Abstract
The structure and stability of membrane proteins can vary widely in different detergents and this variability has great practical consequences for working with membrane proteins. Nevertheless, the mechanisms that operate to alter the behavior of proteins in micelles are poorly understood and not predictable. Atomic simulations could provide considerable insight into these mechanisms. Building protein-micelle complexes for simulation is fraught with uncertainty, however, in part because it is often unknown how many detergent molecules are present in the complex. Here, we describe several convenient ways to employ Micelle Builder in CHARMM-GUI to rapidly construct protein-micelle complexes and performed simulations of the isolated voltage-senor domain of voltage-dependent potassium-selective channel and an antimicrobial peptide papiliocin with varying numbers of detergents. We find that once the detergent number exceeds a threshold, protein-detergent interactions change very little and remain very consistent with experimental observations. Our results provide a platform for future studies of the interplay between protein structure and detergent properties at the atomic level.
Keywords: protein-detergent interactions, voltage-dependent potassium-selective channel, papiliocin
Graphical abstract
1. Introduction
Micelles are commonly used as cell membrane mimetics to solubilize membrane proteins [1–5]. However, it is often difficult to select detergents for particular experiments, because the effects of protein-detergent interactions on protein structure and dynamics are poorly understood. To study such effects at the atomic level, one can build various protein/micelle systems and perform molecular dynamics (MD) simulations [6, 7]. Since detergents can self-assemble to form micelles around a protein, the self-assembling simulations can be applied by placing a certain amount of detergents in a large water box containing a protein [8–10]. Nevertheless, self-assembly simulations require long relaxation time (depending on protein and detergent types), which are computationally expensive. To avoid long relaxation times, one can build a preassembled model of a protein/micelle complex. To simplify and automate the process of building protein-micelle complex model systems for MD simulations, Cheng et al recently developed Micelle Builder (http://www.charmm-gui.org/input/micelle) in CHARMM-GUI [11, 12] following the framework of Membrane Bilayer Builder [13–15]. Using Micelle Builder, a user can choose homogeneous detergents or mixtures of different detergents to build a micelle system with or without a membrane protein. However, the estimation of exact ratios of protein to detergents is challenging in many cases. Even with the known experimental molar ratio of detergent to protein, there could be discrepancy between the experimental molar ratio and the ratio of the protein-associated detergents to protein, because the distribution of detergents is not uniform at nm-scale in solvent and some detergents can dynamically associate to and dissociate from protein/micelle complexes. In addition, it is mostly unknown how detergents solubilize and arrange themselves around a protein.
In this study, we tested two strategies for the construction of micelles around a protein. First, to address the protein/detergent ratio problem, we built multiple protein-micelle systems with different numbers of detergents. Since the membrane-spanning region provides additional hydrophobic surface area for detergent assembly, the number of detergents per micelle should be more than the aggregation number of a detergent-only micelle. In a second strategy, to determine the preferred location and to cover the membrane protein in a micelle, we inserted its transmembrane segments in the center of the micelle. For a protein whose transmembrane segments are undefined, the whole protein can be placed in the center of a micelle to make sure that most protein residues have a chance to interact with the micelle. To avoid the protein unfolding due to the initial non-native-like environment, constraints can be applied to maintain the protein structure at the beginning of the simulations. The protein and detergents are then allowed to adjust to each other and form a favorable assembled complex during the simulation.
To test the protein-micelle systems built according to these two strategies, we employed two distinctively different systems for which there are experimental data for validation. The first is the isolated voltage-senor domain (VSD) of voltage-dependent potassium-selective (Kv) channel (KvAP VSD) [16–20]. The VSD consists of four transmembrane helices (S1–S4; Fig. 1A). The structure of the KvAP VSD in dihetanoylphosphatidylcholine (DHPC) micelles has been extensively characterized by NMR spectroscopy [16], which serves as a useful validation for the simulation results. In this study, we used the different numbers of DHPC detergents to solvate KvAP VSD in the simulations. We find that when the number of detergents gets above a threshold in the system, the resulting systems show conserved properties consistent with the experimental ones. The second system is papiliocin, a small antimicrobial peptide, whose structure was determined in dodecylphosphocholine (DPC) micelles by NMR (Fig. 1F) [21]. It was found to bind to the surface of the micelles rather than become entirely engulfed by the detergents. Papiliocin is proved to be an interesting test case because when we started with the structure buried in the micelle, the protein moved to the surface during the simulations, in conformations that are consistent with experimental observations.
Fig. 1.
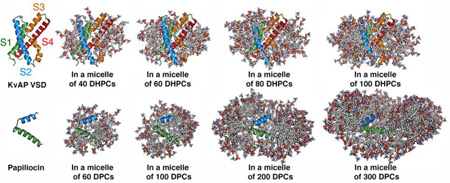
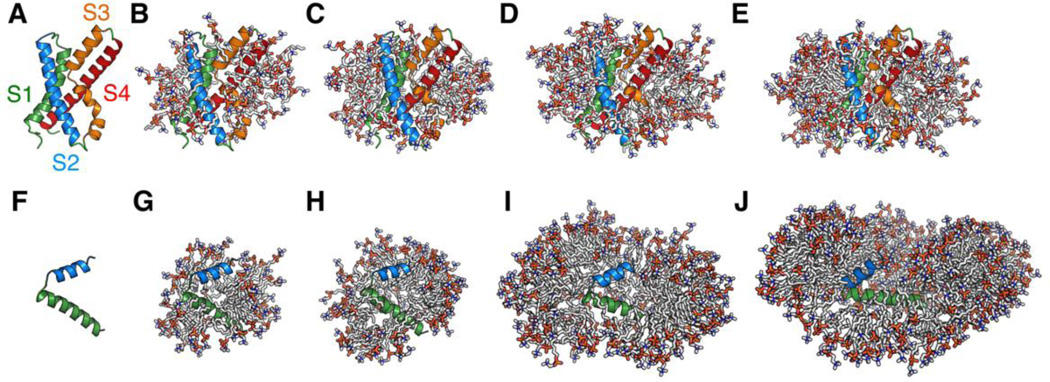
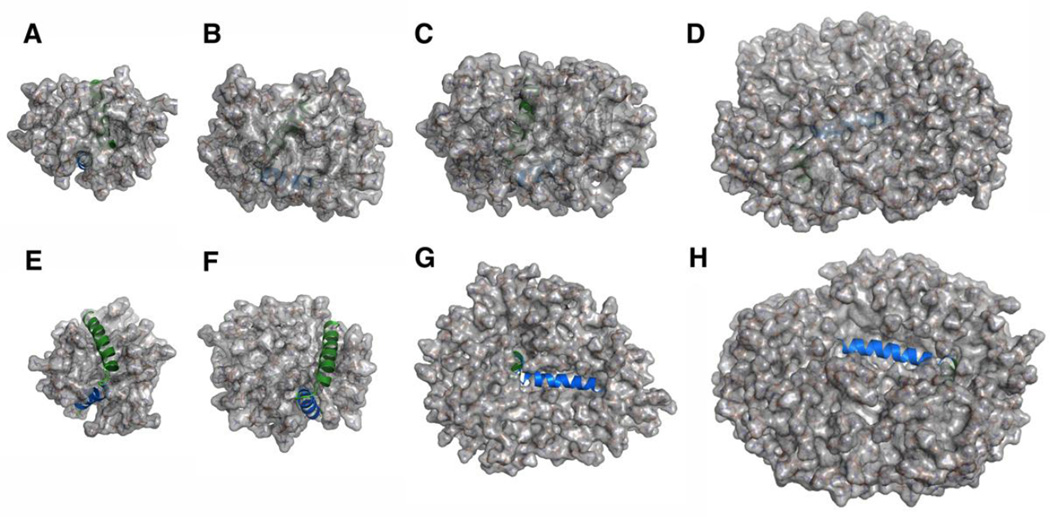
Structures of proteins and protein-micelle complexes. (A) Structure of KvAP VSD (PDB:1ORS). (B–E) Snapshots showing KvAP VSD in the micelles of (B) 40, (C) 60, (D) 80, and (E) 100 DHPC molecules. (F) Structure of papiliocin (PDB:2LA2). (G–J) Snapshots showing papiliocin in the micelles of (G) 60, (H) 100, (I) 200, and (J) 300 DPC molecules. The protein is presented in cartoon and the detergents are shown in sticks. Ions, water, and detergents blocking the view of proteins are omitted for clarity.
2. Material and methods
2.1 Simulations of KvAP VSD
The structure of KvAP VSD solubilized within DHPC micelles (PDB:2KYH) [16] is highly similar to its crystal structure (PDB:1ORS) [18] complexed with an antibody fragment. Since the crystal structure has a higher resolution, we used the crystal structure as a starting structure to build various VSD-DHPC micelle systems. The aggregation number of DHPC in a DHPC-only micelle is 35 [22], but a protein-micelle complex system generally needs more detergent molecules due to the increased hydrophobic surface area on the protein. The Micelle Builder module [11] in CHARMM-GUI (www.charmm-gui.org) [12] was used to insert the VSD structure into four micelle systems consisting of 40, 60, 80, and 100 DHPC molecules, called DHPC40, DHPC60, DHPC80 and DHPC100, respectively. Snapshots are shown in Fig. 1B–E. The transmembrane segments (S1, S2, S3, and S4 in Fig. 1A) were placed in the center of each micelle system. All system used 150 mM KCl. Each system was replicated and assigned with different initial velocities to generate five independent simulations, resulting in a total of 20 simulations. The CHARMM36 force field [23–25] and a TIP3P water model [26] were used. All calculations were performed in an NPT (constant particle number, pressure, and temperature) ensemble [27] at 318.15 K using NAMD 2.9 [28] (with the NAMD input scripts generated by CHARMM-GUI [29]). The particle mesh Ewald algorithm [30] was applied to calculate electrostatic forces, and the van der Waals interactions were smoothly switched off at 10–12 Å by a force-switching function [31]. A time step of 2 fs was used in all simulations. After equilibration, a 100-ns production run was performed for each system.
2.2 Simulations of papiliocin
The structure of papiliocin determined in DPC micelles (PDB:2LA2) [21] was used as a starting point. The aggregation number of DPC in a DPC-only micelle is 54 [32]. Thus, Micelle Builder was used to insert papiliocin into four micelle systems of 60, 100, 200, and 300 DPC molecules in 150 mM KCl solution, called DPC60, DPC100, DPC200, and DPC300, respectively. Snapshots are shown in Fig. 1G–J. Papiliocin consists of an N-terminal helix (residue Lys3–Lys21) and a C-terminal helix (residue Ala25- Val36). The amidated C terminus (CT2) patch was used for the last residue Lys37 through CHARMM-GUI PDB Reader [33]. Since the transmembrane segment of papiliocin is not well defined, the whole structure was placed in the center of micelle in each system. Papiliocin is a flexible peptide adopting different structures in different environments. Thus, backbone restraints were applied on papiliocin to maintain its initial secondary structures for the first 50-ns in DPC60, DPC100, and DPC200 systems. After equilibration, a 50-ns production run was performed for each of these systems without restraints. For the DPC300 system, only short 10-ns production runs were performed with the same secondary backbone restraints on papiliocin to examine any difference between DPC200 and DPC300 systems. Each system was replicated and assigned with different initial velocities to generate five independent simulations. All calculations were performed in an NPT ensemble [27] at 303 K using NAMD 2.9 [28] with the CHARMM force field [23–25]. The simulation protocol is the same as in the simulation of KvAP VSD.
2.3 Paramagnetic 5-doxylstearic acids titration in papiliocin
The location of papiliocin in DPC micelles was examined by measuring the effect of 5-doxylstearic acids on the proton signal of papiliocin. Papiliocin was dissolved at 1.0 mM in 0.50 ml of 9:1 (v/v) H2O/D2O 20 mM phosphate buffer, pH 5.9 containing 300 mM DPC. After recording the TOCSY spectrum in the absence of the 5-doxylstearic acids, the 5-doxylstearic acids were titrated into the samples to yield a final concentration of 5 mM spin-labeled acids, and the TOCSY spectra with a mixing time of 20 ms were recorded at 298K on a Bruker 800 MHz spectrometer (Bruker, Rheinstetten, Germany) at the Korean Basic Science Institute at Ochang. The intensity reductions of TOCSY spectra caused by spin-labeled lipids were measured by comparing the intensities of the TOCSY cross-peaks in the presence and absence of the spin-labeled lipids.
3. Results and Discussion
3.1 Binding of DHPC detergents to KvAP VSD
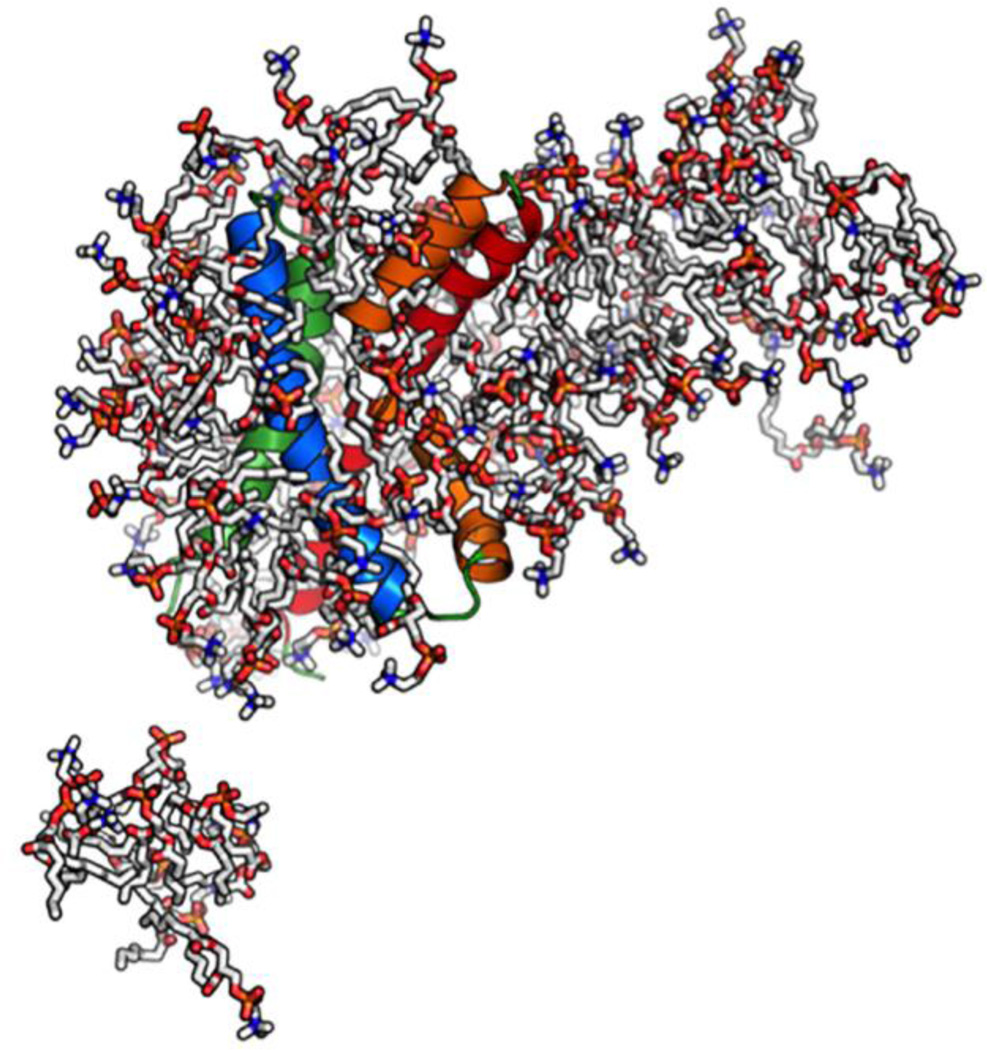
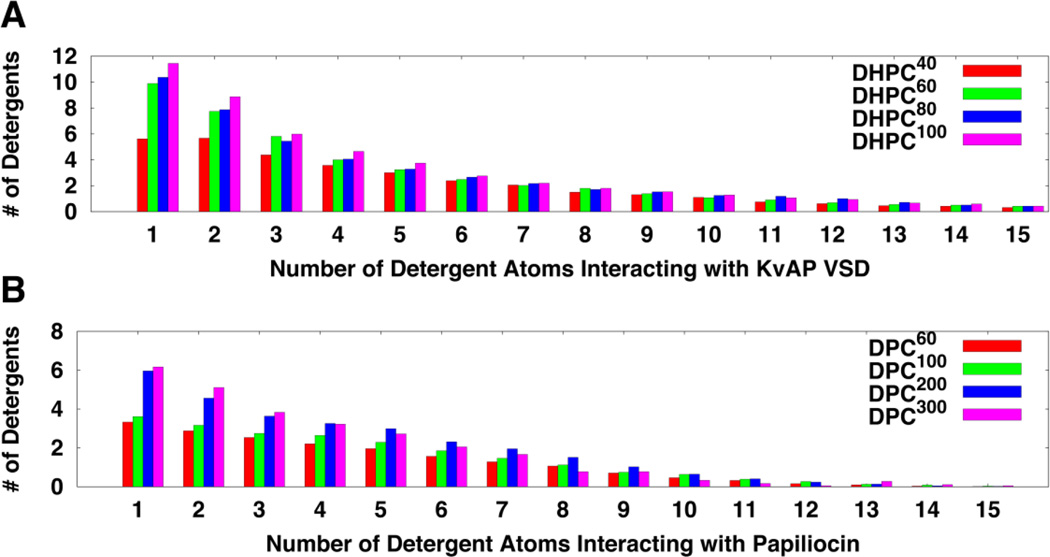
As shown in Fig. 2, some detergent molecules dissociate from a protein-micelle complex and float around during simulations, suggesting that this system is “saturated”. In fact, such dissociation was observed in all systems. In particular, there were 2~4 detergent molecules frequently dissociating from micelles in DHPC60, DHPC80, and DHPC100 systems. The number of detergents in a protein-micelle complex varies in different systems due to different initial micelle sizes (Table 1). However, the amount of detergents that are in direct contact with protein is less varied in different systems. In this study, when a detergent has any heavy atom within 4 Å from any protein heavy atom, the detergent is considered to be in a direct “contact” with protein. As shown in Fig. S1 A–D, the number of direct-contact detergents has small standard errors over five replicated simulations in all systems, suggesting the convergence of these micelle simulations. In addition, the amounts of detergents in direct contact with KvAP VSD are similar in DHPC60, DHPC80, and DHPC100 systems (Table 1 and Fig. S1). These detergents can be further classified according to the number of their interacting atoms. As shown in Fig. 3A, DHPC40 has on average 5 detergent molecules that have one atom interacting with the protein, while DHPC60, DHPC80, and DHPC100 have more than 10 detergent molecules. Notably, DHPC60, DHPC80, and DHPC100 systems show similar interaction patterns, indicating that the micelle of 60 DHPC detergents is interacting with KvAP VSD in a similar way as the larger micelles do. That is, 60 DHPC detergent molecules are sufficient for solubilizing KvAP VSD and from the protein perspective, increasing the number of detergent molecules beyond 60 is of little consequence. Thus, DHPC60 system is used as a representative for further analysis. Indeed, all properties including the protein structure and dynamics in DHPC80 and DHPC100 are very similar to those in DHPC60 (data not shown).
Fig. 2.
A snapshot of KvAP VSD in a micelle of 100 DHPC molecules. The protein is presented in cartoon and the detergents are shown in sticks. Ions and water are omitted for clarity.
Table 1.
Detergent behavior in KvAP VSD/DHPC systems.
| System | Number of detergents in protein/micelle complex |
Number of detergents in direct contact with protein |
|---|---|---|
| DHPC40 | 40±1 | 34±2 |
| DHPC60 | 58±1 | 43±3 |
| DHPC80 | 78±1 | 47±3 |
| DHPC100 | 96±4 | 49±3 |
Fig. 3.
Histogram of protein-contact detergents classified by the number of detergent atoms in direct contact with (A) KvAP VSD and (B) papiliocin in different systems.
3.2 Interactions between KvAP VSD and DHPC60 micelle
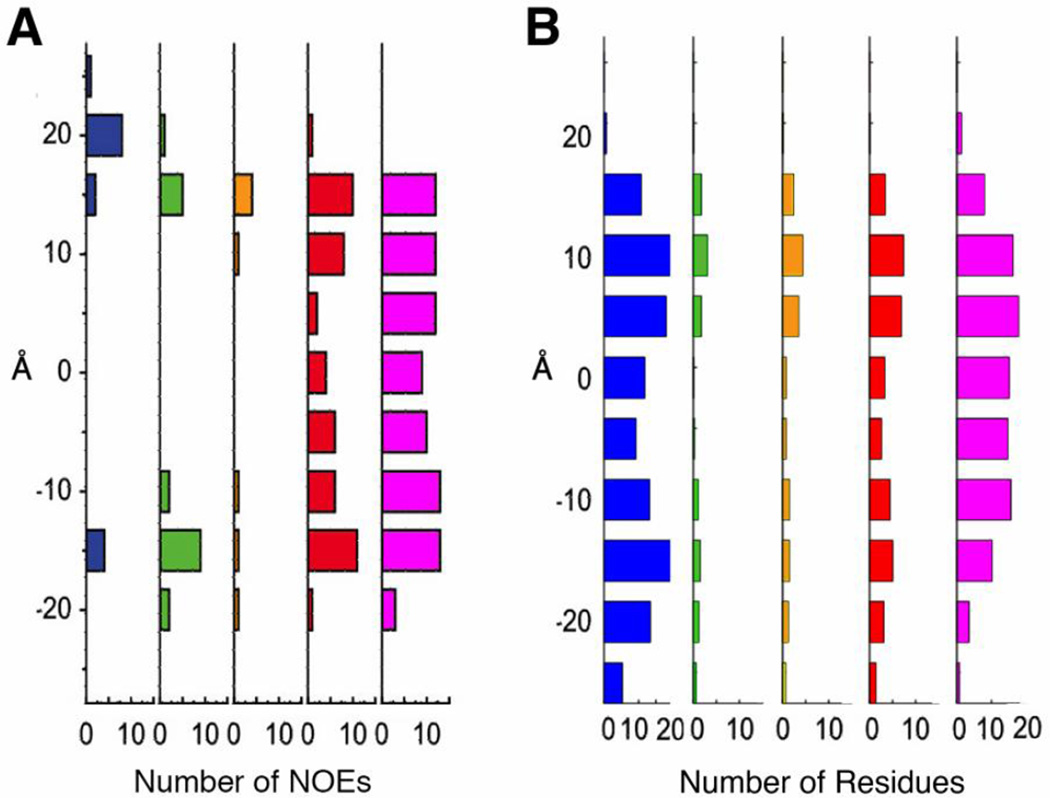
Butterwick et al. [16] employed NMR experiments to carefully characterize the structure of the KvAP VSD-DHPC micelle complex. They report the histograms of NOE cross-peaks from KvAP VSD to water, DHPC choline headgroup atoms, DHPC glycerol backbone atoms, and DHPC aliphatic carbons along the transmembrane axis (Fig. 4A). To compare our protein-micelle simulations to the experimental observations, we calculated the number of transmembrane residues of KvAP VSD interacting with water and different DHPC detergent atoms along the transmembrane axis. As shown in Fig. 4B, the histograms of interactions between protein and detergents (choline headgroup, glycerol backbone, and aliphatic carbons) calculated from DHPC60 simulations are highly consistent with experimental observations of Butterwick et al. [16]. However, we observed water accessing to the center of VSD from both intra- and extracellular sides (Fig. 5). In particular, a few water molecules frequently form hydrogen bonds with Asp62 near the center of VSD during the simulations. The presence of these water-filled crevices and the Asp62-associated water molecule are supported by the NMR data (secondary chemical shifts for amide protons and NOE) of VSD solubilized in DPC/LDOA micelles by Shenkarev et al [17].
Fig. 4.
Histograms of interactions between KvAP VSD transmembrane residues and system components. Interactions with water are shown in blue, DHPC choline headgroup in green, DHPC glycerol backbone in orange, DHPC first two aliphatic carbons in red, and DHPC last four aliphatic carbons in magenta). (A) Butterwick et al. reported a histogram of NOE cross-peaks from KvAP VSD transmembrane segments to water and detergents along KvAP VSD transmembrane axis (the Y-axis). (B) Histogram of transmembrane residues interacting with water and detergents along the transmembrane axis (the Y-axis) in DHPC60 system.
Fig. 5.
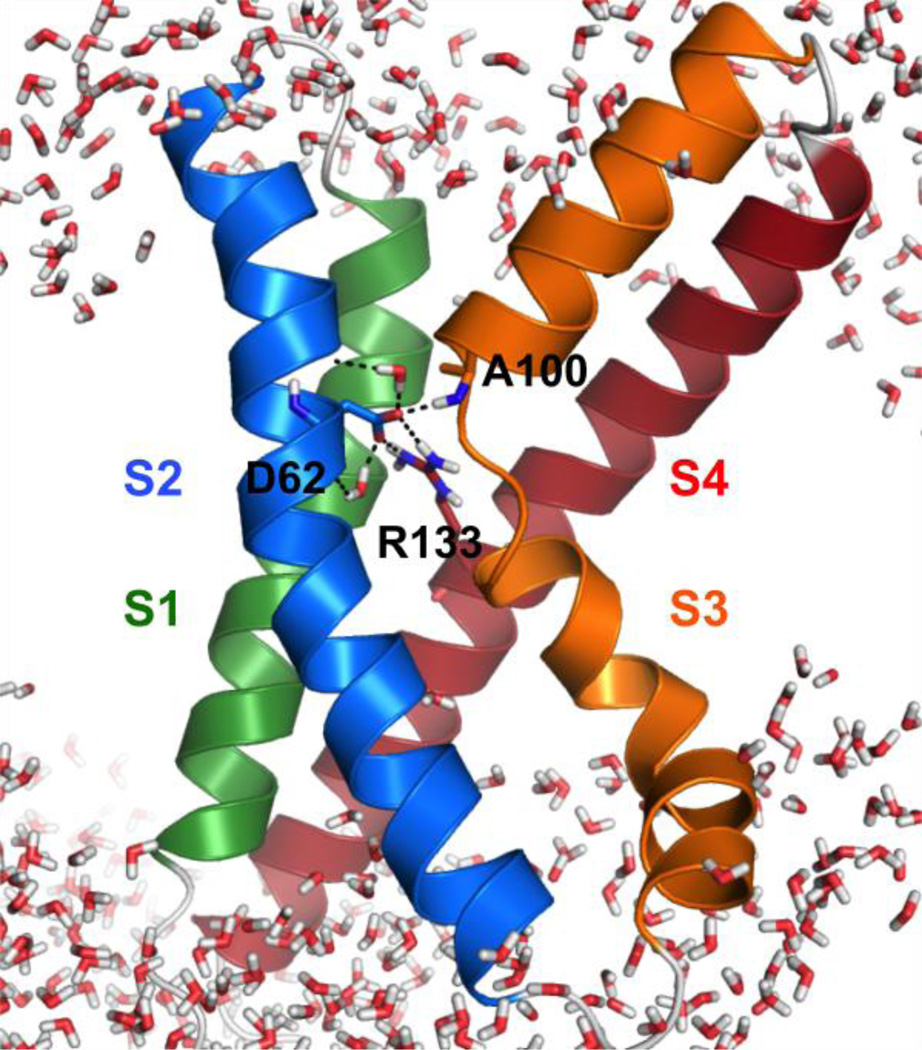
A snapshot of KvAP VSD in a micelle of 60 DHPC molecules. The protein is presented in cartoon. Residues Asp62, Ala100, Arg133, and water molecules are shown in sticks. The black dashed lines represent salt bridges and hydrogen bonds. Ions and detergents are omitted for clarity.
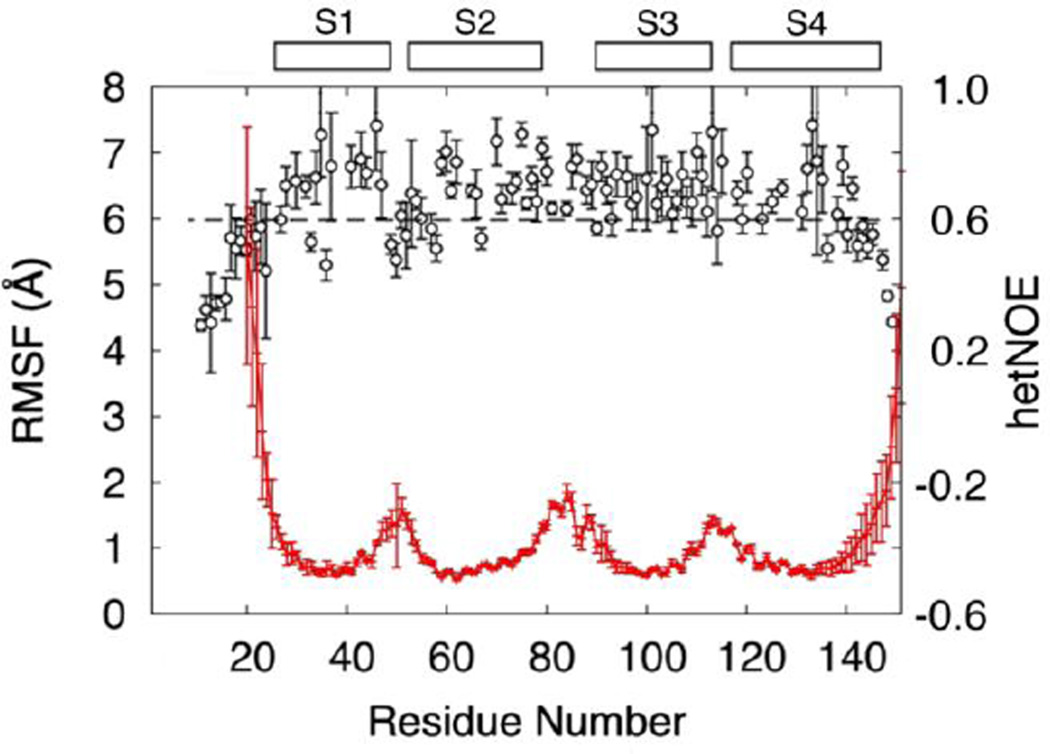
In DHPC60 system, the VSD adopts a stable structure and variations in structural flexibility are broadly consistent with experimental observations. During the simulations, the transmembrane helices maintain a backbone root-mean-squared deviation (RMSD) of 1.9 ± 0.2 Å from PDB:1ORS. Fig. 6 compares the per-residue backbone root-mean-squared fluctuations (RMSF) with experimentally observed 1H-15N heteronuclear NOE (hetNOE) measurements. The low hetNOEs (< 0.6) at the C-terminus and N-terminus as well as the loop regions imply a high degree of flexibility, consistent with the high RMSF values seen in these regions. The large hetNOEs in the transmembrane helices implies a larger structural rigidity in agreement with the reduced RMSF values seen in our simulations.
Fig. 6.
Comparison of flexibility observed in the simulations with hetNOE values. Per-residue backbone RMSF values for KvAP VSD in DHPC60 system are shown in red. Per-residue 1H-15N heteronuclear NOE (hetNOE) values are shown in black. The locations of transmembrane helices S1–S4 are indicated by the boxes. The dashed line indicates a low hetNOE value of 0.6.
3.3 Interactions between papiliocin and DPC micelles
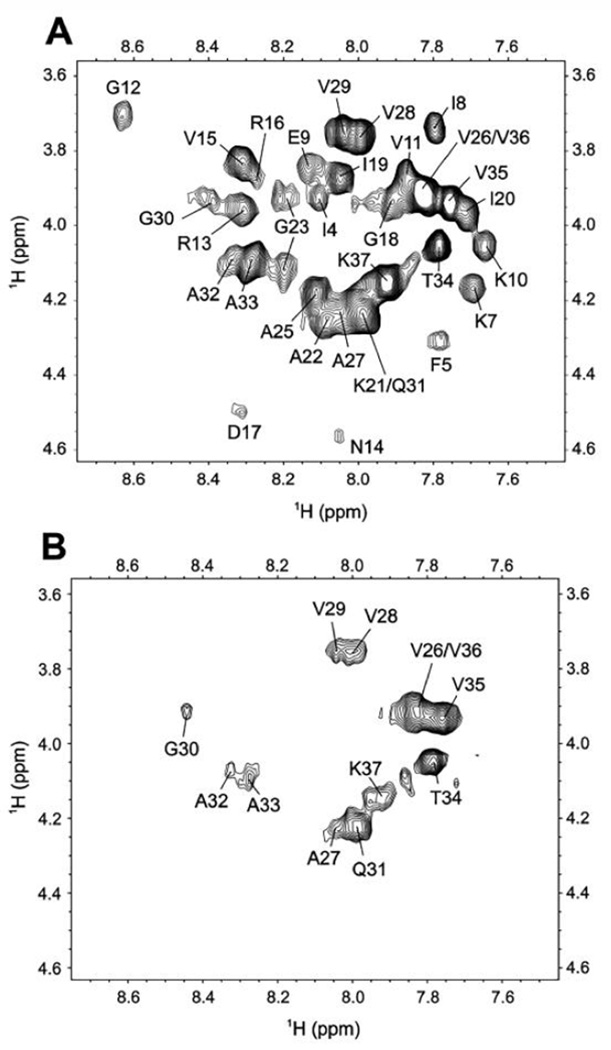
Papiliocin consists of an N-terminal amphipathic helix and a C-terminal hydrophobic helix, linked by a short hinge region. To investigate how papiliocin associates with DPC micelles, we examined the paramagnetic relaxation effects resulting from the presence of spin labels integrated into the DPC micelle. The micelle-integrating spin-label 5-doxylstearic acid is known to cause peak broadening of residues that are close to the head group and the micelle–water interface. As shown in Fig. 7, the intensities of the most of the residues at the N-terminal helix region are significantly affected by the presence of 5-doxylstearic acid and signal attenuations for residues at the C-terminal helix region were much stronger than those in the N-terminal helix region. These results indicate that the N-terminal helix lies on the surface of the micelles, while the C-terminal helix may be buried in the DPC micelles and have hydrophobic interactions with the DPC acyl chains.
Fig. 7.
Membrane-inserted structure of papiliocin in DPC micelle probed by 5-doxylstearic acid. (A) TOCSY spectra of 1mM papiliocin in the absence of 5-doxylstearic acid and (B) TOCSY spectra in the presence of 5-doxylstearic acid. 5-doxylstearic acid was added at a concentration of 1/60 spin label/DPC.
At the beginning of all simulations, papiliocin was buried in each micelle complex, but it eventually moved to the micelle-water interface (Fig. 8). In DPC60 and DPC100 systems, the C-terminal helix region becomes exposed to water, while DPC200 and DPC300 have this region inserted in the micelle. Compared to DPC200 and DPC300, DPC60 and DPC100 have fewer detergent molecules associating with papiliocin when simulations converged (Fig. S1 E–H and Fig. 3B). Therefore, micelles of 60–100 DPC molecules may have insufficient detergent molecules to completely engulf papiliocin. DPC200 and DPC300 systems have similar detergent interaction patterns, suggesting that the DPC200 system is sufficient to solubilize papiliocin. Thus, the DPC200 system is used as a representative in the following analysis.
Fig. 8.
Progress of papiliocin simulations. Initial (A–D) and equilibrated (E–H) structures of papiliocin in DPC micelles of (A, E) 60, (B, F) 100, (C, G) 200, and (D, H) 300 detergents. The protein is presented in green and detergents are shown in sticks and gray surface. Ions and water are omitted for clarity.
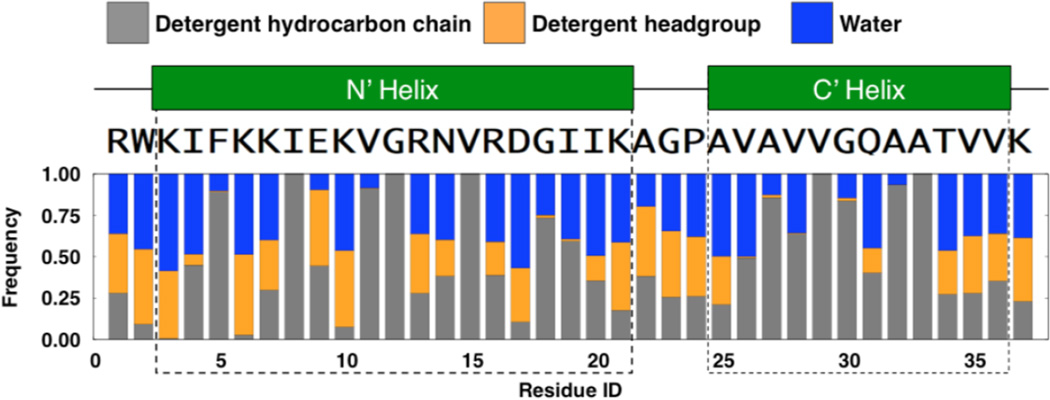
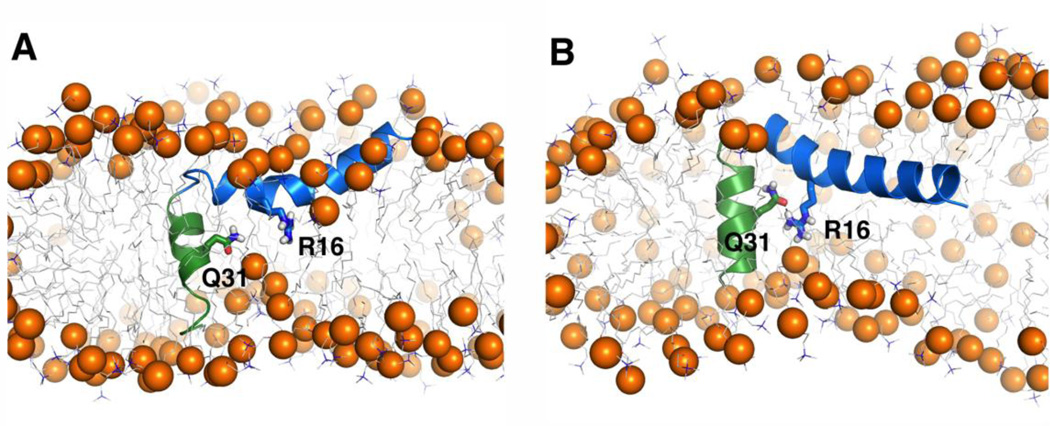
In DPC200 system, the N-terminal helix lies on the micelle surface and the C-terminal helix inserts into the micelle (Fig. 8G), supporting the experimental observations from the TOCSY spectra mentioned above. The interaction profiles between each protein residue and system components (water and detergents) of the DPC200 system were calculated to further characterize how micelles interact with papiliocin at the atomic level (Fig. 9). In the N-terminal helix, the hydrophobic residues (Phe5, Ile8, Val11, Gly12, and Val15) frequently interact with detergent hydrocarbon chains on one side, while the polar or charged residues (Lys3, Lys6, Lys7, Glu9, Lys10, Arg13, Asn14, Arg16, Asp17, and Lys20) on the other side interact with the detergent headgroups and water, thereby stabilizing the orientation of this amphipathic helix on the micelle surface. In the C-terminal helix, Val29, Ala32 and Ala33 frequently interact with the detergent hydrocarbon chains, which helps drive insertion of this short helix in the micelle. However, a polar residue Gln31 in the middle of C-terminal helix and a charged residue Arg16 in the N-terminal helix play roles in exposing the C-terminus to polar interacting partners. As shown in Fig.10A, Arg16 and Gln31 can interact with the detergent headgroups, and therefore induce a local curvature on the micelle surface. In particular, Arg16 directly interacts with the detergent headgroups in 91.8% of all simulation trajectories. As a consequence, water molecules have a chance to access C-terminal helix residues Thr34-Val36 and Gly30-Ala31 (Fig. 9). Nevertheless, less than 10% of the surface area of the C-terminal helix is exposed to water on average in the simulations. Moreover, in 42.0% of the DPC200 simulation trajectories, the side-chain carbonyl oxygen of Gln31 can form a hydrogen bond with the side-chain nitrogen of Arg16 (Fig. 10B). In these structures, the average interaction frequency between Gln31 and detergent headgroups is only ~5%. However, in the structures excluding the Arg16-Gln31 hydrogen bond, the interaction frequency between Gln31 and detergent headgroups is ~65%. That is, Arg16 maybe able to stabilize Gln31 in the hydrophobic core of micelle and reduce its access to the water-micelle interface. Arg16 side chain is mostly buried during the simulations in DPC200 system, while Arg16 is on the micelle surface in DPC100 system. Nevertheless, the frequency of hydrogen bonding interaction between Arg16 and Gln31 is about 20% in DPC100 system. Therefore, burying Arg16 in the hydrophobic micelle center may stabilize the hydrogen bonding of Arg16 and Gln31, but it is not a necessary condition.
Fig. 9.
Interactions between papiliocin residues and various components in DPC200 system. The graph shows the frequency with which any heavy atom of each residue is found within 4 Å of detergent hydrocarbon chain, detergent head groups, and water. The green rectangles indicate the N-terminal helix and the C-terminal helix.
Fig. 10.
Interactions of Arg16 and Gln31. (A) Representative snapshots showing the interactions between protein residues (Arg16 and Gln31) and detergent headgroups. (B) Snapshot showing the hydrogen bond between the side-chain carbonyl oxygen of Gln31 and the side-chain nitrogen of Arg16. The protein is shown in cartoon presentation and Arg16 and Gln31 residues are shown in sticks. The detergent hydrocarbon chains are shown in gray lines and the headgroup phosphorus is presented in spheres. Ions, water and detergents blocking the view of proteins are omitted for clarity.
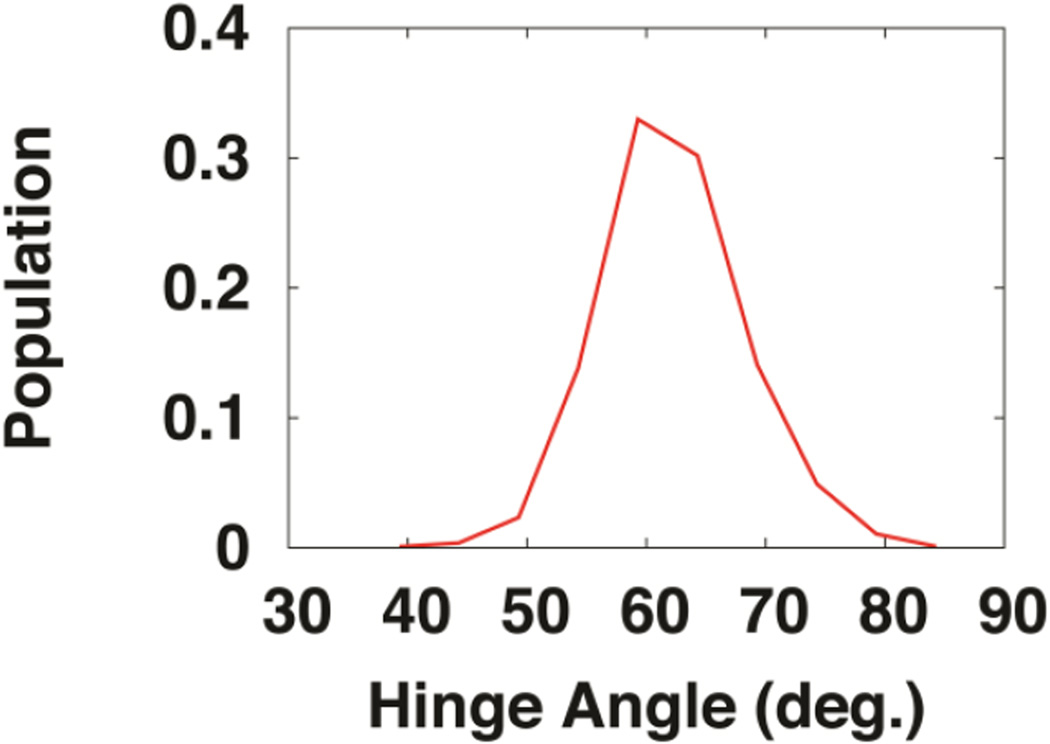
Papiliocin has a large backbone RMSD of 4.4 ± 0.7 Å from PDB:2LA2 in simulations, while the RMSDs of individual N- and C-terminal helices are ~0.5 Å. Since the loop in the protein is short, the large overall RMSD is due to the flexible relative orientation between the N- and C-terminal helices, similar to Pf1 and fd coat proteins [34, 35]. As shown in Fig. 11, the hinge angle between the N- and C-terminal helices ranges from 40° to 85°, which is highly consistent with the previous experimental observations reporting a hinge angle ranging from 45° to 80° [13]. With respect to the micelle, the conserved protein-detergent interactions stabilize the orientations of both N- and C-terminal helices, and therefore help define the structure of papiliocin. Additionally, as discussed above, residues Arg16 and Gln31 can form a hydrogen bond, which may also contribute to stabilizing the structure in micelles.
Fig. 11.
Distribution of hinge angle between N-terminal and C-terminal helices in DPC200 micelle system.
4. Conclusions
This work shows that the number of detergents in contact with protein is not significantly changed by adding more detergents, when the protein structure is maintained and a protein-micelle system is “saturated” as in most experimental conditions. Thus, when the ratio of protein and detergents is unknown, to find an optimal MD simulation system, one can build multiple protein-micelle systems with detergent numbers exceeding the micelle aggregation number. The KvAP VSD simulation system was chosen to test this idea and produced results consistent with experimental data, supporting the utility of this strategy.
To build a reasonable initial preassembly model of a protein-micelle complex, one can insert the transmembrane segments of the protein into the center of the micelle. In the case of a protein whose transmembrane segments are undefined, the whole protein can be placed in the micelle center to maximize the chance of the protein interacting with the micelle. By inserting papiliocin in the DPC micelle center as an initial model, we were able to obtain a protein/micelle complex showing good agreement with experimental observations. Taking advantage of all-atom MD simulations, we are also able to observe how papiliocin is solubilized in micelles at the atomic level. Conserved interactions between papiliocin and detergents are observed, which not only locate papiliocin in the micelle, but also determine its structure. Our results suggest that protein micelle complex simulations hold promise for understanding how different detergents can stabilize or destabilize protein structure.
Supplementary Material
Highlights.
Practical strategies are introduced to perform molecular simulations of various protein-micelle complexes.
Illustrations of the strategies are given for simulations of the isolated voltage-senor domain of voltage-dependent potassium-selective channel and papiliocin with varying numbers of detergents.
Protein-detergent interactions are converged once the detergent number exceeds a threshold.
The protein initially placed in the micelle center can adjust to the detergents and form a favorable assembled complex during the simulation.
Acknowledgements
This research was supported by grants from the National Science Foundation (MCB- 1157677 and DBI-1145987 to WI), XSEDE Resources (MCB070009 to WI), National Institutes of Health (RO1 GM063919 to JUB), the National Research Foundation of Korea (2013R1A1A2058021 to YK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Columbus L, Lipfert J, Klock H, Millett I, Doniach S, Lesley SA. Expression, purification, and characterization of Thermotoga maritima membrane proteins for structure determination. Protein Sci. 2006;15:961–975. doi: 10.1110/ps.051874706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eshaghi S, Hedren M, Nasser MIA, Hammarberg T, Thornell A, Nordlund P. An efficient strategy for high-throughput expression screening of recombinant integral membrane proteins. Protein Sci. 2005;14:676–683. doi: 10.1110/ps.041127005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanders CR, Sonnichsen F. Solution NMR of membrane proteins: practice and challenges. Magn Reson Chem. 2006;44:S24–S40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 4.Berger BW, Garcia RY, Lenhoff AM, Kaler EW, Robinson CR. Relating surfactant properties to activity and solubilization of the human adenosine a3 receptor. Biophysical journal. 2005;89:452–464. doi: 10.1529/biophysj.104.051417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiener MC. A pedestrian guide to membrane protein crystallization. Methods. 2004;34:364–372. doi: 10.1016/j.ymeth.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 6.Cheng X, Im W. NMR observable-based structure refinement of DAP12-NKG2C activating immunoreceptor complex in explicit membranes. Biophysical journal. 2012;102:L27–L29. doi: 10.1016/j.bpj.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patargias G, Bond PJ, Deol SS, Sansom MS. Molecular dynamics simulations of GlpF in a micelle vs in a bilayer: conformational dynamics of a membrane protein as a function of environment. J Phys Chem B. 2005;109:575–582. doi: 10.1021/jp046727h. [DOI] [PubMed] [Google Scholar]
- 8.Bockmann RA, Caflisch A. Spontaneous formation of detergent micelles around the outer membrane protein OmpX. Biophysical journal. 2005;88:3191–3204. doi: 10.1529/biophysj.105.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khelashvili G, LeVine MV, Shi L, Quick M, Javitch JA, Weinstein H. The membrane protein LeuT in micellar systems: aggregation dynamics and detergent binding to the S2 site. J Am Chem Soc. 2013;135:14266–14275. doi: 10.1021/ja405984v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abel S, Dupradeau F, Marchi M. Molecular Dynamics Simulations of a Characteristic DPC Micelle in Water. Journal of Chemical Theory and Compution. 2012;8:4610–4623. doi: 10.1021/ct3003207. [DOI] [PubMed] [Google Scholar]
- 11.Cheng X, Jo S, Lee HS, Klauda JB, Im W. CHARMM-GUI Micelle Builder for Pure/Mixed Micelle and Protein/Micelle Complex Systems. J Chem Inf Model. 2013;53:2171–2180. doi: 10.1021/ci4002684. [DOI] [PubMed] [Google Scholar]
- 12.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. Journal of computational chemistry. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 13.Jo S, Kim T, Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PloS one. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophysical journal. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu EL, Cheng X, Jo S, Rui H, Song KC, Davila-Contreras EM, Qi Y, Lee J, Monje-Galvan V, Venable RM, Klauda JB, Im W. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. Journal of computational chemistry. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butterwick JA, MacKinnon R. Solution Structure and Phospholipid Interactions of the Isolated Voltage-Sensor Domain from KvAP. J Mol Biol. 2010;403:591–606. doi: 10.1016/j.jmb.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shenkarev ZO, Paramonov AS, Lyukmanova EN, Shingarova LN, Yakimov SA, Dubinnyi MA, Chupin VV, Kirpichnikov MP, Blommers MJJ, Arseniev AS. NMR Structural and Dynamical Investigation of the Isolated Voltage-Sensing Domain of the Potassium Channel KvAP: Implications for Voltage Gating. J Am Chem Soc. 2010;132:5630–5637. doi: 10.1021/ja909752r. [DOI] [PubMed] [Google Scholar]
- 18.Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 19.Lee SY, Lee A, Chen J, MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 21.Kim JK, Lee E, Shin S, Jeong KW, Lee JY, Bae SY, Kim SH, Lee J, Kim SR, Lee DG, Hwang JS, Kim Y. Structure and function of papiliocin with antimicrobial and anti-inflammatory activities isolated from the swallowtail butterfly, Papilio xuthus. The Journal of biological chemistry. 2011;286:41296–41311. doi: 10.1074/jbc.M111.269225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Columbus L, Lipfert J, Jambunathan K, Fox DA, Sim AYL, Doniach S, Lesley SA. Mixing and Matching Detergents for Membrane Protein NMR Structure Determination. J Am Chem Soc. 2009;131:7320–7326. doi: 10.1021/ja808776j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klauda JB, Venable RM, Freites JA, O'Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Pastor RW. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. Journal of computational chemistry. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 26.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 27.Feller SE, Zhang YH, Pastor RW. Computer-Simulation of Liquid/Liquid Interfaces .2. Surface-Tension Area Dependence of a Bilayer and Monolayer. J Chem Phys. 1995;103:10267–10276. [Google Scholar]
- 28.Kumar S, Huang C, Zheng G, Bohm E, Bhatele A, Phillips JC, Yu H, Kale LV. Scalable molecular dynamics with NAMD on the IBM Blue Gene/L system. Ibm J Res Dev. 2008;52:177–188. [Google Scholar]
- 29.Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y, Jo S, Pande VS, Case DA, Brooks CL, MacKerell AD, Klauda JB, Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. Journal of Chemical Theory and Compution. 2015 doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A Smooth Particle Mesh Ewald Method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 31.Steinbach PJ, Brooks BR. New Spherical-Cutoff Methods for Long-Range Forces in Macromolecular Simulation. Journal of computational chemistry. 1994;15:667–683. [Google Scholar]
- 32.Tieleman DP, van der Spoel D, Berendsen HJC. Molecular dynamics simulations of dodecylphosphocholine micelles at three different aggregate sizes: Micellar structure and chain relaxation. J Phys Chem B. 2000;104:6380–6388. [Google Scholar]
- 33.Jo S, Cheng X, Islam SM, Huang L, Rui H, Zhu A, Lee HS, Qi Y, Han W, Vanommeslaeghe K, MacKerell AD, Jr, Roux B, Im W. CHARMM-GUI PDB manipulator for advanced modeling and simulations of proteins containing nonstandard residues. Advances in protein chemistry and structural biology. 2014;96:235–265. doi: 10.1016/bs.apcsb.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng X, Jo S, Marassi FM, Im W. NMR-based simulation studies of Pf1 coat protein in explicit membranes. Biophysical journal. 2013;105:691–698. doi: 10.1016/j.bpj.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng X, Jo S, Qi Y, Marassi FM, Im W. Solid-State NMR-Restrained Ensemble Dynamics of a Membrane Protein in Explicit Membranes. Biophysical journal. 2015;108:1954–1962. doi: 10.1016/j.bpj.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.