Summary
Smads are nuclear-shuttling transcriptional mediators of transforming growth factor-β (TGF-β) signaling. While their essential nuclear roles in gene regulation during development and carcinogenesis are well established, whether they have important cytoplasmic functions remains unclear. Here we report that Smad2 is a critical determinant of mitochondrial dynamics. We identified mitofusin 2 (Mfn2) and Rab and Ras interactor 1 (RIN1) as new Smad2 binding partners required for mitochondrial fusion. Unlike TGF-β-induced Smad2/3 transcriptional responses underlying mitochondrial fragmentation and apoptosis, inactive cytoplasmic Smad2 rapidly promotes mitochondrial fusion by recruiting RIN1 into a complex with Mfn2. We demonstrate that Smad2 is a key scaffold, allowing RIN1 to act as a GTP exchange factor (GEF) for Mfn2-GTPase activation to promote mitochondrial ATP synthesis and suppress superoxide production. These results reveal functional implications between Smads and mitochondrial dysfunction in cancer, metabolic, and neurodegenerative disorders.
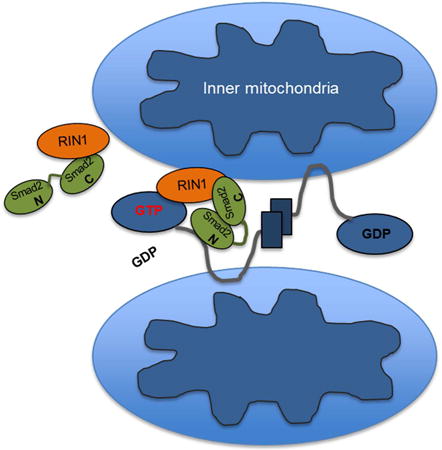
Introduction
Receptor-activated Smads (R-Smads: Smad2/3 and Smad1/5/8) represent an important subset of substrates for the type I serine/threonine receptor kinases, ALK5/TβRI and bone morphogenetic protein (BMP) receptors. Upon phosphorylation, R-Smads translocate to the nucleus to control numerous genes in a highly context-dependent manner (Massague, 2012; Massague et al., 2005; Samanta and Datta, 2012). R-Smads also regulate microRNA expression and processing in the nucleus by directly engaging key components of the microRNA processing machinery (Blahna and Hata, 2012).
The duration and intensity of R-Smad activation are key determinants of TGF-β signaling specificity and versatility (Derynck and Zhang, 2003; Massague, 2012; Massague et al., 2005). In addition to type I receptors, a growing number of negative regulators including kinases, phosphatases, and cytoplasmic-anchoring proteins ensure proper transcriptional control (Derynck and Zhang, 2003; Massague, 2012). Interestingly, despite numerous cytoplasmic binding partners that have been identified in the past two decades, whether R-Smads have any distinct functional roles in the cytoplasm remains unclear.
Mitochondria are highly dynamic, interconnected organelles that continuously undergo fusion and fission (Lackner, 2014; Liesa et al., 2009; Ranieri et al., 2013; Zorzano et al., 2009). The crucial balance between these opposing forces determine the overall mitochondrial architecture, regulated by dynamin related proteins (DRPs), which are a family of large GTPases that rely on GTP-dependent self-oligomerization or scission of mitochondrial membranes (Lackner, 2014; Liesa et al., 2009; Ranieri et al., 2013; Zorzano et al., 2009). Arguably, less is known about the fusion process, although at least three major proteins are involved in the mammalian system: mitofusins 1 and 2 (Mfn), which are DRP mediators of outer-membrane fusion, and optic atrophy protein 1 (OPA1) that orchestrates the inner membrane fusion and maintains membrane potential. Structurally homologous Mfn1 and Mfn2 proteins have their N-and C-termini exposed to the cytoplasm, and form homomeric and heteromeric complexes to induce mitofusion. Recent studies demonstrate several posttranslational mediators of Mfn turnover including PTEN-induced kinase 1 (PINK1)/Parkin induced ubiquitination and proteasomal degradation (Chen and Dorn, 2013) as well as phosphorylation-induced proteasomal degradation in stress-induced apoptosis (Leboucher et al., 2012).
Modulation of GTPase activity is another key mechanism by which Mfn is regulated. Structural studies show that their catalytic domains require efficient GDP to GTP nucleotide exchange in order to initiate the trans-oligomerization process necessary for membrane fusion (Lackner, 2014; Liesa et al., 2009; Ranieri et al., 2013; Zorzano et al., 2009). Indeed, more than 40 inactivating human mutations on Mfn have been identified, many of which reside along the GTPase domain (Honda et al., 2005; Liesa et al., 2009; Ranieri et al., 2013). However, despite the critical importance of the Mfn2-GTPase, how its catalytic function is regulated remains less clear, and no direct modulators (negative or positive) have been characterized.
Here our work defines a new paradigm for Smad function. We demonstrate that Smad2 can act in a transcription-independent manner to regulate mitochondrial dynamics and metabolic functions.
Results
Smad2 Interacts with Mfn1 and Mfn2
As Smad2 and 3 represent canonical transcriptional effectors of TGF-β signaling, we screened for Smad2 interactomes by mass spectrometry. Immunoprecipitation following serum deprivation allowed for enrichment of inactive Smad2 generally localized in the cytoplasm. Among a number of potential hits from the mass spectrometry/proteomics analysis, we noted a mitochondria outer-membrane GTPase, Mfn2 (Figure S1A, B) (Hales and Fuller, 1997; Knott et al., 2008; Lackner, 2014; Ranieri et al., 2013; Santel and Fuller, 2001; Suen et al., 2008). Subsequent co-immunoprecipitation studies confirmed a strong interaction between Smad2 and Mfn2 in both overexpression and endogenous systems (Figure 1A and B). Smad2 also associated with Mfn1 albeit in a less robust manner (Figure 1C). Surprisingly, Smad3 failed to co-precipitate with either Mfn1 or Mfn2 despite the high degree of structural homology to Smad2 (Figure 1A, C). Two other prominent Smad2 binding partners did not associate with Mfn2; SARA, a Smad2 cytoplasmic-retention protein; and Smad4, a co-Smad that facilitates phosphorylated R-Smads to translocate to the nucleus (Shi et al., 1997; Tsukazaki et al., 1998; Wu et al., 2001) (Figure S2A, B). Similarly, Smad2 failed to interact with dynamin-related protein 1 (Drp1), a GTPase that associates with the mitochondrial outer-membrane to induce mitochondrial fission (Figure S2C) (Smirnova et al., 2001).
Figure 1. Inactive Smad2 interacts with Mfn2 and enhances mitochondrial fusion.
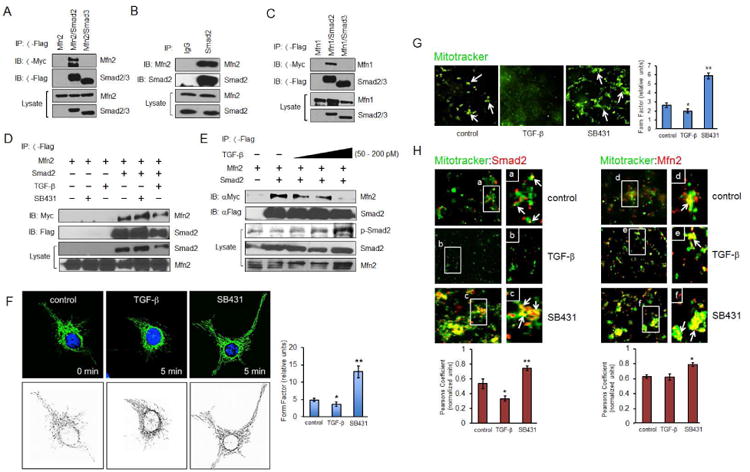
(A) COS-7 cells expressing Myc-Mfn2, Flag-Smad2, and Flag-Smad3 were immunoprecipitated with Flag antibody. (B) COS-7 cell lysate was immunoprecipitated with either IgG control or total Smad2 antibody. (C) COS-7 cells expressing Myc-Mfn1, Flag-Smad2, or Flag-Smad3 were immunoprecipitated with Flag antibody. (D) COS-7 cells expressing Myc-Mfn2 with or without Flag-Smad2 were treated with SB431542 (30 μM) or TGF-β1 (200 pM) for 30 min prior to co-immunoprecipitation. (E) COS-7 cells expressing Myc-Mfn2 with or without Flag-Smad2 were treated with increasing TGF-β1 concentrations (0, 50, 100, 200 pM) for 20 min. (F) HeLa cells were treated with SB431542 (30 μM) or TGF-β1 (200 pM) for 5 min prior to fixation, then imaged for Mfn2 immunofluorescence staining (upper three panels) and quantified using form factor by outlining the mitochondrion area of each cell (lower three panels). Quantification was based on data collected from at least 30 cells per condition from a representative experiment (out of at least three independent experiments). Error bars represent SEM and type 2 T-test results show relative to control: *p<0.05, **p=0.01. (G) COS-7 cells were subjected to serum deprivation for 3 h prior to treatment with MitoTracker (green) for 1 h and SB431542 (30 μM) or TGF-β1 and (200 pM) for 30 min. The cells were then fractionated and isolated mitochondria visualized with MitoTracker. Results shown are representative of 3 independent experiments. 20 to 30 images in the same treatment group from each experiment were analyzed and quantified using form factor. (H) COS-7 cells were subjected to serum deprivation for 3 h prior to treatment with MitoTracker for 1 h and SB431542 (30 μM) or TGF-β1 and (200 pM) for 30 min. Then fractionated mitochondria were labeled for Smad2 (red) or Mfn2 (red). Results are representative of 3 independent experiments. At least 20-30 images in the same treatment group from each experiment were quantified for co-localization using ImageJ and Pearsons coefficient analyses.
Inactive Smad2/Mfn2 Promotes Mitochondrial Fusion
TGF-β-induced ALK5 receptor activation causes Smad2 C-terminal phosphorylation and subsequent nuclear translocation (Massague, 2012; Massague and Chen, 2000; Massague et al., 2005). To determine the effects of TGF-β on Mfn2-binding, cells expressing Smad2 and Mfn2 were treated with either TGF-β or ALK5 inhibitor (SB431542). In both endogenous and overexpression systems, the Smad2/Mfn2 complex increased with SB431542, whereas TGF-β produced the opposite effect (Figure 1D and Figure S2D). Their dissociation also proved TGF-β concentration-dependent, resulting in complete abrogation at saturating concentration (200 pM) (Figure 1E). As before, Mfn1 bound weaker to Smad2 than Mfn2 and was devoid of responsiveness to TGF-β and ALK5 inhibition (Figure S2E). Given the critical role of Mfn2 in mitofusion, we tested how the Smad2/Mfn2 complex affects mitochondrial dynamics with a brief TGF-β or SB431542 treatment. Imaging of the mitochondria with Mfn2, mitotracker, and MTCO1 upon SB431542-treatment showed prominent fusion within 5 min whereas TGF-β attenuated this process in multiple cell types (Figure 1F, S2F, G).
Next, a cell fractionation was performed to visualize endogenous Smad2 and Mfn2 co-localization within isolated mitochondria. Imaging with mitotracker revealed a striking inverse correlation in the overall size of mitochondrial aggregates upon TGF-β and SB431542 treatment relative to control (Figure 1G). Importantly, a parallel experiment showed a robust increase in endogenous Smad2 and Mfn2 co-localization with isolated mitochondrial clusters upon SB431542 but not TGF-β treatment (Figure 1H and graph).
To gauge the overall kinetics of mitofusion, we performed live imaging on cells expressing a mitochondrial fluorescence reporter and observed rapid extension of mitochondrial networks upon treatment with SB431542 (Figure 2A). Given these dynamics, we next determined whether Smad2 is a key determinant by generating Smad2 knockdown stable clones in multiple cell types (sh-Smad2). At basal state, sh-Smad2 cells had greater mitochondrial fragmentation relative to control and further failed to respond to SB431542, suggesting that inactive Smad2 is crucial for mitofusion (Figure 2B). Similar results were obtained in other cell types with stable Smad2 knockdowns including in HeLa, mouse embryonic endothelial cells (MEEC) (Figure S3A-C) and A549 (data not shown), demonstrating that Smad2-dependent mitofusion is a general phenomenon.
Figure 2. Smad2 is required for efficient mitochondrial fusion.
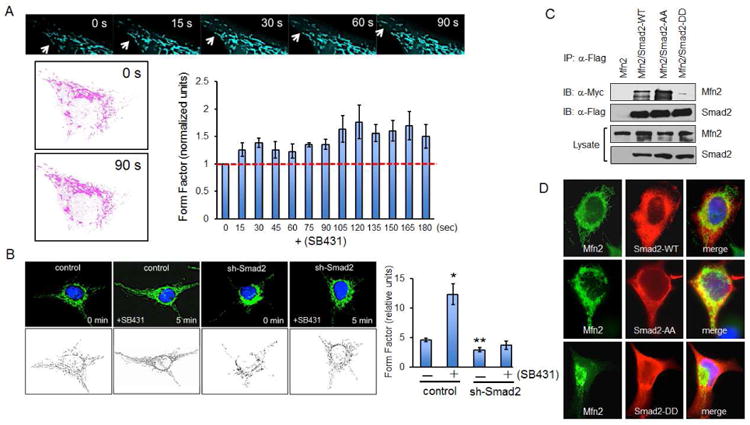
(A) MEECs expressing Turquoise2-Mito were serum starved for 4 h prior to fluorescence live cell confocal imaging. Cells were treated with SB431542 (30 μM) and imaged for 3 to 5 min. Pink outlines indicate the mitochondria at time 0 and 90 s. Graph represents the averaged form factor values for three cells at each time point. Error bars represent SEM. (B) HeLa control and Smad2 knockdown stables (sh-Smad2) were serum deprived for 3 h, then treated with SB431542 (30 μM) for 5 min prior to Mfn2 immunofluorescence staining. Graph is a representative form factor quantification of four independent experiments. Each condition shows the averaged form factor value (20 to 30 cells). Error bars represent SEM and type 2 T-test results show relative to control: *p<0.01; **p<0.03. (C) COS-7 cells expressing Myc-Mfn2, Flag-Smad2, Flag-Smad2-AA (constitutively inactive) and Flag-Smad2-DD (constitutively active) were immunoprecipitated with Flag antibody. (D) COS-7 cells expressing Mfn2, Flag-Smad2, Flag-Smad2-AA and Flag-Smad2-DD were co-stained with Mfn2 and Flag antibody. Immunofluorescence studies are representative data of at least three independent experiments.
In a reciprocal experiment, Mfn2-null (Mfn2-/-) mouse embryonic fibroblasts (MEFs) were used in the presence or absence of Smad2 depletion to test whether Smad2 mediates mitofusion primarily by targeting Mfn2 and not Mfn1. Previous studies have shown that loss of Mfn2 leads to mitochondrial fragmentation that are heterogenous in size and accumulate in perinuclear regions (Chen et al., 2003; Eura et al., 2003). Consistent with these reports, mitotracker and MTCO1 staining in Mfn2-/- MEFs showed a fragmented and perinuclear mitochondria irrespective of Smad2 expression (Figure S3D, E). Notably, neither TGF-β or SB431542 treatment had any impact on their mitochondrial morphology, indicating that the Mfn2/Smad2 complex is critical for TGF-β-based regulation of mitochondrial dynamics (Figure S3D, E).
As SB431542 inhibits Smad2 C-terminal phosphorylation, we tested whether its phosphorylation status modulates the Mfn2/Smad2 interaction by comparing the wild type (Smad2-WT), a nonphosphorylatable inactive (Smad2-AA) and constitutively active mutant (Smad2-DD). Co-immunoprecipitation results showed a far greater Mfn2 interaction with Smad2-AA than Smad2-WT but no interaction with Smad2-DD (Figure 2C). Consistent with this finding, Smad2-WT and Smad2-AA expression led to more extensive mitofusion than Smad2-DD (Figure 2D), suggesting that overall, inactive cytoplasmic Smad2 interacts with Mfn2 to promote mitochondrial fusion.
The Smad2 NH2-Domain Interacts with Mfn2 intermediate Coiled-coil region
To determine the critical structural determinants of the Mfn2/Smad2 complex, a panel of six Mfn2 deletion mutants was generated, comprising a GTPase domain truncation and five serial truncations distal to the GTPase containing several highly conserved regions (Figure 3A: M1-M6) (Honda et al., 2005). Smad2 immunoprecipitation efficiently pulled down M2, M3, M4 and M6 relative to the wild type (Mfn2-WT), indicating that the GTPase domain (M4: residues 80-270) and C-terminal regions including the Mfn2 dimerization domain are largely dispensable (M2, M3, M6: residues spanning 390 to 752) (Figure 3B). However, M1, a mutant harboring the largest deletion (residues 270 to 748) failed to interact with Smad2, whereas M5, which lacks residues 270 to 590, bound poorly (Figure 3B). Hence, the conserved structural region between M1 and M2 (residues 270 to 390) comprises the primary Smad2-binding element, although an auxiliary binding site exists within residues 590 to 748 to further enhance this interaction (Figure 3B). To determine whether Smad2-binding directly correlates with mitofusion, each mutant was expressed in COS7 and co-immunostained for Myc and Mfn2 (Figure S4A). Relative to control (vector), the WT expectedly increased mitofusion whereas most of the mutants had negligible impact with the exception of M4 (Figure S4A, B). Surprisingly, M4 expression was properly localized to the mitochondrial membrane and modestly enhanced fusion despite the GTPase deletion, likely via transoligomerization with neighboring endogenous Mfn2 (Figure S4A, B). Several mutants (M1-M3) failed to express along the normal mitochondrial membranes due to the absence of the transmembrane domain (TM), although their ectopic expression did not affect the overall mitochondrial morphology (Figure 3 and S4A, B). Still, the presence of the TM domain failed to properly target M5 to the mitochondrial network, suggesting that additional factors including Smad2 may be important mediators of Mfn2 activity and localization. In any event, as many of these mutants were able to bind Smad2 but failed to enhance mitofusion, the results indicated that the Smad2/Mfn2 interaction is critical but not sufficient for mitofusion.
Figure 3. Structural determinants of Mfn2 and Smad2 Interaction.
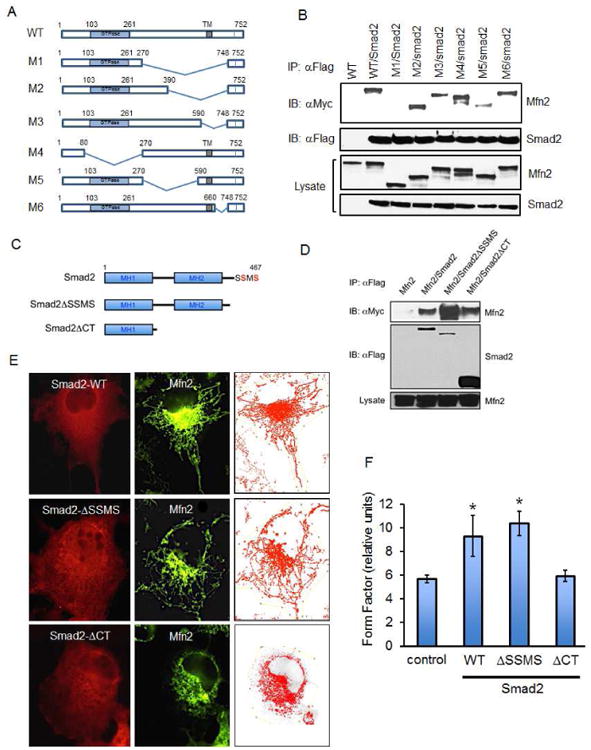
(A) Schematic of Mfn2-WT and deletion mutants M1-M6. (B) Western analysis of co-immunoprecipitation for Mfn2-WT and mutants (M1-M6) upon immunoprecipitation of Flag-Smad2 in COS-7 cells. (C) Schematic representation of Smad2 C-terminal phosphorylation motif (SSMS in red), and truncation mutants lacking the motif in SmadΔSSMS and Smad2ΔCT. (D) HeLa cells expressing Myc-Mfn2 with or without Flag-Smad2, Flag-Smad2ΔSSMS or Flag-Smad2ΔCT were immunoprecipitated with Flag antibody. (E-F) COS-7 cells expressing Flag-Smad2, Smad2ΔSSMS, or Smad2ΔCT were co-stained with Flag and Mfn2 antibodies. All western blots and immunofluorescence studies are representative data of at least three independent experiments and quantified using form factor analysis. Error bars represent SD and type 2 T-test results show relative to control: *p<0.0015.
To further understand the mechanisms underlying Smad2-dependent fusion events, we turned to the Smad2-binding elements. Like all receptor Smads, Smad2 consists of two globular domains, a highly conserved DNA-binding N-terminal Mad-homology 1 (MH1) domain, followed by a regulatory linker-region, and a C-terminal MH2 domain that can associate with ALK5, Smad4, and numerous transcription factors (Figure 3C) (Derynck and Zhang, 2003; Massague, 2012; Massague and Chen, 2000; Massague et al., 2005). We tested for Mfn2 interaction with Smad2 mutants either lacking the entire C-terminal half (Smad2ΔCT), or a short C-terminal truncation of the key phosphorylation sites responsible for transcriptional activity (Smad2ΔSSMS) (Figure 3C). The interaction between Mfn2 and Smad2ΔCT was similar to that of Smad2-WT while Smad2ΔSSMS had even greater affinity (Figure 3D). However, unlike the WT and Smad2ΔSSMS, Smad2ΔCT failed to enhance mitofusion over basal control levels, suggesting that the Smad2 C-terminal region contains an additional structural determinant involved in mitofusion (Figures 3E, F).
Smad2 Recruits RIN1 to Mediate Mitochondrial Fusion
To elucidate the functional role of the Smad2 C-terminal domain in mitofusion, we focused on yet another Smad2-binding candidate identified from our mass spectrometry interactome analysis- Ras and Rab Interactor 1 (RIN1), a Ras effector with reported guanine nucleotide exchange factor (GEF) function (Figure S4C) (Balaji et al., 2014; Deininger et al., 2008; Han et al., 1997; Tall et al., 2001; Wang et al., 2002). Since the Smad2/Mfn2 interaction itself was insufficient for enhanced mitofusion (Figures 3B, E and S4A, B), we hypothesized that Smad2 may recruit RIN1 into a complex to modulate Mfn2 function. We first tested whether RIN1 forms a trimeric complex with Smad2/Mfn2 by immunoprecipitating for endogenous RIN1 in cells expressing Mfn2 alone, Smad2 alone, or Mfn2 and Smad2 together (Figure 4A). Here, Mfn2 and Smad2 each interacted with RIN1 independently, although Mfn2 interaction was enhanced when Smad2 was co-expressed (Figure 4A: lanes 2 versus 4). Two additional biochemical experiments further demonstrated the formation of a trimeric complex, first at the endogenous level where immunoprecipitation of RIN1 resulted in co-precipitation of Smad2 and Mfn2 (Figure 4B), and second, by testing their direct interaction using purified Mfn2, Smad2 and RIN1 generated in the baculovirus/SF21 insect cell system (Figure S4D, E). The RIN1/Mfn2 interaction was greater upon Smad2ΔSSMS expression compared to Smad2-WT, again suggesting that the predominantly cytoplasmic-localized and transcriptionally inactive Smad2 augments RIN1/Mfn2 interaction (Figure S5A). Further validating the specificity of this novel trimeric complex, we found that RIN1 interacted with Mfn2 but not with Drp1, a similar GTPase localized to the mitochondrial outer-membrane (Figure S5B).
Figure 4. RIN1 acts as a GEF for Mfn2 GTP-binding.
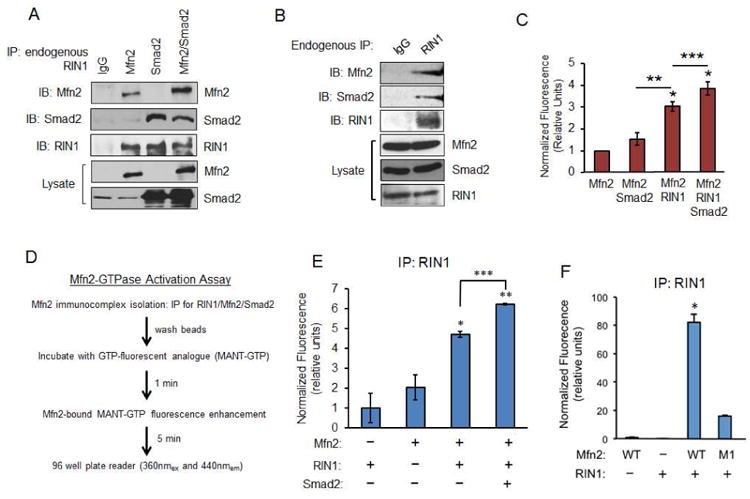
(A) COS-7 cells expressing Myc-Mfn2, Flag-Smad2, or together were immunoprecipitated with IgG control or RIN1 antibody, then immunobloted for Mfn2, Smad2, and RIN1. (B) COS-7 cell lysate was immunoprecipitated with either IgG control or RIN1 antibody and immunoblotted for endogenous Mfn2, RIN1 and Smad2. (C) Graph represents MANT-GTP emission upon binding purified Mfn2 (0.5 μM) alone, with purified Smad2 (0.5 μM), or Mfn2, Smad2 and RIN1 (0.5 μM each). Control MANT-GTP (2.5 μM) emission was subtracted from each condition, then normalized to Mfn2 alone plus MANT-GTP (2.5 μM). Error bars represent SE from three independent experiments (**p< 0.0008; ***p=0.01). (D) Immunocomplex-based MANT-GTP assay protocol. (E) Graph represents MANT-GTP emission of RIN1 complexed with Mfn2 or Mfn2/Smad2. Error bars represent the SE from three independent experiments normalized to RIN1 alone control (*p<0.0004, **p<0.001, ***p=0.01). (F) Graph represents MANT-GTP emission of RIN1 immunocomplex with Mfn2-WT or M1 in COS-7 cells. Error bars represent the SE from three independent experiments normalized to Mfn2 alone control (*p<0.001).
RIN1 acts as a Novel Mfn2 GEF
A number of recent studies demonstrated the importance of post-translational regulation of Mfn2 expression (Amiott et al., 2009; Chen and Dorn, 2013; Glauser et al., 2011; Leboucher et al., 2012). Despite these advances, the molecular mechanisms underlying the Mfn2-GTPase function remain elusive (Escobar-Henriques and Anton, 2013; Hales and Fuller, 1997; Hermann et al., 1998; Santel and Fuller, 2001). Given the evidence demonstrating Smad2 and RIN1 as new Mfn2 binding partners, we devised a fluorescence-based Mfn2-GTPase assay to test whether RIN1 acts as a guanine nucleotide exchange factor (GEF) for Mfn2. Here, we exploited the chemical properties of MANT-GTP, a GTP analogue that markedly increases fluorescence emission upon selectively binding to a GTPase. As the release of GDP in favor of GTP-binding activates most GTPase containing proteins, we used this molecular phenomenon as a potential read-out for RIN1-induced Mfn2-GTPase activation. Using purified proteins generated from SF21 insect cells we first compared the MANT-GTP binding properties of Mfn2 alone, or in a complex with either Smad2 or RIN1 (Figure 4C). Here, incubation of Mfn2 with MANT-GTP resulted in minimal fluorescence enhancement relative to the probe control, indicating that Mfn2 by itself cannot efficiently bind the GTP analogue. Likewise, MANT-GTP emission was not markedly enhanced in a complex with Smad2, suggesting that Smad2 does not act as a GEF (Figure 4C). However, MANT-GTP emission increased 3 to 4-fold upon Mfn2 incubation with RIN1, and was further enhanced in a complex with Smad2/RIN1 (Figure 4C). These results supported our hypothesis that RIN1 is a GEF for Mfn2-GTPase and that Smad2 functions as an important molecular scaffold.
To fully test our above hypothesis, we developed an immunocomplex-based MANT-GTP assay using various Mfn2 and Smad2 mutants (Figure 4D). We validated this system first by immunoprecipitating for RIN1 in the presence or absence of Mfn2 and Smad2 co-expression and found a remarkably similar MANT-GTP emission pattern as before using purified proteins (Figure 4E). Next, we assessed how the RIN1 GEF activity is affected by disrupting the Mfn2/Smad2 interaction by comparing Mfn2-WT versus M1, which we previously characterized as unable to bind Smad2 but still retains the conserved GTPase domain (Figure 3B). Interestingly, the MANT-GTP emission for M1 was significantly weakened compared to Mfn2-WT, indicating that Smad2 is necessary for efficient RIN1-GEF activity (Figure 4F).
As Mfn2 had previously been shown to interact with RAS (Chen et al., 2014; Chen et al., 2004), and given the versatile roles of RIN1 in regulating substrates like Rab5 and RAS, we investigated how these two prominent GTPases relate to the RIN1/Mfn2 complex and influence Mfn2 MANT-GTP binding properties. In co-immunoprecipitation studies, we first tested the degree of interaction between Mfn2 and RIN1 in the presence of RAS overexpression. Under our experimental conditions, RAS did not alter the Mfn2/RIN1 interaction, nor did it impact MANT-GTP binding (Figure S6A, B). However, there existed a potential competition for binding between RIN1 and Rab5 for Mfn2, as we observed an efficient interaction between the Rab5 immunoprecipitate and Mfn2 that was abolished when RIN1 was co-expressed (Figure S6C). Notably, a Rab5 immunocomplex-based MANT-GTP assay showed that Rab5 does not bind the GTP analogue itself, but does so for Mfn2 (Figure S6D). Consistent with our biochemical data, RIN1 expression largely blocked Rab5 from promoting Mfn2-MANT-GTP binding (Figure S6D).
Smad2 is an Essential Scaffold for Mfn2/RIN1 Complex
To determine whether Smad2 plays a crucial role in recruiting and scaffolding RIN1 into a complex with Mfn2, the interaction between RIN1 and Mfn2 was examined in sh-Smad2 stable cells. Consistent with the previous MANT-GTP assay results (Figure 4), Smad2 depletion significantly reduced the RIN1/Mfn2 interaction (Figure 5A). Accordingly, sh-Smad2 cells displayed similarly reduced MANT-GTP emission irrespective of RIN1 (Figure 5B), suggesting that Smad2 is a critical mediator of RIN1-based Mfn2-GTPase activation. To define this molecular complex, Smad2 structural binding elements for RIN1 were examined. Interestingly, RIN1 interacted with Smad2-WT and Smad2ΔSSMS but not Smad2ΔCT (Figure 5C), demonstrating that the N- and C-terminal domains are binding modules for Mfn2 and RIN1, respectively (Figures 3D and 5C). And as expected, both Smad2-WT and Smad2ΔSSMS dramatically enhanced the MANT-GTP emission whereas Smad2ΔCT did not (Figures 3D and 5D). Hence, these results strongly suggested that Smad2 promotes Mfn2-GTPase activity by recruiting RIN1 into a molecular complex. Functionally, ectopic RIN1 expression in control cells, whether acting on endogenous or overexpressed Mfn2, substantially increased mitofusion, whereas sh-Smad2 cells (HeLa, MEECs and A549s) exhibited a rather fragmented mitochondrial profile that did not change even in the presence of ectopic RIN1 and Mfn2 expression (Figures 6A, B and S5C). Taken together, these findings demonstrate that Smad2 is a critical scaffold for RIN1-induced Mfn2-GTPase activation and mitofusion (Figure 6C).
Figure 5. RIN1 requires Smad2 scaffolding action for Mfn2 GTP binding.
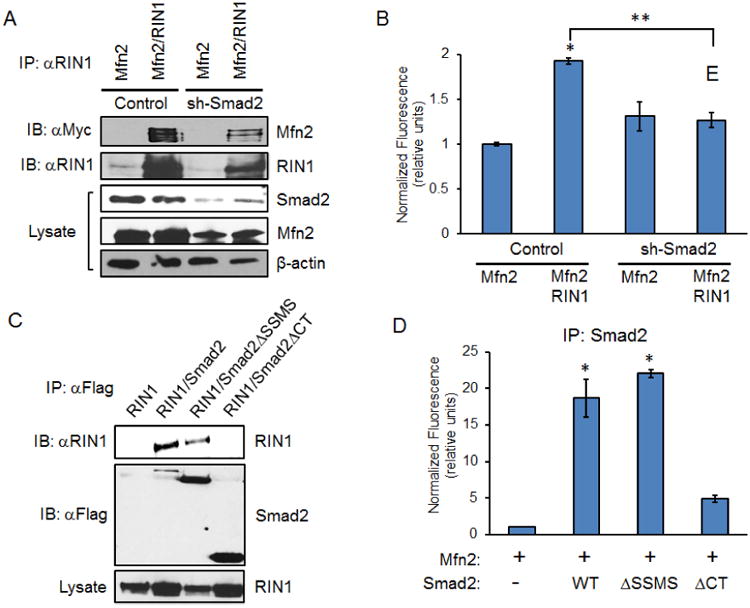
(A) HeLa control and sh-Smad2 stables expressing Myc-Mfn2 or Myc-Mfn2 and RIN1 were immunoprecipitated with RIN1 antibody. (B) Graph shows MANT-GTP emission of RIN1 immunocomplex in control HeLa and sh-Smad2 expressing Myc-Mfn2, or Myc-Mfn2 and RIN1. Error bars represent the SEM from three independent experiments normalized to Mfn2 alone control (*p<0.0001, **p=0.0001). (C) HeLa cells expressing RIN1 along with Flag-Smad2-WT, Smad2ΔSSMS, or Smad2ΔCT were immunoprecipitated with Flag antibody. (D) Graph shows MANT-GTP emission of Smad2 immunocomplex. HeLa cells expressing Mfn2, Mfn2 and Smad2-WT, Mfn2 and Smad2ΔSSMS, or Mfn2 and Smad2ΔCT were immunoprecipitated with Flag antibody. Error bars represent the SEM of a representative data set collected out of three independent experiments normalized to Mfn2 alone control (*p<0.02).
Figure 6. Smad2 and RIN1 are key determinants of mitochondrial fusion.
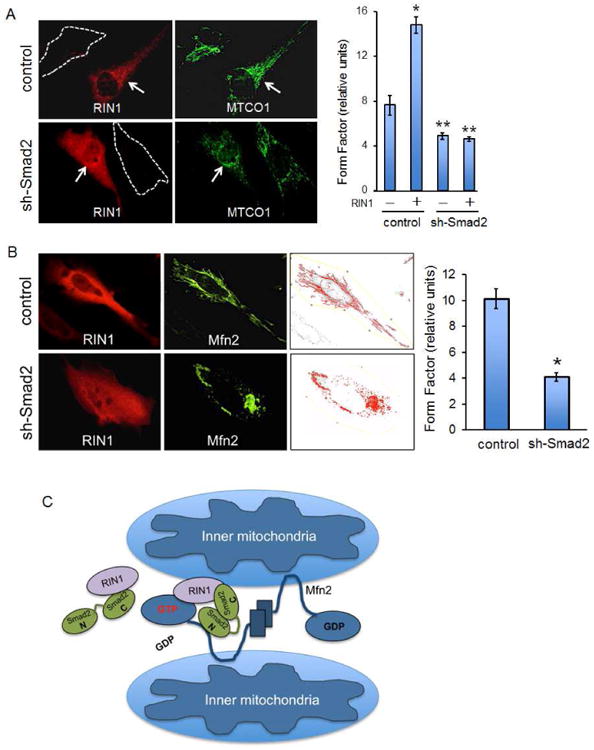
(A) Control MEEC and sh-Smad2 stables that are untransfected (white dashed lines) or expressing RIN1 (red) are co-stained for MTCO1 (green). Graph represents averaged form factor values for each condition based on a representative experiment in which least 30 cells were analyzed. Error bars represent SEM and type 2 T-test shows relative to control: *p<0.001, **p<0.015. (B) Control MEEC and sh-Smad2 stables were transfected with Mfn2 and RIN1, then co-stained with RIN1 and Myc antibodies. Graph represents averaged form factor values based on at least 30 cells of a representative experiment. Relative to control: *p<0.02. (C) Model for the Smad2/RIN1/Mfn2 trimeric complex during mitochondrial outer-membrane fusion.
Bioenergetically Protective Role of Inactive Smad2
Mitochondrial fusion generally facilitates protective effects through even spreading of metabolites, mitochondrial gene products and maximum ATP production (Murphy, 2009). To evaluate the bioenergetic roles of Smad2 in mitochondrial fusion, we monitored the overall respiratory activity of various cell types by ATP synthesis measurements (Figure 7A). First, in COS7 cells we found a modest decrease in ATP synthesis upon a brief treatment with TGF-β, whereas a marked enhancement was observed upon exposure to SB431542 (Figure 7A). A similar pattern was observed in MEECs although in sh-Smad2 MEEC stables ATP synthesis was dramatically reduced irrespective of TGF-β or SB431542 treatment (Figure 7B). Conversely, as mitochondrial superoxide anion (O2-) concentration increases with mitochondrial fragmentation, we examined whether Smad2-mediated fusion has a protective role against superoxide production. As expected, we observed differential responses where TGF-β generally increased and SB431542 decreased O2- production (Figure 7C). Likewise, the same pattern was observed in control MEECs whereas sh-Smad2 stables displayed constitutively higher levels of O2- production (Figure 7D).
Figure 7. Inactive Smad2 promotes normal mitochondrial functions.
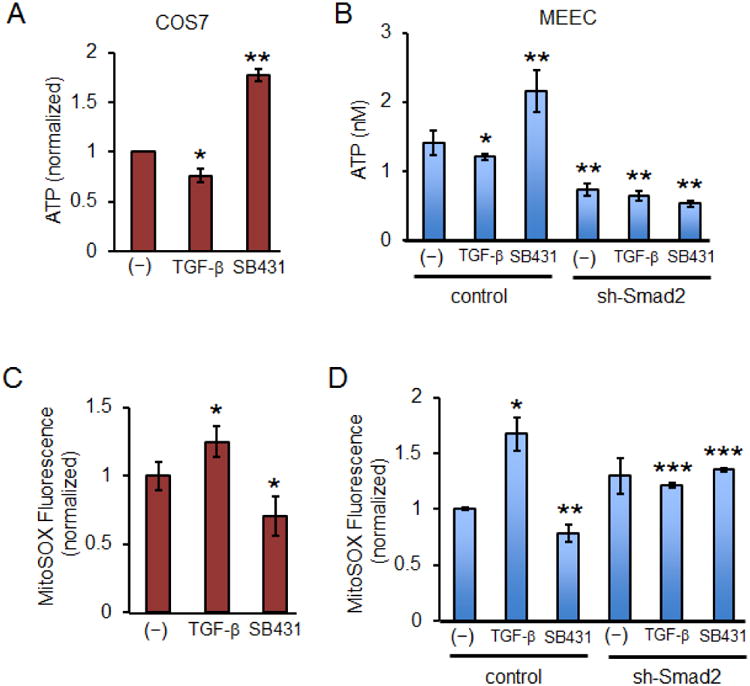
(A) Graph represents ATP concentrations in COS-7 cells upon treatment with SB431542 (30 μM) or TGF-β1 (200 pM) for 30 min. Each condition was normalized to the control ATP. Error bars represent SEM and type 2 T-test shows relative to control: *p<0.05; **p<0.02. (B) Graph represents ATP concentrations in MEEC control and sh-Smad2 stables upon treatment with SB431542 (30 μM) or TGF-β1 (200 pM) for 30 min. Calculations were based on the ATP standard curve. Error bars represent SEM and type 2 T-test shows relative to control: *p=0.027; **p<0.015. (C) Graph represents mitochondrial superoxide measurements. COS-7 cells were treated with SB431542 (30 μM) or TGF-β1 (200 pM) for 30 min. Mitochondrial superoxide was measured by MitoSOX Red mitochondrial superoxide fluorescence indicator and normalized to control. Error bars represent SEM and type 2 T-test shows relative to control: *p<0.04. (D) Graph represents mitochondrial superoxide in MEEC control and sh-Smad2 stables upon serum deprivation for 3 h, then treated with SB431542 (30 μM) or TGF-β1 (200 pM) for 30 min. Error bars represent SEM and type 2 T-test shows relative to control: *p=0.05; **p=0.027; ***p<0.001.
In parallel experiments, Mfn2-/- MEFs were used to test the importance of the Mfn2/Smad2 complex in modulating mitochondrial function (Figure S7). As expected, Mfn2 knockout abrogated the effects of SB431542 in enhancing ATP synthesis irrespective of Smad2 expression (Figure S7A). However, we observed a modest reduction in ATP upon TGF-β treatment in Mfn2-/- MEFs but not Smad2 depleted Mfn2-/- MEFs, suggesting that Smad2 may still promote ATP synthesis through interaction with Mfn1 (Figure S7A). In the case of superoxide production, TGF-β and SB431542 completely failed to regulate O2- levels in Mfn2-/-MEFs and noted only a slight overall increase upon Smad2 depletion likely due to further enhanced mitofission (Figure S7B). Taken together, these results confirm overall that the Mfn2/Smad2 complex is a critical determinant of mitochondrial function.
Discussion
Non-transcriptional Smad2 Regulation of Mitochondrial Morphology
TGF-β regulation of the mitochondria is best characterized in the context of apoptosis, mediated by the activation of caspase proteases and loss of mitochondrial membrane potential (Herrera et al., 2001a; Herrera et al., 2001b; Ribeiro et al., 1999). Specific TGF-β gene targets affecting this process have also been defined including expression of such proteins as death-associated protein kinase (DAP-kinase) as an immediate early response to Smad2, Smad3, and other TGF-β-inducible factors (Jang et al., 2002). While these mechanisms represent a growing body of work demonstrating a central role for TGF-β/Smad pathways in mitochondrial fragmentation, our work provides compelling new evidence for how these primary effectors of TGF-β signaling can actively participate in miotfusion.
Relationship Between Smad2 Function and Mfn2 Mutations in Human Disease
Most mutations that cause Mfn2 dysfunction reside within the GTPase and the coiled-coil intermediate segment (Liesa et al., 2009). Charcot-Marie-Tooth type 2A (CMT2A) is a peripheral neuropathy caused by Mfn2 point mutations, affecting sensory and motor neurons of the distal extremities, and many other neurodegenerative and metabolic diseases have been linked to Mfn2 defects (Honda et al., 2005; Liesa et al., 2009). As the major Smad2-binding region of Mfn2 encompasses the same highly conserved coiled-coil region (Figure 3A and B: residues 270 to 390), our data raises important functional questions as to whether some Mfn2-related diseases involve changes in the ability for Smad2 to bind Mfn2 harboring deleterious mutations. Similarly, mutations for Smad2 and Smad4 have been identified in colon, lung, cervix and hepatocellular carcinomas, with some resulting in the loss of tumor suppressor functions (Hata et al., 1997; Samanta and Datta, 2012). Aside from their transcriptional effects, long-term studies are underway to define the underlying roles of the Mfn2/Smad2 complex in human diseases.
Identification of an Mfn2-GTPase Modulator
An extensive network of kinases, phosphatases, and ubiquitin machinery has now been characterized as crucial regulators of mitochondrial dynamics. In the case of Mfn2, many posttranslational modulators have been identified (Liesa et al., 2009). Many of them exert their influences as ubiquitin-ligases, including the mitochondrial ubiquitin-ligase membrane-associated ring CH (MARCH)-V, Parkin and its associated serine-threonine protein kinase PINK1 (Chen and Dorn, 2013; Glauser et al., 2011; Liesa et al., 2009). Stress-induced JNK activation has also been shown to promote Mfn2 phosphorylation-induced ubiquitination and proteosomal degradation mediated by the E3 ligase Huwe1 (Leboucher et al., 2012). Furthermore, the key apoptosis-related proteins Bak, BCLX, and BCL2 have all been shown to interact with Mfn2, although we found no presence of these components associated with the Mfn2/Smad2/RIN1 complex in our mass spectrometry analysis. While we cannot definitively exclude the possibility that Smad2 promotes mitochondrial fusion by indirectly regulating the apoptotic machinery, our data strongly supports an active role for Smad2 where it recruits RIN1 to the mitochondrial outer-membrane to help catalyze the GTP nucleotide exchange.
The binding and hydrolysis of GTP are essential at distinct steps of mitochondrial outer-membrane fusion. Studies in yeast and mammalian systems have shown that nucleotide binding facilitates mitofusins to adopt a dimeric cis conformation whereas GTP hydrolysis induces the tethering of opposing or trans organelles to drive membrane fusion (Anton et al., 2011; Cohen et al., 2011; Ishihara et al., 2004). Although our data clearly supports the role of RIN1/Smad2 in enhancing GTP binding, whether this molecular complex participates in subsequent hydrolysis remains unclear. However, our kinetic measurements showed enhanced MANT-GTP emission within seconds of binding the Mfn2/Smad2 or Mfn2/Smad2/RIN1 complex and persisted over the course of 30 min with gradual decline (data not shown). This observation is consistent with previous findings in which hydrolysis-deficient forms of Mfn2 actively promoted mitochondrial fusion, suggesting that Mfn2 does not act as a classical dynamin type GTPase (Neuspiel et al., 2005).
In addition, the role of Rab5 in Mfn2-GTPase function hints at a new area of investigation since we show that RIN1 and Rab5 likely compete for Mfn2 binding (Figure S6). Whether RIN1 sequesters Rab5 away from Mfn2, or RIN1/Rab5 complex prevents their GEF activity toward Mfn2 constitute some of the preliminary questions that need to be addressed.
Smad2-based regulation of Mfn1 and Mfn2 in Mitochondrial Dynamics
While Mfn1 and Mfn2 are localized throughout the mitochondrial network in all cell types and have redundant functions in mitochondrial fusion, our data demonstrates the greater Smad2 affinity for Mfn2 than Mfn1 (Figure 1 and S1). This preferential binding may be particularly relevant in areas of high metabolic demand such as the brain and muscle. While our present study primarily based on epithelial and endothelial cells, a more focused approach involving tissue-specific investigations on Smad2/Mfn2 dynamics will be crucial to better understand the functional relationship between Smad2 and mitochondria-related disorders. Still, our work strongly supports a general case where a pool of inactive Smad2 is quite necessary for optimal mitochondrial fusion, ATP synthesis, and suppression of superoxide production (Figure 7).
In conclusion, our work defines an important non-transcriptional role for Smad2 in the cytoplasm. In contrast to the established TGF-β/Smad2/3 transcriptional responses that cause mitochondrial fragmentation, our findings define inactive Smad2 as a critical mediator of mitochondrial fusion. Collectively, our work provides new pathophysiological implications for Smad2 function in cancer, metabolic disorders, and neurodegenerative disease.
Experimental Procedures
Antibodies
Antibodies used in this study include Mfn1, Mfn2, Smad2, p-Smad2, Smad4, and Drp1 (all from Cell Signaling), Myc, Flag, and β-actin (all from Sigma), RIN1, SARA, Rab5, and H-Ras (all from Santa Cruz Biotechnology).
Plasmids
The following constructs were obtained from Addgene: Myc-Mfn2, Myc-Mfn1, and Drp1 were gifts from Dr. David Chan. Flag-Smad2ΔCT was a gift from Dr. Joan Massague. Flag-Smad2ΔSSMS was a gift from Dr. Rik Derynck. Turquoise2-Mito was a gift from Dr. Dorus Gadella, and Flag-SARA was a gift from Dr. Jeff Wrana. Flag-Smad2, Flag-Smad3, and Flag-Smad4 were gifts from Dr. Kohei Miyazono. Flag-Smad2-AA and Flag-Smad2-DD were gifts from Dr. Gerard Blobe. RIN1 was a gift from Dr. John Colicelli. Mfn2 deletion mutants were generated by PCR amplification using Myc-Mfn2-WT as a template, and forward and reverse primers complementary to the sequences near the defined regions of deletions. M1 mutant contained a deletion between nucleotide sequences 810G and 2242T; M2 mutant deletion was between 1170C and 2242T; M3 mutant deletion was between 1770A and 2242T; M4 mutant deletion was between 240G and 811G, M5 deletion was between 810G and 1771C; and M6 deletion was between 1980C and 2242T.
Cell culture and transfection
COS-7, Hela, A549, and MEFs were cultured in DMEM (GIBCO) supplemented with 10% FBS (HyClone). MEECs were maintained in MCDB-131 (GIBCO) supplemented with 15% FBS, 2 mM L-glutamine and endothelial cell growth supplement (Sigma). A549 cells were maintained in F12K (GIBCO) with 10% FBS. A549 and HeLa sh-Smad2 knockdown stables were generated first by shRNA vector transfection, selected in puromycin (1-2 mg/mL), then colonies were isolated and biochemically validated for Smad2 knockdown. Transfection was performed using Lipofectamine 2000 (Invitrogen). Standard immunoprecipitation and immunoblotting were prepared in lysis buffer (20 mM HEPES, pH 7.4, 150 mM NaCl, 2 mM EDTA, 10 mM NaF, 10 % (w/v) glycerol, 1% NP-40) supplemented with protease inhibitors (Sigma) and phosphatase inhibitors (Sigma).
Generation of recombinant baculoviruses and proteins
Genes coding for full length Mfn2, RIN1 and Smad2 were cloned in a pFastBac1 vector (Life Technologies). Mfn2 was cloned between the EcoRI and Hind III restriction site from a DNA fragment produced by PCR using the wild type gene as template, amplified with 5′ and 3′ hexahistidine-tagged primer (5′ GC GAA TTC GCC ATG GGT ATG CAC CAC CAC CAC CAC CAC TCC CTG CTC TTT TCT CGA 3′and CCC AAG CTT CTA TCT GCT GGG CTG CAG). RIN1 was cloned between EcoRI and XhoI restriction site from a DNA fragment produced by PCR using the wild type gene as template, amplified with 5′ and 3′ hexahistidine-tagged primer (5′ GC GAA TTC GCC ATG GGT ATG CAC CAC CAC CAC CAC CAC GAA AGC CCT GGA GAG TCA3′ and CCG CTC GAG CTA CTC CTC TGC TGC CCG). Smad2 was cloned between the EcoRI and Hind III restriction site from a DNA fragment produced by PCR using the wild type gene as template, amplified with 5′ and 3′ hexahistidine-tagged primer (5′ GC GAA TTC GCC ATG GGT ATG CAC CAC CAC CAC CAC CAC TCG TCC ATC TTG CCA TTC and CCC AAG CTT TTA TGA CAT GCT TGA GCA). Once constructed, each plasmid was transformed into DH10Bac competent cells for production of recombinant bacmids by using Bac-to-Bac Baculovirus Expression System. Transfection was performed in SF21 insect cell line by using Cellfectin II (Invitrogen). Purified recombinant viruses were amplified with three rounds of infection in SF21 cells grown at 27°C using a multiplicity of infection (MOI) of 0.1. Viral supernatants were harvested 48-72 h post infection. SF21 cells were infected with baculovirus at 2 MOI. 48 hours post-infection, cells were harvested by centrifugation at 1000g for 3 min, gently washed with resuspension buffer (20mM HEPES at pH 7.4, 0.5 M NaCl, 250 mM sucrose and protease inhibitors [5 μg/ml aprotinin, 5 μg/ml leupeptin, 2 μM pepstatin A, 1mM PMSF]). Cells were then subjected to protein purification protocol as previously described (Lee and Koland, 2005).
Immunoprecipitation assays
Cells were washed with PBS, lysed on ice with lysis buffer for 20 min (20 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM NaF, 1% NP-40) prior to centrifugation at 13,000 rpm for 15 min. Supernatants were incubated with appropriate antibodies and agarose G or protein A agarose for 4 to 6 h at 4 C. Immunoprecipitates were then pelleted and washed three times then stored in 2× sample buffer prior to western blot analyses.
Immunofluorescence Studies
Cells grown on coverslips were fixed with 4% paraformaldehyde, permeabilized in 0.1% Triton X-100 in PBS for 5 min, then blocked with 5% bovine serum albumin (BSA) in PBS for 20 min. All primary and fluorescently conjugated secondary antibodies and DAPI were incubated at room temperature for 1 h unless noted otherwise. Mitochondrial morphology assessment was based on calculating form factor, an average of isolated mitochondrial particles in a region of interest (ROI). Raw images obtained from confocal and immunofluorescence microscopy were binarized and quantified based on Image J using Mito Morphology Macro (Dagda et al., 2009) and the equation: Pm2/4π*Am (where Pm is the length of mitochondrial outline and Am is the area of mitochondrion) (Mortiboys et al., 2008).
Fluorescence Live Cell Confocal Imaging
MEECs were plated in glass chamber slides (Lab-Tek) and transfected with Turquoise2-Mito. 24 h later, cells were monitored via time-lapse confocal microscopy analysis (Visitech Infinity 3 hawk 2D Array). Cells were maintained in a chamber at 37°C equilibrated with 5% CO2, and images were recorded every 15 s.
Mfn2-GTPase Activation assay
Cells were immunoprecipitated in GTPase lysis buffer (20 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM NaF, 1% Nonidet NP-40) with appropriate antibodies, then washed and resuspended with GTPase lysis buffer. The immunocomplexes were incubated with MANT-GTP at 2.5 μM final concentration, and mixed for one min prior to transferring each immunocomplex into quadruplicates in a 96 well fluorescence plate. Additional mixing was performed in three 30 sec bursts in the fluorescence 96-well plate reader (Biotek) at 25 C prior to fluorescence measurements at 360 nmex and 440 nmem using bottom fluorescence area scan (5×5). Control fluorescence emission was subtracted from the experimental readings prior to relative normalization of data to fluorescence emission of Mfn2, Smad2, or RIN1 alone. For MANT-GTP assays using purified proteins, each at 0.5 μM final concentration, were incubated with MANT-GTP (2.5 μM) for 1 min prior to reading at 360 nmex and 440 nmem in a 96 well fluorescence plate.
Measurement of mitochondrial ROS
Mitochondrial superoxide production was measured using the MitoSOX Red mitochondrial superoxide indicator fluorescent probe (Invitrogen). Cells grown in 96 well plates were washed twice with PBS and subsequently incubated for 15 min with Mitosox Red (5 μM) at 37 C. After the incubation, fluorescence was measured with a microplate reader set to 510 nm excitation 580nm emission wavelengths.
ATP synthesis assay
Cellular ATP synthesis was measured with an ATP Determination Kit (Invitrogen). Briefly, cells were re-suspended in a reaction buffer, mixed with the ATP standard solution in a 96 well plate, and measured for luciferase luminescence at 560 nm. The ATP content in the samples was calculated from the standard curve.
Mitochondrial isolation
Mitochondrial fractions were prepared by differential centrifugation as previously described [Garg and Hu 2007]. Briefly, cells were suspended in a homogenizing buffer containing 250 mM sucrose, 5 mM HEPES, 5 mM EDTA, and protease inhibitors (Sigma). Homogenization was performed on ice for two 15 sec homogenization cycles. The homogenate was centrifuged at 1000 g for 10 min at 4 C to remove nuclei and debris. The mitochondrial fractions were prepared by centrifugation at 8500 × g for 20 min at 4°C. The pellet containing the mitochondrial fraction was gently resuspended in the homogenizing buffer and further centrifuged at 8500 g for 20 min at 4 °C. The isolated mitochondria were resuspended and plated on coverslips for immunofluorescence analyses.
Biotin-Streptavidin pull-down assay
Purified Mfn2 or Smad2 (0.5 μM) protein was biotinylated for 15 min on ice with Sulfo-NHS-SS-Biotin (Thermo Scientific). The reaction was neutralized by adding a quenching solution and then exhaustively dialyzed (20 mM Hepes, pH 7.4, 150 mM NaCl). Dialyzed Mfn2 or Smad2 was conjugated to a streptavidin agarose matrix, then incubated with RIN1 and either Smad2 or Mfn2 for 30 min. The streptavidin agarose-conjugated protein complexes were washed 3 times, then resolved on SDS-PAGE and immunoblotted with Mfn2, Smad2 and RIN1 antibodies.
Supplementary Material
Highlights.
- Smad2 is a critical mediator of mitochondrial dynamics and function
- Inactive Smad2 forms a complex with mitofusin2
- Smad2 scaffolds RIN1 into a complex with mitofusin2
- RIN1 is a GEF for mitofusin2 activation
Kumar et al. demonstrate a nontranscriptional role for Smad2 in regulating mitochondrial function. Inactive cytoplasmic Smad2 promotes Mfn2-induced mitochondrial fusion by recruiting RIN1 that acts as a GEF for Mfn2-GTPase activation.
Acknowledgments
We would like to thank Dr. John Colicelli (UCLA) for the RIN1 expression construct, Dr. Kohei Miyazono (University of Tokyo) for the Smad2, 3, 4 constructs. We acknowledge the Mass Spectrometry and Proteomics Facility at The Ohio State University for their technical assistance. This work was supported in part by NIH CA178443 awarded to N.Y.L., The Ohio State University College of Pharmacy, Davis Heart Lung Research Institute and James Comprehensive Cancer Center for internal funding.
Footnotes
Author Contributions N.Y.L. conceived and designed the study, S.K., C.C.P., and N.S. conducted the experiments. N.Y.L., K.M., K.R.H., and S.K. wrote the manuscript.
Author Information Authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amiott EA, Cohen MM, Saint-Georges Y, Weissman AM, Shaw JM. A mutation associated with CMT2A neuropathy causes defects in Fzo1 GTP hydrolysis, ubiquitylation, and protein turnover. Molecular biology of the cell. 2009;20:5026–5035. doi: 10.1091/mbc.E09-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton F, Fres JM, Schauss A, Pinson B, Praefcke GJ, Langer T, Escobar-Henriques M. Ugo1 and Mdm30 act sequentially during Fzo1-mediated mitochondrial outer membrane fusion. Journal of cell science. 2011;124:1126–1135. doi: 10.1242/jcs.073080. [DOI] [PubMed] [Google Scholar]
- Balaji K, French CT, Miller JF, Colicelli J. The RAB5-GEF function of RIN1 regulates multiple steps during Listeria monocytogenes infection. Traffic. 2014;15:1206–1218. doi: 10.1111/tra.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blahna MT, Hata A. Smad-mediated regulation of microRNA biosynthesis. FEBS letters. 2012;586:1906–1912. doi: 10.1016/j.febslet.2012.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. The Journal of cell biology. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Dasgupta A, Ding J, Indig FE, Ghosh P, Longo DL. Role of mitofusin 2 (Mfn2) in controlling cellular proliferation. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2014;28:382–394. doi: 10.1096/fj.13-230037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Guo X, Ma D, Guo Y, Li Q, Yang D, Li P, Qiu X, Wen S, Xiao RP, et al. Dysregulation of HSG triggers vascular proliferative disorders. Nature cell biology. 2004;6:872–883. doi: 10.1038/ncb1161. [DOI] [PubMed] [Google Scholar]
- Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MM, Amiott EA, Day AR, Leboucher GP, Pryce EN, Glickman MH, McCaffery JM, Shaw JM, Weissman AM. Sequential requirements for the GTPase domain of the mitofusin Fzo1 and the ubiquitin ligase SCFMdm30 in mitochondrial outer membrane fusion. Journal of cell science. 2011;124:1403–1410. doi: 10.1242/jcs.079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Cherra SJ, 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. The Journal of biological chemistry. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger K, Eder M, Kramer ER, Zieglgansberger W, Dodt HU, Dornmair K, Colicelli J, Klein R. The Rab5 guanylate exchange factor Rin1 regulates endocytosis of the EphA4 receptor in mature excitatory neurons. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12539–12544. doi: 10.1073/pnas.0801174105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Escobar-Henriques M, Anton F. Mechanistic perspective of mitochondrial fusion: tubulation vs. fragmentation. Biochimica et biophysica acta. 2013;1833:162–175. doi: 10.1016/j.bbamcr.2012.07.016. [DOI] [PubMed] [Google Scholar]
- Eura Y, Ishihara N, Yokota S, Mihara K. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J Biochem. 2003;134:333–344. doi: 10.1093/jb/mvg150. [DOI] [PubMed] [Google Scholar]
- Glauser L, Sonnay S, Stafa K, Moore DJ. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. Journal of neurochemistry. 2011;118:636–645. doi: 10.1111/j.1471-4159.2011.07318.x. [DOI] [PubMed] [Google Scholar]
- Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Han L, Wong D, Dhaka A, Afar D, White M, Xie W, Herschman H, Witte O, Colicelli J. Protein binding and signaling properties of RIN1 suggest a unique effector function. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:4954–4959. doi: 10.1073/pnas.94.10.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A, Lo RS, Wotton D, Lagna G, Massague J. Mutations increasing autoinhibition inactivate tumour suppressors Smad2 and Smad4. Nature. 1997;388:82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. The Journal of cell biology. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera B, Alvarez AM, Sanchez A, Fernandez M, Roncero C, Benito M, Fabregat I. Reactive oxygen species (ROS) mediates the mitochondrial-dependent apoptosis induced by transforming growth factor (beta) in fetal hepatocytes. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2001a;15:741–751. doi: 10.1096/fj.00-0267com. [DOI] [PubMed] [Google Scholar]
- Herrera B, Fernandez M, Alvarez AM, Roncero C, Benito M, Gil J, Fabregat I. Activation of caspases occurs downstream from radical oxygen species production, Bcl-xL down-regulation, and early cytochrome C release in apoptosis induced by transforming growth factor beta in rat fetal hepatocytes. Hepatology. 2001b;34:548–556. doi: 10.1053/jhep.2001.27447. [DOI] [PubMed] [Google Scholar]
- Honda S, Aihara T, Hontani M, Okubo K, Hirose S. Mutational analysis of action of mitochondrial fusion factor mitofusin-2. Journal of cell science. 2005;118:3153–3161. doi: 10.1242/jcs.02449. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Eura Y, Mihara K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. Journal of cell science. 2004;117:6535–6546. doi: 10.1242/jcs.01565. [DOI] [PubMed] [Google Scholar]
- Jang CW, Chen CH, Chen CC, Chen JY, Su YH, Chen RH. TGF-beta induces apoptosis through Smad-mediated expression of DAP-kinase. Nature cell biology. 2002;4:51–58. doi: 10.1038/ncb731. [DOI] [PubMed] [Google Scholar]
- Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nature reviews Neuroscience. 2008;9:505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner LL. Shaping the dynamic mitochondrial network. BMC biology. 2014;12:35. doi: 10.1186/1741-7007-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leboucher GP, Tsai YC, Yang M, Shaw KC, Zhou M, Veenstra TD, Glickman MH, Weissman AM. Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Molecular cell. 2012;47:547–557. doi: 10.1016/j.molcel.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NY, Koland JG. Conformational changes accompany phosphorylation of the epidermal growth factor receptor C-terminal domain. Protein Science. 2005;14:2793–2803. doi: 10.1110/ps.051630305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa M, Palacin M, Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiological reviews. 2009;89:799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- Massague J. TGFbeta signalling in context. Nature reviews Molecular cell biology. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J, Chen YG. Controlling TGF-beta signaling. Genes & development. 2000;14:627–644. [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes & development. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Mortiboys H, Thomas KJ, Koopman WJ, Klaffke S, Abou-Sleiman P, Olpin S, Wood NW, Willems PH, Smeitink JA, Cookson MR, et al. Mitochondrial function and morphology are impaired in parkin-mutant fibroblasts. Ann Neurol. 2008;64:555–565. doi: 10.1002/ana.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuspiel M, Zunino R, Gangaraju S, Rippstein P, McBride H. Activated mitofusin 2 signals mitochondrial fusion, interferes with Bax activation, and reduces susceptibility to radical induced depolarization. The Journal of biological chemistry. 2005;280:25060–25070. doi: 10.1074/jbc.M501599200. [DOI] [PubMed] [Google Scholar]
- Ranieri M, Brajkovic S, Riboldi G, Ronchi D, Rizzo F, Bresolin N, Corti S, Comi GP. Mitochondrial fusion proteins and human diseases. Neurology research international. 2013;2013:293893. doi: 10.1155/2013/293893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro A, Bronk SF, Roberts PJ, Urrutia R, Gores GJ. The transforming growth factor beta(1)-inducible transcription factor TIEG1, mediates apoptosis through oxidative stress. Hepatology. 1999;30:1490–1497. doi: 10.1002/hep.510300620. [DOI] [PubMed] [Google Scholar]
- Samanta D, Datta PK. Alterations in the Smad pathway in human cancers. Frontiers in bioscience. 2012;17:1281–1293. doi: 10.2741/3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. Journal of cell science. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- Shi Y, Hata A, Lo RS, Massague J, Pavletich NP. A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature. 1997;388:87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Molecular biology of the cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes & development. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall GG, Barbieri MA, Stahl PD, Horazdovsky BF. Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Developmental cell. 2001;1:73–82. doi: 10.1016/s1534-5807(01)00008-9. [DOI] [PubMed] [Google Scholar]
- Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- Wang Y, Waldron RT, Dhaka A, Patel A, Riley MM, Rozengurt E, Colicelli J. The RAS effector RIN1 directly competes with RAF and is regulated by 14-3-3 proteins. Molecular and cellular biology. 2002;22:916–926. doi: 10.1128/MCB.22.3.916-926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JW, Fairman R, Penry J, Shi Y. Formation of a stable heterodimer between Smad2 and Smad4. The Journal of biological chemistry. 2001;276:20688–20694. doi: 10.1074/jbc.M100174200. [DOI] [PubMed] [Google Scholar]
- Zorzano A, Liesa M, Palacin M. Mitochondrial dynamics as a bridge between mitochondrial dysfunction and insulin resistance. Archives of physiology and biochemistry. 2009;115:1–12. doi: 10.1080/13813450802676335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


