Abstract
Communication between acute myeloid leukemia (AML) and the bone marrow microenvironment is known to control disease progression. Therefore, regulation of AML cell trafficking and adhesion to the bone marrow is of significant interest. In this study, we demonstrate that differential expression of the membrane scaffold CD82 modulates the bone marrow homing of AML cells. By combining mutational analysis and super-resolution imaging, we identify membrane protein clustering by CD82 as a regulator of AML cell adhesion and bone marrow homing. Cluster analysis of super-resolution data indicates that N-linked glycosylation and palmitoylation of CD82 are both critical modifications that control the microdomain organization of CD82 as well as the nanoscale clustering of associated adhesion protein, N-cadherin. We demonstrate that inhibition of CD82 glycosylation increases the molecular packing of N-cadherin and promotes the bone marrow homing of AML cells. In contrast, we find that inhibition of CD82 palmitoylation disrupts the formation and organization of N-cadherin clusters and significantly diminishes bone marrow trafficking of AML. Taken together, these data establish a mechanism where the membrane organization of CD82, through specific post-translational modifications, regulates N-cadherin clustering and membrane density, which impacts the in vivo trafficking of AML cells. As such, these observations provide an alternative model for targeting AML where modulation of protein organization within the membrane may be an effective treatment therapy to disrupt the bone marrow homing potential of AML cells.
Keywords: Tetraspanin, CD82, N-cadherin leukemia, bone marrow homing, super-resolution imaging
Introduction
AML, the most common acute leukemia affecting adults, is characterized by an increase of immature myeloid blasts in the bone marrow that results from a loss of normal differentiation and proliferation of hematopoietic stem/progenitor cells (HSPCs) (1). Multiple subtypes of AML exist with a range of aggressiveness and treatment sensitivity (2). One sign of disease aggressiveness is the ability of AML cells to home to the bone marrow and displace HSPCs (3). Homing requires multiple steps including the ability to respond to a chemotactic gradient, extravasation, and adhesion to specialized niches within the bone marrow. In fact, adhesion-mediated interactions between AML cells and the bone marrow play an important role in disease progression and chemoresistance (4–7). Therefore, identifying the molecules and mechanisms that mediate AML-bone marrow adhesion and homing are fundamental to the development of future therapeutic treatments.
Recently, an AML protein profile was identified for a subpopulation of leukemic blasts, the leukemia stem cells (LSCs). This mass spectrometry study found an enrichment of specific adhesion-related proteins including CD44, integrin α6, CD47 and CD82 on LSCs (8). An alternative AML screen also identified the upregulation of CD82 in LSCs where it was suggested to modulate AML adhesion to the bone marrow (9). Following its initial cloning (10–12), the tetraspanin CD82 (or Kai1) was described as a metastasis suppressor in solid tumors (13). Tetraspanins are evolutionarily conserved membrane proteins present in most eukaryotes that function as mediators of cell adhesion, trafficking, and cell signaling (14). Through their ability to associate in cis with other tetraspanins, cell adhesion molecules, and signaling receptors, tetraspanins form tetraspanin-enriched microdomains (TEMs) (15, 16). Formation of TEMs enables tetraspanins to serve as molecular organizers for membrane proteins (15). Our recent work identified a role for CD82 in the homing of human HSPCs, which we linked to the membrane organization of CD82 and associated adhesion and signaling molecules (17). Currently, basic questions concerning the formation and regulation of TEMs and their modulation of adhesion receptors, which specifically impact bone marrow homing, still remain.
N-cadherin is a classical cadherin that interacts homophilically with cadherins on neighboring cells to form adherence junctions, which mechanically link cells and relay signaling information from the extracellular environment (18, 19). While the function of N-cadherin remains controversial for HSPCs (20–22), its role in the regulation of specific leukemias is more evident. In AML, the LSC compartment that expresses N-cadherin is relatively resistant to chemotherapy treatments and highly enriched following chemotherapy (23). Subsequent studies suggest that N-cadherin expression facilitates LSCs to initiate and induce AML development (24). In combination, these data indicate that N-cadherin participates in the protection of LSCs and the relapse of AML; therefore, the regulation of N-cadherin function in AML is of significant interest.
The dynamic regulation of cadherin-mediated adhesiveness is thought to involve modulation of cadherin density arrangement on the cell surface (25). Moreover, clustering of cell surface cadherins is known to modify cadherin-mediated adhesion and signal transduction, but the mechanism of cadherin clustering is poorly understood (26). Combining super-resolution imaging, CD82 mutational analysis, and in vivo functional studies, we utilize a multiscale approach that identifies CD82 as a regulator of AML cell adhesion and bone marrow homing. Our work establishes a mechanism where the membrane organization of CD82, which is dependent upon specific post-translational modifications, regulates N-cadherin clustering and membrane density. We demonstrate that the spatial regulation of N-cadherin by CD82 leads to functional in vivo consequences for AML cell behavior.
Results/Discussion
CD82 expression increases AML cell homing to the bone marrow and modulates N-cadherin mediated adhesion
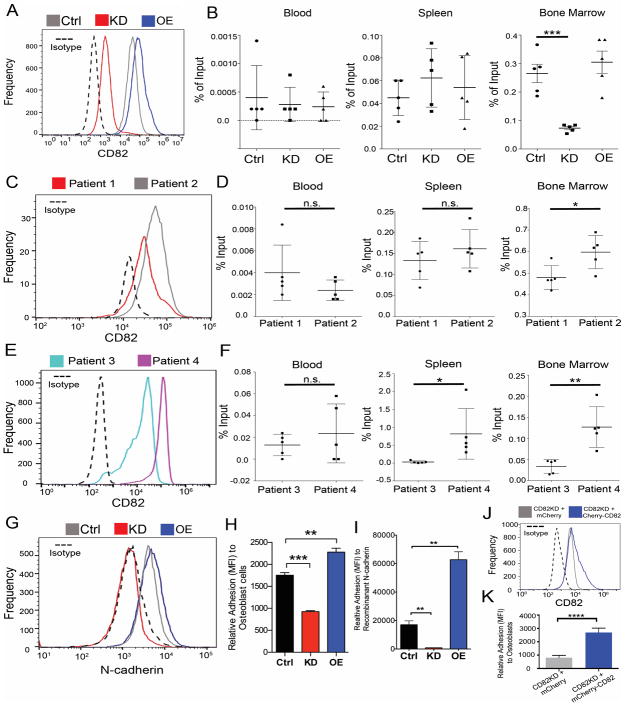
To gain mechanistic insight into how CD82 affects bone marrow homing, we used the previously described control, CD82 overexpression (CD82OE), and CD82 knock down (CD82KD) human KG1a cells (Fig. 1A) (27) to monitor changes in AML cell homing using NSG mice. Sixteen hours following injection, we detected no difference in AML cell localization to the spleen or blood (Fig. 1B). However, when we analyzed the bone marrow, we identified a marked reduction in bone marrow homing of the CD82KD cells along with a modest increase in the bone marrow homing of CD82OE cells when compared to control cells. Therefore, CD82 expression can modify the in vivo trafficking of AML cells. To further evaluate this finding, we compared the homing capacity of primary human AML cells with differential CD82 expression (Fig. 1C, E). Consistent with the cell line data, we find that AML cells with higher CD82 expression display improved bone marrow homing when compared to AML cells with lower expression of CD82 (Fig. 1D, F). The combined cell line and primary AML cell data suggest that CD82 expression modulates AML cell homing to the bone marrow microenvironment, which is an indicator of aggressive AML.
Figure 1. CD82 expression regulates homing to the bone marrow and adhesion to niche components.
(A) Flow cytometry analysis of CD82 surface expression using previously described CD82OE, CD82KD and control KG1a cell lines(27) (ATCC; CCL-246.1). Cells were characterized using Alexa Fluor 647 anti-human CD82 (clone ASL-24, BioLegend). Data was acquired using an Accuri flow cytometer C6 (BD Bioscience) and analyzed with FlowJo X software (Tree Star, Inc). (B) Bone marrow homing of CD82OE, KD or Ctrl KG1a cells. Cells were labeled with CFSE according to manufactures protocol. After labeling, 1×106 cells were injected i.v. into female NOD.Cg-PrkdcscidIl2rgtm1wjl/SzJ (NSG) mice 8–12 weeks of age. NSG mice were housed and bred at the Animal Research Facility under specific pathogen free conditions at the University of New Mexico Health Sciences Center (Albuquerque, NM). All procedures were approved by the University of New Mexico Institutional Animal Care and Use Committee and carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals. 16 hours after injection the blood, spleen and bone marrow were harvested. A single cell suspension was generated and red blood cells were lysed with ACKs buffer (15M NH4Cl, 10mM KHCO3, 0.1mM EDTA). Cells were treated with Fc block then stained for human-CD45 and analyzed by flow cytometry for CFSE and huCD45 (Clone HI30, Biolegend) double positive cells. Percent input was calculated based on number of double positive events multiplied by total tissue cell number divided by the number of cells injected all multiplied by 100 (n = 5 mice). (C, E) Flow cytometry analysis of CD82 on the surface of primary AML cells. (D, F) Tissue harvest from 8–10 week old male and female NSG mice 16 hrs after i.v. injection of CFSE labeled primary AML cells (1×106 cells) using protocol described above (n = 5 mice/patient sample). AML patient samples were deidentified and obtained from the UNM Health Science center (HSC) cell bank. Flow cytometry analysis of (G) N-cadherin (Clone 8C11, Biolegend) surface expression on Ctrl, CD82KD or CD82OE KG1a cells. (G) Fluorescence based cell adhesion assay using Ctrl, CD82KD, CD82OE cells. Cells were labeled with 2 μM calcein (Invitrogen) and allowed to adhere to (H) SaOS-2 osteoblastic cells (ATCC) or (I) purified N-cadherin (R&D Systems) for one hour. Non-adherent cells were removed by washing and remaining fluorescent cells were measured by using synergyH1 plate reader (Biotek) and analyzed with the Gen5 2.00.18 plate reader software (n=3 replicates). (J) Flow cytometry analysis for CD82 following the nucleofection of mCherry or the mCherry-CD82 vectors into the CD82KD cells. (K) Osteoblastic cell adhesion analysis (as previously described) for CD82KD cells upon CD82 reintroduction. For all graphs, mean is displayed with error bars denoting S.D., all variances were determined to be similar; no randomization or blinding methods were used; statistics were performed using two-sided unpaired t-test. (* p < .05, ** p < .01, *** p < .001).
Bone marrow homing of AML cells requires a series of complex steps involving a combination of cell migration and adhesion signaling. The chemokine receptor, CXCR4, with its ligand, stromal derived factor-1 (SDF-1), is the major receptor signaling pathway used for bone marrow homing by HSPCs (28) and various types of leukemic cells (29). While functional interactions between tetraspanins and CXCR4 signaling were shown previously (30), we did not detect any CXCR4 expression differences between the control, CD82OE, and CD82KD cells (Suppl. Fig. 1A, B). Additional analysis of cell migration toward SDF-1 illustrates no difference in the migratory behavior of these cells in a transwell assay (data not shown). Therefore, these data suggest that the observed changes in bone marrow homing are not likely due to CD82-mediated effects on the CXCR4 homing signal.
Next, we turned to evaluate whether CD82 expression may affect AML cell adhesion within the bone marrow by screening the cell lines for expression changes in the cadherin family of cell-cell adhesion molecules (18, 19). While we were unable to detect differences in the expression of E-cadherin and P-cadherin (Suppl. Fig. 1C, D), the surface expression of N-cadherin was significantly reduced in the CD82KD cells (Fig. 1G). Recently, N-cadherin enrichment was identified on the surface of LSCs, which was proposed to enable the cell adhesion of AML cells to the bone marrow (23, 24). Therefore, we used a fluorescence-based adhesion assay to measure changes in cell adhesion to osteoblasts and purified N-cadherin. Consistent with the homing experiments, we find that CD82KD results in a decrease in cell adhesion to osteoblastic cells as well as purified N-cadherin, whereas CD82OE cells display an increase in cell adhesion (Fig. 1H, I). Furthermore, the reintroduction of CD82 back into the CD82KD cells recovered the reduced adhesion phenotype (Fig. 1J, K). Together, these data implicate a specific role for N-cadherin in CD82-mediated AML cell adhesion.
CD82 membrane clustering is altered by glycosylation and palmitoylation status
A distinct feature of tetraspanins is their ability to associate with other tetraspanins, cell adhesion molecules and signaling receptors, thereby serving as molecular facilitators for membrane proteins (15, 16). Therefore, the mechanism by which CD82 regulates AML cell adhesion and homing is likely to be dependent upon its ability to form higher order protein complexes in the cell membrane. Moreover, the regulation of TEM formation and stability is of significant interest. Previously, our group and others showed that the palmitoylation of the membrane proximal cysteines of CD82 promotes the oligomerization and dynamic reorganization of proteins into microdomains (27, 31–33). Furthermore, cell surface glycosylation, which can alter protein-protein interactions, also regulates the membrane organization of proteins. The glycosylation of membrane bound proteins is perturbed in many cancers and can be regulated by oncogenic factors (34–36). Recently the membrane glycosylation of CD82 was shown to play a role in cell adhesion and motility in specific cancers (37, 38). To evaluate how palmitoylation and glycosylation of CD82 affect its membrane organization and the aggressive potential of AML, two constructs were generated where: 1) the membrane proximal cysteines were mutated to serine, preventing palmitoylation (Palm-CD82) (27, 39), and 2) the three N-linked glycosylation sites were mutated to glutamine, inhibiting glycosylation (Ngly-CD82) (Fig. 2A). These constructs were stably transfected into KG1a cells and Figure 2B indicates that the Ngly-CD82 and Palm-CD82 cells express similar CD82 surface levels as the CD82OE cells. Interestingly, both mutants contain intracellular CD82, which may further suggest changes in CD82 protein trafficking that are regulated by these post-translational modifications.
Figure 2. Palmitoylation of CD82 is critical for CD82 membrane organization.
(A) Cartoon of CD82 highlighting N-linked glycosylation and palmitoylation sites. Using the mCherry-CD82 plasmid(27), three N-linked glycosylation sites on CD82, Asparagine 129, 157 and 198 were mutated individually to glutamine using a QuickChange II site-directed mutagenesis kit (Agilent) according to the manufacture’s instructions (Ngly-CD82). All mutations were confirmed by DNA sequence analysis (ACGT Inc.). CD82 palmitoylation mutant was generated as previously described (Palm-CD82) (27). (B) Flow cytometry analysis of CD82 surface expression on CD82OE, Ngly-CD82, and Palm-CD82 cells (clone ASL-24, BioLegend). (C–F) Reconstructed dSTORM images of CD82 distribution on each cell line. The previously described labeling, imaging, and fitting protocols were followed (27, 48). (G) Hopkins analysis of CD82 cellular membrane organization on ctrl (n=8), CD82OE (n=11), Palm-CD82 (n=13) and Ngly-CD82 (n=12 cells) using reconstructed dSTORM images was performed using SuperCluster Matlab software from the UNM Spatiotemporal Modeling Center. (Code availability: http://stmc.health.unm.edu/tools-and-data/index.html). The reconstructed dSTORM images were also analyzed with the DBSCAN algorithm to generate DBSCAN images (C–F zooms), which represent clustered CD82 localizations in color and non-clustered CD82 localizations in gray. A 6 × 6 μm box was examined for clustering using an epsilon value of 100 nm and an n value of 10 localizations. Quantification of (H) CD82 cluster diameter and (I) CD82 cluster area based on DBSCAN analysis (ctrl n=3 cells n=315 clusters, CD82OE n=4 cells n=711 clusters, Ngly-CD82 n=5 cells n=772 clusters and Palm-CD82 n=3 cells n=356 clusters). (J–L) dSTORM imaging and DBSCAN analysis for CD82 cluster area and diameter was performed on four primary AML samples (P1 n=4 cells n=130 clusters, P2 n=3 cells n=74 clusters, P3 n=3 cells n=75 clusters, P4 n=3 cells n= 84 clusters). The mean is displayed with S.E.M.; statistics were performed using one-way ANOVA, post-hoc two-sided unpaired t-test with Welch’s correction for groups with unequal standard deviations. (** p < .01, *** p < .001, **** p < .0001).
Next, we assessed how these CD82 mutations affect the membrane organization of the CD82 scaffold. To measure differences in microdomain organization between control, CD82OE, Ngly-CD82 and Palm-CD82 cells, we used the super-resolution imaging technique, direct stochastic optical reconstruction microscopy (dSTORM) (40). Super-resolution imaging allows us to quantify changes in CD82 membrane organization at the level of individual molecules on the nanometer scale (Fig. 2C–F). Initially, the reconstructed dSTORM images were analyzed using the Hopkins index, which determines the extent to which CD82 is present in a random distribution on the cell surface (41, 42). Consistent with our visual observations, we find that each of the CD82 expressing AML cells has a Hopkins index that is significantly higher than what would be expected for a random distribution of molecules (0.5), demonstrating that CD82 is not randomly distributed, but organized into membrane clusters (Fig. 2G).
The CD82 dSTORM images were also analyzed using the density-based spatial clustering of applications with noise clustering algorithm (DBSCAN) (Fig. 2C–F, zoom) as previously described (43). From these measurements, we determined that CD82OE cells have an increased CD82 cluster diameter and area with respect to control cells, which is likely due to the increased expression of CD82 (Fig. 2H, I). Interestingly, the CD82 cluster size quantified for both the Ngly-CD82 and Palm-CD82 cells indicates an even further increase in CD82 cluster diameter and area when compared to CD82OE cells (Fig. 2H, I). Measurements of the Palm-CD82 cells detect the most significant increase in CD82 cluster size and decrease in CD82 cluster organization, which is consistent with previous work demonstrating the importance of the palmitoylation sites in the lateral packing of CD82 (27, 33). Previous work from our lab identified smaller CD82 cluster sizes in the Palm-CD82 cells using pair-auto correlation function analysis, which is an averaged radial cluster measurement. In contrast, the DBSCAN algorithm enables the quantification of larger scale clusters of varying shapes and sizes, which is what we find for CD82. As for the N-glycosylation mutation, the effects on CD82 cluster size are more modest, however we do detect an increase in CD82 cluster diameter and area. We also imaged and analyzed the CD82 cluster area and diameter in primary AML cells. Consistent with the cell line data, Fig. 2J–L further illustrate the differentiation clustering of CD82 in primary patient samples. In combination, these data illustrate that while the CD82OE, Ngly-CD82 and Palm-CD82 cells all have similar CD82 surface expression, the Ngly- and Palm- mutations change the CD82 membrane distribution into larger ordered CD82 clusters. Therefore, these specific post-translational modifications regulate the membrane organization of CD82, which may in turn modulate protein-protein interactions important for bone marrow homing and adhesion.
N-cadherin clustering is regulated by CD82 membrane organization
Next, we set out to determine whether the described changes in CD82 membrane organization affect the expression and distribution of N-cadherin. First, we confirmed that N-cadherin surface expression is consistent between the CD82OE, Ngly-CD82, and Palm-CD82 cell lines (Fig. 3A). Next, we performed confocal immunofluorescence imaging to analyze N-cadherin distribution in the cells. Figure 3B illustrates that both CD82 and N-cadherin are localized to the plasma membrane in each of the cells except for the CD82KD cells, which have reduced expression levels of CD82 and a punctate distribution of N-cadherin. In addition to the change in N-cadherin distribution upon CD82KD, a reduction in N-cadherin expression is observed, which is consistent with the flow cytometry data (Fig. 1F). Moreover, double staining of primary AML cells suggests a similar surface expression profile for CD82 and N-cadherin (Fig. 3C). To further assess potential protein-protein interactions between CD82 and N-cadherin, we completed co-immunoprecipitation experiments using Brij lystates. The ability of CD82 to pull down N-cadherin in this mild detergent (Fig. 3D) suggests that CD82 and N-cadherin are present in a protein complex.
Figure 3. CD82 interacts with N-cadherin on the plasma membrane.
(A) Flow cytometry analysis of N-cadherin surface expression on CD82OE, Ngly-CD82, and Palm-CD82 cells (Clone 8C11, BioLegend). (B) Confocal immunofluorescence imaging of CD82 and N-cadherin. Cells were fixed in 4% PFA then blocked and permeabilized with PBS + 1.0 % BSA + 0.1% tween 20. Alexa Fluor 647-conjugated anti-human CD82 (Clone ASL-24, BioLegend) and anti-human N-cadherin (clone 32/N-cadherin, BD Bioscience) antibodies were diluted 1:500 in permeabilization buffer and added to the sample overnight at 4°C. Cells were washed and then Alexa Fluor 488-goat-anti-mouse secondary antibody (Invitrogen) was added to the cells for 1hr at room temperature. Following PBS washes, cells were imaged by laser scanning confocal microscopy with a Zeiss Axiovert 100M inverted microscope (LSM 510) system using excitation wavelengths of 488 or 633 nm and a 63X 1.2 N.A. oil immersion objective. Image analysis was performed using the Zeiss LSM 510 software and Image J (NIH, Bethesda, MD). (C) Double surface expression analysis by flow cytometry for CD82 and N-cadherin on primary AML cells. (D) Co-immunoprecipitation of CD82 and N-cadherin. Co-immunoprecipitations were performed using BRIJ O10 cell lysates incubated with CD82 antibody (Clone B-L2, Abcam) or control IgG antibody (Santa Cruz Biotechnology) and then immunoprecipitated using protein A/G Beads (Santa Cruz Biotechnology). Western blots were performed as previously described (24) using the N-cadherin antibody (32/N-Cadherin, BD Biosciences).
Surface clustering of N-cadherin can trigger signaling events, which promote cell adhesion (25). Furthermore, the regulatory mechanism of cadherin clustering is a critical aspect of cadherin adhesion since the adhesive capacity of individual cadherins is negligible (26). Therefore, the lateral association between cadherin receptors is a prerequisite for the formation of adhesive dimers (44). To quantify how changes in CD82 membrane organization affect the nanoscale organization of N-cadherin, we again used dSTORM (Fig. 4A–D). Analysis of the N-cadherin dSTORM images with the DBSCAN algorithm (Fig. 4E–H) suggests that N-cadherin cluster size and diameter is significantly decreased in Ngly-CD82 and Palm-CD82 cells when compared to control and CD82OE cells (Fig. 4I, J). More importantly, Palm-CD82 cells display a marked decrease in the number of N-cadherin clusters when compared to CD82OE or control cells (Fig. 4K). Additional analysis of Palm-CD82 cells also identified that the majority of the N-cadherin molecules are distributed diffusely throughout the membrane and not localized to organized clusters (Fig. 4L). Thus, the palmitoylation of CD82 and its lateral assembly significantly affects the formation of N-cadherin adhesive protein complexes. Interestingly, we also find that the Ngly-CD82 cells demonstrate a significant increase in the density or molecular confinement of N-cadherin molecules into a cluster (Fig. 4M), which is predicted to modulate N-cadherin function. We find that N-glycosylation of CD82 maintains N-cadherin clusters at approximately 80 nm. However, when the N-linked glycosylation sites on CD82 are mutated, the average size of N-cadherin clusters shrinks to approximately 65 nm, which leads to an increase in the molecular confinement of N-cadherin in each cluster. Together, these data suggest that while palmitoylation of CD82 regulates N-cadherin assembly into clusters, N-glycosylation of CD82 affects the nanoscale packing of N-cadherin. Therefore, in addition to N-cadherin expression, the regulation of N-cadherin membrane organization by CD82 may also be an important regulatory mechanism for controlling N-cadherin function and subsequent behavior of AML.
Figure 4. CD82 regulates N-cadherin cell membrane organization and AML homing.
(A–D) Reconstructed dSTORM images of N-cadherin distribution on each cell line. (E–H) DBSCAN images of N-cadherin clustering generated from DBSCAN analysis from the highlighted white boxes from the reconstructed dSTORM images. Clustered N-cadherin localizations are displayed in color and non-clustered N-cadherin localizations are in gray. An epsilon value of 50 nm and an n value of 30 localizations were used to examine N-cadherin clustering. Quantification of (I) N-cadherin cluster diameter, (J) N-cadherin cluster area, (K) number of N-cadherin clusters, (L) percent N-cadherin localizations clustered, and (M) density of N-cadherin in a cluster based on DBSCAN analysis (ctrl n=6 cells n=716 clusters, CD82OE n=6 cells n=768 clusters, Ngly-CD82 n=8 cells n=677 clusters, Palm-CD82 n=6 cells n=467 clusters). Bone marrow homing analysis of CFSE labeled ctrl, (N) Ngly-CD82 and (O) Palm-CD82 cells injected i.v. into 8–12 week old female NSG mice and analyzed as described in figure 1. (P) Ngly-CD82 cells were pretreated with 40 μg of N-cadherin blocking antibody (GC-4: Sigma) or IgG control (Santa Cruz Biotechnology) for 30 min at 37°C prior to i.v. injection into 8–10 week old male and female NSG mice (n=4). 16 hours following injection, homing analysis was completed as previously described. (Q) Working model of how CD82 post-translational modifications regulate N-cadherin protein organization and confinement, thereby contributing to functional differences in adhesion and homing. For super-resolution data, means are shown with S.E.M.; for homing data, means are shown with S.D,. Data was analyzed using one-way ANOVA followed by a two-sided unpaired t-test with Welch’s correction for groups with unequal standard deviations; no randomization or blinding methods were used. For homing studies, one animal was excluded based upon Grubbs outlier test (alpha =.05). (* p<.05, ** p < .01, *** p < .001).
Molecular scale organization of CD82 alters the bone marrow homing capacity of AML cells
The lateral assembly of cadherins in the membrane can stimulate signaling events and promote cell adhesion (25). Therefore, we assessed whether the CD82-mediated changes in N-cadherin clustering affect the homing of AML cells into the bone marrow. We injected the Ngly-CD82, Palm-CD82 and control cells into NSG mice to measure potential differences in bone marrow homing. Interestingly, we detect a significant increase in the ability of the Ngly-CD82 cells to home to the bone marrow when compared to control cells, while the Palm-CD82 cells display a substantial decrease in bone marrow homing (Fig. 4N, O). Analysis of the blood and spleen for Ngly-CD82 and Palm-CD82 cell localization identified no differences. To assess the role of N-cadherin in the enhanced homing of the Ngly-CD82 cells, we pretreated the cells with the N-cadherin blocking antibody (GC-4) prior to injection. Fig. 4P shows a disruption in Ngly-CD82 cells homing when N-cadherin is inhibited. Together these data demonstrate that CD82 and its post-translational modifications regulate N-cadherin cluster size, organization, and density, which modulate AML bone marrow homing.
While protein expression plays a critical role in AML (45), our study suggests that protein organization can be equally important. We define a pathway by which CD82 regulates bone marrow homing of AML cells through the membrane clustering of N-cadherin (Fig. 4Q). Establishment of AML within the bone marrow has extremely poor patient outcomes and we speculate that N-cadherin clustering may serve as a valuable marker to predict the aggressive behavior of AML. In addition, these findings provide an alternative model for targeting AML where modulation of protein organization within the membrane may be an effective treatment to dislodge AML cells from the protective environment of the bone marrow. Although N-cadherin is a focus of this study, we propose that N-cadherin will most likely model other adhesive proteins expressed on the cell surface such as selectins and integrins. In fact, CD82 regulation of specific integrin organization has been previously described in a variety of cellular systems (27, 46, 47).
In summary, these observations strengthen the significance of tetraspanin-mediated membrane organization within a complex multi-step process such as bone marrow homing. Moreover, we reason that CD82 serves as to regulate cellular behavior by modulating the topological distribution of protein networks on the cell membrane. It is plausible that this regulation ultimately leads to more robust signaling and adhesive potential that can be harnessed in disease states such as AML where cancer stem cells have a greater fitness advantage over normal HSPCs. Together, these data suggest that membrane clustering of proteins can regulate the aggressive potential of AML cells and may serve as a novel therapeutic target for future disease treatments.
Supplementary Material
Flow cytometry analysis of (A) CXCR4 (Clone 12G5, BD Bioscience), (C) E-cadherin (clone 36/E-cadherin, BD Bioscience) and (D) P-cadherin (clone 56/P-cadherin BD Bioscience) surface expression on Ctrl, CD82KD or CD82OE KG1a cells. (B) Western blot analysis of total CXCR4 protein expression (clone Ab-2 (1-14), Calbiochem) on Ctrl, CD82KD or CD82OE KG1a cells with actin (clone AC-74, Sigma) used as the loading control.
Acknowledgments
We would like to acknowledge all of the funding sources that made this work possible. This includes funding from an NIH R01 HL122483-01A1 to J.M.G, a New Mexico INBRE grant subaward to J.M.G (NIH P20 GM103451), pilot funding from the University of New Mexico Cancer Center (NIH P30CA118100), an American Cancer Society Institutional Research Grant, a Post-Doctoral Training Fellowship to K.D.M. (NIH T32 HL007736), Graduate Student Training Fellowships to C.M.T. from the NM Spatiotemporal Modeling Center (NIH P50 GM085273) and (NIH F31 HL124977), and a Graduate Student Training Fellowship to C.S.R. (NIH T32 HL007736). We also thank the NM Spatiotemporal Modeling Center for supporting the Super-Resolution Imaging Core Facility (NIH P50 GM085273) and the University of New Mexico Cancer Center (NIH P30CA118100) for use of the Keck-UNM Small Animal Models and Imaging Shared Resource. This work was also supported through faculty start-up funds to J.M.G. from the University of New Mexico Department of Pathology. Finally, we would like to acknowledge the technical assistance of Rebecca J. Dodd and Dr. I-Ming Chen, University of New Mexico Health Sciences Center. In addition, we would like to acknowledge the assistance of Dr. Ravi Majeti, Division of Hematology, Institute for Stem Cell Biology and Regenerative Medicine, and Cancer Institute, Stanford University and the Stanford University Division of Hematology Tissue Bank for samples.
Footnotes
Conflict-of-interest disclosure: The authors declare no conflict of interest.
Supplemental Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Machida U, Kami M, Hirai H. Hematopoietic stem-cell transplantation for acute leukemia. The New England journal of medicine. 1999;340(10):810. author reply 1–2. Epub 1999/03/13. [PubMed] [Google Scholar]
- 2.Guzman ML, Allan JN. Concise review: Leukemia stem cells in personalized medicine. Stem Cells. 2014;32(4):844–51. doi: 10.1002/stem.1597. Epub 2013/11/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konopleva M, Konoplev S, Hu W, Zaritskey AY, Afanasiev BV, Andreeff M. Stromal cells prevent apoptosis of AML cells by up-regulation of anti-apoptotic proteins. Leukemia. 2002;16(9):1713–24. doi: 10.1038/sj.leu.2402608. Epub 2002/08/30. [DOI] [PubMed] [Google Scholar]
- 4.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nature medicine. 2006;12(10):1167–74. doi: 10.1038/nm1483. Epub 2006/09/26. [DOI] [PubMed] [Google Scholar]
- 5.Gibson LF. Survival of B lineage leukemic cells: signals from the bone marrow microenvironment. Leukemia & lymphoma. 2002;43(1):19–27. doi: 10.1080/10428190210188. Epub 2002/03/23. [DOI] [PubMed] [Google Scholar]
- 6.Bradstock KF, Gottlieb DJ. Interaction of acute leukemia cells with the bone marrow microenvironment: implications for control of minimal residual disease. Leukemia & lymphoma. 1995;18(1–2):1–16. doi: 10.3109/10428199509064917. Epub 1995/06/01. [DOI] [PubMed] [Google Scholar]
- 7.Zhang B, Li M, McDonald T, Holyoake TL, Moon RT, Campana D, et al. Microenvironmental protection of CML stem and progenitor cells from tyrosine kinase inhibitors through N-cadherin and Wnt-beta-catenin signaling. Blood. 2013;121(10):1824–38. doi: 10.1182/blood-2012-02-412890. Epub 2013/01/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonardi F, Fusetti F, Deelen P, van Gosliga D, Vellenga E, Schuringa JJ. A proteomics and transcriptomics approach to identify leukemic stem cell (LSC) markers. Molecular & cellular proteomics: MCP. 2013;12(3):626–37. doi: 10.1074/mcp.M112.021931. Epub 2012/12/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishioka C, Ikezoe T, Furihata M, Yang J, Serada S, Naka T, et al. CD34(+)/CD38(−) acute myelogenous leukemia cells aberrantly express CD82 which regulates adhesion and survival of leukemia stem cells. International journal of cancer Journal international du cancer. 2013;132(9):2006–19. doi: 10.1002/ijc.27904. Epub 2012/10/12. [DOI] [PubMed] [Google Scholar]
- 10.Gil ML, Vita N, Lebel-Binay S, Miloux B, Chalon P, Kaghad M, et al. A member of the tetra spans transmembrane protein superfamily is recognized by a monoclonal antibody raised against an HLA class I-deficient, lymphokine-activated killer-susceptible, B lymphocyte line. Cloning and preliminary functional studies. J Immunol. 1992;148(9):2826–33. Epub 1992/05/01. [PubMed] [Google Scholar]
- 11.Imai T, Fukudome K, Takagi S, Nagira M, Furuse M, Fukuhara N, et al. C33 antigen recognized by monoclonal antibodies inhibitory to human T cell leukemia virus type 1-induced syncytium formation is a member of a new family of transmembrane proteins including CD9, CD37, CD53, and CD63. J Immunol. 1992;149(9):2879–86. Epub 1992/11/01. [PubMed] [Google Scholar]
- 12.Lebel-Binay S, Gil ML, Lagaudriere C, Miloux B, Marchiol-Fournigault C, Quillet-Mary A, et al. Further characterization of CD82/IA4 antigen (type III surface protein): an activation/differentiation marker of mononuclear cells. Cellular immunology. 1994;154(1):468–83. doi: 10.1006/cimm.1994.1092. Epub 1994/04/01. [DOI] [PubMed] [Google Scholar]
- 13.Dong JT, Lamb PW, Rinker-Schaeffer CW, Vukanovic J, Ichikawa T, Isaacs JT, et al. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science. 1995;268(5212):884–6. doi: 10.1126/science.7754374. Epub 1995/05/12. [DOI] [PubMed] [Google Scholar]
- 14.Boucheix C, Rubinstein E. Tetraspanins. Cellular and molecular life sciences: CMLS. 2001;58(9):1189–205. doi: 10.1007/PL00000933. Epub 2001/10/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemler ME. Targeting of tetraspanin proteins--potential benefits and strategies. Nature reviews Drug discovery. 2008;7(9):747–58. doi: 10.1038/nrd2659. Epub 2008/09/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bassani S, Cingolani LA. Tetraspanins: Interactions and interplay with integrins. The international journal of biochemistry & cell biology. 2012;44(5):703–8. doi: 10.1016/j.biocel.2012.01.020. Epub 2012/02/14. [DOI] [PubMed] [Google Scholar]
- 17.Larochelle A, Gillette JM, Desmond R, Ichwan B, Cantilena A, Cerf A, et al. Bone marrow homing and engraftment of human hematopoietic stem and progenitor cells is mediated by a polarized membrane domain. Blood. 2012;119(8):1848–55. doi: 10.1182/blood-2011-08-371583. Epub 2012/01/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeichi M. Morphogenetic roles of classic cadherins. Current opinion in cell biology. 1995;7(5):619–27. doi: 10.1016/0955-0674(95)80102-2. Epub 1995/10/01. [DOI] [PubMed] [Google Scholar]
- 19.Kemler R. From cadherins to catenins: cytoplasmic protein interactions and regulation of cell adhesion. Trends in genetics: TIG. 1993;9(9):317–21. doi: 10.1016/0168-9525(93)90250-l. Epub 1993/09/01. [DOI] [PubMed] [Google Scholar]
- 20.Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425(6960):841–6. doi: 10.1038/nature02040. Epub 2003/10/24. [DOI] [PubMed] [Google Scholar]
- 21.Bromberg O, Frisch BJ, Weber JM, Porter RL, Civitelli R, Calvi LM. Osteoblastic N-cadherin is not required for microenvironmental support and regulation of hematopoietic stem and progenitor cells. Blood. 2012;120(2):303–13. doi: 10.1182/blood-2011-09-377853. Epub 2012/05/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenbaum AM, Revollo LD, Woloszynek JR, Civitelli R, Link DC. N-cadherin in osteolineage cells is not required for maintenance of hematopoietic stem cells. Blood. 2012;120(2):295–302. doi: 10.1182/blood-2011-09-377457. Epub 2012/02/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhi L, Wang M, Rao Q, Yu F, Mi Y, Wang J. Enrichment of N-Cadherin and Tie2-bearing CD34+/CD38−/CD123+ leukemic stem cells by chemotherapy-resistance. Cancer letters. 2010;296(1):65–73. doi: 10.1016/j.canlet.2010.03.021. Epub 2010/05/07. [DOI] [PubMed] [Google Scholar]
- 24.Qiu S, Jia Y, Xing H, Yu T, Yu J, Yu P, et al. N-Cadherin and Tie2 positive CD34(+)CD38(−)CD123(+) leukemic stem cell populations can develop acute myeloid leukemia more effectively in NOD/SCID mice. Leukemia research. 2014;38(5):632–7. doi: 10.1016/j.leukres.2014.03.007. Epub 2014/04/08. [DOI] [PubMed] [Google Scholar]
- 25.Hong S, Troyanovsky RB, Troyanovsky SM. Binding to F-actin guides cadherin cluster assembly, stability, and movement. The Journal of cell biology. 2013;201(1):131–43. doi: 10.1083/jcb.201211054. Epub 2013/04/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson WJ. Regulation of cell-cell adhesion by the cadherin-catenin complex. Biochemical Society transactions. 2008;36(Pt 2):149–55. doi: 10.1042/BST0360149. Epub 2008/03/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Termini CM, Cotter ML, Marjon KD, Buranda T, Lidke KA, Gillette JM. The membrane scaffold CD82 regulates cell adhesion by altering alpha4 integrin stability and molecular density. Molecular biology of the cell. 2014 doi: 10.1091/mbc.E13-11-0660. Epub 2014/03/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aiuti A, Webb IJ, Bleul C, Springer T, Gutierrez-Ramos JC. The chemokine SDF-1 is a chemoattractant for human CD34+ hematopoietic progenitor cells and provides a new mechanism to explain the mobilization of CD34+ progenitors to peripheral blood. The Journal of experimental medicine. 1997;185(1):111–20. doi: 10.1084/jem.185.1.111. Epub 1997/01/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaitseva L, Murray MY, Shafat MS, Lawes MJ, MacEwan DJ, Bowles KM, et al. Ibrutinib inhibits SDF1/CXCR4 mediated migration in AML. Oncotarget. 2014;5(20):9930–8. doi: 10.18632/oncotarget.2479. Epub 2014/10/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida T, Kawano Y, Sato K, Ando Y, Aoki J, Miura Y, et al. A CD63 mutant inhibits T-cell tropic human immunodeficiency virus type 1 entry by disrupting CXCR4 trafficking to the plasma membrane. Traffic. 2008;9(4):540–58. doi: 10.1111/j.1600-0854.2007.00700.x. Epub 2008/01/10. [DOI] [PubMed] [Google Scholar]
- 31.Berditchevski F, Odintsova E, Sawada S, Gilbert E. Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. The Journal of biological chemistry. 2002;277(40):36991–7000. doi: 10.1074/jbc.M205265200. Epub 2002/07/12. [DOI] [PubMed] [Google Scholar]
- 32.Yang X, Kovalenko OV, Tang W, Claas C, Stipp CS, Hemler ME. Palmitoylation supports assembly and function of integrin-tetraspanin complexes. The Journal of cell biology. 2004;167(6):1231–40. doi: 10.1083/jcb.200404100. Epub 2004/12/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou B, Liu L, Reddivari M, Zhang XA. The palmitoylation of metastasis suppressor KAI1/CD82 is important for its motility- and invasiveness-inhibitory activity. Cancer research. 2004;64(20):7455–63. doi: 10.1158/0008-5472.CAN-04-1574. Epub 2004/10/20. [DOI] [PubMed] [Google Scholar]
- 34.Dwivedi C, Dixit M, Hardy RE. Plasma sialyltransferase as a tumor marker. Cancer detection and prevention. 1988;11(3–6):191–6. Epub 1988/01/01. [PubMed] [Google Scholar]
- 35.Swindall AF, Londono-Joshi AI, Schultz MJ, Fineberg N, Buchsbaum DJ, Bellis SL. ST6Gal-I protein expression is upregulated in human epithelial tumors and correlates with stem cell markers in normal tissues and colon cancer cell lines. Cancer research. 2013;73(7):2368–78. doi: 10.1158/0008-5472.CAN-12-3424. Epub 2013/01/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seales EC, Jurado GA, Singhal A, Bellis SL. Ras oncogene directs expression of a differentially sialylated, functionally altered beta1 integrin. Oncogene. 2003;22(46):7137–45. doi: 10.1038/sj.onc.1206834. Epub 2003/10/17. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Zhang W, Zhao J, Zhang L, Liu M, Yan G, et al. N-Glycosylation pattern of recombinant human CD82 (KAI1), a tumor-associated membrane protein. Journal of proteomics. 2012;75(4):1375–85. doi: 10.1016/j.jprot.2011.11.013. Epub 2011/11/30. [DOI] [PubMed] [Google Scholar]
- 38.White A, Lamb PW, Barrett JC. Frequent downregulation of the KAI1(CD82) metastasis suppressor protein in human cancer cell lines. Oncogene. 1998;16(24):3143–9. doi: 10.1038/sj.onc.1201852. Epub 1998/07/22. [DOI] [PubMed] [Google Scholar]
- 39.Mazurov D, Heidecker G, Derse D. The inner loop of tetraspanins CD82 and CD81 mediates interactions with human T cell lymphotrophic virus type 1 Gag protein. The Journal of biological chemistry. 2007;282(6):3896–903. doi: 10.1074/jbc.M607322200. Epub 2006/12/15. [DOI] [PubMed] [Google Scholar]
- 40.Heilemann M, van de Linde S, Schuttpelz M, Kasper R, Seefeldt B, Mukherjee A, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47(33):6172–6. doi: 10.1002/anie.200802376. Epub 2008/07/23. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Leiderman K, Pfeiffer JR, Wilson BS, Oliver JM, Steinberg SL. Characterizing the topography of membrane receptors and signaling molecules from spatial patterns obtained using nanometer-scale electron-dense probes and electron microscopy. Micron. 2006;37(1):14–34. doi: 10.1016/j.micron.2005.03.014. Epub 2005/08/06. [DOI] [PubMed] [Google Scholar]
- 42.Mattila PK, Feest C, Depoil D, Treanor B, Montaner B, Otipoby KL, et al. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity. 2013;38(3):461–74. doi: 10.1016/j.immuni.2012.11.019. Epub 2013/03/19. [DOI] [PubMed] [Google Scholar]
- 43.Ester M, Kreigel H, Sander J, Xu X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. 2nd International Conference on Knowledge Discovery and Data Mining; 1996. [Google Scholar]
- 44.Chitaev NA, Troyanovsky SM. Adhesive but not lateral E-cadherin complexes require calcium and catenins for their formation. The Journal of cell biology. 1998;142(3):837–46. doi: 10.1083/jcb.142.3.837. Epub 1998/08/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, Zhuo J, Duan Y, Shi B, Chen X, Zhang X, et al. Construction of protein profile classification model and screening of proteomic signature of acute leukemia. International journal of clinical and experimental pathology. 2014;7(9):5569–81. Epub 2014/10/23. [PMC free article] [PubMed] [Google Scholar]
- 46.Malik FA, Sanders AJ, Jiang WG. KAI-1/CD82, the molecule and clinical implication in cancer and cancer metastasis. Histology and histopathology. 2009;24(4):519–30. doi: 10.14670/HH-24.519. Epub 2009/02/19. [DOI] [PubMed] [Google Scholar]
- 47.Miranti CK. Controlling cell surface dynamics and signaling: how CD82/KAI1 suppresses metastasis. Cellular signalling. 2009;21(2):196–211. doi: 10.1016/j.cellsig.2008.08.023. Epub 2008/09/30. [DOI] [PubMed] [Google Scholar]
- 48.Huang F, Schwartz SL, Byars JM, Lidke KA. Simultaneous multiple-emitter fitting for single molecule super-resolution imaging. Biomedical optics express. 2011;2(5):1377–93. doi: 10.1364/BOE.2.001377. Epub 2011/05/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow cytometry analysis of (A) CXCR4 (Clone 12G5, BD Bioscience), (C) E-cadherin (clone 36/E-cadherin, BD Bioscience) and (D) P-cadherin (clone 56/P-cadherin BD Bioscience) surface expression on Ctrl, CD82KD or CD82OE KG1a cells. (B) Western blot analysis of total CXCR4 protein expression (clone Ab-2 (1-14), Calbiochem) on Ctrl, CD82KD or CD82OE KG1a cells with actin (clone AC-74, Sigma) used as the loading control.