Abstract
Evolution of osmoregulatory systems is a key factor in the transition of species between fresh- and saltwater habitats. Anopheles coluzzii and An. merus are stenohaline and euryhaline malaria vector mosquitoes belonging to a larger group of sibling species, the Anopheles gambiae complex, which radiated in Africa within the last 2 million years. Comparative ecological genomics of these vector species can provide insight into the mechanisms that permitted the rapid radiation of this species complex into habitats of contrasting salinity. Here we use RNA-Seq to investigate gene expression differences between An. coluzzii and An. merus after briefly exposing both young and old larval instars of each species to either saltwater (SW) or freshwater (FW). Our examination aims to identify candidate genes and pathways responsible for the greater SW tolerance of An. merus. Our results are congruent with the ability of gene induction to mediate salinity tolerance, with both species showing increasing amounts of differential gene expression between SW and FW as salt concentrations increase. Besides ion transporters such as AgAE2 that may serve as effectors for osmoregulation, we also find mitogen activated protein kinases that may serve in a phosphorylation signaling pathway responding to salinity, and report potential cross-talk between the mosquito immune response and osmoregulation. This study provides a key step towards applying the growing molecular knowledge of these malaria vectors to improve understanding of their ecological tolerances and habitat occupancy.
Keywords: Anopheles merus, c-Jun N-terminal kinase, gene expression reaction norm, osmoregulation, saltwater tolerance, transporters
Introduction
Transitions between freshwater (FW) and saltwater (SW) systems can represent dramatic shifts, requiring adaptation to sudden fluxes of water and ions. Salinity has been described as the most important physical environmental influence on species’ distributions (Whitehead et al. 2011). Within fishes, salinity tolerance differs both among closely related species and within populations of the same species, with killifish and sticklebacks representing two well-studied examples (Jones et al. 2012; Kozak et al. 2013). Differences in saltwater tolerance have been implicated in the stable coexistence of sibling species in cryptic species complexes and local adaptation of aquatic invertebrates such as rotifers (Montero-Pau & Serra 2011; Scheuerl & Stelzer 2013). Rapid evolution of enzymes (V-type H+ ATPase and Na+/K+ ATPase) in the copepod Eurytema affinis has been linked with the capacity of this invasive species to move from marine to FW systems (Lee et al. 2011). Ability to cross the osmotic gradient between such FW and SW systems is central to the biogeography, population dynamics, and local adaptation of a variety of organisms.
Freshwater systems are thought to be the ancestral habitat for the Culicidae family of mosquitoes, with ~95% of species inhabiting FW, but the lack of phylogenetic clustering of species occupying SW suggests that there have been multiple acquisitions (and/or losses) of salinity tolerance (Bradley 1987; Clements 1992). Both SW and FW species co-occur repeatedly in different species complexes across genus Anopheles, including 3 non-sister species tolerant of SW (An. merus, An. melas, and An. bwambae) out of a group of 8 morphologically indistinguishable species in the An. gambiae complex (Fontaine et al. 2015; White et al. 2011). Distinct molecular mechanisms for osmoregulation may apply, even among the three SW tolerant sibling species, as An. bwambae requires high mineral content to complete normal development (Harbach et al. 1997; White et al. 2011), while both An. merus and An. melas larvae can develop in either fresh- or brackish waters. Determining the role of salinity tolerance in local adaptation and ecological speciation of anophelines requires detailed phenotypic and molecular analyses of the trait.
Physiological studies suggest a key role of protein localization in anopheline osmoregulation. In FW, mosquitoes absorb ions from the external medium with their anal papillae, and in the rectum further resorption of ions takes place (Bradley 2008). In tolerant anophelines, SW exposure is mediated by osmoregulation and ion excretion (Bradley 2008; Smith et al. 2008). With antibody stains, Linser and colleagues demonstrated that the location of expression of Na+/K+-ATPase of FW species does not change regardless of water salinity, while in SW-tolerant anophelines (An. albimanus, An. merus) it is up-regulated in the dorsal anterior rectum (DAR) and down-regulated in the non-DAR (Smith et al. 2010; Smith et al. 2008; White et al. 2013). The authors propose that this disrupts coordination of Na+/K+-ATPase with V-ATPase, ultimately transforming the role of non-DAR cells from ion resorption to production of hyper-osmotic urine. However, ability of the larvae to shift Na+/K+-ATPase and survive transfers from FW to saline media declines after ~1 d, with >80% of 48 h-old An. merus larvae placed in SW failing to reach pupation (Smith et al. 2010; Smith et al. 2008; White et al. 2013).
Research on the genetic basis of mosquito osmoregulation is in its infancy. Rheault and colleagues (2007) describe two mechanisms mediating ion transport, the first consisting of electroneutral ion exchange (e.g., by Na+/H+-exchangers) energized via ATP hydrolysis, as by Na+/K+-ATPase. Second, Na+/nH+ antiporters can use voltage gradients established by H+-ATPases to influence ion transport (Rheault et al. 2007). Bioinformatic and physiological analyses have suggested the presence of both mechanisms in anophelines, with reports of anopheline electrophoretic antiporters AgNHA1 and AgNHA2 (Rheault et al. 2007; Xiang et al. 2012), as well as anion exchangers such as AgAE2 (Linser et al. 2012). Likely, an integrated network of signaling systems regulates their expression. In fish, osmoregulation is proposed to result from the combined action of osmosensors that detect osmotic stress and signal transduction pathways that respond, along with effectors such as ion transporters that act to restore homeostasis (Kültz 2012). Mitogen-activated protein kinase (MAPK) pathways are associated with osmotic stress signal transduction in taxa ranging from fish species (Whitehead et al. 2013) to plants (Cowan & Storey 2003), and the high-osmolarity glycerol (HOG) pathway is a MAPK signaling pathway associated with osmotic stress in yeast (Li et al. 2012). Thus similar themes in osmoregulatory responses are seen throughout diverse organisms, and may be shared by mosquitoes.
To further understanding of anopheline SW tolerance, we began with QTL mapping from a cross of euryhaline An. merus and stenohaline An. coluzzii, using 2387 markers distributed genome-wide (Smith et al. 2015). We identified six genomic regions associated with osmoregulation: one on chromosome arm 2R, one encompassing the centromere of chromosome 2, two on arm 3R, and two on the X chromosome. Our results suggested the potential for multiple interacting loci, and perhaps different yet fungible combinations of alleles, to contribute to SW survival. However, these six QTL intervals were too large to allow inference of specific genes involved in the trait. Here we expand these findings with RNA-Seq to associate mRNA expression levels of individual genes with SW exposure. We hypothesize that inducible gene expression is associated with osmoregulation, influencing the overall greater tolerance of SW by An. merus compared to An. coluzzii. To address this hypothesis we compare transcriptomic responses of young (18 h post-hatch) An. merus and An. coluzzii transferred for 6 h from FW into one of six solutions, (FW, 10, 20, 30, 40, and 50% the salinity of seawater). We also investigated the response of older (66 h) larvae, which cannot survive a transition to SW (White et al. 2013).
Materials and Methods
Cultures and saltwater exposure
Colonies of the Mali-NIH strain of Anopheles coluzzii, formerly known as An. gambiae “M” form (Coetzee et al. 2013), and the An. merus MAF strain were maintained at the University of Notre Dame insectary as in (White et al. 2013), with minor modifications. Strains were obtained from MR4 (MRA-1156 and MRA-860, MR4, ATCC® Manassas, Virginia). To minimize maternal effects from prior SW exposure, larvae of both the SW-tolerant species An. merus as well as the FW species An. coluzzii were reared in FW (reverse-osmosis water) for ≥2 generations prior to the experiment. Except during the 6 h experimental exposures, larvae were fed daily until pupation with 2:1 w/w ground mixture of Koi fish food to active dry yeast. Saltwater used in experimental assays consisted of sea salts (~98.6% NaCl). In an initial experiment in 2012, species-specific near-lethal salinity levels were chosen for SW concentrations based on the chronic exposure bioassays of White et al. (2013). Survivorship through pupation declines at ~20% the salinity of seawater for An. coluzzii, while for An. merus survivorship declines at ~60% (White et al. 2013). Thus for An. merus the salinity was set slightly below that inducing substantial mortality, at 50% salinity (15.85 g/L sea salts, defining seawater as 31.7 g/L salt), and for An. coluzzii we used 20% salinity (6.34 g/L). Subsequently in 2014, we repeated the experiment exposing each species to a gradient of 0%, 10%, 20%, 30%, 40%, and 50% the salinity of seawater, as detailed below.
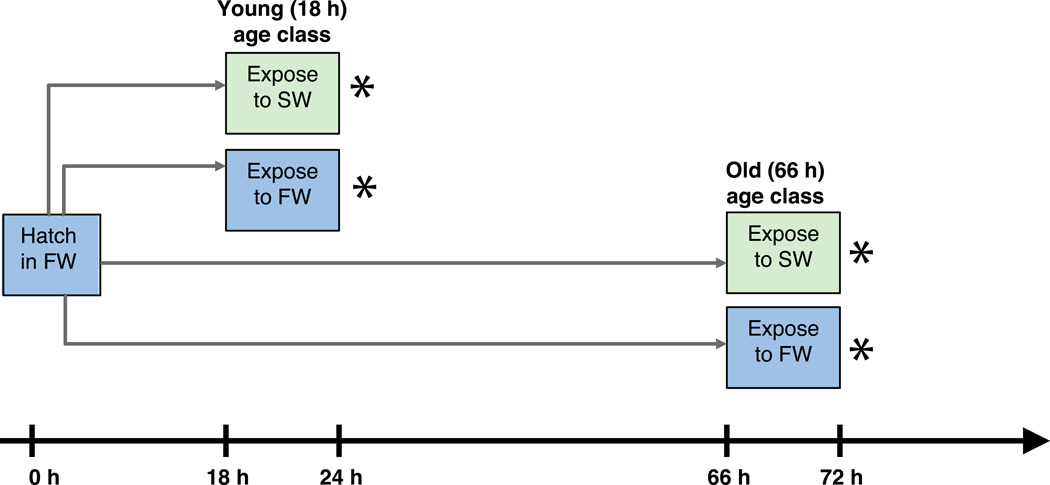
In both years, experiments were performed separately but simultaneously on An. merus and An. coluzzii (Fig. 1). Cages of adults received blood meals to stimulate egg production. Synchronous hatching was induced in FW, following (White et al. 2013). This procedure requires a very brief (~6 min.) exposure of eggs to air, and hatching in FW rather than beginning experimental exposure in SW prevents uncontrolled increases in salinity from evaporative water loss. Neonates were placed in small plastic containers (~12.5 cm2) at densities of 50 larvae / 200 mL FW; all 50 larvae were kept together for the remainder of the experiment as a pooled replicate sample. At 18 h post-hatch (approximately 6–7 a.m.), experimental exposures were begun. All larvae in each replicate container were transferred to a new container with 200 mL of one of six solutions: water of 0%, 10%, 20%, 30%, 40%, or 50% salinity. Exposure lasted 6 h, after which time ~35–50 animals from each container were pooled into 1.5 mL eppendorf tubes, snap-frozen in liquid nitrogen, and stored at −80°C until RNA extraction. In 2012 the experiment only tested FW vs. a single saline water concentration of 50% for An. merus, and 20% for An. coluzzii. However in addition to 18 h larvae, the identical protocol was performed on sibling larvae that remained in FW until 66 h post-hatch, and then were transferred to FW or SW for 6 h. With two salinities (FW or SW), two age classes of exposure, and two species, we had eight separate treatments replicated four times apiece, for a total of 32 samples in 2012. In the 2014 experiment we only assayed 18 h larvae, but used the gradient of salinities of 0–50% in 10% increments. Thus with two species, six salinities, and three replicates (12 treatments), we produced 36 samples in 2014.
Figure 1.
Schematic representation of experimental design. Exposures were performed separately, but simultaneously, for An. coluzzii and An. merus. Each species was exposed to SW (or FW) for 6 h, followed by immediate preservation for RNA extraction as designated by *. In 2012, two ages were tested by starting exposure at age 18 h post-hatch or age 66 h post-hatch. In 2014, only one age was tested, starting exposure at 18 h post-hatch.
No mortality was observed for An. coluzzii at salinities <50% during the short 6 h exposure period, nor for An. merus at any salinity. Yet roughly 1/3rd of the animals in the An. coluzzii samples at 50% salinity died. Due to the impossibility of separating the tiny live and dead first instar larvae quickly enough to avoid prolonging exposure beyond 6 h, all An. coluzzii larvae were preserved for library preparation. As discussed below, results for An. coluzzii at salinities ≥30% were treated with caution as some of this gene expression data may reflect impending mortality.
Library preparation and sequencing
All samples of ~35–50 pooled larvae were used to construct separate mRNA-Seq libraries. Total RNA was extracted and DNA removed with the RNeasy® Mini Kit and RNase-Free DNAse Set following the manufacturer’s instructions (QIAGEN). Construction and barcoding of libraries was performed with the TruSeq® RNA Sample Preparation kit v2 (Illumina) from 600 ng of total RNA per sample. For sequencing we pooled one replicate of each of the eight treatments per lane, with a total of four lanes for the 32 libraries in 2012. Pooled libraries were submitted to the Princeton University Sequencing Core Facility for sequencing on the Illumina HiSeq (V3 chemistry), with 101 base pair single read sequencing. This protocol was repeated in 2014 with all 36 libraries made by the same individual (HAU) using the same eight barcodes. However, HiSeq sequencing was performed by the Beijing Genomics Institute at the University of California Davis facility on five lanes, with 101 base pair paired read sequencing.
Statistical and network analyses
Preliminary filtering included application of Illumina’s default Chastity filter, demultiplexing, and visualization using either the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html) or FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to ensure reads were of high quality. The program Trimmomatic (Lohse et al. 2012) was used for trimming adapters and filtering based on quality scores (default settings). Reads were then mapped to the An. gambiae PEST genome with the software Bowtie2 (Langmead & Salzberg 2012) and TopHat2 (Kim et al. 2013) using the most current geneset downloaded from VectorBase (Giraldo-Calderón et al. 2015), consisting of AgamP3.7 for the 2012 experiment and AgamP4.2 for the 2014 data. Numbers of uniquely mapped reads were counted with HTSeq using the default “union” setting and excluding multi-mapped reads (Anders et al. 2015). We obtained a mean (±1 SE) of ≥6.26 (±0.86) million uniquely mapped reads per treatment in 2012, and a mean of ≥14.0 (±0.6) million uniquely mapped reads per treatment in 2014.
Mapping An. merus reads to the An. gambiae PEST genome could have missed detection of some genes, although these species are very closely related. However we chose this reference genome because the An. merus reference assembly was not well annotated at the time of analysis. Because of the difference in the year of the experiment and the use of paired end data in 2014, statistical analyses of differential gene expression for the 2012 and 2014 data were performed separately. We focus our study on the more comprehensive 2014 salinity experiment, unless otherwise noted. However, 66 h larvae were only assayed in 2012, and hence analysis of ontogenetic effects on SW tolerance is derived from this dataset. Overall patterns and major conclusions for treatments tested in either year are similar for both years. In both 2012 and 2014, An. merus at 50% salinity differentially expresses far more genes than An. coluzzii at 20% salinity. Statistically there is little evidence for an interaction effect of year and water type on gene expression. For An. coluzzii, merely seven genes were significant for a two-way interaction effect of year and water salinity (20% vs. 0%). For An. merus, only four genes were significant for the interaction of year and water salinity (50% vs. 0%).
Finally, raw library counts were normalized with the TMM approach (Robinson & Oshlack 2010) and tested for differential expression using generalized linear model likelihood ratio tests with edgeR (McCarthy et al. 2012). Specific contrasts tested are in Table S1, S2 (Supporting information). We only retained genes with >1 count per million in the same number of libraries as we had replicates, thereby filtering out genes with extremely low expression. This resulted in analysis of 10,905 genes in 2014 and 11,025 in 2012. We refined our list of significantly differentially expressed genes by focusing on those with FDR <0.05 that also showed a log2 fold change <-2 or >2 between SW and FW. Although small-effect changes missed by applying the fold change threshold may be important, we assumed most of the critical genes whose induced expression influences saltwater tolerance would show larger changes. For An. coluzzii, log2 fold change values ranged from −10.47 to 9.44, while for An. merus values ranged from −6.96 to 7.98.
We used DAVID (Huang et al. 2008) to cluster genes in similar functional categories (default settings), focusing on clusters of ≥5 genes that were significantly enriched (enrichment score >1.3, corresponding to a P-value <0.05). This tool incorporates information from multiple functional annotations to group similar genes, and determine enrichment relative to a background set. Here, DAVID was used to identify functional enrichment of differentially expressed genes, relative to a background of all genes passing our filters for low expression. Manual scans of VectorBase gene descriptions for the members of each significant DAVID cluster were used to confirm functional identity of the cluster.
Based on DAVID clusters, five key functional groups emerged. We assessed the norm of reaction for each functional category by computing the number of genes up- and down-regulated across salinities. We used Gene Ontology (GO) membership as reported in VectorBase to identify membership in these five groups. First we designated all genes with GO terms of transport, carrier, channel, antiporter, symporter, or exchanger as transport genes. Second, a list generated by Rob Waterhouse was used to identify immune-related genes, as in (Cassone et al. 2014; Waterhouse et al. 2013). Third, a combined list of potential cytoskeleton and cuticle components was made of GO terms including cuticle, microtubule, cytoskeleton, cytoskeletal, chitin, and chitinase. Fourth, a list of cell cycle genes was made of GO terms including cell cycle, meiosis, meiotic, mitotic, mitosis, cell division, anaphase, checkpoint, cytokinesis, and cyclin (metaphase, telophase, and interphase either were not found, or were combined with other terms). Fifth, a list of sensory genes was made of genes with GO terms of odorant, olfactory, and sensory. Although assignment of gene functional categories from homology approaches such as this is inevitably somewhat subjective, it permitted a coarse assessment of how not only numbers but also functional categories of genes expressed changes across a salinity gradient.
Construction of co-expression modules for network analysis was performed with WGCNA version 1.34 (Langfelder & Horvath 2008). This analysis grouped genes into modules based on their showing similar patterns of increasing or decreasing expression across treatments (species identity as An. coluzzii or An. merus, by water salinity as FW or SW). Data was pooled from both 2012 and 2014 experiments to increase the number of samples, which can increase reliability of such network analyses. Prior to implementing WGCNA the TMM normalized data was log transformed, and effects of year of the experiment were removed using the combat algorithm of the SVA package v. 3.12.0 (Leek et al. 2012). Only the young 18 h data from 2012 was used for network analysis, to avoid the influence of age on gene expression overwhelming differences from salinity and species identity. Large transcriptomic changes are known to occur during larval development (Cassone et al. 2014; Koutsos et al. 2007). Prior work has shown that roughly half of An. coluzzii larvae do not survive to pupation at salinities of 20% and most die at ≥30% salinity; in contrast An. merus survives salinities from 0–50% (White et al. 2013). Thus we questioned whether data from higher salinities for An. coluzzii would be representative of typical gene expression networks, as it is not known how long after exposure to high salinities the expression data reflects impending death. Hence salinities of 30–50% were excluded for An. coluzzii.
Co-expression signed networks were made of positively correlated genes, using data for all genes except those removed due to low expression (genes with ≤1 count per million in the same number of libraries as we had replicates). Thus networks include differentially expressed genes as well as those that do not change across salinities. Construction of networks was based on non-parametric Spearman rank correlations of gene expression levels; other parameters were kept at default settings. We used a soft-thresholding power of 13, yielding an r2 of 0.829 in plots of scale-free topology model fit. Log2 fold change values based on the 2014 dataset alone were plotted for all genes in each module using the “matplot” function in R. These plots provided visual confirmation that the identified modules did represent groups of genes with similar expression shifts across salinities.
Additionally, we assigned regulatory impact factors (RIF) to genes in a differential co-expression analysis, implementing the method of (Hudson et al. 2012) with a custom R script adapting it to RNA-Seq data (Script S1, Supporting information). The RIF statistic is reported as a z-score that ranks genes with potential regulatory roles, based on the difference between two conditions in the regulatory gene’s correlation to differentially expressed genes. This correlation is weighted by the average abundance of the differentially expressed genes, and by the difference in their level of expression (here, TMM normalized mRNA counts) between the two conditions. Because we anticipate transcriptomic networks vary between species, we focus on the difference in Spearman correlation between them, with the two species identities as our two conditions. Similar to our concern in the WGCNA networks, we did not want any subversion of typical gene interactions due to the mortality of An. coluzzii at high salinity. Yet the RIF analysis focuses on differential correlations between species, so we omitted data from salinities ≥30% for both An. merus and An. coluzzii. Expression levels are taken from the 2014 dataset, averaged across 0%, 10%, and 20% salinity.
The RIF analysis was designed to test for regulatory roles of genes such as transcription factors, which may correlate with and influence differentially expressed genes even if they themselves are not differentially expressed. Here we employed RIF twice. First in following the original design of Hudson et al. (2012), we generated a list of potential transcription factors using all genes in the AgamP4.2 geneset with a Gene Ontology category including the word “transcription.” This yielded 416 genes, and is notably a liberal list of possible transcription factors and other genes that may influence transcription. Second, we applied the RIF analysis to test for a relationship between differentially expressed genes, and genes we previously localized to QTL peak regions (Smith et al. 2015). These QTL peaks were identified by mapping genomic loci associated with saltwater tolerance in backcross larvae, which were generated by mating parental An. merus with F1 hybrids of An. merus and An. coluzzii. In this second formulation of the RIF analysis, the transcription factor gene list was replaced with the genes within all six QTL peaks, based on the genomic coordinates reported in (Smith et al. 2015).
Results and Discussion
Transcriptomic responses to saltwater vs. freshwater
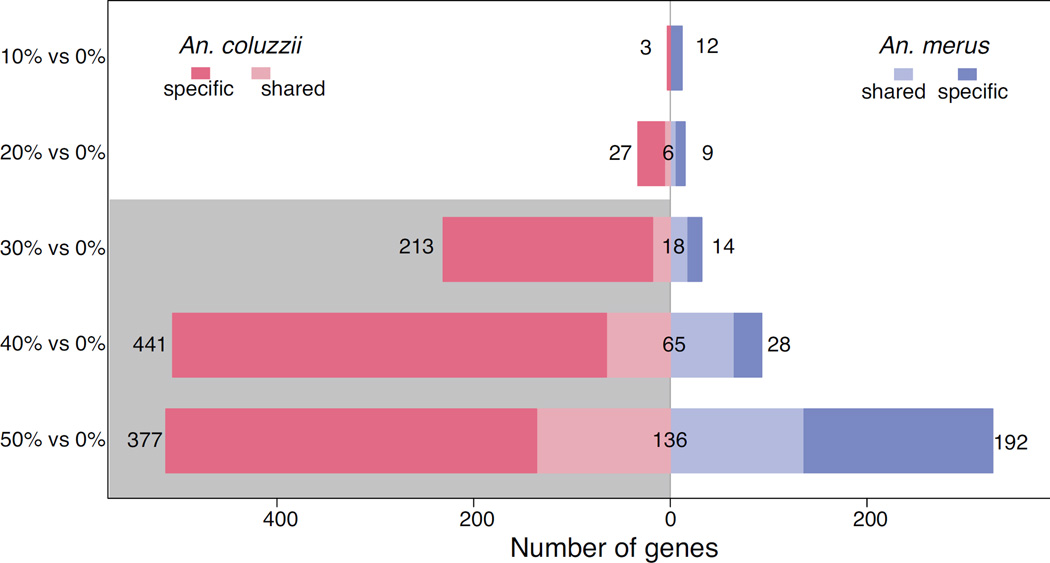
Data support our overall hypothesis that inducible gene expression occurs in response to transfer from FW to SW (Fig. 2, Table S1–S2, Supporting information). We assessed the reaction norm of gene expression for young larvae of the SW tolerant An. merus and the obligate FW species An. coluzzii transferred from FW (0%), back into FW or to one of five salt levels: 10%, 20%, 30%, 40%, and 50% the salinity of seawater. Comparing exposure of 10% salinity to FW, An. merus significantly down-regulates 10 genes in saltwater and up-regulates two genes, while An. coluzzii only differentially expresses 3 genes, all down-regulated in SW. At all other salinities, An. coluzzii shows more differential expression than An. merus (Fig. 2).
Figure 2.
Number of genes differentially expressed by An. merus and An. coluzzii at 10% SW relative to FW; 20% SW relative to FW; 30% SW relative to FW; 40% SW relative to FW; and 50% SW relative to FW. Bars show number of genes only significantly induced by SW in each species (specific), along with genes induced by both species (shared). Lethal salinities for An. coluzzii are shaded gray.
The greater induction by An. merus than its sibling species at 10% salinity could be important, revealing greater sensitivity to small increases in ion concentrations. Yet the species only exhibited differential expression of nine more genes than An. coluzzii. Moreover in a two-way test of genes with differential induction between species (interaction of species and water salinity), only two genes were significant (AGAP002331, a sodium-independent sulfate anion transporter, and AGAP013746, a gene of unknown function with no GO terms or description in VectorBase at the time of writing). Each species induces a distinct set of genes, sometimes with little overlap (Fig. 2). Indeed at 10% salinity there is no overlap in genes differentially expressed by the two species. Anopheles merus differentially expresses some genes between SW and FW that do not change in An. coluzzii, and vice versa, for all salinities tested.
Analysis of genes uniquely induced by one species reveals a few interesting genes. Allantoinase (AGAP000239) shows inconsistent results in An. coluzzii with up-regulation at 30% and down-regulation at 50%, whereas in An. merus it is up-regulated in all salt levels ≥10%. Allantoinase is involved in nitrogenous waste processing in mosquitoes (Esquivel et al. 2014). This response may reflect increased nitrogen processing from catabolism, as energy is consumed by the SW response. Another gene of interest is the Solute carrier family 4 member 1 (AGAP006968), designated as AgAE2 in the Slc-4-like anion transport family (Linser et al. 2012). We find that it is up-regulated by An. merus at 20–40% SW, and also up-regulated at 10% and 50% although it did not meet our fold change threshold (FDR <0.05, but log2 fold changes of ~1.98 and ~1.67, respectively). The gene is neither up- nor down-regulated by An. coluzzii in any SW vs. FW comparison. Considering this induction by An. merus and lack of induction in the FW species An. coluzzii, AgAE2 represents a strong candidate for an ion transport effector gene that contributes to SW tolerance in An. merus.
Transcriptomic norm of reaction of larval anophelines transferred to saltwater
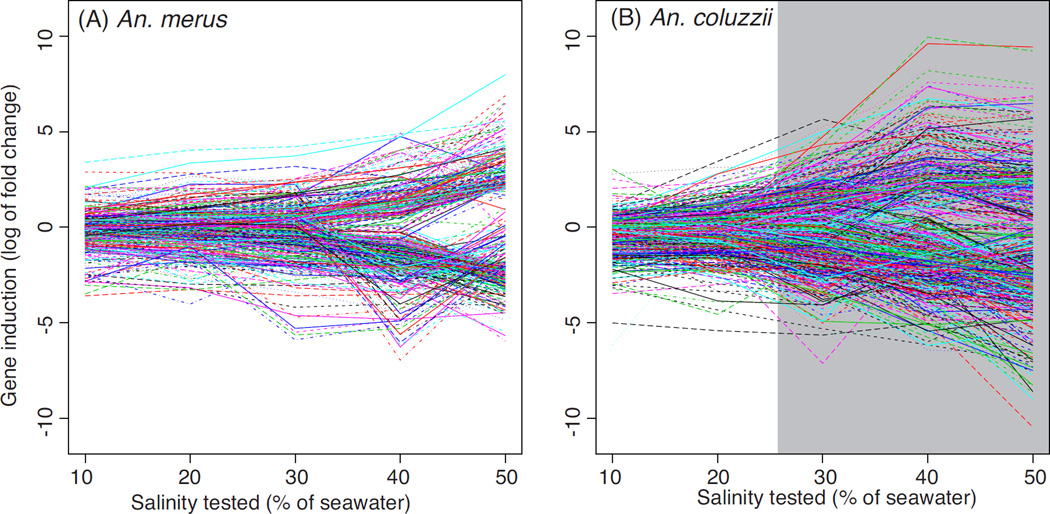
For both species, the number of up- and down-regulated genes tends to increase as salinity increases. Neither species shows strong induction until higher salinities, ≥30% for An. coluzzii and 50% for An. merus (Fig. 2, Fig. 3). Rather than linear, the reaction norm of gene expression vs. salinity stress appears closer to exponential. For An. merus the most striking change is seen between the 158 genes differentially expressed at 40% salinity, almost tripling to reach 464 genes differentially expressed at 50% salinity. Down-regulation consistently exceeds up-regulation of expression across species and salinities.
Figure 3.
Plot of log2 fold change in SW relative to FW for all differentially expressed genes in An. merus (A) and An. coluzzii (B). Positive log2 fold change values reflect an increase in gene expression in SW relative to FW, while negative values represent a decrease. Each line depicts a single gene. Lethal salinities for An. coluzzii are shaded gray.
Spearman rank correlations were computed to relate gene expression (averaged across replicate libraries) to salinity levels. Correlations were computed separately for each species (Table S3, Supporting information). Of those genes with very strong correlations (rho >0.8, P <0.05), 15 are correlated positively to salinity for An. merus and negatively for An. coluzzii, while another 15 genes show the opposite relations to salinity (positive for An. coluzzii, negative for An. merus) (Table 1). Of particular interest is GTPase Kras (AGAP002219), a gene correlated positively to salinity for An. merus and negatively for An. coluzzii. This gene is an upstream part of the ERK signaling pathway (Horton et al. 2011), which is one of the three primary MAPK families (together with the JNK and p38 pathways) responsible for transmitting external signals and stressors into cellular responses (Cowan & Storey 2003). The ERK pathway is known for roles in development as well as immunity, and in mosquitoes can be induced by the Plasmodium parasite (Cowan & Storey 2003; Horton et al. 2011). Conversely, the eukaryotic translation initiation factor 3 subunit E (AGAP006944) is correlated negatively with salinity in An. merus and positively in An. coluzzii. Decreasing expression of the eukaryotic translation initiation factor 3 subunit E gene in An. merus, could reflect a tradeoff towards down-regulating protein production for growth, until osmotic homeostasis is restored.
Table 1.
Spearman correlations between gene expression and salinity. Positive correlation coefficients (rho) indicate gene expression increases as salinity increases. Genes included in this table are those with strong correlations to salinity within each species, but with a different direction of correlation in An. merus vs. An. coluzzii.
| GeneID |
An. merus FDR |
An. merus rho |
An. coluzzii FDR |
An. coluzzii rho |
Gene description |
|---|---|---|---|---|---|
| AGAP001150 | 0.013 | 0.943 | 0.021 | −0.943 | |
| AGAP002219 | 0.013 | 0.943 | 0.000 | −1.000 | GTPase KRas |
| AGAP002335 | 0.013 | 0.943 | 0.021 | −0.943 | nucleolysin TIA-1/TIAR |
| AGAP005072 | 0.000 | 1.000 | 0.021 | −0.943 | |
| AGAP007054 | 0.000 | 1.000 | 0.000 | −1.000 | solute carrier family 13 (sodium-dependent dicarboxylate transporter), member |
| AGAP007055 | 0.013 | 0.943 | 0.021 | −0.943 | Sodium-dependent dicarboxylate transporter sdcS |
| AGAP007374 | 0.013 | 0.943 | 0.000 | −1.000 | glucuronosyltransferase |
| AGAP007572 | 0.000 | 1.000 | 0.021 | −0.943 | All-trans/9-cis |
| AGAP008105 | 0.047 | 0.886 | 0.021 | −0.943 | cullin 3 |
| AGAP009102 | 0.047 | 0.886 | 0.000 | −1.000 | |
| AGAP009747 | 0.013 | 0.943 | 0.021 | −0.943 | Gamma-interferon-inducible lysosomal thiol reductase precursor |
| AGAP010143 | 0.013 | 0.943 | 0.021 | −0.943 | |
| AGAP010414 | 0.000 | 1.000 | 0.021 | −0.943 | cytochrome P450 |
| AGAP011790 | 0.000 | 1.000 | 0.000 | −1.000 | Clip-domain serine protease |
| AGAP013410 | 0.013 | 0.943 | 0.021 | −0.943 | |
| AGAP000771 | 0.047 | −0.886 | 0.021 | 0.943 | Vacuolar-sorting protein SNF8 |
| AGAP001738 | 0.013 | −0.943 | 0.021 | 0.943 | Mitochondrial ribosomal protein L1 |
| AGAP001825 | 0.047 | −0.886 | 0.000 | 1.000 | salivary lipase |
| AGAP002945 | 0.013 | −0.943 | 0.000 | 1.000 | bifunctional glutamyl/prolyl-tRNA synthetase |
| AGAP003343 | 0.013 | −0.943 | 0.021 | 0.943 | cytochrome P450 |
| AGAP004029 | 0.000 | −1.000 | 0.021 | 0.943 | DNA excision repair protein ERCC-1 |
| AGAP006166 | 0.013 | −0.943 | 0.021 | 0.943 | |
| AGAP006668 | 0.013 | −0.943 | 0.021 | 0.943 | squid |
| AGAP006944 | 0.013 | −0.943 | 0.021 | 0.943 | Eukaryotic translation initiation factor 3 subunit E |
| AGAP008336 | 0.013 | −0.943 | 0.021 | 0.943 | protein phosphatase methylesterase 1 |
| AGAP008876 | 0.013 | −0.943 | 0.021 | 0.943 | ATP synthase mitochondrial F1 complex assembly factor 2 |
| AGAP009505 | 0.013 | −0.943 | 0.000 | 1.000 | T-complex protein 1, delta subunit |
| AGAP010292 | 0.047 | −0.886 | 0.021 | 0.943 | guanine nucleotide exchange factor VAV |
| AGAP011696 | 0.000 | −1.000 | 0.021 | 0.943 | Duplex-specific nuclease |
| AGAP012533 | 0.047 | −0.886 | 0.021 | 0.943 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 6 |
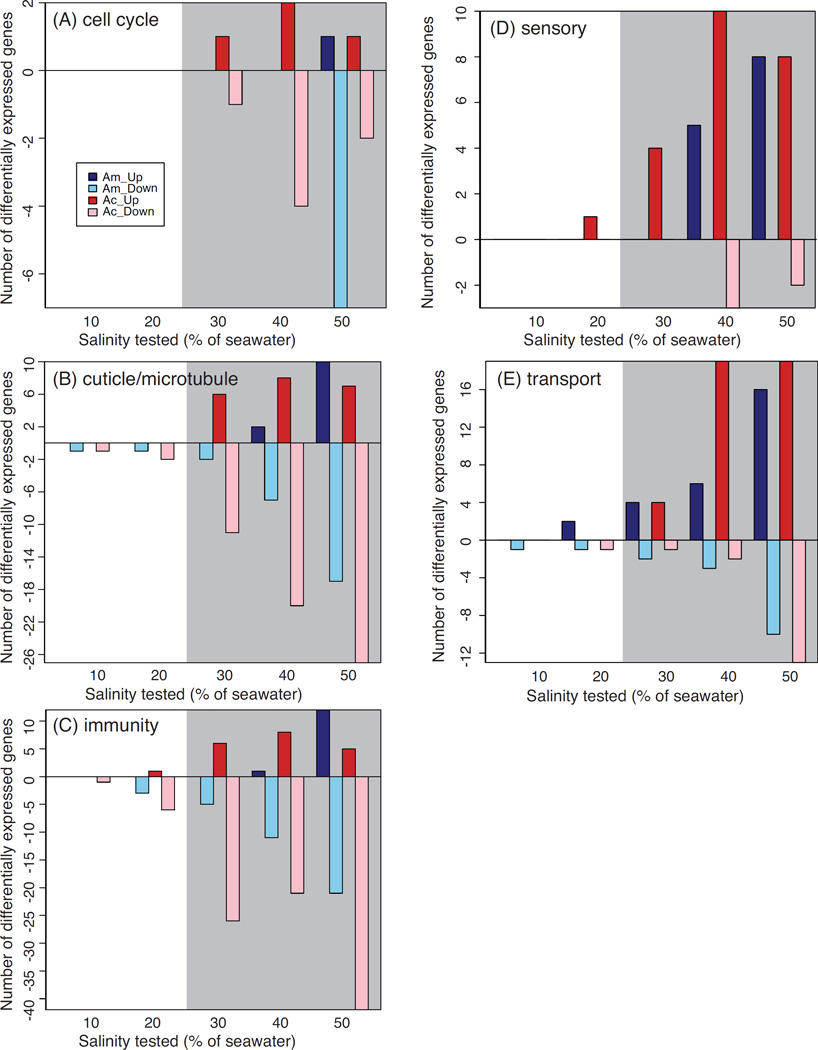
To gain additional insight into transcriptional responses, we focused on the induction of genes potentially involved in similar processes, aided by the DAVID clustering algorithm. Clusters identified by DAVID were summarized into five functional categories: immunity, transport, sensory, cell cycle, and cuticle/cytoskeleton (Fig. 4, Table 2, Table S4–S5, Supporting information). At 10% salinity, insufficient numbers of genes are induced to recover functional clusters in either species. At 20%, both species induce genes associated with immune stress, such as caspase, prophenoloxidase, and fibrinogen. For An. merus, transport represents the category most strongly differentially expressed at low salinity. More transport genes are up- than down-regulated at and above 20% salinity, in contrast to the overall trend of greater gene down-regulation in SW. For sensory genes such as gustatory and odorant receptors, both species show more up- than down-regulation, although sensory genes are not induced by An. merus until ≥40% salinity. An. merus cell cycle genes are not induced until 50% salinity, a sub-lethal stress level for An. merus. In An. coluzzii, cell cycle genes are not differentially expressed until lethal salinity concentrations are reached (≥30%). The cuticle/cytoskeleton genes are not robustly induced in An. merus until salinities reach 40–50%, with a slightly earlier induction at ~30% for An. coluzzii.
Figure 4.
Plots of number of differentially expressed (A) cell cycle, (B) cuticle and microtubule, (C) immunity, (D) sensory, and (E) transport genes. Positive values reflect genes up-regulated in SW vs. FW; negative values correspond to down-regulation in SW relative to FW. Bars correspond to expression in An. merus (Am, light and dark blue) and An. coluzzii (Ac, light and dark red). Lethal salinities for An. coluzzii are shaded gray.
Table 2.
Functionally enriched clusters with ≥5 genes and significant enrichment scores >1.3 from DAVID analysis, from tests of the response to FW vs. SW for An. merus age 18 h and 66 h. There were no clusters with ≥5 genes and scores >1.3 for An. merus at 20% vs. 0%. Neither species had enriched clusters at 10% vs. 0%, and the only significantly enriched cluster for An. coluzzii at 20% vs. 0% was peptidase/immune, with 7 genes and a score of 2.31.
| DAVID Cluster | Functional Category | Enrichment Score | Genes in Cluster |
|---|---|---|---|
| 18 h, 30% vs. 0% | |||
| Cluster 1 | serine peptidase, caspase, protease | 2.75 | 10 |
| Cluster 2 | peptidase C14, caspase | 2.75 | 7 |
| 18 h, 40% vs. 0% | |||
| Cluster 1 | peptidase, caspase | 3.58 | 20 |
| Cluster 4 | cuticle | 2.00 | 6 |
| 18 h, 50% vs. 0% | |||
| Cluster 1 | peptidase, CLIP domain serine protease, caspase | 6.01 | 47 |
| Cluster 2 | cuticle | 5.06 | 15 |
| Cluster 3 | fibrinogen | 3.76 | 7 |
| Cluster 4 | cell cycle, mitosis, meiosis, kinase | 2.61 | 15 |
| Cluster 5 | microtubule | 2.17 | 17 |
| Cluster 6 | gustatory receptor, odorant receptor | 1.60 | 13 |
| Cluster 7 | peptidyl-dipeptidase | 1.48 | 8 |
| Cluster 8 | EGF | 1.38 | 8 |
| Cluster 9 | transporter, sugar transporter | 1.34 | 11 |
| 66 h, 50% vs. 0% | |||
| Cluster 1 | peptidase, CLIP domain serine protease, chymotrypsin | 3.36 | 32 |
| Cluster 2 | cellular retinaldehyde binding, alpha tocopherol | 2.37 | 7 |
| Cluster 3 | fibrinogen | 1.98 | 5 |
| Cluster 4 | odorant receptor, gustatory receptor | 1.59 | 19 |
| Cluster 5 | C-type lectin | 1.41 | 6 |
| Cluster 6 | transport, channel, receptor | 1.33 | 32 |
Overall, induction of these functions is not unexpected. Transporters are expected to show induction, with a role in mobilizing ions for excretion, and potentially mobilizing energy reserves. Cuticle composition may be altered to combat osmoregulatory stress by decreasing permeability in SW, although for anophelines this strategy has been better studied in the adult stage (Reidenbach et al. 2014). Development is slowed upon saltwater exposure in both species (White et al. 2013), potentially indicating a life-history tradeoff between growth and stress tolerance and explaining impacts on the cell cycle. It is possible that sensory receptors are utilized in detecting changes in ionic composition or heightening sensitivity in the face of environmental stress. Osmosensing is thought to be an important part of the salinity stress response in fishes (Kültz 2012).
Another intriguing category induced by salt stress is the immune system. Prior evidence exists for cross-talk between immune and other stress-related genes in dipterans (Davies et al. 2012). In mosquitoes, the innate immune system responds to infection and can decrease survival of Plasmodium parasites within the insect (Clayton et al. 2014; Garver et al. 2013). Our results suggest the possibility for salinity stress to inhibit the immune system by down-regulating immunity genes, which would be consistent with a diversion of resources from combatting internal pathogens to maintaining homeostasis under environmental stress. It is not known whether the apparent down-regulation of immunity genes would continue to the adult form, when Plasmodium infection can occur. If it persists, larval stress exposure could have important implications for malaria transmission to humans.
Co-expression networks of genes responding to SW vs. FW
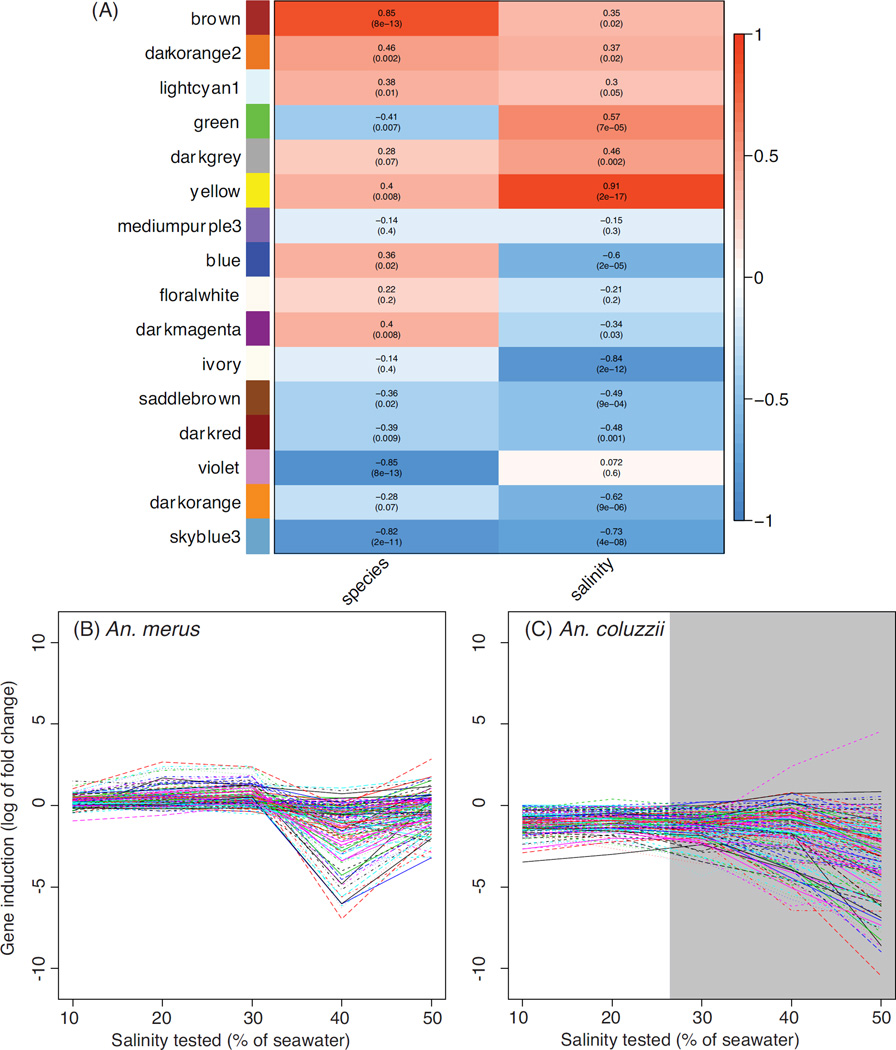
We also performed a weighted correlation network analysis (WGCNA) on all genes tested for differential expression, using both the 2014 and 2012 datasets. This allowed clustering of genes that showed similar expression level changes across species and salinity concentrations (Fig. 5, Table S6, Supporting information). The sixteen modules of co-expressed genes identified by WGCNA were assigned a unique color and assessed for the significance of the relationship to salinity and species. Most modules (N =12) were significantly related to salinity (P <0.05), with 9 remaining significant if a Bonferroni-corrected α of 0.003 (=0.05/16) was imposed. For this analysis, modules with positive correlations for species contain genes with higher expression in An. merus; those with positive correlations for salinity contain genes whose expression increases with increasing salinity (Fig. 5).
Figure 5.
Weighted gene co-expression networks for An. merus and An. coluzzii. Panel (A) shows correlations between each module (rows, named by color) and the traits of species identity (left column) and salinity (right column). Heatmap colors in the left column are red for higher expression in An. merus, and blue for higher expression in An. coluzzii. In the right column, red represents increasing expression as salinity increases, while blue depicts an inverse correlation for the module with salinity. Log2 fold change in SW relative to FW for genes in the darkred module are shown for An. merus (B) and An. coluzzii (C). Lethal salinities for An. coluzzii are shaded gray.
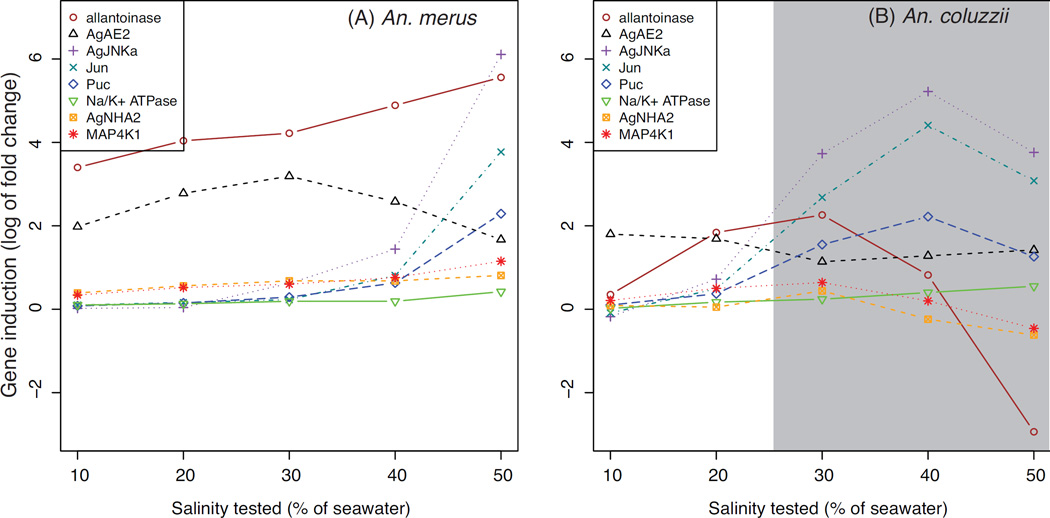
The WGCNA analysis revealed several co-expression modules of interest. The brown module is even more strongly correlated to species identity (rho =0.85, P <0.001) than it is to salinity (rho =0.35, P <0.02). This module contains the Na+/H+ antiporter AgNHA1 (AGAP002093). Another module of particular interest is the yellow module (correlation to species: rho =0.4, P =0.008; salinity: rho =0.91, P <0.001). This module (Fig. 6) contains several osmoregulatory candidates, including AgAE2 and three potential members of a MAPK phosphorylation signaling pathway: AgJNKa (AGAP009460), Jun (AGAP006386), Puc (AGAP004353). This module also includes Na+/K+-ATPase (AGAP002858), and another Na+/H+ antiporter, AgNHA2 (AGAP002324). The yellow module is particularly intriguing considering the proposed role of electrophoretic antiporters and a shift in Na+/K+-ATPase localization in anopheline osmoregulation (Rheault et al. 2007; Smith et al. 2008; Xiang et al. 2012). Co-occurrence of several candidates, and the module’s overall correlation to salinity and species identity, suggests that genes in this module may function in an overall network of signals (e.g., JNK phosphorylation) and effectors (e.g., AgNHA2) facilitating the survival of An. merus in saltwater. The particularly strong induction of allantoinase (AGAP000293) by An. merus could reflect additional catabolism to provide energy for osmoregulatory processes. Alternatively induction of allantoinase may be a byproduct of an overall up-regulation of excretory processes, including nitrogen excretion, to maintain osmotic homeostasis in saline waters. While postprandial nitrogen excretion has been extensively studied in adult females (Coast et al. 2005; Esquivel et al. 2014), further research is needed on the larval stage of anophelines.
Figure 6.
Plot of candidate genes in the yellow WGCNA module, showing log2 fold change in SW relative to FW in An. merus (A) and An. coluzzii (B). Positive log2 fold change values reflect an increase in gene expression in SW relative to FW, while negative values represent a decrease. Lethal salinities for An. coluzzii are shaded gray.
Intramodular connectivity scores from WGCNA gradually declined, rather than being divided into clear groups of either highly connected “hub” genes or non-hub genes with low connectivity. Although our data do not provide strong support for the existence of hubs, one of the most highly connected genes (27th of 1720 genes) in the yellow module is of interest: AGAP010837, identified as MAP4K1 (Horton et al. 2011). This gene is an ortholog of the Drosophila melanogaster gene happyhour, and based on its known role in other systems, Horton et al. (2011) suggest that this kinase may ultimately activate JNK and p38 MAPK signaling. The JNK pathway is our strongest candidate for a phosphorylation signaling cascade induced by SW, as discussed below.
Although little is known of the function of AgJNKa, one can speculate on potential pathways based on those predicted for its neighboring paralog AgJNKb (AGAP009461, not differentially expressed). JNK protein phosphorylates the transcription factor Jun (encoded by AGAP006386, an ortholog of the D. melanogaster Jun-related antigen or Jra) (Garver et al. 2013; Ríos-Barrera & Riesgo-Escovar 2013). However this activity can be halted by the phosphatase Puc, which dephosphorylates JNK in a negative feedback loop (encoded by AGAP004353, an ortholog of the D. melanogaster gene Puckered) (Garver et al. 2013; Martín-Blanco et al. 1998; Ríos-Barrera & Riesgo-Escovar 2013). Up-regulation of all three genes by An. merus in SW genes suggests the JNK signaling pathway may be induced by osmotic stress by An. merus.
In mammals, JNK contributes to regulating mRNA expression of Na+/K+-ATPase subunits (Capasso et al. 2004; Vadász et al. 2012; Wang et al. 2007). Considering the importance of Na+/K+-ATPase protein localization to mosquito SW tolerance, a similar regulatory role of the anopheline JNK is plausible. In general c-Jun N-terminal kinases interact with a variety of transcription factors such as p53, Jun, and NFAT molecules, and were first described as stress-activated protein kinases that mediate responses to several environmental stressors (e.g., ultraviolet radiation and oxidative stress in Drosophila) (Bogoyevitch & Kobe 2006; Ríos-Barrera & Riesgo-Escovar 2013). Thus it is possible that up-regulation of AgJNKa and the downstream transcription factor Jun contributes to regulation of gene expression by An. merus in response to salinity.
Transcription factors and QTL correlated to differentially expressed genes
The RIF statistic assigns a score to a list of input genes, based on the difference between mosquito species in the correlation of those genes to differentially expressed genes. Even if a given input gene does not itself change expression in SW, a strong correlation with genes that are differentially expressed suggests the potential for it to serve as a regulator of the transcriptomic response to salinity. When using a predicted list of transcription factors as input genes and analyzing salinities of 0–20%, 16 received high RIF z-scores >2 (Table 3, Table S7A, Supporting information). Among these is POU domain transcription factor, class 3 (AGAP005878). This gene has the 10th-highest RIF score (Table S7A, Supporting information), with RIF =2.99. The POU homeobox family contains transcription factors, at least some of which are expressed in osmoregulatory tissues and proposed to regulate osmoregulatory genes (Bürglin & Ruvkun 2001; Wang et al. 2012).
Table 3.
List of RIF z-scores for transcription factors.
| GeneID | RIF z-score | Gene description |
|---|---|---|
| AGAP006699 | 6.28 | hairy and enhancer of split, invertebrate |
| AGAP005437 | 4.71 | |
| AGAP012143 | 4.59 | mTERF domain-containing protein, mitochondrial |
| AGAP007985 | 4.40 | homeobox protein aristaless-related |
| AGAP009028 | 3.89 | Teashirt-like protein |
| AGAP009559 | 3.49 | transcription initiation factor TFIIB |
| AGAP004228 | 3.41 | GATA-binding protein 1/2/3 |
| AGAP011436 | 3.27 | |
| AGAP011134 | 3.01 | LIM homeobox protein 5 |
| AGAP005878 | 2.99 | POU domain transcription factor, class 3 |
| AGAP000056 | 2.73 | neuronal PAS domain-containing protein |
| AGAP013275 | 2.62 | |
| AGAP009539 | 2.52 | DNA-directed RNA polymerase I subunit RPA2 |
| AGAP001618 | 2.51 | homeobox protein EMX |
| AGAP011253 | 2.16 | |
| AGAP002958 | 2.05 | transcription initiation factor TFIIA large subunit |
Besides ranking transcription factors with the RIF, we also were interested in exploring the relationship between genomic regions with sequence differences associated with SW tolerance, and gene expression changes. Previously we backcrossed hybrids of An. merus and An. coluzzii to parental An. merus, and related SW tolerance of the backcross progeny to genomic sequences at thousands of loci across the genome (Smith et al. 2015). Based on this QTL mapping experiment, we identified six genomic locations associated with SW tolerance, which altogether encompass 2271 genes. Thus we ran the RIF analysis a second time, using as the input gene list all 2271 genes located within these six QTL. With this implementation, we found 125 genes in the QTL regions with high RIF z-scores of >2 (or <-2). Among these are mitogen-activated protein kinase kinase kinase 7-interacting protein 1 or MAP3K7IP1 (AGAP002953) in the 2Rop inversion with RIF=4.01 and rank 20 (Table S7B, Supporting information), and mitogen-activated protein kinase kinase 3 or MAP2K3 (AGAP000310) on the X chromosome with RIF=3.22 and rank 38. According to the kinome depicted by (Horton et al. 2011), MAP3K7 is part of the JNK pathway, while MAP2K3 is upstream in the p38 pathway.
Ontogenetic impacts on the transcriptomic response to saltwater
Older larvae of An. merus alter expression of more genes than younger larvae, in the single-factor tests of response to 50% SW (423 vs. 351 genes, respectively) (Table S1, Supporting information). Functional clustering of differentially expressed genes reveals similar functional classes between age groups (Table S5, S8, Supporting information), including sensory genes (e.g., odorant and gustatory receptors), and immune genes (e.g., fibrinogens).
The knowledge that larval tolerance of a transfer to SW diminishes at ages of approximately ≥24 h (White et al. 2013) prompted us to examine candidate genes (Table S9, Supporting information) for patterns of greater expression at the younger vs. older age class. For example, expression of AgJNKa and the Jun transcription factor is high in both age classes. Thus, while the JNK pathway may play a role in the osmotic stress response, it likely is not responsible for the unique ability of young An. merus to survive high salinity. However, comparison of age classes reveals AgAE2 to be a strong candidate for the SW tolerance of young larvae. Not only is expression of AgAE2 consistently higher in SW between An. merus and An. coluzzii, but also the expression of this gene barely changes between FW and SW in older An. merus (a mere log2 fold of ~0.14) (Table S9, Supporting information).
Conclusions
By assaying differential expression of larval An. coluzzii and An. merus in SW and FW, we supported our hypothesis that salinity exposure induces differential gene expression, which may in turn influence their different SW tolerances and habitat occupancy. One of the greatest shifts in gene expression was seen for AgJNKa, a mosquito-specific MAPK, while AgAE2 represents one of the most promising ion transport candidate genes up-regulated by young but not old An. merus. Future knockdown or gene silencing assays such as CRISPR/Cas (Sander & Joung 2014, Bono et al. 2015) should be employed to confirm the roles of candidate genes in osmoregulation. It also will be important to investigate tissue-specific changes in gene expression; our use of whole bodies may miss genes that change expression only in one tissue, or change expression in opposite directions among different tissues. Field research is needed to explore how transcriptomic shifts during salinity transitions differ in a natural setting where other biotic (e.g., predation, competition, food resources) and abiotic (e.g., thermal fluctuations, hypoxia, pollution) forces may modulate the response. Bradley (2008) suggests that osmoregulation is an energetically costly strategy for tolerating salinity, but viable in environments with abundant food resources. Once successful laboratory colonies are established for additional members of the An. gambiae complex, it will be interesting to compare transcriptomic responses to a salinity gradient in other SW-tolerant species and explore the potential for adaptive plasticity vs. constitutive expression. Indeed the effect of age on the ability of An. merus to survive SW has been interpreted as a form of developmental plasticity by White et al. (2013).
Despite its limitations, the controlled laboratory system employed here allows for a crucial first step in developing hypotheses of candidate osmoregulatory genes and pathways. Overlap between genes responding to salinity with those associated with immunity reveals that such studies also may provide important insights regarding vectorial capacity, and perhaps differences in malaria transmission between species or habitat types. With climate change and extreme weather events creating the potential for saltwater intrusion and salinization of inland waters (Ramasamy & Surendran 2011), improved understanding of SW-tolerant disease vectors may be particularly timely.
Supplementary Material
Acknowledgments
We gratefully acknowledge the help of Luke Xiao and Marcy Kern in the insectary, the Notre Dame Genomics Core for quality tests during library construction, Paul Howell and MR4 for anopheline eggs (Anopheles gambiae Mali-NIH M form, MRA-860, deposited by T. Lehmann ≫ NJB, and Anopheles merus MAF, MRA-1156, deposited by M. Coetzee), and Dr. Bradley White for discussions that aided project conceptualization. We appreciate the insight of Dr. Antonio Reverter regarding RIFs, and comments by reviewers that improved this manuscript. Funding was provided by National Institutes of Health award 1R21AI101459 to NJB.
Footnotes
Disclaimer
The views and opinions expressed by the authors do not necessarily reflect those of the agencies or institutions with which they are affiliated.
Data Accessibility
Expression levels (raw counts from HTSeq) for each gene identified by their VectorBase gene ID (AGAP + 6 digits for gene identifier), along with links to fastq files, are available from NCBI GEO reference series GSE74820.
Author Contributions
This project was designed by NJB and HAU. Larval water exposure, library construction, and statistical analyses were conducted by HAU; RIF analyses were performed by HAU and CC. The manuscript was drafted by HAU, and edited and approved by NJB, CC, and HAU.
REFERENCES
- Anders S, Pyl PT, Huber W. HTSeq — A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoyevitch MA, Kobe B. Uses for JNK: the many and varied substrates of the c-Jun N-terminal kinases. Microbiology and Molecular Biology Reviews. 2006;70:1061–1095. doi: 10.1128/MMBR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono JM, Olesnicky EC, Matzkin LM. Connecting genotypes, phenotypes and fitness: harnessing the power of CRISPR/Cas9 genome editing. Molecular Ecology. 2015;24:3810–3822. doi: 10.1111/mec.13252. [DOI] [PubMed] [Google Scholar]
- Bradley TJ. Physiology of osmoregulation in mosquitoes. Annual Review of Entomology. 1987;32:439–462. doi: 10.1146/annurev.en.32.010187.002255. [DOI] [PubMed] [Google Scholar]
- Bradley TJ. Saline-water insects: ecology, physiology and evolution. In: Lancaster J, Briers RA, editors. Aquatic Insects: Challenges to Populations (Royal Entomological Society) Oxfordshire, UK: CAB International; 2008. pp. 20–35. [Google Scholar]
- Bürglin TR, Ruvkun G. Regulation of ectodermal and excretory function by the C. elegans POU homeobox gene ceh-6 . Development. 2001;128:779–790. doi: 10.1242/dev.128.5.779. [DOI] [PubMed] [Google Scholar]
- Capasso JM, Rivard CJ, Berl T. The synthesis of the Na/K-ATPase γ subunit is regulated at both the transcriptional and translational levels in Inner Medullary Collecting Duct (IMCD3) cells. American Journal of Physiology - Renal Physiology. 2004;288:F76–F81. doi: 10.1152/ajprenal.00026.2004. [DOI] [PubMed] [Google Scholar]
- Cassone BJ, Kamdem C, Cheng C, et al. Gene expression divergence between malaria vector sibling species Anopheles gambiae and An. coluzzii from rural and urban Yaoundé Cameroon. Molecular Ecology. 2014;23:2242–2259. doi: 10.1111/mec.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton AM, Dong Y, Dimopoulos G. The Anopheles innate immune system in the defense against malaria infection. Journal of Innate Immunity. 2014;6:169–181. doi: 10.1159/000353602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements AN. The Biology of Mosquitoes. London: Chapman & Hall; 1992. Osmotic and ionic regulation; pp. 124–149. [Google Scholar]
- Coast GM, Garside CS, Webster SG, Schegg KM, Schooley DA. Mosquito natriuretic peptide identified as a calcitonin-like diuretic hormone in Anopheles gambiae (Giles) Journal of Experimental Biology. 2005;208:3281–3291. doi: 10.1242/jeb.01760. [DOI] [PubMed] [Google Scholar]
- Coetzee M, Hunt RH, Wilkerson R, et al. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa. 2013;3619:246–274. [PubMed] [Google Scholar]
- Cowan KJ, Storey KB. Mitogen-activated protein kinases: new signaling pathways functioning in cellular responses to environmental stress. Journal of Experimental Biology. 2003;206:1107–1115. doi: 10.1242/jeb.00220. [DOI] [PubMed] [Google Scholar]
- Davies S-A, Overend G, Sebastian S, et al. Immune and stress response ‘cross-talk’ in the Drosophila Malpighian tubule. Journal of Insect Physiology. 2012;58:488–497. doi: 10.1016/j.jinsphys.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Esquivel CJ, Cassone BJ, Piermarini PM. Transcriptomic evidence for a dramatic functional transition of the Malpighian tubules after a blood meal in the Asian tiger mosquito Aedes albopictus . PLoS Neglected Tropical Diseases. 2014;8:e2929. doi: 10.1371/journal.pntd.0002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine MC, Pease JB, Steele A, et al. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science. 2015;437:1258524. doi: 10.1126/science.1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver LS, de Almeida Oliveira G, Barillas-Mury C. The JNK pathway is a key mediator of Anopheles gambiae antiplasmodial immunity. PLoS Pathog. 2013;9:e1003622. doi: 10.1371/journal.ppat.1003622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo-Calderón GI, Emrich SJ, MacCallum RM, et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Research. 2015;43:D707–D713. doi: 10.1093/nar/gku1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbach RE, Townson H, Mukwaya LG, Adeniran T. Use of rDNA-PCR to investigate the ecological distribution of Anopheles bwambae in relation to other members of the An. gambiae complex of mosquitoes in Bwamba County, Uganda. Medical and Veterinary Entomology. 1997;11:329–334. doi: 10.1111/j.1365-2915.1997.tb00418.x. [DOI] [PubMed] [Google Scholar]
- Horton AA, Wang B, Camp L, et al. The mitogen-activated protein kinome from Anopheles gambiae: identification, phylogeny and functional characterization of the ERK, JNK and p38 MAP kinases. BMC Genomics. 2011;12:574. doi: 10.1186/1471-2164-12-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hudson NJ, Dalrymple BP, Reverter A. Beyond differential expression: the quest for causal mutations and effector molecules. BMC Genomics. 2012;13:356. doi: 10.1186/1471-2164-13-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsos AC, Blass C, Meister S, et al. Life cycle transcriptome of the malaria mosquito Anopheles gambiae and comparison with the fruitfly Drosophila melanogaster . Proceedings of the National Academy of Sciences. 2007;104:11304–11309. doi: 10.1073/pnas.0703988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak GM, Brennan RS, Berdan EL, Fuller RC, Whitehead A. Functional and population genomic divergence within and between two species of killifish adapted to different osmotic niches. Evolution. 2013;68:63–80. doi: 10.1111/evo.12265. [DOI] [PubMed] [Google Scholar]
- Kültz D. The combinatorial nature of osmosensing in fishes. Physiology. 2012;27:259–275. doi: 10.1152/physiol.00014.2012. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CE, Kiergaard M, Gelembiuk GW, Eads BD, Posavi M. Pumping ions: rapid parallel evolution of ionic regulation following habitat invasions. Evolution. 2011;65:2229–2244. doi: 10.1111/j.1558-5646.2011.01308.x. [DOI] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SC, Diakov TT, Rizzo JM, Kane PM. Vacuolar H+-ATPase works in parallel with the HOG pathway to adapt Saccharomyces cerevisiae cells to osmotic stress. Eukaryotic Cell. 2012;11:282–291. doi: 10.1128/EC.05198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linser PJ, Neira Oviedo M, Hirata T, et al. Slc4-like anion transporters of the larval mosquito alimentary canal. Journal of Insect Physiology. 2012;58:551–562. doi: 10.1016/j.jinsphys.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Bolger AM, Nagel A, et al. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Research. 2012;40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Blanco E, Gampel A, Ring J, et al. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila . Genes & Development. 1998;12:557–570. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Research. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Pau J, Serra M. Life-cycle switching and coexistence of species with no niche differentiation. PLoS ONE. 2011;6:e20314. doi: 10.1371/journal.pone.0020314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy R, Surendran SN. Possible impact of rising sea levels on vector-borne infectious diseases. BMC Infectious Diseases. 2011;11:18. doi: 10.1186/1471-2334-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidenbach K, Cheng C, Liu F, et al. Cuticular differences associated with aridity acclimation in African malaria vectors carrying alternative arrangements of inversion 2La. Parasites & Vectors. 2014;7:176. doi: 10.1186/1756-3305-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheault MR, Okech BA, Keen SBW, et al. Molecular cloning, phylogeny and localization of AgNHA1: the first Na+/H+ antiporter (NHA) from a metazoan, Anopheles gambiae . Journal of Experimental Biology. 2007;210:3848–3861. doi: 10.1242/jeb.007872. [DOI] [PubMed] [Google Scholar]
- Ríos-Barrera LD, Riesgo-Escovar JR. Regulating cell morphogenesis: The Drosophila Jun N-terminal Kinase pathway. Genesis. 2013;51:147–162. doi: 10.1002/dvg.22354. [DOI] [PubMed] [Google Scholar]
- Robinson M, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature Biotechnology. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuerl T, Stelzer C-P. Patterns and dynamics of rapid local adaptation and sex in varying habitat types in rotifers. Ecology and Evolution. 2013;3:4253–4264. doi: 10.1002/ece3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HA, White BJ, Kundert P, et al. Genome-wide QTL mapping of saltwater tolerance in sibling species of Anopheles (malaria vector) mosquitoes. Heredity. 2015;115:471–479. doi: 10.1038/hdy.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KE, Raymond SL, Valenti ML, Smith PJS, Linser PJ. Physiological and pharmacological characterizations of the larval Anopheles albimanus rectum support a change in protein distribution and/or function in varying salinities. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 2010;157:55–62. doi: 10.1016/j.cbpa.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KE, VanEkeris LA, Okech BA, Harvey WR, Linser PJ. Larval anopheline mosquito recta exhibit a dramatic change in localization patterns of ion transport proteins in response to shifting salinity: a comparison between anopheline and culicine larvae. Journal of Experimental Biology. 2008;211:3067–3076. doi: 10.1242/jeb.019299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadász I, Dada LA, Briva A, et al. Evolutionary conserved role of c-Jun-N-terminal kinase in CO2-induced epithelial dysfunction. PLoS ONE. 2012;7:e46696. doi: 10.1371/journal.pone.0046696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Kawakami K, Gick G. Divergent signaling pathways mediate induction of Na,K-ATPase α1 and β1 subunit gene transcription by low potassium. Molecular and Cellular Biochemistry. 2007;294:73–85. doi: 10.1007/s11010-006-9247-y. [DOI] [PubMed] [Google Scholar]
- Wang J-Q, Hou L, Yi N, Zhang R-F, Zou X-Y. Molecular analysis and its expression of a pou homeobox protein gene during development and in response to salinity stress from brine shrimp, Artemia sinica . Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 2012;161:36–43. doi: 10.1016/j.cbpa.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Tegenfeldt F, Li J, Zdobnov EM, Kriventseva EV. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Research. 2013;41:D358–D365. doi: 10.1093/nar/gks1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, Collins FH, Besansky NJ. Evolution of Anopheles gambiae in relation to humans and malaria. Annual Review of Ecology, Evolution, and Systematics. 2011;42:111–132. [Google Scholar]
- White BJ, Kundert PN, Turissini DA, et al. Dose and developmental responses of Anopheles merus larvae to salinity. The Journal of Experimental Biology. 2013;216:3433–3441. doi: 10.1242/jeb.087189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Roach JL, Zhang S, Galvez F. Genomic mechanisms of evolved physiological plasticity in killifish distributed along an environmental salinity gradient. Proceedings of the National Academy of Sciences. 2011;108:6193–6198. doi: 10.1073/pnas.1017542108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Zhang S, Roach JL, Galvez F. Common functional targets of adaptive micro- and macro-evolutionary divergence in killifish. Molecular Ecology. 2013;22:3780–3796. doi: 10.1111/mec.12316. [DOI] [PubMed] [Google Scholar]
- Xiang MA, Linser PJ, Price DA, Harvey WR. Localization of two Na+- or K+-H+ antiporters, AgNHA1 and AgNHA2, in Anopheles gambiae larval Malpighian tubules and the functional expression of AgNHA2 in yeast. Journal of Insect Physiology. 2012;58:570–579. doi: 10.1016/j.jinsphys.2011.12.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.