene repertoire of Mycobacterium tuberculosis reveal functional overlap of cell envelope lipids in host-pathogen interactions. Cell. Microbiol. 16, 195–213. 10.1111/cmi.12214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.