Abstract
Essential host cofactors in retrovirus replication bind cis-acting sequences in 5’untranslated region (UTR). Although host RBPs are crucial to all aspects of virus biology, elucidating their roles in replication remains a challenge to the field. Here RNA affinity-coupled-proteomics generated a comprehensive, unbiased inventory of human and avian RNA binding proteins (RBPs) co-isolating with 5’UTRs of HIV-1, spleen necrosis virus and Rous sarcoma virus. Applying stringent biochemical and statistical criteria, we identified 185 RBP; 122 were previously implicated in retrovirus biology and 63 are new to the 5’UTR proteome. RNA electrophoretic mobility assays investigated paralogs present in the common ancestor of vertebrates and one hnRNP was identified central node to the biological process-anchored networks of HIV-1, SNV, and RSV 5’ UTR-proteomes. This comprehensive view of the host constituents of retroviral RNPs is broadly applicable to investigation of viral replication and antiviral response in both human and avian cell lineages.
Keywords: Post-transcriptional control, cis-acting replication sequences, virus-host interaction, RNA recognition motif, hnRNPs
Introduction
The biogenesis of retroviral virions requires a collection of host RNA binding proteins (RBPs) responsible for the formation of ribonucleoprotein particles (RNPs). Specific host RBPs are recruited to the viral 5’ terminal untranslated region (UTR) to sculpt RNPs with versatile activity. These cellular proteins catalyze intricate post-transcriptional modifications at the 5’ and 3’ RNA termini, alternative splicing, nuclear export, mRNA translation, and facilitate dimerization of virion precursor RNA. Precisely elucidating the responsible host RBPs that generate different types of retroviral RNPs remains a complex challenge.
To date, host factors comprising the host proteome of retroviral 5’ UTRs are principally identified by genetic studies and evaluation of candidate proteins from host cell lineages. For instance, mutational analyses of the 5’ UTR of avian sarcoma virus led to the discovery of the cis-acting negative regulatory of splicing (NRS) (McNally et al., 1991). Partially complementary to chicken U1 snRNA, NRS was recognized as an antagonist of snRNP U1 at the major splice donor, preventing efficient intron removal; the result is the nuclear export and translation of the unspliced mRNA template (McNally et al., 1991). Likewise, optimization of the 5’ splice site to consensus improves splicing efficiency, but impedes downstream steps in replication that require cis-acting residues within the excised intron (Cobrinik et al., 1987; McNally et al., 1991). Beyond regulating intron retention, host RBPs direct the nuclear export and translation of unspliced mRNA and their assembly into progeny virions [reviewed in (Leblanc et al., 2013)]. Collectively, previous studies established the fundamental principle that retrovirus cis-acting 5’ UTRs and host RBPs manipulate host RNP activities to balance biogenesis of the full repertoire of transcripts required to produce infectious retrovirus particles.
Relatively minimal information is available about the scope of requisite host RBPs and redundancy between host cell lineages. All retroviral 5’ UTRs contain the RU5 region of the proviral RNA as part of the 5’ exon. The precursor RNA (pre-mRNA) is subject to alternative splicing, which fuses the 5’ exon to various downstream open reading frames. Studies of cellular mRNAs have identified splicing-dependent deposition of RNA binding proteins near the 5’ splice site in an RNP designated the exon junction complex (EJC), which facilitates nucleocytoplasmic transport and efficient translation of mature mRNAs (Tange et al., 2004). For retroviruses, the pre-mRNA forms an atypical unspliced RNP that is subject to nucleocytoplasmic transport and is translated to virion structural proteins (Gag and Gag-Pol). Formation of this atypical RNP is due to distal sequences in the 5’ leader, which encompass the dimer initiation sequence (DIS) and the packaging signal (Kuzembayeva et al., 2014).
The 5’ UTR is a central hub of regulatory activity by cis-acting replication sequences. In terms of post-transcriptional gene expression, the present paradigm posits that mutually exclusive RNA conformations expose the translation start codon or the dimer initiation sequence and these conformations are mediated by host RNP rearranging activity (RNPase) (Lu et al., 2011a; Lu et al., 2011b). The cis-acting replication sequences necessary to sculpt distinct RNPs are well-defined, but the cellular components of their cognate RNPs are poorly defined.
Herein, a comprehensive screen of 5’ UTR-host proteomes from human and avian lineages was generated by RNA affinity chromatography and proteomics identification. The results demonstrate overlap in a subset of host RBPs co-isolating with the 5’ UTR of HIV-1, spleen necrosis virus (SNV) and Rous sarcoma virus (RSV). The results provide evidence of partial cofactor redundancy between human and avian host cell lineages and provide candidates for future mechanistic investigations of RNA biology.
2. Materials and Methods
2.1 Isolation of RNA-protein complexes by RNA affinity chromatography
We used PCR to generate DNA templates for in vitro transcription. Each 5’ primer contained the T7 promoter followed by ~20nt of a designated template DNA; the 3’ primers were complementary to the template 3’ boundary. The resulting HIV-1 (NL4-3), SNV (PB101) and RSV (Prague A, pATV8) in vitro transcripts are shown in Figure 1. We used T7 RNA polymerase and RiboMAX™ Large Scale RNA Production System biotin-labeled UTP (bio-11-UTP) to generate biotinylated transcripts, as described (Promega). The reactions were treated with 1 unit of DNAase 1 (Promega) at 37°C for 30 minutes to digest template DNA. The reaction volume was increased to 500 µl with DEPC-treated water and 0.1 volume of 3M NaOAc, mixed with an equivalent volume of phenol:chloroform:isoamyl alcohol (25:24:1) and vigorously vortexed. The aqueous layer was collected by centrifugation, 1 µl glycogen and 2.5 volumes of cold ethanol was added and the sample was incubated at −80°C for 30 min. Precipitates were collected at 12,000×g for 15 min, washed with 70% ethanol, partially air-dried for 5 min, and resuspended in 30 µl DEPC-treated water. These samples were applied to G25 Sepharose (Roche) to remove unincorporated dNTPs, collected, and stored at −80°C. The appropriate size and homogeneity of the biotinylated RNAs was validated by electrophoresis, as described (Hartman et al., 2006).
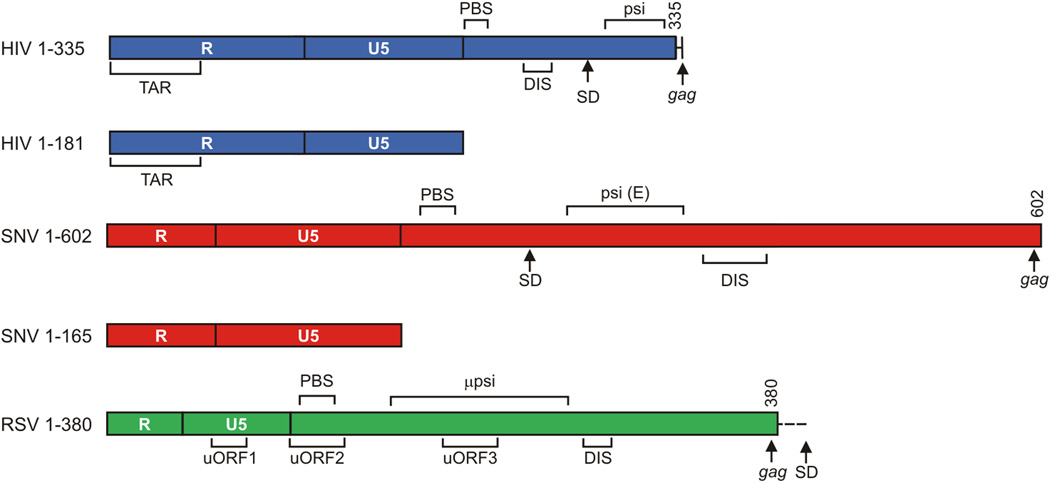
Figure 1.
A: Drawing of the retroviral transcripts used in this study. Numbers indicate nucleotide position relative to the start of the viral RNA. TAR: trans-activation response element; PolyA: Polyadenylation Signal; PBS: Primer Binding Site; 5’ss: 5’ Splice Site; DIS: Dimerization initiation signal; uORF: Upstream Open Reading Frame. µpsi coordinates from pATV8.
B: Immunoblot analysis of candidate 5’ UTR binding proteins relative to negative controls. Evaluation of stringency achieved in the RNA affinity chromatography of Hela cell extract. RNA affinity chromatography was performed on the indicated RNAs and eluates were collected after progressive wash cycles and equivalent volumes were subjected to SDS-PAGE and Western blot (WB) with the indicated antiserum. HeLa Input demonstrates abundant DHX30, YBX1 and GAPDH in the lysate and chemiluminscence indicates DHX30 and YBX1 are enriched relative to MYC 5’ UTR or RNA-free resin (Beads Only) and GAPDH.
A slurry of streptavidin-coated agarose beads was generated in 5-volumes of binding buffer (10 mM HEPES pH 7.6; 5 mM EDTA pH 8.0; 3 mM MgCl2; 40 mM KCl; 5% glycerol; 1% NP40; 2 mM DTT) by inverting the tube 5 times, centrifuging at 1,200×g for 1 min adding 5-volumes of binding buffer. After three repeats, the streptavidin preparation was supplemented with 8 µg biotinylated RNA bait and gently rocked for 1 hour at 4 °C. Each experiment included streptavidin beads incubated in buffer without RNA bait to differentiate bead-binding proteins from those bound to bait RNAs.
After RNA loading, beads were collected by centrifugation at 1,200×g for 1 min at 4 °C and resuspended in 1 ml of biotin blocking solution (2mM). The slurry was gently rocked at room temperature for 5 min to coat the beads and residual biotin eliminated by centrifuging the slurry at 1,200×g for 1 min at 4°C. The supernatant was discarded and the beads incubated for another 5 min in 1 ml binding buffer without RNA bait. The supernatant was discarded and the blocked beads were collected and gently rocked with 300 µg of protein lysate in 200 µl binding buffer at 4°C. Protein lysates had been prepared from either DF1 chicken cells or HeLa human cells by the protocols of (Dignam et al., 1983) for nucleus-enriched lysate or (Manley et al., 1980) for total cell lysate.
After 2 h incubation, beads were collected at 1200×g for 1 min and washed 3 times in 1 ml of binding buffer. Three additional washes served to release low affinity RNA binding proteins. These washes progressed with 40 mM, 100 mM, and 200 mM KCl in 10 mM HEPES pH 7.6; 3 mM MgCl2; 5 mM EDTA pH 8.0; 0.2% glycerol; 2 mM DTT (elution buffer) in 200 µl volumes. Each tube was inverted 5 times, incubated at room temperature for 5 min, and centrifuged at 1200×g for 1 min. The final treatment eluted high affinity RNA binding proteins using 2 M KCl in 200 µl elution buffer. The eluates were dialyzed overnight against binding buffer at 4 °C and after a brief SpeedVac incubation, separated by SDS-PAGE in 8% acrylamide. Proteins were stained by Coomassie Brilliant Blue and the entire lane was harvested in one slice. After digestion by trypsin, the peptides were processed for tandem mass spectrophotometry at Ohio State’s shared services proteomics core. For immunoblotting, the SDS-PAGE was followed by transfer of the proteins to nitrocellulose, incubation with antisera to human YB-1 (Cell Signaling Technology 4202), DHX30 (Bethyl A302-218A), GAPDH (Bethyl A300-639A), and visualization by chemiluminescence detection (Pierce) using Fuji imaging system and Multigauge software.
2.2 Peptide identification from mass spectra
Data were reported in Mascot Generic Format and analyzed by the Paragon algorithm contained in ProteinPilot 4.5 Software (AB SCIEX, Framingham MA). The Paragon Algorithm is a next-generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra (Shilov et al., 2007) and match these peptides against an NCBI-derived protein database. We limited the datasets to peptides matching to chicken or human proteins, and common laboratory contaminants were removed. A custom UNIX program using the awk command (provided by Bruce Stanley, Penn State College of Medicine Proteomics and Mass Spectrometry Core Facility) identified proteins in the RNA-containing sample with an unused score corresponding to an estimated local false-discovery rate (FDR) of ≤5%, as calculated by the Proteomics System Performance Evaluation Pipeline Software (Tang et al., 2008) within ProteinPilot software. The proteins carried forward for downstream analysis were enriched ≥2-fold by unused score relative to bead-only benchmark samples in all replicates; these enriched proteins were designated as candidate 5’ UTR binding proteins.
2.3 Network analysis of the retroviral RNA proteomes
The Database for Annotation, Visualization, and Integrated Discovery (DAVID, version 6.7) was used to assign each protein to one cellular compartment and biological process category. In cases where a protein was assigned to >1 biological process or cellular compartment, we narrowed its assignment to only one category based on the published literature (Supplementary Table 1). Candidate proteins were organized by their Official Gene Name for entries into DAVID 6.7 (http://david.abcc.ncifcrf.gov/) (Huang et al., 2009a, b), and the Homo sapiens species database was probed using query terms GOTERM_CC_ALL and GOTERM_BP_ALL. Categories with a p-value of ≤ 0.05, as determined by modified Fisher’s Exact Test, were considered statistically overrepresented. We used the BioVenn online comparison tool [http://www.cmbi.ru.nl/cdd/biovenn/index.php (Hulsen et al., 2008)] and Venn Diagram Plotter (http://omics.pnl.gov/software/venn-diagram-plotter (Pacific Northwest National Laboratory) to generate Venn Diagrams. Network analysis was performed using the IPA (Ingenuity Systems, www.ingenuity.com) with default settings. Because IPA does not contain Gallus gallus information, Homo sapiens gene names were substituted for the corresponding Gallus gene names. Only categories that were statistically overrepresented in the IPA analysis were chosen for construction of networks.
2.4 Recombinant protein isolation and RNA electromobility shift assay (EMSA)
In vitro transcripts were generated in the RiboMAX large-scale RNA production system (Promega), treated with DNase (Promega), separated on 8% denaturing urea gels, and eluted in passive gel elution buffer (Ambion). RNAs preparations were treated with Trizol-LS (Life Technology) and precipitated in 95% ethanol and 0.3 m NaOAc at −80 °C for 20 min, collected by centrifugation, resuspended in diethyl pyrocarbonate-treated water. HIV-1 RNA was treated to facilitate dimer conformation as described by (Heng et al., 2012).
The N-terminal domain of human DHX9 (N-term, 300 amino acids) or chicken DHX30 (N-term, 280 amino acids) were expressed from pGEx2T in E. coli BL21 (DE3) (GE Healthcare). After induction with 1 mm isopropyl-β-d-thiogalactopyranoside (United States Biochemical Corp.) for 3.5 h at 33°C, cells were resuspended in 50 mM sodium phosphate pH 8 and 300 mM NaCl with 10 µl/ml protease inhibitor cocktail (Sigma) and 1 µl/ml dithiothreitol (1 M) and lysed with an Aminco French pressure cell at 5000 units of pressure. Soluble proteins were collected by centrifugation at 12,000 rpm for 20 min at 4 °C in Sorvall RC5C SS-34 rotor, and incubated with glutathione-Sepharose beads (GE Healthcare) for 2 h at 4 degrees. The beads were washed twice with 10-bead volumes of 50 mM sodium phosphate (pH 8) and 300 mM NaCl and once with 50 mM sodium phosphate (pH 8), 300 mM NaCl and 10% glycerol, and then treated with elution buffer containing 50 mM sodium phosphate pH 8, 300 mM NaCl, 10% glycerol and 10 mM glutathione (reduced).The eluates were dialyzed against 10 mM sodium phosphate (pH 8.0) containing 150 mM NaCl at 4°C for 24 h with three buffer changes, followed by treatment with 1 unit of thrombin to 100 µg of protein for 2 h followed by Heparin column chromatography (GE Healthcare) to remove thrombin and GST from the protein preparation. The desired proteins were eluted with 50 mM sodium phosphate (pH 8) and 800 mM NaCl. The eluted proteins were dialyzed against 10 mM HEPES (pH 7.4) containing 50 mM NaCl at 4°C for 24 h with three buffer changes. The purity of the recombinant proteins was evaluated by SDS PAGE and Coomassie stain. One protein was used for RNA EMSA.
The EMSAs were performed with 1 µM recombinant protein and 1 µM RNA in EMSA binding buffer (10 mM HEPES, pH 7.4; 200 mM KCl 1 mM MgCl2; 1 mM EDTA, pH 8; and 10% glycerol). Control experiments to establish equilibrium conditions varied the incubation time and temperature and RNA and protein concentrations. RNAs diluted in water were placed in boiling water for 2 min and incubated on ice for 5 min. Thereafter, 1 µM N-term DHX30 or N-term DHX9 was added in EMSA binding buffer, producing a total reaction volume of 16 µl. After incubated at room temperature for 1 h, 16 µl of 2X RNA loading buffer (Ambion) was added and the samples were separated on 1% agarose with 0.5% TBE buffer at 4 degrees. Complexes were visualized by ethidium bromide staining of the nucleic acid. EMSAs were repeated with independent preparations of protein and RNA.
3. Results
The 5’ UTR sequences of HIV-1, SNV, and RSV were compared using the Basic Local Alignment Search Tool (BLAST, NCBI) and there was no apparent homology of the primary sequences (data not shown). However, there are important similarities, in that the 5’ UTRs of each retrovirus are highly structured and they serve conserved functions in regulating retroviral gene expression, alternative splicing, genomic RNA and dimer formation (Bolinger et al., 2010; Butsch and Boris-Lawrie, 2002). Therefore, we hypothesized that these seemingly unrelated UTR sequences would bind an overlapping set of host RBPs that are co-opted from the cell to perform regulatory functions that are crucial for retrovirus replication. To test that possibility, we performed a proteomic study to identify cellular proteins that bound each retroviral 5’ UTR and compared the results based on protein identity, domain analysis, and biological role.
3.1 Redundancy is observed among the human and chicken 5’ UTR binding proteins identified between HIV-1, SNV and RSV
A total of eight independent RNA affinity chromatography experiments were performed with human (HeLa) or chicken (DF1) cell lysates and the 5’ UTRs of HIV-1 (1–355 and 1–181), SNV (1–602 and 1–165) or RSV (1–380), which are depicted in Figure 1. As previously validated experimentally (Sharma and Boris-Lawrie, 2012), negative control samples consisted of cell lysates mixed with streptavidin-coated beads without biotinylated RNA. Stringency was introduced by isolating RNA-protein complexes resistant to a salt step gradient buffer (40 mM to 1 M KCl), progressively eliminating proteins that bound with low affinity. After enrichment of high affinity RNA-protein complexes, eluates were collected and subjected to MALDI-TOF tandem mass spectrophotometry. The complete datasets for each 5’ UTR affinity chromatography and mass spectrometry analysis are available in Supplementary files 2–5. The cumulative results of eight experiments identified 185 proteins passing the stringency criterion; the candidate proteins were those enriched 2-fold or greater relative to the negative controls. Immunoblotting experiments were performed to validate the negative controls (Sharma and Boris-Lawrie, 2012). Antisera to DHX30, YBX1 and GAPDH verified their detection in lysate (Supplementary Figure 1). Consistent with peptides identified by our mass spectrometry, DHX30 and YBX1 but not GAPDH were enriched in the HIV-1 and RSV eluates. The results validated enrichment over the background signal present in the RNA-free resin (Beads alone) and nonviral c-myc RNA (MYC).
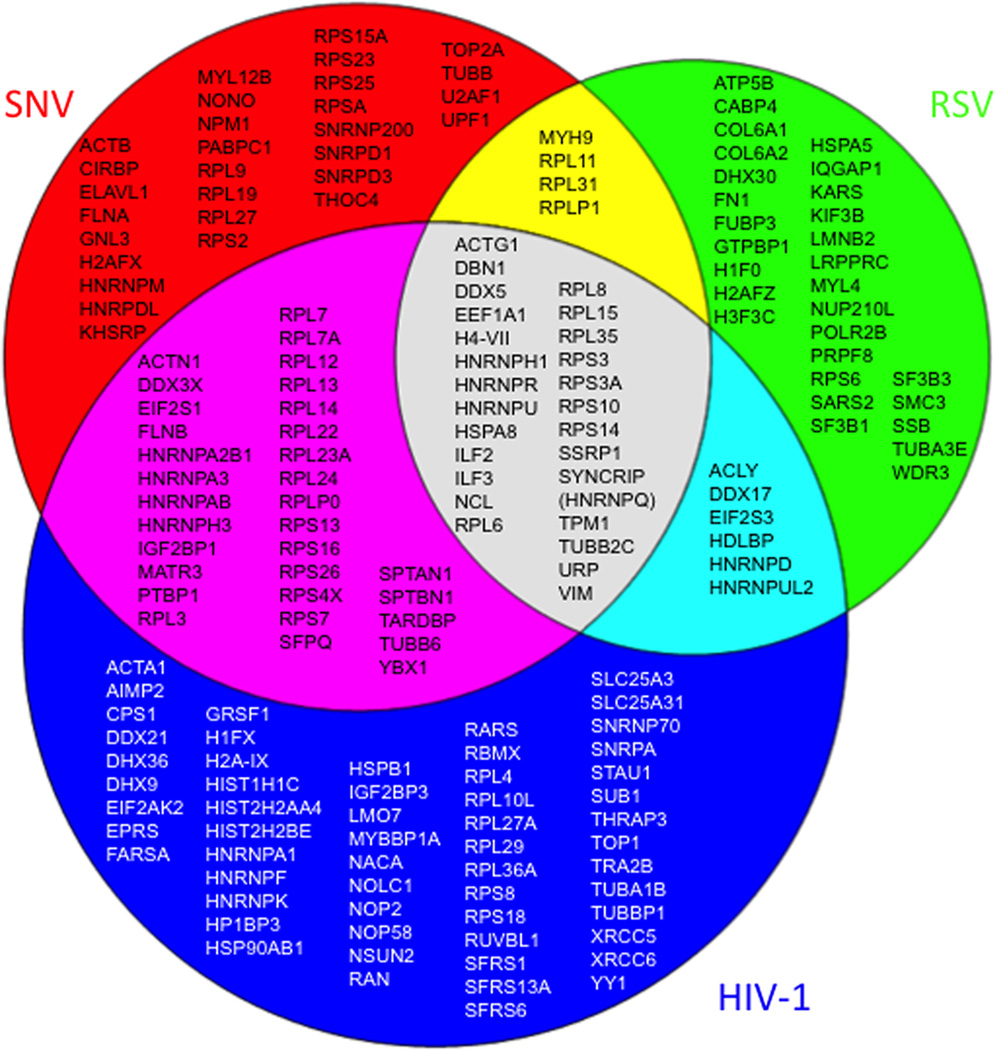
The peptides identified by mass spectrometry revealed a total of 185 enriched proteins, with 117 proteins identified exclusively by one retroviral 5’ UTR; 43 isolated by two of the three biotinylated UTRs; and 25 isolated by all three retroviral 5’ UTRs (Figure 2). Literature searches identified potential or existing roles for 122 of the RNA binding proteins in retrovirus biology (Supplementary Table 1); the remaining 63 were newly identified candidates that will require further validation for discovery of potential functions in retrovirus replication. The collection of the 185 proteins was designated the 5’ UTR proteome.
Figure 2.
Venn diagram of 5’ UTR RBPs identified by RNA affinity chromatography. Summarized are results of 8 independent experiments using the biotinylated transcripts depicted in Figure 1.
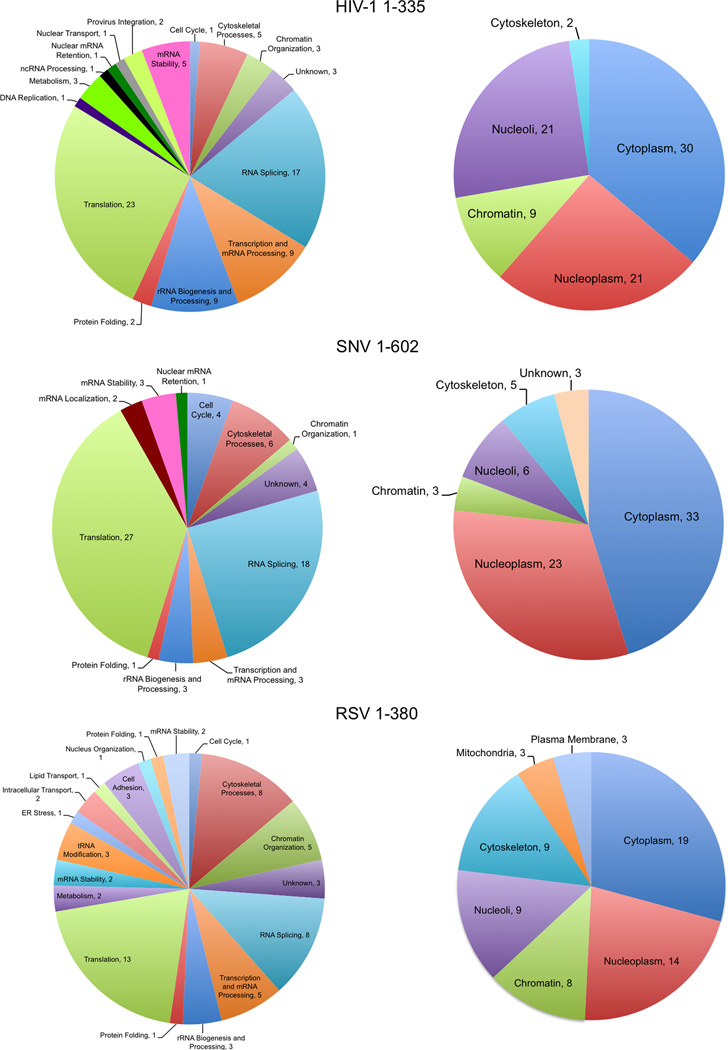
Because retroviruses are completely reliant on cellular machinery for all aspects of their RNA biology, we expected to isolate posttranscriptional regulatory proteins that undergo nucleocytoplasmic transport. In agreement with that hypothesis, the most abundant proteins identified were hnRNPs; double-stranded RNA binding proteins; scaffold and helicase proteins; and ribosome subunits (Table 1). The DAVID statistical tool (Huang et al., 2009a, b) was used to determine the biological processes (gene ontology (GO) terms) that were most highly enriched in the study. The analysis revealed that the GO terms with highest significance for binding to the HIV 5’ UTR were transcription and mRNA processing; translation and ribosome biogenesis; response to unfolded protein; provirus integration; and nucleocytoplasmic transport (Table 2). The classes of enriched factors that bound to the SNV 5’ UTR were similar, except that in addition to transcription/mRNA processing and translation/ribosome biogenesis, the posttranscriptional regulation of gene expression and rRNA processing was also highly represented (Table 3).
Table 1.
DAVID biological process enrichment analysis for human proteins isolated by HIV 5’ UTR (+1–335)
| Identifier Activity (Protein count) |
Fold enrichment (p-value) |
Proteins |
|---|---|---|
| Transcription and mRNA Processing | ||
| GO:0006396 RNA processing (32) |
10 (2.678E-23) |
RPL36A, RPL14, TRA2B, SYNCRIP/HNRNP Q, YBX1, SFRS6, HNRNPA3, DDX17, HNRNPK, RPL7, HNRNPF, HNRNPD, SNRNP70, NSUN2, DHX9, PTBP1, HNRNPA2B1, GRSF1, SFRS13A, SFRS1, HNRNPR, RBMX, HNRNPA1, HNRNPU, RPS7, HNRNPH3, NOP2, NOLC1, RPS14, SNRPA, NOP58, HNRNPH1 |
| GO:0000375 RNA splicing, via transesterification reactions (21) |
24 (3.644E-22) |
DHX9, RPL36A, TRA2B, PTBP1, HNRNPA2B1, SFRS13A, SFRS1, RBMX, HNRNPA1, HNRNPR, HNRNPU, YBX1, HNRNPA3, SFRS6, HNRNPH3, HNRNPK, HNRNPF, HNRNPD, SNRPA, SNRNP70, HNRNPH1 |
| GO:0043489 RNA stabilization (5) |
57 (1.309E-06) |
DHX9, HNRNPD, SYNCRIP/HNRNPQ, HNRNPU, YBX1 |
| GO:0043484 regulation of RNA splicing (4) |
34 (1.934E-04) |
HNRNPF, SFRS13A, RPS13, SNRNP70 |
| GO:0006376 mRNA splice site selection (3) |
37 (2.830E-03) |
SFRS6, SFRS13A, SFRS1 |
| GO:0000245 spliceosome assembly (3) |
16 (1.443E-02) |
SFRS6, SFRS13A, SFRS1 |
| GO:0033119 negative regulation of RNA splicing (2) |
69 (2.837E-02) |
SFRS13A, RPS13 |
| Nucleocytoplasmic Transport | ||
| GO:0006913 nucleocytoplasmic transport (5) |
5.5 (1.237E-02) |
RAN, SFRS13A, NOP58, HNRNPA1, MYBBP1A |
| GO:0050658 RNA transport (4) |
7 (1.821E-02) |
RAN, HNRNPA2B1, SFRS13A, HNRNPA1 |
| GO:0051168 nuclear export (3) |
8.7 (4.646E-02) |
RAN, SFRS13A, HNRNPA1 |
| Translation/Ribosome Biogenesis | ||
| GO:0006412 translation (26) |
13.5 (1.107E-21) |
RPL36A, NACA, RPL14, RPL13, RPL15, RPL35, RPL27A, IGF2BP3, RPL10L, RPL7, RPLP0, RPL3, RPL4, RPL7A, RPL12, EEF1A1, RPL24, RPL23A, RPS8, RPS7, RPL29, RPS18, RPL22, RPS14, RPS13, EIF2AK2 |
| GO:0042254 ribosome biogenesis (10) |
14 (3.090E-08) |
NOP2, NOLC1, RPL14, RPL7, RPS14, RPLP0, NOP58, RPL24, RPL7A, RPS7 |
| GO:0006364 rRNA processing (7) |
13 (1.424E-05) |
NOP2, NOLC1, RPL14, RPL7, RPS14, NOP58, RPS7 |
| GO:0042273ribosomal large subunit biogenesis (3) |
52 (1.420E-03) |
RPL14, RPL7, RPL24 |
| GO:0042257 ribosomal subunit assembly (2) |
115 (1.712E-02) |
RPS14, RPL24 |
| Other | ||
| GO:0006986 response to unfolded protein (4) |
10 (7.861E-03) |
HSP90AB1, HSPB1, EIF2AK2, HSPA8 |
| GO:0019047 provirus integration (2) |
43 (4.500E-02) |
XRCC5, XRCC6 |
Terms presented have p value greater than 0.05 and redundant, less significant terms were excluded.
Overrepresented GO categories were clustered by their relationship to the steps of gene expression (transcription, splicing, and translation).
Table 2.
DAVID biological process enrichment analysis of chicken proteins isolated by SNV 5’ UTR (+1–602)
| Identifier term Activity (Protein count) |
Fold enrichment p-value |
Proteins |
|---|---|---|
| Transcription and mRNA Processing | ||
| GO:0010467 gene expression (n= 32) |
37 9.107E-22 |
RPL19, RPL14, RPL13, SNRPD3, RPL15, SNRPD1, SYNCRIP, RPS2, YBX1, NONO, RPS3A, RPLP0, TARDBP, RPLP1, U2AF1, RPL11, RPL12, PTBP1, HNRNPA2B1, RPS4X, HNRNPR, HNRNPU, HNRPDL, RPS16, RPS14, SNRNP200, THOC4, RPS10, RPL35, RPS15A, HNRNPA3, RPS25, HNRNPM, RPS26, RPL6, RPL31, RPL9, RPL3, PABPC1, RPL7A, RPS23, RPSA, EEF1A1, RPL27, RPL23A, DDX5, RPS7, HNRNPH3, ILF2, RPL22, SFPQ, HNRNPH1 |
| GO:0006396 RNA processing (n= 26) |
10 1.019E-18 |
RPL14, SNRPD3, SNRPD1, SYNCRIP, YBX1, NONO, HNRNPA3, HNRNPM, TARDBP, U2AF1, RPL11, PABPC1, PTBP1, HNRNPA2B1, DDX5, HNRNPR, HNRNPU, RPS7, HNRPDL, HNRNPH3, RPS16, RPS14, SFPQ, SNRNP200, THOC4, HNRNPH1 |
| GO:0008380 RNA splicing (n=20) |
15 3.112E-17 |
SNRPD3, PTBP1, HNRNPA2B1, SNRPD1, SYNCRIP, DDX5, HNRNPR, HNRNPU, YBX1, NONO, HNRNPA3, HNRNPM, HNRNPH3, TARDBP, SFPQ, SNRNP200, U2AF1, THOC4, PABPC1, HNRNPH1 |
| GO:0043489 RNA stabilization n= 6 |
83 5.990E-09 |
ELAVL1, SYNCRIP, IGF2BP1, PABPC1, HNRNPU, YBX1 |
| GO:0022613 ribonucleoprotein complex biogenesis n= 10 |
12 1.652E-07 |
RPL14, RPS16, RPS14, RPLP0, SNRPD3, SNRNP200, SNRPD1, RPL11, RPL7A, RPS7 |
| Translation/Ribosome Biogenesis | ||
| GO:0006412 translation n= 30 |
19 5.445E-30 |
RPL19, RPL14, RPL13, RPL15, RPL35, RPS15A, RPS2, RPS25, RPS26, RPL31, RPS3A, RPL6, RPLP0, RPL9, RPLP1, RPL3, RPL11, RPL12, RPL7A, RPS23, RPSA, EEF1A1, RPL27, RPL23A, RPS4X, RPS7, RPS16, RPL22, RPS14, RPS10 |
| GO:0042254 ribosome biogenesis n= 7 |
12 2.390E-05 |
RPL14, RPS16, RPS14, RPLP0, RPL11, RPL7A, RPS7 |
| Other | ||
| GO:0010608 posttranscriptional regulation of gene expression n= 8 |
8 6.138E-05 |
UPF1, ELAVL1, SYNCRIP, IGF2BP1, PABPC1, RPS4X, HNRNPU, YBX1 |
| GO:0006364 rRNA processing n= 5 |
5 9.465E-04 |
RPL14, RPS16, RPS14, RPL11, RPS7 |
Started with Terms with p value greater than 0.05, removed redundant and less meaningful BP Categories, rounded fold enrichment and p values. Overrepresented GO categories were clustered by their relationship to the steps of gene expression (transcription, splicing, and translation).
Table 3.
DAVID biological process enrichment analysis of chicken proteins isolated by RSV 5’ UTR (+1–380)
| Identifier term Activity (Protein count) |
Fold enrichment and (p-value) |
Proteins |
|---|---|---|
| Transcription and mRNA Processing | ||
| GO:0010467 gene expression (34) |
3 (3.194E-09) |
RPL15, RPL35, SYNCRIP, POLR2B, KARS, SF3B3, RPS3, SF3B1, FUBP3, DDX17, RPL6, RPS3A, RPL31, PRPF8, RPL8, RPLP1, HNRNPD, RPL11, SSRP1, EEF1A1, EIF2S3, SSB, ILF3, RPS6, DDX5, HNRNPR, SARS2, HNRNPU, ILF2, RPS14, WDR3, RPS10, HNRNPH1, LRPPRC |
| GO:0006396 RNA processing (16) |
7 3.512E-09 |
SYNCRIP/HNRNPQ, SSB, RPS6, DDX5, HNRNPR, SF3B3, POLR2B, HNRNPU, SF3B1, DDX17, RPS14, PRPF8, HNRNPD, WDR3, RPL11, HNRNPH1 |
| GO:0008380 RNA splicing (10) |
8 (2.137E-06) |
SF3B1, PRPF8, HNRNPD, SYNCRIP, HNRNPH1, DDX5, HNRNPR, HNRNPU, SF3B3, POLR2B |
| GO:0048255 mRNA stabilization (3) |
47.9 (1.683E-03) |
HNRNPD, SYNCRIP, HNRNPU |
| Translation/Ribosome Biogenesis | ||
| GO:0006412 translation (16) |
12 3.107E-12 |
EEF1A1, RPL15, RPL35, EIF2S3, RPS6, KARS, SARS2, RPS3, RPS3A, RPL6, RPL31, RPS14, RPLP1, RPL8, RPS10, RPL11 |
| GO:0006364 rRNA processing (4) |
10 6.382E-03 |
RPS14, WDR3, RPL11, RPS6 |
| GO:0016072 rRNA metabolic process (4) |
10.0 7.178E-03 |
RPS14, WDR3, RPL11, RPS6 |
| GO:0042254 ribosome biogenesis (4) |
8 1.376E-02 |
RPS14, WDR3, RPL11, RPS6 |
| GO:0006413 translational initiation (3) |
16 1.467E-02 |
RPS3A, EIF2S3, RPS3 |
| GO:0042274 ribosomal small subunit biogenesis (2) |
43.5 4.430E-02 |
RPS14, RPS6 |
| Other | ||
| GO:0034660 ncRNA metabolic process (7) |
7 3.497E-04 |
RPS14, WDR3, RPL11, SSB, RPS6, SARS2, KARS |
| GO:0030705 cytoskeleton-dependent intracellular transport (4) |
18 1.261E-03 |
KIF3B, MYH9, TPM1, LRPPRC |
| GO:0034470 ncRNA processing (5) |
6 7.250E-03 |
RPS14, WDR3, RPL11, SSB, RPS6 |
| GO:0007018 microtubule-based movement (4) |
8.5 1.120E-02 |
KIF3B, TUBB2C, TUBA3E, LRPPRC |
| GO:0006334 nucleosome assembly (3) |
8.5 4.667E-02 |
H1F0, H2AFZ, H3F3C |
Started with Terms with p value greater than 0.05, removed redundant and less meaningful BP Categories, rounded fold enrichment and p values. Overrepresented GO categories were clustered by their relationship to the steps of gene expression (transcription, splicing, and translation).
RSV 5’ UTR binding proteins identified were in these same categories with the addition of proteins involved in noncoding RNA metabolism, cytoskeleton-dependent intracellular transport, microtubule-based movement, and nucleosome assembly (Table 3). Overall, transcription/mRNA processing factors were the most significantly enriched, with more than 30 proteins represented (HIV-1, p=10−23; SNV p=10−22; RSV, p=10−9). Proteins involved in translation were the next most overrepresented functional class (HIV-1, p=10−25; SNV, p=10−19; RSV, p=10−12; >30 proteins), as summarized in Figure 3.
Figure 3.
Biological function and subcellular assignments given for 5’ UTR RBPs by IPA. Evidence for functional qualities of each protein originates from Ingenuity Knowledge.
3.2 Conserved domains are characteristics recapitulated by the human and chicken 5’ UTR RBPs
Using DAVID, we identified conserved domains that were significantly enriched amongst the 5’ UTR binding proteins. As shown in Tables 4, 5, and 6, included are the nucleotide-binding, alpha-beta plait domain (HIV-1, 23-fold enriched, p= 10−25; SNV, 24-fold enriched, p= 10−22; RSV, 7-fold enriched, p=10−3), and the RNA recognition motif (RRM) (HIV-1, 22-fold enriched, p=10−23; SNV, 23-fold enriched, p=10−21; RSV 7.5-fold enriched, p=10−3), which are common in hnRNPs and small nuclear RNPs. HIV-1 and SNV binding proteins were also enriched in the double-stranded RNA binding domain (dsRBD) of α-β-β-β-α topology and the DEAD/DEAH box RNA helicase superfamily 2 domain (Table 7). Numerous RNA helicase family members were identified as binding partners of the HIV-1 UTR (DDX3, DDX5, DDX17, DDX21, DHX9, DHX30, DHX36, and UPF1; see Table 3), which have designated roles in HIV-1 biology (Table 7; (Lorgeoux et al., 2012; Roy et al., 2006).
Table 4.
DAVID domain analysis of human proteins Interacting with HIV 5’ UTR (+1–335)
| Domain identifer Domain term (Protein count) |
Fold enrichment p-value |
Proteins |
|---|---|---|
| IPR012677 Nucleotide-binding, alpha-beta plait (24) |
23 4.13E-25 |
RPL36A, TRA2B, PTBP1, HNRNPA2B1, GRSF1, SFRS13A, SYNCRIP/HNRNPQ, RPL23A, IGF2BP3, SFRS1, RBMX, HNRNPA1, NCL, HNRNPR, HNRNPA3, SFRS6, HNRNPH3, HNRNPF, HNRNPD, SNRPA, SNRNP70, HNRNPH1, MATR3, HNRNPAB |
| IPR000504 RNA recognition motif, RNP-1 (23) |
22 1.12E-23 |
RPL36A, TRA2B, PTBP1, HNRNPA2B1, GRSF1, SFRS13A, SYNCRIP/HNRNPQ, IGF2BP3, SFRS1, RBMX, HNRNPA1, NCL, HNRNPR, HNRNPA3, SFRS6, HNRNPH3, HNRNPF, HNRNPD, SNRPA, SNRNP70, HNRNPH1, MATR3, HNRNPAB |
| IPR012996 Zinc finger, CHHC-type (3) |
203 6.98E-05 |
RPL36A, HNRNPF, HNRNPH1 |
| IPR011545 DNA/RNA helicase, DEAD/DEAH box type, N-terminal (4) |
13 3.25E-03 |
DDX17, DDX21, DHX9, DHX36 |
| IPR018316; IPR017975 Tubulin/FtsZ, 2-layer sandwich domain; Tubulin, conserved site (3; 3) |
28 5.06E-03 |
TUBB, TUBB2C, TUBA1B |
| IPR001159 Double-stranded RNA binding (3) |
28 5.06E-03 |
DHX9, ILF3/NF90, EIF2AK2 |
| IPR003034 DNA-binding SAP (3) |
27 5.53E-05 |
HNRNPUL2, XRCC6, HNRNPU |
| IPR003008 Tubulin/FtsZ, GTPase domain (3) |
25 6.01E-03 |
TUBB, TUBB2C, TUBA1B |
| IPR012956 CARG-binding factor, N-terminal (2) |
203 9.70E-03 |
HNRNPD, HNRNPAB |
| IPR005161; IPR005160 Ku70/Ku80, N-terminal alpha/beta; and C-terminal arm (2; 2) |
203 9.70E-03 |
XRCC5, XRCC6 |
| IPR001650; IPR014021 DNA/RNA helicase, C-terminal; and Helicase, superfamily 1 & 2, ATP- binding (4; 4) |
8 1.53E-02 |
DDX17, DDX21, DHX9, DHX36 |
| IPR014001 DEAD-like helicase, N-terminal (4) |
7.5 1.61E-02 |
DDX17, DDX21, DHX9, DHX36 |
| IPR006561 DZF (2) |
81 2.41E-02 |
ILF2/NF45, ILF3/NF90 |
| IPR006535 HnRNP R and Q splicing factor (2) |
81 2.41E-02 |
SYNCRIP/HNRNPQ, HNRNPR |
| IPR018314 Eukaryotic nucleolar NOL1/Nop2p, conserved site (2) |
58 3.35E-02 |
NOP2, NSUN2 |
Table 5.
DAVID domain analysis of chicken proteins interacting with SNV 5’ UTR (+1–602)
| Identifier term Domain term (Protein count) |
Fold enrichment p-value |
Proteins |
|---|---|---|
| IPR012677 Nucleotide-binding, alpha-beta plait (21) |
24 2.57E-22 |
PTBP1, HNRNPA2B1, SYNCRIP, IGF2BP1, ELAVL1, RPL23A, HNRNPR, NONO, HNRNPA3, HNRPDL, HNRNPM, HNRNPH3, TARDBP, SFPQ, U2AF1, CIRBP, THOC4, PABPC1, HNRNPH1, MATR3, HNRNPAB |
| IPR000504 RNA recognition motif, RNP-1 (20) |
23 7.41E-21 |
PTBP1, HNRNPA2B1, SYNCRIP, IGF2BP1, ELAVL1, HNRNPR, NONO, HNRNPA3, HNRPDL, HNRNPM, HNRNPH3, TARDBP, SFPQ, U2AF1, CIRBP, THOC4, PABPC1, HNRNPH1, MATR3, HNRNPAB |
| IPR002017 Spectrin repeat (3) |
35 3.28E-03 |
SPTBN1, ACTN1, SPTAN1 |
| IPR012975 NOPS (2) |
161 1.22E-02 |
NONO, SFPQ |
| IPR001813 Ribosomal protein 60S (2) |
161 1.22E-02 |
RPLP0, RPLP1 |
| IPR006535 HnRNP R and Q splicing factor (2) |
97 2.02E-02 |
SYNCRIP/HNRNPQ, HNRNPR |
| IPR014837 EF-hand, Ca insensitive (2) |
81 2.42E-02 |
ACTN1, SPTAN1 |
| IPR011545 DNA/RNA helicase, DEAD/DEAH box type, N-terminal (3) |
112 2.57E-02 |
DDX3X, DDX5, SNRNP200 |
| IPR005824 KOW (2) |
54 3.62E-02 |
RPL27, RPS4X |
| IPR002453 Beta tubulin (2) |
44 4.40E-02 |
TUBB, TUBB2C |
Started with terms with p value greater than 0.05, removed redundant domain categories, rounded fold enrichment and p values. Overrrepresented domains listed in order of decreasing statistical significance.
Table 6.
DAVID domain analysis of chicken proteins isolated by RSV 5’ UTR (+1–380)
| Identifier term Domain term (Protein count) |
Fold enrichment p-value |
Proteins |
|---|---|---|
| IPR000504 RNA recognition motif, RNP-1 (6) |
7.5 1.12E-03 |
HNRNPD, SYNCRIP/HNRNPQ, SSB, HNRNPH1, NCL, HNRNPR |
| IPR012677 Nucleotide-binding, alpha-beta plait (6) |
7.4 1.16E-03 |
HNRNPD, SYNCRIP, SSB, HNRNPH1, NCL, HNRNPR |
| IPR004161 Translation elongation factor EFTu/EF1A, domain 2 (3) |
53 1.34E-03 |
GTPBP1, EEF1A1, EIF2S3 |
| IPR000795 Protein synthesis factor, GTP- binding (3) |
47 1.79E-03 |
GTPBP1, EEF1A1, EIF2S3 |
| IPR004087 K Homology (3) |
21 8.35E-03 |
FUBP3, HDLBP, RPS3 |
| IPR006535 HnRNP R and Q splicing factor (2) |
106 1.85E-02 |
SYNCRIP, HNRNPR |
| IPR006561 DZF (2) |
106 1.85E-02 |
ILF2, ILF3 |
| IPR004160 Translation elongation factor EFTu/EF1A, C-terminal (2) |
59 3.30E-02 |
GTBP1, EEF1A1 |
| IPR001023 Heat shock protein Hsp70 (2) |
44 4.38E-02 |
HSPA5, HSPA8 |
Started with terms with p value greater than 0.05, removed redundant domain categories, rounded fold enrichment and p values. Overrrepresented domains listed in order of decreasing statistical significance.
Table 7.
RNA binding proteins in common to 5’ UTRs of HIV-1, SNV, and RSV
| Protein Family | In common to HIV-1, SNV and RSV |
In common to 2 of 3 retroviruses |
|---|---|---|
| Double Stranded RNA Binding Proteins |
ILF2/NF90 | IGF2BP1/IMP1/ZBP1 ILF3/NF45 |
| Helicase Superfamily | DDX5 | DDX3X DDX17 |
| HNRNPs | HNRNPH1 SYNCRIP/HNRNPQ HNRNPR HNRNPU |
HNRNPA2B1 HNRNPA3 HNRNPAB HNRNPD HNRNPH3 HNRNPUL2 PTBP1/HNRNPI |
| RRM and Nucleotide Binding |
NCL | MATR3 SFPQ/PSF TARDBP YXB1 |
| Ribosomal Proteins | RPL6 RPL8 RPL15 RPL35 RPS1 RPS3 RPS3A RPS14 |
RPL3 RPL7 RPL7A RPL11 RPL12 RPL13 |
| Scaffold Proteins | ACTG1 DBN1 TPM1 TUBB2C URP VIM |
ACTN1 |
| Other | SSRP1/FACT H4-VII HSPA8 EEF1A1 |
EIF2S1 EIF2S3 HDLBP/VIGILIN |
Several proteins identified in this study contain the conserved dsRBD of α-β-β -β-α secondary structure: Staufen1, eIF2AK (eukaryotic translation initiation factor 2-alpha kinase 2; PKR); trans-activation responsive DNA-binding protein (TARBP); ILF3 (NF90); DHX9 and its paralog, DHX30. The α-β-β-β-α dsRBD facilitates interaction with retroviral RNA sequences, in particular the 5’ UTR (Agbottah et al., 2007; Fujii et al., 2001; Mouland et al., 2000; Ranji et al., 2011).
Staufen was identified as virion-associated HIV-1 RNA binding protein in proportion with genomic RNA, and was demonstrated to function in virion RNP assembly. Staufen promotes Gag oligomerization and virion RNP encapsidation, processes essential to viral replication (Chatel-Chaix et al., 2004; Ghoujal et al., 2012; Mouland et al., 2000). The isolation of EIF2AK2 (protein kinase R, PKR) is of interest given that NF90/45, TRBP, eIF2α and DHX9 are substrates for phosphorylation by PKR (Burugu et al., 2014). PKR’s activity is central to innate antiviral response by phosphorylating eIF2α and attenuating viral mRNA translation (Burugu et al., 2014). Notably, eIF2α-phosphorylation was not detectable in HIV-1 infected lymphocytes (Sharma et al., 2012), which may be attributable to viral antagonism of PKR activity. In concordance with this notion, eIF2-α has been identified in complex with NF90 and the other PKR substrates isolated in this study (Burckstummer et al., 2006), which potentially squelches PKR activity. This intriguing possibility warrants future investigation.
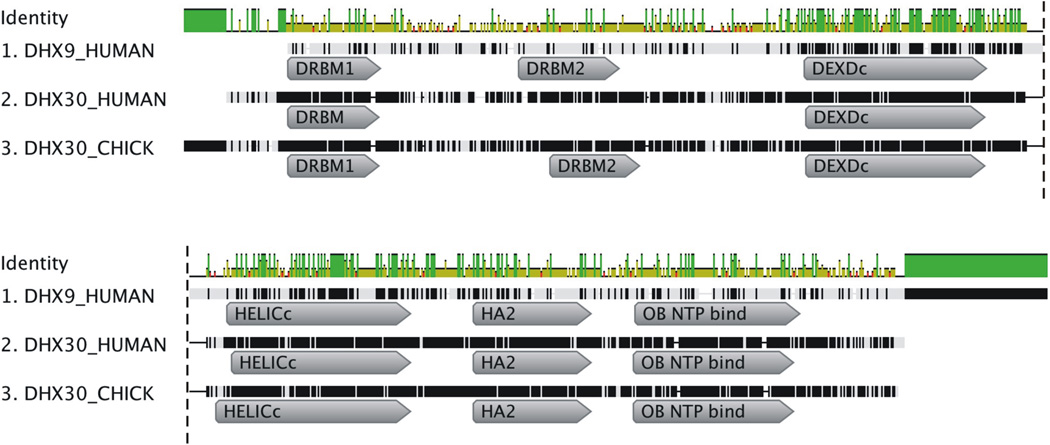
The dsRBD of NF90 binds the HIV-1 5’ LTR (R region) at the Tat transactivation response element (TAR), inhibiting transcriptional activation of HIV-1 (Agbottah et al., 2007; Hoque et al., 2011). DHX9 binding to TAR has been shown (Fujii et al., 2001) although DHX9 interaction with TAR and contiguous 5’ leader sequences is necessary for its post-transcriptional activity (Bolinger et al., 2007) and incorporation into virions (Boeras and Boris-Lawrie, unpublished). In the case of DHX9, two dsRBDs within the N-terminal domain (NTD) are responsible for recognition of the 5’ UTR of many retroviruses, including SNV (Bolinger et al., 2007; Ranji et al., 2011). The dsRBDs tether DHX9’s helicase core to the cognate retrovirus RNA, sculpting an mRNP that facilitates translation into virion proteins (Bolinger et al., 2010; Hartman et al., 2006). The human paralog, DHX30 contains one consensus dsRBD and a second atypical dsRBD (Gleghorn and Maquat, 2014) and alignment of the DHX9 and DHX30 domains reveals partial amino acid similarity (Supplementary Figure 2A). Also important to consider is DHX30 and DHX9 share highly analogous domain content and organization, in addition to the two dsRBDs in the N-terminal domain (N-term), DEIH box helicase, HELICc, HA2, and OBN domains (Figure 4). Notably, the chicken (Gallus gallus) genome does not encode a DHX9 paralog (GenBank). The lack of a DHX9 paralog in chicken, yet the conservation of DHX30 in human and chicken introduced the possibility that DHX30 and DHX9 serve analogous roles in posttranscriptional control of retroviral RNA expression attributable, at least in part, to conservation of dsRBDs of α-β-β -β-α secondary structure in N-terminal domains.
Figure 4.
DHX9 and DHX30 have a very similar overall domain structure. Geneious software version 5.5 (http://www.geneious.com, Kearse et al., 2012) was used to align the amino acid sequences and domain locations of H. sapiens DHX9, H. sapiens DHX30 and G. gallus DHX30. The top row depicts the amino acid identity shared between the three helicases at each position. Identical amino acids shared between the three helicase are depicted by a vertical green line, amino acids shared by two of the helicases are a shorter yellow line, red denotes no shared residues. Domain locations assigned by the NCBI database are shown. Extraneous domains and sequence conflicts were omitted for clarity. DRBM: double stranded RNA binding motif, DEXDc: DEAD-like helicases superfamily, HELICc: Helicase-superfamily C-terminal domain, HA2: Helicase associated domain, OB NTP bind: Oligonucleotide/oligosaccharide binding fold.
3.3 Human DHX9 RNA binding activity is recapitulated by HIV-1 and SNV, but not RSV
To assess the possibility of reciprocal RNA binding activity of DHX9 and DHX30 to the retroviral 5’ UTRs, RNA EMSAs were performed with recombinant DHX30 N-term and DHX9 N-term. The results demonstrated that N-term DHX30 bound to RSV 5’ UTR, providing independent validation of the RNA affinity results (Figure 5A). By comparison, in vitro binding of RSV to DHX9 was not observed; only the SNV and HIV-1 RNA sequences bound DHX9 (also known as RNA helicase A, RHA) (Figures 5A–B). The ability of SNV to bind both DHX30 and DHX9 may be explained by the recent evidence that SNV is actually a mammalian virus that was unintentionally introduced into chickens in the early 20th century (Niewiadomska and Gifford, 2013). However, based on the finding that the DHX30 dsRBDs are highly conserved between human and chicken (Supplementary Figure 2B), it also feasible that DHX30 may play similar roles in RSV and HIV-1 infection. Further experiments will be needed to discriminate between these hypotheses.
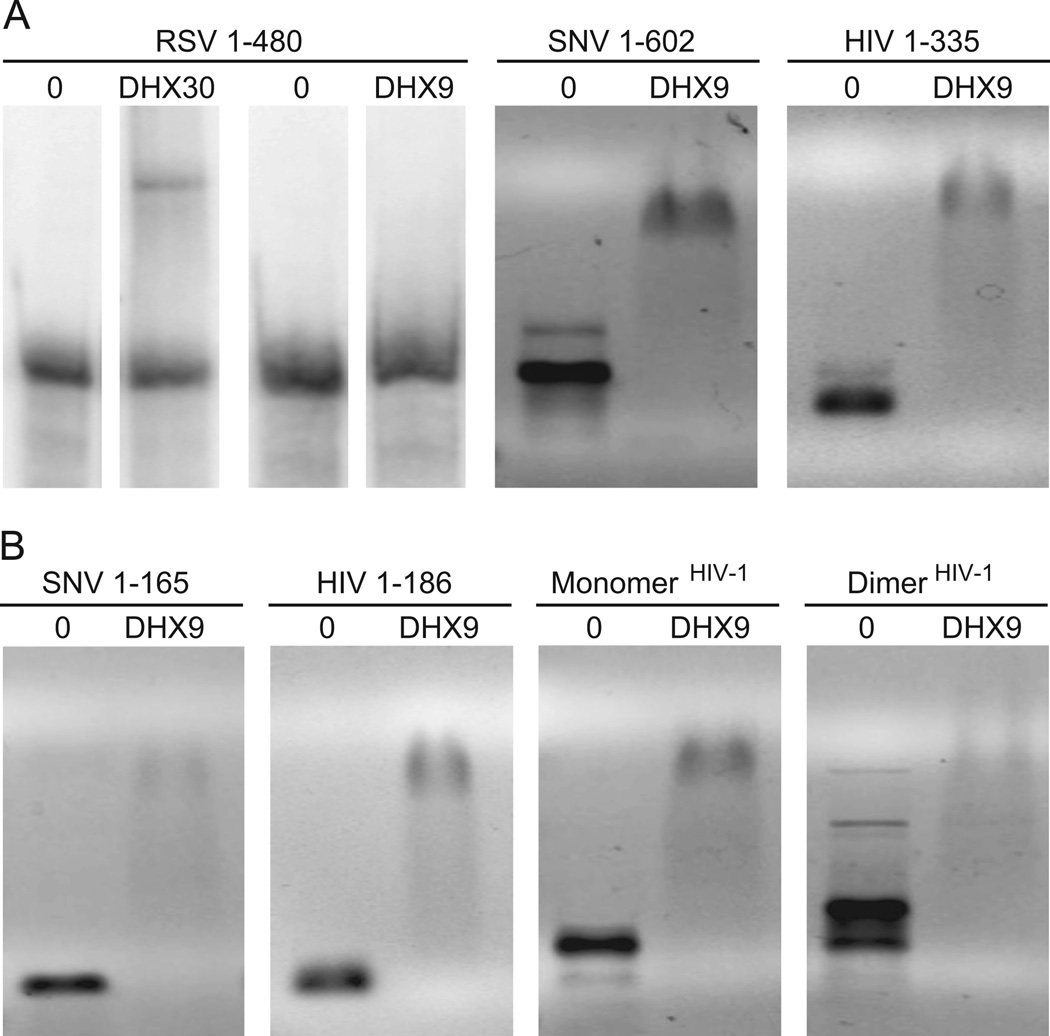
Figure 5.
RNA electrophoretic mobility shift assay of 5’ UTR interaction with DHX30 or DHX9. RSV, SNV or HIV-1 transcripts were incubated on ice for 30 min with recombinant N-terminal RNA binding domains of DHX30 or DHX9 (NTD) and complexes were resolved on chilled 1% agarose gels. Representative EMSAs using 300 nM protein. A) RSV 5’ leader (1–480) is shifted by DHX30 NTD, but not DHX9 NTD. B) SNV and HIV-1 5’ leaders are shifted by DHX9. C) SNV and HIV-1 5’ terminal nts corresponding to RU5 are sufficient for interaction with DHX9. D) DHX9 produces distinct mobility changes of HIV-1 5’ leader RNA preferentially in monomer conformation or dimer conformation.
Because the SNV and HIV-1 transcripts tested contained the DIS, both monomer and dimer conformations of these 5’ leader RNAs were observed (Figure 5B). Moreover, addition of DHX9 N-term shifted both RNAs to a higher molecular mass complex, implicating redundant RNA binding interactions (Figure 5B). The electrophoretic shift was also exhibited by SNV 1–165 and HIV 1–185, which lack the DIS. These results are consistent with our earlier reports that binding sites for DHX9 are present in 5’ terminal nts of in SNV and HIV-1 (RU5) (Bolinger et al., 2010; Hartman et al., 2006). They also suggest DHX9 cis-binding sequences are recapitulated in the three-dimensional structure of the HIV-1 leader, which forms monomer and dimer conformations. We therefore examined whether the three-dimensional structure of the HIV-1 leader would affect DHX9 binding by performing EMSA of DHX9 N-term with HIV-1 RNAs that preferentially adopt a monomer or dimer conformation in solution (Lu et al., 2011a). There was significant mobility shift detected in the lane containing DHX9 N-term with the HIV-1 monomer (Figure 5C). In comparison, the lane containing DHX9 N-term and abundant dimer revealed an electrophoretic shift more heterogeneous in size, indicating RNA structural features distinct to the monomer or the dimer are recognized by DHX9 N-term. Further studies will be needed to investigate the structural basis for DHX9 interactions with HIV-1 and SNV RNA conformers.
3.4 Identifying host constituents of the retroviral 5’UTR nuclear RNP
Several nuclear RBPs were identified reiteratively amongst the 5’ UTRs in the eight RNA affinity isolation experiments (Table 7). The HIV-1 and SNV RNPs were enriched in MATR3, splicing factor SFPQ (PSF), and NONO (Sewer et al.) and all three retroviral UTRs isolated nucleolin (NCL) as a binding factor (Figure 2). The importance of PSF and p54nrb in accumulating an appropriate ratio of unspliced transcript to spliced viral mRNA has been shown in HIV-1 (Zolotukhin et al., 2003). MATR3 binding stabilizes HIV-1 unspliced and incompletely spliced Rev/Rev-responsive element (RRE)-dependent HIV-1 mRNAs (Kula et al., 2011; Yedavalli and Jeang, 2011). PSF, p54nrb and MATR3 are components of nuclear paraspeckles, which retain RNAs in the nucleus during active transcription (Sewer et al., 2002; Zhang and Carmichael, 2001). Upon transcriptional arrest, PSF, p54nrb, MATR3 and NCL redistribute to perinucleoar caps (Sewer et al., 2002; Zhang and Carmichael, 2001). The accumulation of retroviral RNPs in the perinucleolar space has been shown (Levesque et al., 2006; Poole et al., 2005) and may execute an RNP sorting mechanism important to virion RNP assembly.
Another partner of NCL, IGF2BP1 (ZBP1/IMP1), was identified in this study of HIV-1, SNV and RSV RNPs. IMP1 is cotranscriptionally recruited into an IGF2BP1 (ZBP1/IMP1)-dependent mRNP complex on nascent transcripts (Jonson et al., 2007; Oleynikov and Singer, 2003). Liang and colleagues demonstrated that IMP1 (IGF2BP1/ZBP1) interfered with the assembly of virion RNPs and processing of nascent particles on cellular membranes (Zhou et al., 2008). In summary, the isolation of MATR3, PSF, NONO, NCL and Imp1 by RNA affinity chromatography is concordant with their demonstrated roles as nuclear retrovirus cofactors involved in determining the posttranscriptional fate of the viral RNA, and these proteins coincide with the reiterative isolation of additional nucleolar RNA binding proteins in our affinity chromatography experiments (Figure 3). In the case of RSV and HIV-1, a subpopulation of the Gag precursor protein accumulates in nucleoli, raising the possibility that a nuclear pool of Gag could participate in virion RNP assembly (Lochmann et al., 2013).
3.5 HIV-1 and the avian retroviruses isolate orthologous human and chicken hnRNPs
Highly conserved hnRNPs were identified in this study, demonstrating ≥90% amino acid identity between human hnRNPs and chicken hnRNPs or homolog 1-like proteins (Table 8). The family of hnRNP proteins has characteristic RRMs and arginine-glycine-rich basic domains and function as positive and negative regulators of eukaryotic spliceosome activity and splice site selection (Chen and Manley, 2009; Krecic and Swanson, 1999; Mayeda and Krainer, 1992). Several human hnRNPA/B paralogs (A1, A2, A3, AB) are demonstrated to modulate post-transcriptional expression of human, avian, and murine retroviruses. As expected, the SNV 5’ UTR isolated several RNPs of human origin, RSV identified the chicken hnRNP R orthologous with human, and HIV-1 identified ~20 RNPs of chicken origin (Table 7). Notable is hnRNPQ because it was identified by the HIV-1, SNV and RSV 5’ UTRs and from human and chicken cell lineages. Also known as SYNCRIP and the mouse minute virus NS1-associated protein (NASP), hnRNPQ and has been recognized to participate in the regulation of translation and miRNA-mediated repression of specific mRNAs, and influencing the activity of APOBEC1, a homolog of APOBEC3G (Lau et al., 2001; Shimizu et al., 2014; Svitkin et al., 2013). hnRNPQ plays dual roles in HCV, influencing viral RNA replication and translation (Liu et al., 2009). These and studies of HIV-1 ribonucleoprotein complexes suggest investigation of hnRNPQ in retrovirus RNA biology will produce valuable information (Kozak et al., 2006).
Table 8.
Amino acid identity of proteins isolated by 5’ UTRs
| Protein | Percent Identity* |
|---|---|
| Double-Stranded RNA Binding Proteins | |
| ILF2/NF90 | 97 |
| ILF3/NF45 | 87 |
| Helicase Superfamily | |
| DHX9 | no chicken ortholog |
| DHX30 | 78.5 |
| HNRNPs | |
| HNRNPA2B1 | 98 |
| HNRNPA3 | 89 |
| HNRNPAB | 85 |
| HNRNPH1 | 87 |
| HNRNPU | 97 |
| SYNCRIP/HNRNPQ | 97 |
| RRM and Nucleotide Binding Proteins | |
| NCL | 59 |
| SSRP1 | 90 |
| YBX1 | 95 |
| Ribosomal Proteins | |
| RPL3 | 96 |
| RPL7A | 96 |
| RPL12 | 95 |
| RPL13 | 92 |
| RPL15 | 99.5 |
| RPL35 | 98 |
| RPLP0 | 94 |
| RPS13 | 100 |
| RPS14 | 100 |
| Other | |
| ACTG1 | 99 |
| HSPA8 | 98 |
Percent identity at the amino acid level for Homo sapiens and Gallus gallus orthologs (www.genecards.org)
3.6 Overlap identified between 5’ UTR binding proteins and HIV-1 Gag interacting factors
Recently, a carefully controlled proteomic analysis of host interacting partners of the HIV-1 Gag protein was reported (Engeland et al., 2014). Of interest, numerous nuclear, nucleolar, and nucleocytoplasmic shuttling proteins were isolated in the affinity isolation experiments. To identify proteins that could be involved in HIV-1 packaging RNP formation, we compared the proteins we identified as binding to the HIV-1 5’ UTR, which contains the psi packaging sequence, with the list of HIV-1 Gag interacting factors reported by (Engeland et al., 2014) (Table 9). The subset of proteins identified in both the RNA and Gag proteomic studies that were previously demonstrated to be incorporated into HIV-1 virions includes DDX3X, DHX9, EEF1A1, HNRNPD, NCL, and STAU1. Several of these proteins have already been shown to participate in HIV-1 RNP formation, again validating our approach and the results of our study. In addition, there are 19 proteins (excluding co-identified ribosomal proteins) that interact with both Gag and HIV-1 5’ UTR sequences that are associated with genomic RNA processes including dimerization, genome recognition/binding, RNP complex formation and intracellular transport of RNPs. Further studies will be needed to investigate the potential biological roles of these proteins in HIV-1 genomic RNA packaging and virion assembly. In sum, our RNA affinity chromatography experiments identified previously authenticated as well as novel but logical candidates to more thoroughly investigate RNP activities important in retrovirus biology.
Table 9.
Overlap of human proteins isolated by HIV-1 5’ UTR or by HIV-1 Gag binding proteins as reported in (Engeland et al., 2014)
| DDX17 | EEF1A1 | H1FX | IGF2BP1 | RARS | RPL15 | RPL6 | RPS13 | RPS3A |
| DDX21 | EIF2S1 | HNRNPD | IGF2BP3 | RBMX | RPL23A | RPL7 | RPS14 | RPS8 |
| DDX3X | EIF2S3 | HNRNPH1 | MYBBP1A | RPL12 | RPL24 | RPL7A | RPS16 | STAU1 |
| DDX5 | EPRS | HNRNPH3 | NCL | RPL13 | RPL27A | RPL8 | RPS26 | TOP1 |
| DHX9 | GRSF1 | HNRNPK | NOP58 | RPL14 | RPL4 | RPLP0 | RPS3 | YBX1 |
Bold italized proteins were identified components of HIV-1 virions (Engeland et al., 2014)
4. Discussion
Commonalities were observed in the biological process anchored networks identified for HIV-1, SNV, and RSV and orthologous RNA binding proteins
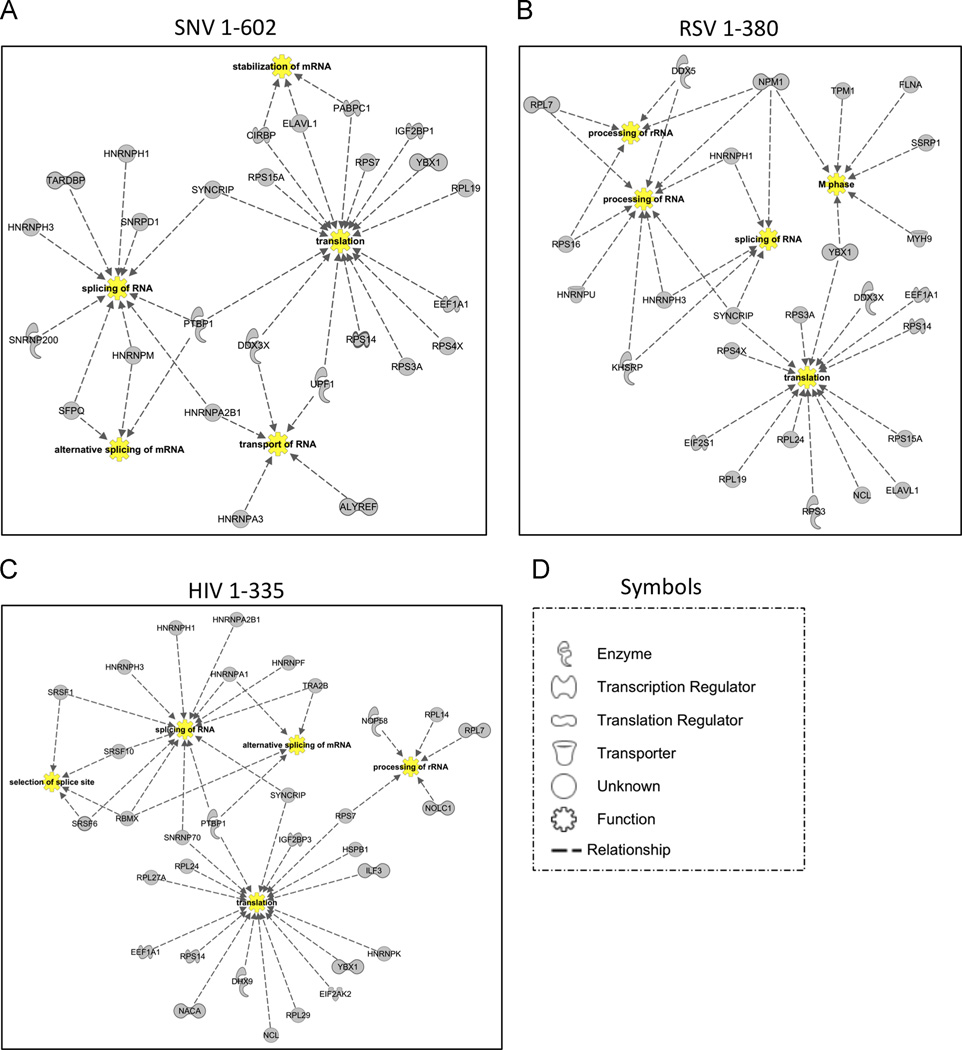
One hnRNP was the central node of the retrovirus networks: SYNCRIP/HNRNPQ (Figure 6). The results implicate orthologous hnRNPQ activities in human and avian cells integrate retroviral pre-mRNA splicing, processing, decay, and translation (Arif et al., 2012; Jurica et al., 2002; Weidensdorfer et al., 2009). Moreover, the co-isolation of ribosomal proteins, RNA synthetases and translation regulator proteins (DHX9, YY1, HuR) is reflective of the crosstalk between nuclear and cytoplasmic RNPs necessary for translation utilization of cellular mRNAs.
Figure 6.
Biological function network analysis for 5’ UTR RBPs by IPA. Evidence for interactions and functional qualities of each protein originate from Ingenuity Knowledge, and biological functions determined to be significantly overrepresented in the gene list by IPA were chosen as nodes to generate the network depicted. The biological process nodes are listed in yellow. Dashed lines indicate a relationship between the protein and the biological process nodes. The shape of each protein in the network indicates the designated activity of that protein (see legend at right).
Several RBP isolated by the HIV-1, SNV, and RSV 5’ UTRs function at the interface of transcriptional and post-transcriptional regulation
NF90/IL3 and NF45/ILF2 form a heterodimeric multifunctional complex regulating transcriptional control, translational control, mRNA processing and localization of cellular genes (Masuda et al., 2013). Initially described as interleukin enhancer binding proteins, NF90/NF45 heterodimers exhibit zinc-finger DNA- and RNA-binding activities that are pivotal to T cell homeostasis, influencing .mitotic control, DNA damage response, and susceptibility to apoptosis (Guan et al., 2008; Reichman et al., 2003; Shamanna et al., 2011; Shamanna et al., 2013). Notably, the 5’ UTRs isolated numerous proteins involved in mitotic control, and this activity coincides with the requirement of cell division for efficient SNV replication (Fritsch and Temin, 1977a; Fritsch and Temin, 1977b).
Also isolated in this study by the HIV-1 5’ UTR, DNA damage response proteins XRCC5 and XRCC6 (Roberts et al., 2010), which bind to and can regulate transcription from the HIV-1 LTR (Manic et al., 2013); downregulation of XRCC5 impairs HIV-1 integration and transcription (Waninger et al., 2004). Furthermore, XRCC6-XRCC5 (Ku p70-p86 dimer) heterodimerize with PRKDC to form the DNA-PK. Activation of DNA-PK during viral integration has a central role in CD4(+) T-cell depletion during HIV-1 productive infection (Cooper et al., 2013).
Kula et al utilized RNA immunoprecipitation and mass spectrometry to identify 32 HIV-1 RNA binding proteins from human osteosarcoma U20S cell line (Kula et al., 2011. In distinction to our study, which has focused on the 5’ UTRs of 3 viruses, their study identified proteins that co-precipitated with intron-containing vector RNA and exon-containing vector RNA, and were not exclusive to the HIV 5’ UTR. Appreciable overlap was recapitulated with RBPs identified by the 5’ UTRs of HIV, SNV and RSV RBPs, including DNA-PK, NF90, PSF, and Matrin 3. Notable distinctions were MOV10 and BAT1/UAP56, as well as CFL1 and SPIN1, which may associate outside of the 5’ UTR to influence nuclear export, RNA trafficking, or other yet-to-be defined activity.
The 5’ UTR RNA binding proteins identified in this study are overwhelmingly representative of the biological processes of RNA processing, splicing and mRNA translation
Their activities in retroviral RNA biology are attributable to combinations of direct and indirect interactions between the viral RNAs, protein adapters, and select proteins that utilize direct RNA binding motifs. The prevalence of physical and functional interconnections between these cellular RNA binding proteins implicates these functional linkages are necessary for retroviral post-transcriptional control. The network analysis provides valuable inferences into the complexity of the virus-host interface. Several of the nuclear RNA binding proteins isolated in this study overlap with RNA binding proteins identified to co-isolate HIV-1 Gag protein in cells and in virion particles (Engeland et al., 2014), provoking speculation that nuclear interactions are necessary to assembly of virion RNPs. Understanding the crosstalk between nuclear and cytoplasmic RNPs that assemble on the viral 5’ UTR may have translational implications for designing more effective regimens to attenuate retroviral infections in human and animals.
Supplementary Material
Cellular proteins are responsible for activity of retroviral 5’ cis-responsive sequences
Comprehensive inventory of high affinity cellular partners of retroviral RNA is needed
Retroviral RNA affinity-coupled-human/avian proteomics is comprehensive and unbiased
Higher-order structure of HIV-1 and SNV 5’ UTRs are similar, RSV is distinct
Nonreciprocal DXH9-5’ UTR binding explains human restriction of avian sarcoma virus
Acknowledgments
We thank Bruce Stanley, PhD, Penn State College of Medicine Proteomics and Small Molecule Mass Spectrometry Core Facility, for help with data analysis using Protein Pilot. This research was performed with support by the National Institutes of Health’s National Institutes of General Medical Sciences P50GM103297 to LJP and KBL; and National Cancer Institute P01CA16058 to The Ohio State University Comprehensive Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplemental Tables 1–6.
Contributor Information
Matthew Stake, Email: mss31@psu.edu.
Deepali Singh, Email: deepali.gbu@gmail.com.
Gatikrushna Singh, Email: singh.758@osu.edu.
J. Marcela Hernandez, Email: hernandez.1111@osu.edu.
Leslie Kaddis, Email: rjk297@psu.edu.
Leslie J. Parent, Email: lparent@psu.edu.
Kathleen Boris-Lawrie, Email: boris-lawrie.1@osu.edu.
References
- Agbottah ET, Traviss C, McArdle J, Karki S, St Laurent GC, 3rd, Kumar A. Nuclear Factor 90(NF90) targeted to TAR RNA inhibits transcriptional activation of HIV-1. Retrovirology. 2007;4:41. doi: 10.1186/1742-4690-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif A, Chatterjee P, Moodt RA, Fox PL. Heterotrimeric GAIT complex drives transcript-selective translation inhibition in murine macrophages. Mol Cell Biol. 2012;32:5046–5055. doi: 10.1128/MCB.01168-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolinger C, Sharma A, Singh D, Yu L, Boris-Lawrie K. RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res. 2010;38:1686–1696. doi: 10.1093/nar/gkp1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolinger C, Yilmaz A, Hartman TR, Kovacic MB, Fernandez S, Ye J, Forget M, Green PL, Boris-Lawrie K. RNA helicase A interacts with divergent lymphotropic retroviruses and promotes translation of human T-cell leukemia virus type 1. Nucleic Acids Res. 2007;35:2629–2642. doi: 10.1093/nar/gkm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- Burugu S, Daher A, Meurs EF, Gatignol A. HIV-1 translation and its regulation by cellular factors PKR and PACT. Virus Res. 2014;193:65–77. doi: 10.1016/j.virusres.2014.07.014. [DOI] [PubMed] [Google Scholar]
- Butsch M, Boris-Lawrie K. Destiny of unspliced retroviral RNA: ribosome and/or virion? J Virol. 2002;76:3089–3094. doi: 10.1128/JVI.76.7.3089-3094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L, Clement JF, Martel C, Beriault V, Gatignol A, DesGroseillers L, Mouland AJ. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol Cell Biol. 2004;24:2637–2648. doi: 10.1128/MCB.24.7.2637-2648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobrinik D, Katz R, Terry R, Skalka AM, Leis J. Avian sarcoma and leukosis virus pol-endonuclease recognition of the tandem long terminal repeat junction: minimum site required for cleavage is also required for viral growth. J Virol. 1987;61:1999–2008. doi: 10.1128/jvi.61.6.1999-2008.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Garcia M, Petrovas C, Yamamoto T, Koup RA, Nabel GJ. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature. 2013;498:376–379. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeland CE, Brown NP, Borner K, Schumann M, Krause E, Kaderali L, Muller GA, Krausslich HG. Proteome analysis of the HIV-1 Gag interactome. Virology. 2014;460–461:194–206. doi: 10.1016/j.virol.2014.04.038. [DOI] [PubMed] [Google Scholar]
- Fritsch E, Temin HM. Formation and structure of infectious DNA of spleen necrosis virus. J Virol. 1977a;21:119–130. doi: 10.1128/jvi.21.1.119-130.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch EF, Temin HM. Inhibition of viral DNA synthesis in stationary chicken embryo fibroblasts infected with avian retroviruses. J Virol. 1977b;24:461–469. doi: 10.1128/jvi.24.2.461-469.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii R, Okamoto M, Aratani S, Oishi T, Ohshima T, Taira K, Baba M, Fukamizu A, Nakajima T. A Role of RNA Helicase A in cis-Acting Transactivation Response Element-mediated Transcriptional Regulation of Human Immunodeficiency Virus Type 1. J Biol Chem. 2001;276:5445–5451. doi: 10.1074/jbc.M006892200. [DOI] [PubMed] [Google Scholar]
- Ghoujal B, Milev MP, Ajamian L, Abel K, Mouland AJ. ESCRT-II’s involvement in HIV-1 genomic RNA trafficking and assembly. Biol Cell. 2012;104:706–721. doi: 10.1111/boc.201200021. [DOI] [PubMed] [Google Scholar]
- Gleghorn ML, Maquat LE. ‘Black sheep’ that don’t leave the double-stranded RNA-binding domain fold. Trends Biochem Sci. 2014;39:328–340. doi: 10.1016/j.tibs.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, Altan-Bonnet N, Parrott AM, Arrigo CJ, Li Q, Khaleduzzaman M, Li H, Lee CG, Pe’ery T, Mathews MB. Nuclear factor 45 (NF45) is a regulatory subunit of complexes with NF90/110 involved in mitotic control. Mol Cell Biol. 2008;28:4629–4641. doi: 10.1128/MCB.00120-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat Struct Mol Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- Heng X, Kharytonchyk S, Garcia EL, Lu K, Divakaruni SS, LaCotti C, Edme K, Telesnitsky A, Summers MF. Identification of a minimal region of the HIV-1 5’-leader required for RNA dimerization, NC binding, and packaging. J Mol Biol. 2012;417:224–239. doi: 10.1016/j.jmb.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Shamanna RA, Guan D, Pe’ery T, Mathews MB. HIV-1 replication and latency are regulated by translational control of cyclin T1. J Mol Biol. 2011;410:917–932. doi: 10.1016/j.jmb.2011.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. Bmc Genomics. 2008:9. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonson L, Vikesaa J, Krogh A, Nielsen LK, Hansen T, Borup R, Johnsen AH, Christiansen J, Nielsen FC. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics. 2007;6:798–811. doi: 10.1074/mcp.M600346-MCP200. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. Rna. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak SL, Marin M, Rose KM, Bystrom C, Kabat D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J Biol Chem. 2006;281:29105–29119. doi: 10.1074/jbc.M601901200. [DOI] [PubMed] [Google Scholar]
- Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- Kula A, Guerra J, Knezevich A, Kleva D, Myers MP, Marcello A. Characterization of the HIV-1 RNA associated proteome identifies Matrin 3 as a nuclear cofactor of Rev function. Retrovirology. 2011;8:60. doi: 10.1186/1742-4690-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzembayeva M, Dilley K, Sardo L, Hu WS. Life of psi: how full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology. 2014;454–455:362–370. doi: 10.1016/j.virol.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau PP, Chang BH, Chan L. Two-hybrid cloning identifies an RNA-binding protein, GRY-RBP, as a component of apobec-1 editosome. Biochem Biophys Res Commun. 2001;282:977–983. doi: 10.1006/bbrc.2001.4679. [DOI] [PubMed] [Google Scholar]
- Leblanc J, Weil J, Beemon K. Posttranscriptional regulation of retroviral gene expression: primary RNA transcripts play three roles as pre-mRNA, mRNA, and genomic RNA. Wiley Interdiscip Rev RNA. 2013;4:567–580. doi: 10.1002/wrna.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque K, Halvorsen M, Abrahamyan L, Chatel-Chaix L, Poupon V, Gordon H, DesGroseillers L, Gatignol A, Mouland AJ. Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic. 2006;7:1177–1193. doi: 10.1111/j.1600-0854.2006.00461.x. [DOI] [PubMed] [Google Scholar]
- Liu HM, Aizaki H, Choi KS, Machida K, Ou JJ, Lai MM. SYNCRIP (synaptotagmin-binding, cytoplasmic RNA-interacting protein) is a host factor involved in hepatitis C virus RNA replication. Virology. 2009;386:249–256. doi: 10.1016/j.virol.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochmann TL, Bann DV, Ryan EP, Beyer AR, Mao A, Cochrane A, Parent LJ. NC-mediated nucleolar localization of retroviral gag proteins. Virus Res. 2013;171:304–318. doi: 10.1016/j.virusres.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorgeoux RP, Guo F, Liang C. From promoting to inhibiting: diverse roles of helicases in HIV-1 Replication. Retrovirology. 2012;9:79. doi: 10.1186/1742-4690-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, Divakaruni SS, LaCotti C, Barton S, Tummillo D, Hosic A, Edme K, Albrecht S, Telesnitsky A, Summers MF. NMR detection of structures in the HIV-1 5’-leader RNA that regulate genome packaging. Science. 2011a;334:242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Heng X, Summers MF. Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol. 2011b;410:609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manic G, Maurin-Marlin A, Laurent F, Vitale I, Thierry S, Delelis O, Dessen P, Vincendeau M, Leib-Mosch C, Hazan U, Mouscadet JF, Bury-Mone S. Impact of the Ku complex on HIV-1 expression and latency. PLoS One. 2013;8:e69691. doi: 10.1371/journal.pone.0069691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley JL, Fire A, Cano A, Sharp PA, Gefter ML. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980;77:3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Kuwano Y, Nishida K, Rokutan K, Imoto I. NF90 in posttranscriptional gene regulation and microRNA biogenesis. Int J Mol Sci. 2013;14:17111–17121. doi: 10.3390/ijms140817111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- McNally MT, Gontarek RR, Beemon K. Characterization of Rous sarcoma virus intronic sequences that negatively regulate splicing. Virology. 1991;185:99–108. doi: 10.1016/0042-6822(91)90758-4. [DOI] [PubMed] [Google Scholar]
- Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol. 2000;74:5441–5451. doi: 10.1128/jvi.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomska AM, Gifford RJ. The extraordinary evolutionary history of the reticuloendotheliosis viruses. PLoS Biol. 2013;11:e1001642. doi: 10.1371/journal.pbio.1001642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH. Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr Biol. 2003;13:199–207. doi: 10.1016/s0960-9822(03)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole E, Strappe P, Mok HP, Hicks R, Lever AM. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and FRET. Traffic. 2005;6:741–755. doi: 10.1111/j.1600-0854.2005.00312.x. [DOI] [PubMed] [Google Scholar]
- Ranji A, Shkriabai N, Kvaratskhelia M, Musier-Forsyth K, Boris-Lawrie K. Features of double-stranded RNA-binding domains of RNA helicase A are necessary for selective recognition and translation of complex mRNAs. J Biol Chem. 2011;286:5328–5337. doi: 10.1074/jbc.M110.176339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichman TW, Parrott AM, Fierro-Monti I, Caron DJ, Kao PN, Lee CG, Li H, Mathews MB. Selective regulation of gene expression by nuclear factor 110, a member of the NF90 family of double-stranded RNA-binding proteins. J Mol Biol. 2003;332:85–98. doi: 10.1016/s0022-2836(03)00885-4. [DOI] [PubMed] [Google Scholar]
- Roberts SA, Strande N, Burkhalter MD, Strom C, Havener JM, Hasty P, Ramsden DA. Ku is a 5’-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy BB, Hu J, Guo X, Russell RS, Guo F, Kleiman L, Liang C. Association of RNA helicase a with human immunodeficiency virus type 1 particles. J Biol Chem. 2006;281:12625–12635. doi: 10.1074/jbc.M510596200. [DOI] [PubMed] [Google Scholar]
- Sewer MB, Nguyen VQ, Huang CJ, Tucker PW, Kagawa N, Waterman MR. Transcriptional activation of human CYP17 in H295R adrenocortical cells depends on complex formation among p54(nrb)/NonO, protein-associated splicing factor, and SF-1, a complex that also participates in repression of transcription. Endocrinology. 2002;143:1280–1290. doi: 10.1210/endo.143.4.8748. [DOI] [PubMed] [Google Scholar]
- Shamanna RA, Hoque M, Lewis-Antes A, Azzam EI, Lagunoff D, Pe’ery T, Mathews MB. The NF90/NF45 complex participates in DNA break repair via nonhomologous end joining. Mol Cell Biol. 2011;31:4832–4843. doi: 10.1128/MCB.05849-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamanna RA, Hoque M, Pe’ery T, Mathews MB. Induction of p53, p21 and apoptosis by silencing the NF90/NF45 complex in human papilloma virus-transformed cervical carcinoma cells. Oncogene. 2013;32:5176–5185. doi: 10.1038/onc.2012.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Boris-Lawrie K. Determination of host RNA helicases activity in viral replication. Methods Enzymol. 2012;511:405–435. doi: 10.1016/B978-0-12-396546-2.00019-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Yilmaz A, Marsh K, Cochrane A, Boris-Lawrie K. Thriving under stress: selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog. 2012;8:e1002612. doi: 10.1371/journal.ppat.1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Nishitsuji H, Marusawa H, Ujino S, Takaku H, Shimotohno K. The RNA-editing enzyme APOBEC1 requires heterogeneous nuclear ribonucleoprotein Q isoform 6 for efficient interaction with interleukin-8 mRNA. J Biol Chem. 2014;289:26226–26238. doi: 10.1074/jbc.M114.563221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Yanagiya A, Karetnikov AE, Alain T, Fabian MR, Khoutorsky A, Perreault S, Topisirovic I, Sonenberg N. Control of translation and miRNA-dependent repression by a novel poly(A) binding protein, hnRNP-Q. PLoS Biol. 2013;11:e1001564. doi: 10.1371/journal.pbio.1001564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WH, Shilov IV, Seymour SL. Nonlinear fitting method for determining local false discovery rates from decoy database searches. J Proteome Res. 2008;7:3661–3667. doi: 10.1021/pr070492f. [DOI] [PubMed] [Google Scholar]
- Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Waninger S, Kuhen K, Hu X, Chatterton JE, Wong-Staal F, Tang H. Identification of cellular cofactors for human immunodeficiency virus replication via a ribozyme-based genomics approach. J Virol. 2004;78:12829–12837. doi: 10.1128/JVI.78.23.12829-12837.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidensdorfer D, Stohr N, Baude A, Lederer M, Kohn M, Schierhorn A, Buchmeier S, Wahle E, Huttelmaier S. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. Rna. 2009;15:104–115. doi: 10.1261/rna.1175909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedavalli VS, Jeang KT. Matrin 3 is a co-factor for HIV-1 Rev in regulating post-transcriptional viral gene expression. Retrovirology. 2011;8:61. doi: 10.1186/1742-4690-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Rong L, Lu J, Pan Q, Liang C. Insulin-like growth factor II mRNA binding protein 1 associates with Gag protein of human immunodeficiency virus type 1, and its overexpression affects virus assembly. J Virol. 2008;82:5683–5692. doi: 10.1128/JVI.00189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin AS, Michalowski D, Bear J, Smulevitch SV, Traish AM, Peng R, Patton J, Shatsky IN, Felber BK. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol Cell Biol. 2003;23:6618–6630. doi: 10.1128/MCB.23.18.6618-6630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.