Abstract
We have previously shown that potentially pathogenic isolates of Staphylococcus epidermidis occur at high incidence in ready-to-eat food. Now, within 164 samples of ready-to-eat meat products we identified 32 S. epidermidis isolates. In 8 isolates we detected the genes encoding for staphylococcal enterotoxins, but in 7 S. epidermidis isolates these genes were not stable over passages. One isolate designated 4S was shown to stably harbour sec and sel genes.In the genome sequence of S. epidermidis 4S we identified 21,426-bp region flanked by direct-repeats, encompassing sec and sel genes, corresponding to the previously described composite staphylococcal pathogenicity island (SePI) in S. epidermidis FRI909. Alignment of S. epidermidis 4S and S. epidermidis FRI909 SePIs revealed 6 nucleotide mismatches located in 5 of the total of 29 ORFs. Genomic location of S. epidermidis 4S SePI was the same as in FRI909. S. epidermidis 4S is a single locus variant of ST561, being genetically different from FRI909. SECepi was secreted by S. epidermidis 4S to BHI broth ranging from 14 to almost 36 μg/mL, to milk ranging from 6-9 ng/mL, to beef meat juice from 2-3 μg/mL and to pork meat juice from 1-2 μg/mL after 24 and 48 hours of cultivation, respectively. We provide the first evidence that S. epidermidis occurring in food bears an element encoding an orthologue to S. aureus SEC, and that SECepi can be produced in microbial broth, milk and meat juices. Regarding that only enterotoxins produced by S. aureus are officially tracked in food in EU, the ability to produce enterotoxin by S. epidermidis pose real risk for food safety.
Keywords: Staphylococcus epidermidis, staphylococcal pathogenicity island, staphylococcal enterotoxin, SEC, food safety
1. Introduction
Genus Staphylococcus comprises 52 species and 28 subspecies (http://www.bacterio.net) of gram-positive, non-motile bacteria that frequently occur as commensal colonizers of the mucocutaneous membranes of the warm-blooded animals and human (Becker et al., 2014; Fitzgerald and Penadés, 2008; Wertheim et al., 2005). With regard to the ability to coagulate rabbit plasma staphylococci fall into coagulase-positive (CPS) and coagulase-negative (CNS) groups, however this division does not reflect heterogeneity in the pathogenicity and habitat preferences within taxon. For decades, most of research was focused on coagulase-positive Staphylococcus aureus; hence, its virulence factors, population structure, pathogen-host interactions, and ability to cause life-threatening infections remain characterised to the greatest extent (Bergdoll, 1989; Lowy, 1998). S. aureus is also well known for its ability to evoke food poisoning due to the secretion of heat stable enterotoxins that may express superantigenic activity (Hennekinne et al., 2012; Le Loir et al., 2003). Several CNS species (e.g. S. carnosus, S. xylosus, and S. equorum) are widely applied in food industry, exerting positive impact on fermentation processes and sensory characteristics of meat products (Nilsen and Rødbotten, 2007). Recent research highlighted the need of studies on involvement of CNS in human and animal disease. Enterotoxigenic CNS strains were already isolated from cases of human clinical infections (Ataee et al., 2011; de Cuhna et al., 2007; Vasconcelos et al., 2011), and foodstuffs (Even et al., 2010; Marín et al., 1992; Rall et al., 2010; Rodríguez et al., 1996; Zell et al., 2008). CNS isolates endowed with enterotoxigenic properties were also isolated from either healthy or diseased animals (Adesiyun and Usman, 1983; Park et al., 2011; Unal and Cinar, 2012; Valle et al., 1990). To date, the only well characterized enterotoxigenic CNS is S. epidermidis FRI909 strain isolated in the 1960’s from a human clinical case (Madhusoodanan et al., 2011). FRI909 was shown to harbour sec and sel genes on an element similar to S. aureus pathogenicity island (hence designated SePI), and to express SEC and SEL. As it was first described by Park et al. (2011), some enterotoxin genes seem to occur in CNS in unstable form (Piette and Verschraegen, 2009). The significance of these genes for food safety, and public health remains unknown (Podkowik et al., 2013).
Among CNS Staphylococcus epidermidis has gained the greatest attention, so far. Involvement of S. epidermidis in serious hospital infections especially in device-associated cases has been proven (Otto, 2004, 2009; Ziebuhr, 2001).Our previous studies have shown high incidence of S. epidermidis in ready-to-eat (RTE) meat products, and have confirmed significant prevalence of potentially pathogenic isolates among them (Podkowik et al., 2012a, 2012b). Therefore we aimed to describe incidence and characteristics of enterotoxigenic S. epidermidis isolates derived from RTE meat products.
2. Materials and methods
2.1. Isolation and identification of S. epidermidis
One hundred and sixty four samples of ready-to-eat porcine, bovine and chicken meat products were screened for presence of staphylococci. The samples were taken during a thirteen-month period from seven randomly selected supermarkets in Wrocław, Poland. Staphylococci from food samples were cultured on Giolitti-Cantoni enrichment broth (Thermo Fisher Scientific Inc., Waltham, MA, USA) and then subcultured onto Baird-Parker agar (Thermo Fisher Scientific Inc.). One isolate per product was taken for further analyses. The Staphylococcus epidermidis isolates were identified by API Staph ID 32 (bioMerieux, Marcy l'Etoile, France), and additional tests for catalase, clumping factor and coagulase were done. Simultaneously, tuf (Martineau et al., 2001) and 16S rDNA genes (primers from htpp://rdna4.ridom.de) were partially sequenced.
2.2. Preparation of bacterial DNA
Two millilitres of bacterial cell suspension from an overnight culture grown in brain-heart infusion (BHI) broth (Thermo Fisher Scientific Inc.) were centrifuged for 5 min at 12,000 × g and suspended in 100 μL of 100 mM Tris-HCl buffer, pH 7.4, containing 10 μg of lysostaphin (A&A Biotechnology, Gdynia, Poland). After 30-minute incubation at 37 °C, 10 μL of 10% SDS was added and the sample was incubated for another 30 min at 37 °C. Two hundred μL of 5 M guanidine hydrochloride were added and the sample was mixed and incubated at room temperature for 10 min. The DNA was extracted by phenol and chloroform, precipitated with ethanol, and dissolved in water.
2.3. Detection of enterotoxin genes
Detection of SEs genes (sea-see, seg, seh, sei, selj, sek, sem, seo, tst1, sel, sen, sep, seq, ser, selu) was performed with the use of primers and cycling conditions described by Park et al. (2011). The only modification was setting separate reactions for detection of every SE gene. All the amplicons in the size close to expected were sequenced. Sequencing was performed with the BigDye Terminator ready-reaction cycle sequencing kit (Applied Biosystems, Foster City, CA, USA). S. aureus reference strains FRI913, A900322, FRI1151m, and CCM5757 served as PCR controls (Table 1).
Table 1.
Reference S. aureus strains used for enterotoxin genes detection.
| Strain | Enterotoxin gene content |
|---|---|
| FRI913 | sea, sec, see, sek, sel, tst |
| FRI137 | sec, seh, sel, sem, sen, seo, seg, sei, selu |
| CCM5757 | seb, sek |
| A900322 | sep, sem, sen ,seo, seg, sei, |
| FRI1151m | sed, selj, ser |
2.4. Assessment of enterotoxin genes stability in S. epidermidis isolates
Enterotoxigenic S. epidermidis isolates were cultured on Columbia Agar with 5% sheep blood (BTL, Łódź, Poland) at 37 °C. The bacteria were subcultured every 24 hours. The entire procedure comprised 10 consecutive passages. A wire loop of bacterial cells (several randomly selected colonies) was taken from each passage for further DNA isolation and analysis of enterotoxin gene content. During the cultivation colony morphology was assessed focussing on potential colony heterogeneity. The stability of S. epidermidis enterotoxin genes was also assessed following freezing at −80 °C in 30% glycerol. In addition, enterotoxigenic S. epidermidis 4S isolate was subjected to 10 consecutive passages in liquid microbiological medium. In précis, 100 μL of 24-hour S. epidermidis planktonic culture was transferred everyday into 5 mL of fresh BHI broth and grown at 37 °C with agitation (230 rpm). Each day 500 μL of 24-hour bacterial culture was harvested and centrifuged, and then the pellet was used for DNA isolation. PCR for respective enterotoxin genes was performed on the DNA from each bacterial passage. The same procedure was performed in altered temperature or shaking velocity (20 °C and 80 rpm, respectively).
2.5. Genome sequencing and analysis of S. epidermidis4S isolate
An indexed, paired-end sequencing library was prepared for S. epidermidis 4S isolate using a Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA, USA) and cleaned with AMPure XP beads (Agencourt Bioscience, Beverly, MA, USA). The library was sequenced on an IlluminaMiSeq instrument using a 600 cycle Reagent Kit v3 (Illumina), which yielded 2.47 million paired-end reads for this isolate. CLC Genomics Workbench v7 was used to assemble the reads de novo after trimming and filtering for base quality of Q13 (equivalent to base error probability of ≤0.05), number of ambiguities ≤2, and length ≥15 bp. The assembly resulted in 321 contigs ranging in size from 500 bp to 70,462 bp with an N50 of 14,093 bp. Multilocus sequence type (ST) was determined from the genome sequence by extracting the appropriate gene fragments and comparing with the sequences deposited in the international MLST database (sepidermidis.mlst.net). S. epidermidis 4S contigs were also aligned against S. epidermidisFRI909 SePI sequence (WGS accession number AENR00000000). The sequence gaps between contigs comprising SePI were filled by Sanger dideoxy sequencing, and the resulting SePI sequence was submitted to GenBank and given accession number KT845956.
2.6. Cloning, expression, and purification of rSEC from S. epidermidis 4S isolate
The region encoding mature SEC was PCR-amplified from S. epidermidis 4S DNA using the Prime STAR HS DNA Polymerase (Takara Bio Inc., Otsu, Shiga, Japan) according to the protocol: 98°C for 2 min; followed by 30 cycles at 98 °C for 10 s; 55 °C for 10 s; 72 °C for 7 s. The forward cloning primer CATGCCATGGGAGAGAGTCAACCAGACC and the reverse cloning primer CGGCTCGAGTCCATTCTTTGTTGTAAG carried the restriction sites for NcoI and XhoI. The product was purified from agarose gel, digested with XhoI and NcoI (Thermo Fisher Scientific Inc.), ligated into the pET-22b plasmid vector (Merck, Kenilworth, NJ, USA)., and introduced using calcium chloride into NovaBlue Escherichia coli cells (Merck). The plasmid was purified using NucleoSpin Plasmid (Macherey-Nagel, Düren, Germany) and sequenced. The plasmid containing the intact sequence of the respective region of sec was transformed into E. coli Rosetta cells (Merck). Expression was performed using the IPTG induction protocol. Briefly, 500mL of freshly inoculated bacterial culture in LB medium was incubated in 37°C, at 230rpm, until OD600of 0.7, then IPTG (Sigma-Aldrich, St. Louis, MO, USA) was added to a final concentration of 0.5mM.After 3h of incubation bacterial cells were harvested by centrifugation for 10 min at 5,000 × g in 4 °C. Purification of recombinant SEC (rSEC) was performed on His-Select cobalt affinity gel (Sigma-Aldrich), with on-column refolding. Briefly, the bacterial pellet was lysed in 20 mM Tris-Cl (pH 8.0), containing 50 mM NaH2PO4, 150 mM NaCl, 8 M urea, and 5% glycerol. The lysate was centrifuged for 45 min at 16,000 × g, and the supernatant was applied on a column pre-equilibrated with the lysis buffer. The column was washed with lysis buffer, followed by washes with 20 mM Tris-Cl (pH 8.0), 50 mM NaH2PO4, 150 mM NaCl, and 5% glycerol containing decreasing concentrations of urea, from 6 M to 0 M. The protein was eluted with 250 mM imidazole in 20 mM Tris-Cl (pH 8.0), 50 mM NaH2PO4, 150 mM NaCl, and 5% glycerol. Protein concentration was measured using the Bradford reagent (Sigma-Aldrich). The purity of rSEC preparation was checked with sodium dodecyl sulfate– polyacrylamide gel electrophoresis (SDS-PAGE).
2.7. Culture of S. epidermidis 4S in BHI broth, milk and meat juices
Frozen stock culture of S. epidermidis 4S was resuscitated by plating on brain heart infusion (BHI) agar supplemented with 1% yeast extract (YE) and incubated at 37 °C overnight. Single colony was transferred to 5 mL of BHI broth with 1%YE in test tube for 18 hours (37 °C, 230 rpm). One hundred microliters of overnight culture was inoculated into fresh BHI broth with 1% YE and grown in the same conditions and incubated for 2 hours. Next, cells pelleted from 1 mL of culture were washed twice with phosphate-buffered saline (PBS) to remove residual media by centrifugation at 12,000 × g for 5 min. The cells were then suspended in 1 mL of PBS. We used sterilized liquid milk (UHT 0.0% fat milk, Mlekpol, Grajewo, Poland), and meat juices obtained from pork and beef meats according to Rantsiou et al. (2012). All used media were confirmed to be bacteriologically negative and determined to be SEC-free based on the ELISA described below. BHI broth with 1% YE and milk inoculated with PBS-washed cells was suspended in 1 mL of PBS to give an optical density at 600 nm (OD600) of 0.02. Cultures were conducted at 37 °C with agitation (230 rpm). Samples for SEC detection were collected after 24 hours of growth, 1.5 mL of each medium was harvested, centrifuged for 5 min at 12,000 × g and supernatant stored at −20 °C until analysed.
2.8. Growth curve determination in S. epidermidis 4S
Growth curve was determined for S. epidermidis 4S isolate cultivated in BHI broth supplemented with 1% of yeast extract and milk, beef and pork meat juices at 37°C with agitation (230rpm). Cell counts at seven time points were assessed by seeding consecutive 10-fold dilutions of cultures on BHI-agar.
2.9. RNA extraction and Real-time PCR
Bacterial pellets were suspended in 70 μL 100 mM Tris-HCl (pH 7.4), containing 28 U/mL of lysostaphin and incubated for 15 min at 37 °C. RNA was extracted with TRI Reagent® (Sigma-Aldrich) according to manufacturer’s instructions. RNA was dissolved in 50 μL of water and quantified by measuring A260 and A280. One μg of RNA was treated with RNAse-free DNase I (Sigma-Aldrich) in order to eliminate residual genomic DNA. cDNA was synthesized using random hexamers and SuperScript III® (Life Technologies, Carlsbad, CA, USA) following manufacturer’s instructions. Primers used in real-time PCR, SECfor: 5’-CTTGTATGTATGGAGGAATAACAA-3’ and SECrev: 5’-TGCAGGCATCATATCATACCA-3’ were taken from Monday et al. (1999). Primers for rpoBgene, used for normalization of cDNA, were as follows: Rpofor 5’-CTACAAAACCAATTCCGTATCG-3’ and Rporev: 5’-TTAATTGTTGAGGTGTGATAGAC-3’. Real-time quantitative PCR was carried out in iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA), using iQ™ Sybr® Green Supermix (Bio-Rad). The reaction mixture contained 1 μL of template cDNA, 0.5 μM of each primer, 10 μL of iQSybr Green Supermix and water up to 20 μL. The reaction protocol was: 95 °C, 30 s; 35 repeats of 95 °C, 10 s, 55 °C, 15 s, and 72 °C, 15 s. Specificity of PCR was evaluated by melt curve analysis in a temperature range from 90 to 65 °C performed for each reaction. Residual DNA contamination was checked in each RNA sample by running no-RT controls. PCR efficiencies for each primer pair were determined on genomic S. aureus DNA from respective reference strains by running serial 5-fold dilutions of the template. Determined efficiencies were taken into account when calculating relative transcript levels according to Pfaffl (2001). Two or three independent cultures were performed for each strain.
2.10. Western blotting
The protein concentrations in culture supernatants were measured using the Bradford reagent (Sigma-Aldrich), and 5μg of protein was loaded per well in SDS-PAGE. Western immunoblots were probed with rabbit anti-SEC antibodies (Acris, Herford, Germany). To detect the primary antibody, a conjugate of protein A–horseradish peroxidase (Thermo Fisher Scientific Inc.) was used. The blots were developed using the ECL Lumi-Light substrate (Roche, Basel, Switzerland), according to the manufacturer’s instructions.
2.11. Sandwich ELISA
Culture supernatants from reference S. aureus strains and S. epidermidis 4S were pre-incubated with 20% normal rabbit serum, in order to bind protein A, and diluted 5 times in PBS containing 0.01% Tween-20. ELISA was performed according to the protocol described by Freed et al. (1982), with modifications. Rabbit polyclonal anti-SEC antisera were from Acris. Instead of coupling of the horseradish peroxidase (HRP) to the antibody, the antibody was biotinylated with biotin N-hydroxysuccinimide ester (Sigma-Aldrich). Biotinylated antibody was detected with conjugate of HRP-streptavidin (Sigma-Aldrich). 3.3′, 5.5′-Tetramethylbenzidine (Sigma-Aldrich) was used as a substrate for HRP. The specificity of the ELISA was established using culture supernatants of S. aureus reference strains FRI913, A900322, FRI1151m, CCM5757 as controls for enterotoxins SEA, SEB, SEC, SED, SEE, SEG, SEI, SElJ, SEK, SEL, SEM, SEN, SEO, SEP, and SER. The concentration of the enterotoxin in samples was measured with rSECepi from S. epidermidis 4S as a standard, using a 4-parameter logistic curve fit. Data analysis was carried out using GraphPad Prism software (GraphPad Software Inc., La Jolla, CA, USA).
3. Results
3.1. Incidence of S. epidermidis in ready-to-eat meat products
In 164 samples of ready-to-eat porcine, bovine and chicken meat products obtained from seven randomly selected supermarkets in Wrocław, Poland 32 isolates S. epidermidis were identified.
3.2. Detection and stability of enterotoxin genes in S. epidermidis
In 8 of 32 studied S. epidermidis isolates enterotoxin genes were detected. The seh gene was detected in 2 isolates and seq in 4 isolates. Two isolates harboured more than one enterotoxin gene, i.e., in one of them the sec together with sel was detected, and in another one sen, seq together with selu genes were found.
The eight S. epidermidis isolates positive for enterotoxin genes were subjected to sequential passages in order to assess SEs genes stability. In 7 S. epidermidis isolates a gradual loss of PCR signal for enterotoxin genes was observed, and the signal was completely lost after 4-5 passages on solid medium. This phenomenon comprised all of the detected enterotoxin genes. No difference in colony morphology was observed. Both colony types were tested for the presence of enterotoxin genes using colony-PCR. Additionally, the species affiliation of analysed colonies was corroborated aiming to exclude culture contamination. No contamination by additional species was detected in these analyses. Resuscitation of 30% glycerol stock cultures stored in −80°C revealed loss of PCR signal for enterotoxin genes in all the 7 S. epidermidis isolates (bacterial stocks were resuscitated by plating on BHI agar supplemented with 1% YE, and incubated at 37 °C overnight). Reexamination of these 7 S. epidermidis isolates from dozen or so days old colonies on solid medium, obtained during primary isolation, also yielded negative results for enterotoxin genes.
Only one isolate of 8 analysed, namely the S. epidermidis 4S isolate was found to possess stable enterotoxin sec and sel genes, with no loss of PCR signal throughout all 10 passages on solid media. This enabled us to perform liquid media passages. Similarly, no loss of PCR signal for enterotoxin genes was observed in this experiment. Storing of stock cultures in 30% glycerol in −80 °C did not influence S. epidermidis 4S enterotoxin gene content.
3.3. Genotyping of S. epidermidis 4S isolate
Sequence analysis of the 7 loci used for MLST of S. epidermidis yielded a previously unknown allele pattern: arcC49, aroE3, gtr9, mutS5, a new allele at pyrR, tpiA4, and yqiL4. The new pyrR allele resulted in a new sequence type (ST) for S. epidermidis 4S that was a single locus variant of ST561.
Previously, the 437 STs in the international MLST database for S. epidermidis were clustered into 6 genetic clusters (GCs) using Bayesian methods. An updated analysis of 578 STs shows that ST561 is assigned to GC4. In addition, a subset of 7 SNPs that can rapidly assign isolates to GCs (Tolo and Robinson, manuscript in preparation) places both ST561 and S. epidermidis 4S into GC4 with >95% confidence.
3.4. Sequence analysis of S. epidermidis 4S genomic island
In the genome sequence of S. epidermidis 4S we identified a region comprising 21,426 bp (GenBank Accession number GenBank KT845956), flanked by direct-repeat sequences of 73 bp (DR2) at the 5’end and 15 bp (DR1) and the 3’ end. This genomic island encompassed two superantigen genes, i.e., sec and sel, and corresponds to the previously described composite staphylococcal pathogenicity island (SePI) in S. epidermidis. Alignment of S. epidermidis 4S and S. epidermidis FRI909 SePIs revealed 6 nucleotide mismatches located in 5 of the total of 29 ORFs. Five of them resulted in amino acid substitution (Table 2).Analysis of sequences located downstream and upstream of DRs defining the ends of SePI revealed that in S. epidermidis 4S SePI is inserted in the same region as in FRI909.
Table 2.
Nucleotide mismatches and corresponding amino acid alternations in S. epidermidis 4S vs. S. epidermidis FRI909.
| ORFa | Predicted function |
Nucleotide | Mismatch positionb |
Amino acid | Strand | ||
|---|---|---|---|---|---|---|---|
| 4S | FRI909 | 4S | FRI909 | ||||
| 2 | Integrase | A | G | 21,012 | I | V | + |
| 11 | Transposase | T | G | 13,206 | D | A | − |
| 15 | Hypothetical S. aureus protein |
C | T | 9,125 | D | N | − |
| 20 | Hypothetical S. aureus protein |
T | C | 6,202 | P | P | − |
| 20 | Hypothetical S. aureus protein |
T | C | 6,099 | T | A | − |
| 22 | Hypothetical protein |
G | T | 4,439 | W | L | + |
S. epidermidis 4S sec gene was analysed for homology to known sec gene sequences. This search revealed its identity to sec gene from S. epidermidis FRI909 strain. Comparison of translated coding sequence of S. epidermidis SECepi indicated that it is most related to S. aureus SEC3, having 3 amino-acid substitutions in signal peptide and 9 in mature enterotoxin and most distant from SECovine, having 8 amino-acid substitutions in signal peptide and 18 in mature enterotoxin (Table 3).
Table 3.
Comparison of predicted 266 amino acid sequences of SEC1, SEC2, SEC3,SECovine, SECbovine, and SECepi
| Enterotoxin | Strain | Variable positions in predicted amino acid SEC sequences | aa difference | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 4 | 5 | 7 | 9 | 15 | 20 | 21 | 22 | 34 | 37 | 41 | 43 | 47 | 49 | 53 | 58 | 66 | 81 | 86 | 99 | 102 | 121 | 133 | 154 | 160 | 192 | 218 | 243 | 245 | 249 | total | sp | mp | ||
| SEC3 |
S. aureus
Mu3 |
- | - | - | I | - | - | - | S | T | - | - | - | - | - | - | - | - | - | S | K | - | K | - | G | - | V | - | N | M | N | T | 12 | 3 | 9 |
| SEC2 |
S. aureus
Tokyo12381 |
N | S | R | I | C | - | L | - | T | T | E | - | - | - | - | - | - | M | S | K | - | K | - | G | - | - | - | N | M | N | T | 18 | 7 | 11 |
| SEC1 | S. aureus | N | S | R | I | C | - | L | - | T | T | - | A | K | - | E | V | - | - | S | K | G | K | - | G | - | - | - | N | M | N | T | 21 | 7 | 14 |
| SECovine |
S. aureus
ED133 |
N | S | R | I | C | S | L | - | T | T | E | A | K | T | E | V | R | - | S | K | - | K | C | G | - | - | S | N | M | N | T | 26 | 8 | 18 |
| SECbovine |
S. aureus
RF122 |
N | S | R | I | C | - | L | - | T | T | E | A | K | T | E | V | R | - | S | K | - | K | - | G | K | - | - | N | M | N | T | 24 | 7 | 17 |
| SECepi |
S.
epidermidis 4S/FRI909 |
Y | R | L | V | R | A | I | F | I | M | D | S | E | L | G | Y | H | K | N | N | D | N | Y | S | N | I | N | S | I | K | M | - | - | - |
|
Signal peptide |
|||||||||||||||||||||||||||||||||||
sp - signal peptide, mp - mature protein
Accession numbers (GenBank): SEC3 - S. aureus Mu3: AP009324.1 (complete genome), SEC2 - S. aureus Tokyo12381: AB860418.1 (SaPITokyo12381, complete sequence), SEC1: KF386012.1 (enterotoxin type C1 precursor gene), SECovine- S. aureus ED133: NC_017337.1 (complete genome), SECbovine- S. aureus RF122: NC_007622.1 (complete genome)
3.5. Expression of sec gene in S. epidermidis 4S
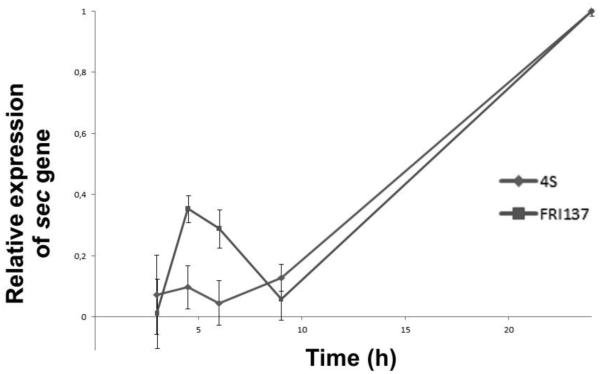
qRT-PCR was performed aiming to determine growth phase-dependent profile of SECepi expression. Results were compared with SEC expression profile in S. aureusFRI137 strain, in which sec gene is known to remain under control of the agr quorum sensing system. As expected SECepi expression increased towards early stationary phase of growth, reflecting the SEC expression pattern observed in FRI137 (Figure 1).
Figure 1.
RNA expression of S. epidermidis 4S secepi and S. aureus FRI137 sec genes.
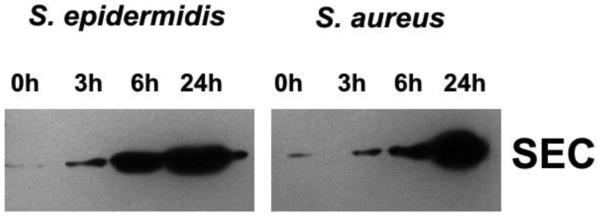
Using Western blotting SECepi was detected in culture supernatants from S. epidermidis 4S. Its expression profile at 3 hour, 6 hour, and 24 hour of cultivation has been compared to SEC produced by S. aureus reference strains FRI137 and FRI913(SEC1 and SEC3, respectively). In all tested strains SEC was detectable at 3 hour of culture, corresponding to early exponential phase of growth (OD600 0.4- 0.5), and SEC concentration increased with time (Figure 2).
Figure 2.
Western blotting of culture supernatants collected from S. epidermidis 4S and S. aureus FRI137 grown for 0, 3, 6 and 24 hours in BHI broth. Five-μg portions of protein were loaded per well in SDS-PAGE. Western immunoblots were probed with rabbit anti-SEC antibodies (Acris, Herford, Germany).
Supernatants from 24 and 48-hour cultures of S. epidermidis 4S, FRI137 and FRI913 reference S. aureus strains, and wild, food sampled S. aureus isolate designated 175 grown in BHI broth, milk, beef and pork meat juices were tested with ELISA for SEC production. S. aureus FRI137 (0.1-2 μg/mL) and FRI913 (85-332 ng/mL) strains were shown to secrete to milk several dozen-fold more SEC than S. epidermidis 4S (6-9 ng/mL) and S. aureus 175 (1-2 ng/mL) did. On the other hand, BHI (14-36 μg/mL) and beef meat juice (2-3 μg/mL) production of SEC exhibited by S. epidermidis 4S exceeded SEC amounts elaborated by FRI137 (0.3-30 μg/mL in BHI and 9-15 ng/mL in beef juice), FRI913 (4-12 μg/mL in BHI and 0.5-2 μg/mL in beef juice), and S. aureus 175 (236-371 ng/mL in BHI and 112-601 ng/mL in beef juice). S. epidermidis 4S secreted more SEC to pork meat juice (1-2 μg/mL) than S. aureus FRI137 (27-43 ng/mL) and 175 (133-263 ng/mL), and less than S. aureus FRI913 strain (3-6 μg/mL) (Table 4). Analysis of S. epidermidis 4S growth in milk indicated it can reach high cell numbers (8.9 log CFU/mL at 24 hour), if compared to tested S. aureus strains, but it did not result in high production of SEC by S. epidermidis 4S. In turn, despite low cell numbers of S. epidermidis 4S in beef meat juice, reaching 6.7 log CFU/mL at 24 hour it could produce the highest amount of SEC (Table 4).
Table 4.
SEC production and bacterial growth in BHI broth, milk, beef and pork meat juices assessed with ELISA.
| BHI broth | Milk | Beef meat juice | Pork meat juice | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |||||||||
| Isolate | SEC [ng/mL] |
Log CFU/ mL |
SEC [ng/mL] |
Log CFU/ mL |
SEC [ng/mL] |
Log CFU/ mL |
SEC [ng/mL] |
Log CFU/ mL |
SEC [ng/mL] |
Log CFU/ mL |
SEC [ng/mL] |
Log CFU/ mL |
SEC [ng/mL] |
Log CFU/ mL |
SEC [ng/mL] |
Log CFU/ mL |
| 4S | 14,001 ± 2,651 |
9.0 ± 0.2 |
35,626 ± 7,892 |
9.4 ± 0.1 |
6 ± 1 | 8.9 ± 0.7 |
9 ± 2 | 7.8 ± 0.1 |
2,283 ± 192 |
6.7 ± 0.3 |
2,979 ± 211 |
7.7 ± 0.2 |
1,256 ± 44 |
8.4 ± 0.2 |
1,877 ± 121 |
7.3 ± 0.2 |
| 175* | 236 ± 47 |
9.0 ± 0.2 |
371 ± 36 |
9.5 ± 0.4 |
2 ± 1 | 8.5 ± 0.3 |
8 ± 1 | 7.8 ± 0.3 |
112 ± 18 |
6.0 ± 0.1 |
601 ± 2 | 7.9 ± 0.2 |
133 ± 8 | 7.5 ± 0.3 |
263 ± 18 |
8.4 ± 0.1 |
| FRI137 | 296 ± 41 |
8.4 ± 0.5 |
30,087 ± 621 |
9.3 ± 0.4 |
131 ± 15 |
8.6 ± 0.4 |
2,319 ± 76 |
9.3 ± 0.3 |
9 ± 1 | 9.1 ± 0.3 |
15 ± 1 | 9.1 ± 0.4 |
27 ± 2 | 8.4 ± 0.2 |
43 ± 3 | 7.3 ± 0.3 |
| FRI913 | 4,134 ± 315 |
9.7 ± 0.2 |
11,969 ± 3,793 |
9.8 ± 0.1 |
85 ± 10 | 9.0 ± 0.1 |
332 ± 62 |
8.7 ± 0.1 |
471 ± 25 |
9.5 ± 0.1 |
2,428 ± 237 |
9.1 ± 0.1 |
3,210 ± 243 |
8.7 ± 0.1 |
5,736 ± 232 |
9.7 ± 0.1 |
wild-type S. aureus isolate derived from food of animal origin. Positive for sec, sel, sep, enterotoxin genes.
4. Discussion
According to the EC Regulation 1441/2007 on microbiological criteria for foodstuffs, selected food products are being examined for the presence of SEA-SEE. The underlying condition for routine food control is the presence of S. aureus exceeding 105 CFU/g (http://eur-lex.europa.eu). For this, the food safety cannot be guaranteed when enterotoxins are produced by staphylococcal species other than S. aureus. So far, data concerning S. epidermidis counts in food products remains obscure. Most studies focus rather on CNS species distribution in food specimens than on their counts. In the study performed by da Cuhna et al., (2006) S. epidermidis counts were assessed in bakery goods, milk and sandwiches. Bacteria numbers oscillated from 1.3 × 103 CFU/g in bakery products, and 3.2 × 104 CFU/mL in milk, to 7.2 × 104 CFU/g in ready to eat sandwiches. In these analyses S. epidermidis was the most predominant species, accounting for 40% of all CNS, followed by S. xylosus (20%), S. warneri (20%), S. saccharolyticus (15%), and S. hominis (5%). It is cumbersome to establish clear relation of staphylococcal loads and food safety risk associated with enterotoxin production. Complexity of the problem can be illustrated using data from analyzes of milk- associated S. aureus isolates. Surveys on bulk milk samples in Hungary and Switzerland indicated low levels of enterotoxigenic S. aureus in milk ranging from 101 CFU/mL to 103 CFU/mL (Peles et al., 2007; Stephan et al., 2002). Moreover, since production of some enterotoxins starts at relatively high bacterial counts some authors argue for low risk associated with SEC-production by S. aureus in milk (Hunt et al., 2014). However, some data indicate low starting S. aureus counts may easily multiply in favorable conditions reaching densities posing risk of SE secretion. Analysis of large SFP outbreak in Brazil due to the cheese and unpasteurized milk showed that S. aureus starting from 103 CFU/g were able to produce SEA, SEB, and SEC (Simeao do Carmo et al., 2007). In Norway a SEH-producing S. aureus found at 103 CFU/mL in product containing raw milk was able to secrete SEH at amounts reaching 0.3μg/mL (Jorgensen et al., 2005).
The role of enterotoxigenic coagulase-negative staphylococci as food safety hazard remains a matter of debate. Moreover, some data on enterotoxin production by CNS seem to be controversial (Podkowik et al., 2013).
Results of this study provide the first evidence of the stable sec gene in S. epidermidis isolate derived from food. We also confirm SECepi transcriptional and translational expression in both laboratory medium and food matrix. To date, the only CNS identified to carry stable sec gene was S. epidermidis FRI909, a strain recovered from human source (Madhusoodanan et al., 2011). Although the data are still scarce, it seems that S. epidermidis harbouring stable enterotoxin genes are not frequent in food products or from human infections. Within 32 S. epidermidis cultures isolated by us from 164 food samples only one stably harboured sec. Recent study by Madhusoodanan et al. (2011) indicate even lower frequency of enterotoxigenic S. epidermidis, since no such isolates were found within more than 200 human bacteraemia isolates.
Current status of knowledge is still not sufficient to describe the genetic processes allowing S. aureus and other staphylococci to exchange mobile genetic elements and stably maintain them. The enterotoxin genes detected in some CNS isolates seem to be unstable. According to a phenomenon observed by Park et al. (2011) intensity of PCR amplicons in enterotoxigenic bovine CNS becomes weak or even completely disappear after several passages of the culture. These observations raise a possibility that some genetic elements harboring superantigen-related genes in CNS behave differently than those known from S. aureus. We suppose that one of the likely explanations for instability of SE genes in CNS may be inability of the certain mobile genetic elements (MGE) bearing these genes to integrate into host chromosome. That aspect was debated over with regard to Staphylococcus aureus Pathogenicity Islands (SaPIs) in S. aureus isolates (Novick et al., 2010). It was also experimentally unveiled that MGE instability occurs when problems with segregation to the daughter cells exist (Úbeda et al., 2007).
SEC is among the most common enterotoxins recovered from food poisoning outbreaks worldwide (Kérouanton et al., 2007; Kitamoto et al., 2009). The sec gene encoding enterotoxin C was reported as the most frequently occurring within S. aureus recovered from milk-related environments (Gutierrez et al., 1982; Scherrer et al., 2004). On the basis on their antigenic properties SECs fall into several serological variants (SEC1, SEC2, SEC3, SECovine, SECbovine) that display high sequence similarity (Marr et al., 1993). Data on the distribution of SEC subtypes in S. aureus related to food-borne outbreaks are scarce; however, in the study performed by Hsiao and colleagues (2003) SEC3 was shown to dominate within S. aureus isolates derived from food-borne outbreaks in Taiwan. Nucleotide sequence of our secepi gene shares 100% identity with S. epidermidis FRI909 sec, and it seems to be unique to S. epidermidis as the alignment with all available S. aureus sec sequences reveals multiple nucleotide substitutions located in sequences encoding both signal peptide and mature protein. Alignment of translated 239-amino acid sequence of mature SECepi with known S. aureus orthologues showed its highest similarity with SEC3 with 9 amino acids different, and highest disparity with SECovine, with 18 amino acid substitutions in mature protein.
Some amino acid substitutions identified in SECepi are located in regions that are conservative among known S. aureus SEC (Table 3). However, based on current knowledge it cannot be definitely resolved whether these mutations can alter biological activity of SECepi if referred to S. aureus SECs. Data on structure/function relations in SE family in many cases are still insufficient to precisely identify residues involved in mitogenic activity of SEs (Zhang and Rogers, 2013). Moreover, site-specific mutagenesis revealed substantial differences within SEs in term of epitopes responsible for mitogenic activity (Briggs et al., 1997).
A disulphide loop comprising from 9 to 19 amino-acids was reckoned to be crucial in SE-mediated emesis (Le Loir et al., 2003). It was identified between 93-110 position in mature SEC3 (Leder et al., 1998). None of cysteine in that region is substituted in SECepi. Only one mutation appears in SECepi, namely G106S, in conserved region in known S. aureus SEC.
Staphylococcal pathogenicity islands belong to a group of phage-inducible chromosomal islands (PICIs) occurring in gram-positive bacteria (Novick et al., 2010). Most of them carry genes for one or more superantigenic toxins, besides the subset of genes that allows them to take the advantage of the temperate phages they parasitize (Maiques et al., 2007; Tormo et al., 2008). SaPIs horizontal transfer has already been unequivocally proven (Lindsay et al., 1998). Genome sequencing of S. epidermidis 4S isolate revealed that it carries 21,426-bp fragment flanked by DR sequences. This element is almost identical to the composite genomic island designated SePI, previously described by Madhusoodanan et al. (Madhusoodanan et al., 2011) in S. epidermidis FRI909 isolate. We have noticed only 6 nucleotide differences in 5 ORF’s; however, two of them were located in genes encoding for proteins involved in classical excision-replication-integration pathway, i.e. integrase and transposase (Maiques et al., 2007). Up to date multiple reports on CNS possessing genes homologous to S. aureus enterotoxins have appeared (as reviewed by Podkowik et al. (2013)), nonetheless the knowledge about their genetic context is still in its infancy. It may be noteworthy that SePI in both sequenced S. epidermidis isolates is larger than most of S. aureus SaPIs, that were shown to average 14-17 kb in length (Novick and Subedi, 2007), what may be a reason for the fail in attempts to mobilize SePI, as shown by Madhusoodanan et al. (2011). Dwelling on this issue aforementioned researchers postulate that disruption of ORFs implicated in formation of small SePI phage particles caused by the insertion of composite SePI elements resulted in inability of SePI-specific transfer (SPST) induction. However, possibility of classical generalized transduction is not excluded, as after mitomycin C treatment the full sized phage particles were observed in culture lysates (in this experiment PCR for toxin genes performed on phage DNA yielded negative results). The primary pending question that should be posed is what factors enabled the stabilization of SePI in S. epidermidis chromosome.
The global catalogue of S. epidermidis diversity, as sampled by the international MLST database, currently consists of less than 600 sequence types (STs). Our isolate represents a previously unknown ST (allelic profile: 49-3-9-5-NEW-4-4) that is similar to ST561 (allelic profile: 49-3-9-5-8-4-4). By clustering methods, both ST561 and S. epidermidis 4S are assigned to the same genetic cluster, GC4. S. epidermidis FRI909 belongs to a different, unique ST (allelic profile: 28-16-NEW-5-3-22-56) that has some similarities to ST441 (allelic profile: 28-16-5-5-3-19-49). This ST441 is represented by S. epidermidis strain AK18W isolated in 2012 from bovine milk in Bangalore, India (http://sepidermidis.mlst.net/).By clustering methods, both ST441 and S. epidermidis FRI909 are also assigned to GC4. Thus, despite the different MLST allelic profiles, the analysis of nucleotides places both SEC-producing strains into the same genetic cluster, GC4. The question of the normal habitat of GC4 isolates requires a close investigation, considering that some of its isolates produce high levels of SEC under certain conditions.
qRT-PCR, western blotting and ELISA that we have performed unambiguously confirmed that SePI-located sec gene is expressed in S. epidermidis 4S isolate. Based on qRT-PCR data for S. epidermidis 4S, with high post-exponential sec transcript increase, we reckon that SECepi production remains under control of the same global regulator (agr) as the S. aureus SEC.
SEC production was assayed in culture supernatants of studied strains after 24 and 48h cultivation in BHI broth, milk and meat juices. The choice of milk as a food model was due to the fact that S. aureus is a common pathogen contaminating milk-related environments, and that milk was already used in studies examining SE production (Valihrach et al., 2013, 2014). Thus far, the production of SEC in milk was reported by Valihrach and colleagues (2014), indicating a low SEC production in milk by 14 S. aureus isolates studied if compared with SEC production in laboratory media. This remains consistent with our data obtained for S. epidermidis 4S and food-derived S. aureus 175 isolate. On the contrary, our reference S. aureus strains, i.e., FRI137 and FRI913, elaborated several dozen-fold higher amounts of SEC in milk compared to S. epidermidis 4S isolate. However, our results on production of various enterotoxins in milk and microbial broths by a number of food-derived S. aureus strains allowed us to classify S. aureus FRI137 and FRI913 as enterotoxin hyper-producers (authors’ unpublished data). It can be thus possible that some S. aureus strains can secrete less enterotoxin to milk than to microbial media, and this is also the case of our S. epidermidis isolate. On the contrary, examination of microbial broth culture supernatants revealed that S. epidermidis 4S is able to elaborate several fold higher amounts of SEC than isolates studied by Valihrach et al. (2014), and even more than S. aureus FRI137, FRI913, and 175. Since enterotoxigenic S. epidermidis was isolated from meat product SEC production was also assessed in meat juices. We revealed that S. epidermidis 4S secreted less SEC to meat juices than to BHI, but much more than to milk. It produced highest amounts of SEC in beef juice as compared to tested S. aureus strains. In pork meat juice S. epidermidis 4S was able to produce almost 2 μg/mL of SEC after 48-hour culture.
In most cases SEs produced by S. aureus in food involved in SFP outbreaks were detected between 5 and 100 ng/mL (Bergdoll, 1989; Evenson et al., 1988). Data presented by Jay (1992) indicate that even 1 ng/g of SE in food is enough to cause food poisoning symptoms.
This implies that amounts of SEC secreted by S. epidermidis 4S to milk (running up to 9 ng/mL after 48 hour) and especially to meat juices (almost 3 μg/mL after 48 hours) are sufficient to act as food safety hazard.
We demonstrated that S. epidermidis occurring in food bears a genetic element encoding an orthologue to S. aureus SEC. SECepi can be produced both in microbial broth, meat juices and in milk. Regarding that only enterotoxins produced by S. aureus are officially tracked in food in EU the ability of enterotoxin production by S. epidermidis pose real risk for food safety.
Highlights.
S. epidermidis 4S isolate from food stably harbor sec and sel genes
enterotoxin genes are on staphylococcal pathogenicity island in S. epidermidis 4S
genomic location of S. epidermidis 4S SePI is the same as in S. epidermidis FRI909
S. epidermidis 4S is genetically different from S. epidermidis FRI909
SECepi is secreted by S. epidermidis 4S to milk, meat juice and to microbial broth
5. Acknowledgements
Project was financially supported by National Science Centre, Poland, on the basis of decision DEC-2012/05/N/NZ7/01945. Genome sequencing of strain 4S was supported in part by National Institutes of Health grant GM080602 (to D.A.R.) and QIA international grant (to K.S.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- Adesiyun AA, Usman B. Isolation of enterotoxigenic strains of staphylococci from dogs. Veterinary Microbiology. 1983;8:459–468. doi: 10.1016/0378-1135(83)90040-8. [DOI] [PubMed] [Google Scholar]
- Ataee RA, Mehrabi-Tavana A, Izadi M, Hosseini SM, Ataee MH. Bacterial meningitis: a new risk factor. Journal of Research in Medical Sciences. 2011;16:207–210. [PMC free article] [PubMed] [Google Scholar]
- Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clinical Microbiology Reviews. 2014;27:870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergdoll M. In: Staphylococcus aureus. Doyle MP, editor. Foodborne Bacterial Pathogens; Marcel Dekker, New York: 1989. pp. 463–524. [Google Scholar]
- Briggs C, Garcia C, Zhang L, Guan L, Gabriel JL, Rogers TJ. Mutations affecting the superantigen activity of staphylococcal enterotoxin B. Immunology. 1997;90:169–175. doi: 10.1046/j.1365-2567.1997.00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cuhna M, Calsolari RA, Júnior JP. Detection of enterotoxin and toxic shock syndrome toxin 1 genes in Staphylococcus, with emphasis on coagulase-negative staphylococci. Microbiology and Immunology. 2007;51:381–390. doi: 10.1111/j.1348-0421.2007.tb03925.x. [DOI] [PubMed] [Google Scholar]
- da Cuhna M, Peresi E, Calsolari RA, Júnior JP. Detection of enterotoxins genes in coagulase-negative staphylococci isolated from foods. Brazilian Journal of Microbiology. 2006;37:70–74. [Google Scholar]
- Even S, Leroy S, Charlier C, Zakour NB, Chacornac JP, Lebert I, Jamet E, Desmonts MH, Coton E, Pochet S, Donnio PY, Gautier M, Talon R, Le Loir Y. Low occurrence of safety hazards in coagulase-negative staphylococci isolated from fermented foodstuffs. International Journal of Food Microbiology. 2010;139:87–95. doi: 10.1016/j.ijfoodmicro.2010.02.019. [DOI] [PubMed] [Google Scholar]
- Evenson ML, Hinds MW, Bernstein RS, Bergdoll MS. Estimation of dose of staphylococcal enterotoxin A from a large outbreak of staphylococcal human food poisoning involving chocolate milk. International Journal of Food Microbiology. 1988;7:311–316. doi: 10.1016/0168-1605(88)90057-8. [DOI] [PubMed] [Google Scholar]
- Fitzgerald JR, Penadés JR. Staphylococci of Animals. In: Lindsay JA, editor. Staphylococcus: Molecular Genetics. Caister Academic Press; Norfolk: 2008. pp. 255–269. [Google Scholar]
- Freed RC, Evenson ML, Reiser RF, Bergdoll MS. Enzyme-linked immunosorbent assay for detection of staphylococcal enterotoxins in foods. Applied and Environmental Microbiology. 1982;44:1349–1355. doi: 10.1128/aem.44.6.1349-1355.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez LM, Menes I, Garcia ML, Moreno B, Bergdoll MS. Characterization and enterotoxigenicity of staphylococci isolated from mastitic ovine milk in Spain. Journal of Food Protection. 1982;45:1282–1286. doi: 10.4315/0362-028X-45.14.1282. [DOI] [PubMed] [Google Scholar]
- Hennekinne JA, de Buyser ML, Dragacci S. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiology Reviews. 2012;36:815–836. doi: 10.1111/j.1574-6976.2011.00311.x. [DOI] [PubMed] [Google Scholar]
- Hsiao MH, Chen TR, Tsen HY. Novel PCR primers for specific detection of C1, C2 and C3 enterotoxin genes in Staphylococcus aureus. Journal of Food and Drug Analysis. 2003;11:338–343. [Google Scholar]
- http://rdna4.ridom.de 3/6/2016.
- http://eur-lex.europa.eu 3/6/2016.
- http://sepidermidis.mlst.net 3/6/2016.
- http://www.bacterio.net 3/6/2016.
- Hunt K, Butler F, Jordan K. Factors affecting staphylococcal enterotoxin C bovine production in milk. International Dairy Journal. 2014;39:41–46. [Google Scholar]
- Jay JM. Indicators of Food Microbial Quality and Safety. In: Jay JM, editor. Modern Food Microbiology. Chapman & Hall; New York: 1992. pp. 413–433. [Google Scholar]
- Jørgensen HJ, Mathisen T, Løvseth A, Omoe K, Qvale KS, Loncarevic S. An outbreak of staphylococcal food poisoning caused by enterotoxin H in mashed potato made with raw milk. FEMS Microbiology Letters. 2005;252:267–72. doi: 10.1016/j.femsle.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Kérouanton A, Hennekinne JA, Letertre C, Petit L, Chesneau O, Brisabois A, De Buyser ML. Characterization of Staphylococcus aureus strains associated with food poisoning outbreaks in France. International Journal of Food Microbiology. 2007;115:369–375. doi: 10.1016/j.ijfoodmicro.2006.10.050. [DOI] [PubMed] [Google Scholar]
- Kitamoto M, Kito K, Niimi Y, Shoda S, Takamura A, Hiramatsu T, Akashi T, Yokoi Y, Hirano H, Hosokawa M, Yamamoto A, Agata N, Hamajima N. Food poisoning by Staphylococcus aureus at a university festival. Japanese Journal of Infectious Diseases. 2009;62:242–243. [PubMed] [Google Scholar]
- Le Loir Y, Baron F, Gautier M. Staphylococcus aureus and food poisoning. Genetics and Molecular Research. 2003;2:63–76. [PubMed] [Google Scholar]
- Leder L, Llera A, Lavoie PM, Lebedeva MI, Li H, Sékaly RP, Bohach GA, Gahr PJ, Schlievert PM, Karjalainen K, Mariuzza RA. A mutational analysis of the binding of staphylococcal enterotoxins B and C3 to the T cell receptor beta chain and major histocompatibility complex class II. The Journal of Experimental Medicine. 1998;187:823–833. doi: 10.1084/jem.187.6.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Molecular Microbiology. 1998;29:527–543. doi: 10.1046/j.1365-2958.1998.00947.x. [DOI] [PubMed] [Google Scholar]
- Lowy FD. Staphylococcus aureus infections. The New England Journal of Medicine. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- Madhusoodanan J, Seo KS, Remortel B, Park JY, Hwang SY, Fox LK, Park YH, Deobald CF, Wang D, Liu S, Daugherty SC, Gill AL, Bohach GA, Gill S,R. An enterotoxin-bearing Pathogenicity Island in Staphylococcus epidermidis. Journal of Bacteriology. 2011;8:1854–1862. doi: 10.1128/JB.00162-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiques E, Úbeda C, Tormo MA, Ferrer MD, Lasa I, Novick RP, Penadés JR. Role of staphylococcal phage and SaPI integrase in intra- and interspecies SaPI transfer. Journal of Bacteriology. 2007;189:5608–5616. doi: 10.1128/JB.00619-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín ME, de la Rosa MC, Cornejo I. Enterotoxigenicity of Staphylococcus strains isolated from Spanish dry-cured hams. Applied and Environmental Microbiology. 1992;58:1067–1069. doi: 10.1128/aem.58.3.1067-1069.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr JC, Lyon JD, Roberson JR, Lupher M, Davis WC, Bohach GA. Characterization of novel type C staphylococcal enterotoxins: biological and evolutionary implications. Infection and Immunity. 1993;61:4254–4262. doi: 10.1128/iai.61.10.4254-4262.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau F, Picard F, Ke D, Paradis S, Roy P, Ouellette P, Bergeron M. Development of a PCR assay for identification of staphylococci at genus and species levels. Journal of Clinical Microbiology. 2001;39:2541–2547. doi: 10.1128/JCM.39.7.2541-2547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monday SR, Bohach GA. Use of Multiplex PCR to detect classical and newly described pyrogenic toxin genes in Staphylococcal isolates. Journal of Clinical Microbiology. 1999;37:3411–3414. doi: 10.1128/jcm.37.10.3411-3414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen A, Rødbotten M. Sensory Attributes. General Considerations. In: Toldrá F, editor. Handbook of Fermented Meat and Poultry. Blackwell Publishing; Iowa: 2007. pp. 197–203. [Google Scholar]
- Novick RP, Christie GE, Penadés JR. The phage-related chromosomal islands of Gram-positive bacteria. Nature Reviews Microbiology. 2010;8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Subedi A. The SaPIs: mobile pathogenicity islands of Staphylococcus. Chemical Immunology and Allergy. 2007;93:42–57. doi: 10.1159/000100857. [DOI] [PubMed] [Google Scholar]
- Otto M. Virulence factors of the coagulase-negative staphylococci. Frontiers in Bioscience. 2004;9:841–863. doi: 10.2741/1295. [DOI] [PubMed] [Google Scholar]
- Otto M. Staphylococcus epidermidis- the 'accidental' pathogen. Nature Reviews Microbiology. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Fox L, Seo K, McGuire M, Park Y, Rurangirwa F, Sischo W, Bohach G. Detection of classical and newly described staphylococcal superantigen genes in coagulase-negative staphylococci isolated from bovine intramammary infections. Veterinary Microbiology. 2011;147:149–154. doi: 10.1016/j.vetmic.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peles F, Wagner M, Varga L, Hein I, Rieck P, Gutser K, Keresztúri P, Kardos G, Turcsányi I, Béri B, Szabó A. Characterization of Staphylococcus aureus strains isolated from bovine milk in Hungary. International Journal of Food Microbiology. 2007;118:186–193. doi: 10.1016/j.ijfoodmicro.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette A, Verschraegen G. Role of coagulase-negative staphylococci in human disease. Veterinary Microbiology. 2009;134:45–54. doi: 10.1016/j.vetmic.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Podkowik M, Bystroń J, Bania J. Genotypes, antibiotic resistance and virulence factors of staphylococci from ready-to-eat food. Foodborne Pathogens and Disease. 2012a;9:91–93. doi: 10.1089/fpd.2011.0962. [DOI] [PubMed] [Google Scholar]
- Podkowik M, Park JY, Seo KS, Bystroń J, Bania J. Enterotoxigenic potential of coagulase-negative staphylococci. International Journal of Food Microbiology. 2013;163:34–40. doi: 10.1016/j.ijfoodmicro.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podkowik M, Bystroń J, Bania J. Prevalence of antibiotic resistance genes in staphylococci isolated from ready-to-eat meat products. Polish Journal of Veterinary Sciences. 2012b;15:233–237. doi: 10.2478/v10181-011-0139-z. [DOI] [PubMed] [Google Scholar]
- Rall VL, Sforcin JM, de Deus MF, de Sousa DC, Camargo CH, Godinho NC, Galindo LA, Soares TC, Araújo Júnior JP. Polymerase chain reaction detection of enterotoxins genes in coagulase-negative staphylococci isolated from Brazilian Minas cheese. Foodborne Pathogens and Disease. 2010;7:1121–1123. doi: 10.1089/fpd.2009.0478. [DOI] [PubMed] [Google Scholar]
- Rantsiou K, Greppi A, Garosi M, Acquadro A, Mataragas M, Cocolin L. Strain dependent expression of stress response and virulence genes of Listeria monocytogenes in meat juices as determined by microarray. International Journal of Food Microbiology. 2012;152:116–122. doi: 10.1016/j.ijfoodmicro.2011.08.009. [DOI] [PubMed] [Google Scholar]
- Rodríguez M, Núñez F, Córdoba JJ, Bermúdez E, Asensio MA. Gram-positive, catalase-positive cocci from dry cured Iberian ham and their enterotoxigenic potential. Applied and Environmental Microbiology. 1996;6:1897–1902. doi: 10.1128/aem.62.6.1897-1902.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer D, Corti S, Muehlherr JE, Zweifel C, Stephan R. Phenotypic and genotypic characteristics of Staphylococcus aureus isolates from raw bulk-tank milk samples of goats and sheep. Veterinary Microbiology. 2004;101:101–107. doi: 10.1016/j.vetmic.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Simeao do Carmo L, Souza Dias R, Roberto, Linardi V, Jose de Sena M, Aparecidados Santos D, Eduardo de Faria M, Castro Pena E, Jett M, Guilherme Heneine L. Food poisoning due to enterotoxigenic strains of Staphylococcus present in Minas cheese and raw milk in Brazil. Food Microbiology. 2002;19:9–14. [Google Scholar]
- Stephan R, Buehler K, Lutz C. Prevalence of genes encoding enterotoxins, exfoliative toxins and toxic shock syndrome toxin 1 in Staphylococcus aureus strains isolated from bulk-tank milk samples in Switzerland. Milchwissenschaft. 2002;57:502–504. [Google Scholar]
- Tormo MA, Ferrer MD, Maiques E, Úbeda C, Selva L, Lasa I, Calvete JJ, Novick RP, Penadés JR. Staphylococcus aureus pathogenicity island DNA is packaged in particles composed of phage proteins. Journal of Bacteriology. 2008;190:2434–2440. doi: 10.1128/JB.01349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Úbeda C, Barry P, Penadés JR, Novick RP. A pathogenicity island replicon in Staphylococcus aureus replicates as an unstable plasmid. Proceedings of National Academy of Sciences of USA. 2007;104:14182–14188. doi: 10.1073/pnas.0705994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal N, Cinar OD. Detection of stapylococcal enterotoxin, methicillin-resistant and Panton-Valentine leukocidin genes in coagulase-negative staphylococci isolated from cows and ewes with subclinical mastitis. Tropical Animal Health and Production. 2012;44:369–375. doi: 10.1007/s11250-011-0032-x. [DOI] [PubMed] [Google Scholar]
- Valihrach L, Alibayov B, Demnerova K. Production of staphylococcal enterotoxin C in milk. International Dairy Journal. 2013;30:103–107. [Google Scholar]
- Valihrach L, Alibayov B, Zdenkova K, Demnerova K. Expression and production of staphylococcal enterotoxin C is substantially reduced in milk. Food Microbiology. 2014;44:54–59. doi: 10.1016/j.fm.2014.05.020. [DOI] [PubMed] [Google Scholar]
- Valle J, Gomez-Lucia E, Piriz S, Goyache J, Orden JA, Vadillo S. Enterotoxin production by staphylococci isolated from healthy goats. Applied and Environmental Microbiology. 1990;56:1323–1326. doi: 10.1128/aem.56.5.1323-1326.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos NG, Pereira VC, Araújo Júnior JP, da Cunha M.deL. Molecular detection of enterotoxins E, G, H and I in Staphylococcus aureus and coagulase-negative staphylococci isolated from clinical samples of newborns in Brazil. Journal of Applied Microbiology. 2011;111:749–762. doi: 10.1111/j.1365-2672.2011.05076.x. [DOI] [PubMed] [Google Scholar]
- Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. The role of nasal carriage in Staphylococcus aureus infections. The Lancet Infectious Diseases. 2005;12:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- Zell C, Resch M, Rosenstein R, Albrecht T, Hertel C, Götz F. Characterization of toxin production of coagulase-negative staphylococci isolated from food and starter cultures. International Journal of Food Microbiology. 2008;127:246–251. doi: 10.1016/j.ijfoodmicro.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Zhang L, Rogers TJ. Assessment of the functional regions of the superantigen staphylococcal enterotoxin B. Toxins. 2013;5:1859–1871. doi: 10.3390/toxins5101859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr W. Staphylococcus aureus and Staphylococcus epidermidis: emerging pathogens in nosocomial infections. Contributions to Microbiology. 2001;8:102–107. doi: 10.1159/000060402. [DOI] [PubMed] [Google Scholar]