Abstract
Francisella (F.) philomiragia is a Gram-negative bacterium with a preference for brackish environments that has been implicated in causing bacterial infections in near-drowning victims. The purpose of this study was to characterize the ability of F. philomiragia to infect cultured mammalian cells, a commonly used invertebrate model, and, finally, to characterize the ability of F. philomiragia to infect BALB/c mice via the pulmonary (intranasal) route of infection. This study shows that F. philomiragia infects J774A.1 murine macrophage cells, HepG2 cells and A549 human Type II alveolar epithelial cells. However, replication rates vary depending on strain at 24 h. F. philomiragia infection after 24 h was found to be cytotoxic in human U937 macrophage-like cells and J774A.1 cells. This is in contrast to the findings that F. philomiragia was non-cytotoxic to human hepatocellular carcinoma cells, HepG2 cells and A549 cells. Differential cytotoxicity is a point for further study. Here, it was demonstrated that F. philomiragia grown in host-adapted conditions (BHI, pH 6.8) is sensitive to levofloxacin but shows increased resistance to the human cathelicidin LL-37 and murine cathelicidin mCRAMP when compared to related the Francisella species, F. tularensis subsp. novicida and F. tularensis subsp. LVS. Previous findings that LL-37 is strongly upregulated in A549 cells following F. tularensis subsp. novicida infection suggest that the level of antimicrobial peptide expression is not sufficient in cells to eradicate the intracellular bacteria. Finally, this study demonstrates that F. philomiragia is lethal in two in vivo models; Galleria mellonella via hemocoel injection, with a LD50 of 1.8 × 103, and BALB/c mice by intranasal infection, with a LD50 of 3.45 × 103. In conclusion, F. philomiragia may be a useful model organism to study the genus Francisella, particularly for those researchers with interest in studying microbial ecology or environmental strains of Francisella. Additionally, the Biosafety level 2 status of F. philomiragia makes it an attractive model for virulence and pathogenesis studies.
Keywords: Francisella philomiragia, murine model, mammalian cells, Galleria mellonella, pulmonary infection, intranasal
Introduction
Members of the Francisella genus are small, non-motile, Gram-negative coccobacilli of the gamma-proteobacteria class (Sjostedt, 2007). Francisella philomiragia was first identified in an ailing muskrat located in Utah approximately 50 years ago, following the discovery of its related species, F. tularensis (Hollis et al., 1989). Mistakenly characterized as Yersinia philomiragia due to its 24% genomic homology with Y. pestis and similarities in morphology, it took 30 years to re-categorize the species as a member of the Francisella genus (Hollis et al., 1989).
Francisella philomiragia has an affinity for aquatic environments which may increase its host species potential (Anda et al., 2001; Tarnvik et al., 2004). The natural range of F. philomiragia reflects its preference for aquatic environments as it is found near bodies of water, particularly brackish or salt water in the mainland United States (Hollis et al., 1989; Whipp et al., 2003; Berrada and Telford, 2010; Siddaramappa et al., 2012; Whitehouse et al., 2012). F. philomiragia may exist naturally by forming biofilms on exposed surfaces of the environment and infecting the aquatic amoeba, Acanthamoeba castellanii (Verhoeven et al., 2010).
Virulence factors in F. philomiragia have not been well studied in this species, but likely include proteins encoded by the Francisella Pathogenicity Island (FPI) and phospholipase C (Zeytun et al., 2012), similar to other members of the Francisella genus (Nano and Schmerk, 2007; Dai et al., 2010).
A related species, F. noatunensis (formerly named F. philomiragia noatunensis), is pathogenic to many fish and mollusk species, which inflicts negative economic and health effects on fisheries (Kay et al., 2006; Ostland et al., 2006; Mauel et al., 2007; Mikalsen and Colquhoun, 2009). Mikalsen et al. (2009) previously asserted that F. philomiragia subsp. noatunensis is a fish pathogen that is not lethal to mice and does not pose a threat to human health (Mikalsen et al., 2009). However, in the time since that publication, F. noatunensis has been elevated to species level, which leaves the ability of F. philomiragia to infect mice in question and untested (Mikalsen and Colquhoun, 2009; Cowley and Elkins, 2011).
Near-drowning victims are susceptible to numerous bacterial infections due to the direct inoculation of the bacteria into the lungs (Ender and Dolan, 1997; Relich et al., 2015). F. philomiragia infections have been reported in otherwise healthy individuals via direct lung exposure resulting from near-drowning experiences in brackish or salty water or immunocompromised individuals with contact to contaminated water or fish (Wenger et al., 1989; Ender and Dolan, 1997; Cora et al., 2010; Kreitmann et al., 2015). Despite differences in genomic sequences (88% homologous to F. tularensis subsp. LVS and 84% to F. tularensis subsp. novicida and SchuS4; Zeytun et al., 2012; Davenport et al., 2014; Johnson et al., 2015), slightly different plasmids (Le Pihive et al., 2009), and reports that F. philomiragia does not cause disease in mice (Mikalsen et al., 2009), some of these near drowning victims infected by F. philomiragia develop a severe pneumonic infection. This prompted further investigation on the similarity of F. philomiragia to F. tularensis subsp. novicida and subsp. LVS, strains related to virulent F. tularensis subsp. SchuS4, and whether it may be an opportunistic pathogens in humans. This comparison was achieved through the use of in vitro experiments using cell models involved in tularemia infections (macrophages, lungs, and liver) and in vivo animal infection models (G. mellonella and BALB/c mice).
Materials and Methods
Bacterial Strains
Francisella tularensis Live Vaccine Strain (LVS; ATCC 29684), F. tularensis subsp. novicida (ATCC 15482), and F. philomiragia (ATCC 25015) were obtained from the American Type Culture Collection (Manassas, VA, USA). All bacterial strains were streaked onto Chocolate II Agar (GC II Agar with Hemoglobin and IsoVitaleXTM, BD 221267) and single colonies were inoculated into Brain Heart Infusion (BHI pH 6.8) broth (TekNova, Hollister, CA, USA).
Tissue Culture Cells
Murine macrophages, J774A.1 (ATCC TIB-67), human hepatocellular carcinoma cells, HepG2 (ATCC HB-8065), and human Type II alveolar epithelial cells, A549 (ATCC CCL-185), were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen #10566-016) supplemented with 10% fetal bovine serum (FBS) as per the manufacturer’s recommendations. Human U937 macrophage-like cells (ATCC CRL-1593.2), were cultured in RPMI 1640 media with 2 mM L-glutamine and 10% FBS as per the manufacturer’s recommendations (Lonza # 3163826). U937 cells were differentiated from monocytes to macrophages as instructed by manufacturer.
Infection Protocol for A549, J774A.1, HepG2, and U937 Cells
Cells were infected at a multiplicity of infection (MOI) of approximately 500, as previously described (Han et al., 2008; Hegedus et al., 2008; Ahmad et al., 2010), with a 2-h preinfection and 1-h gentamicin pulse. Briefly, cells were seeded (105/well) in a 48-well plate and allowed to attach overnight. After verifying successful cell attachment, culture media was gently removed and rinsed twice with culture media. Francisella strains were grown to mid-logarithmic phase, collected by centrifugation (10 min at 4000 × g, 4°C), washed three times with 1x phosphate buffered saline (PBS), and diluted in serum-free DMEM to a verified bacterial concentration (CFU/mL). Dilutions of bacteria were used to infect each cell line at MOI = 500. Sets of three wells were prepared for each condition (n = 3). Characteristically, Francisella infects host cells inefficiently, despite its infectivity via multiple routes in animals and humans. Therefore, the standard MOI of 500 CFU was used to infect cells with the Francisella strains in order to achieve infection of most of the cells (Lai et al., 2001; Lai and Sjostedt, 2003). Cells were then incubated with bacteria at 37°C, 5% CO2. After a 2-h incubation, well media was gently aspirated, washed twice with PBS, and treated with 50 μg/mL gentamicin in serum-free DMEM for 1 h to kill extracellular bacteria. Following the gentamicin pulse, cell media was gently aspirated, replaced with DMEM supplemented with 10% FBS and 5 μg/mL gentamicin, and allowed to incubate for 24 h at 37°C, 5% CO2. (Lai et al., 2001; Han et al., 2008; Ahmad et al., 2010) Cells were lysed and plated on Chocolate agar for CFU determination.
Cytotoxicity Assay of Mammalian Tissue Cultured Cells Infected with Francisella
PrestoBlue Cell Viability Reagent (A-13261, Life Technologies, Carlsbad, CA, USA) was used according to the manufacturer’s protocol. This reagent functions by using the reducing environment of the cell’s cytosol to determine cell viability. The reagent contains a cell-permeable compound, which is blue in color. When added to viable cells, it encounters the reducing environment and modifies the reagent to become a red fluorescent, which can be detected by fluorescence or absorbance measurements. Briefly, reagent was added to infected cells 24 h post gentamicin-pulse at a 1:10 ratio. The reagent was incubated with cells at 37°C for 2 h. Fluorescence was measured at excitation and emission spectra of 560 and 590 nm, respectively. Three wells were used per condition (n = 3). Data was averaged and a no-cell well was subtracted as background. Data was then plotted using GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA, USA) to reflect the cytotoxic effect of Francisella species on eukaryotic cell lines.
EC50 Antimicrobial Assays
Peptides used in this study were custom synthesized by ChinaPeptides Company (Shanghai, China) and had purities of ≥95% based on chromatographic analysis of the purified peptides. Antimicrobial activity (EC50) assays of the antibiotic control levofloxacin, human cathelicidin LL-37, and murine cathelicidin mCRAMP were performed against F. philomiragia, F. tularensis subsp. novicida, and F. tularensis subsp. LVS as previously described (Amer et al., 2010). Briefly, 1 × 105 CFU/well of Francisella species were grown in BHI (pH 6.8), added to a sterile 96-well plate and incubated with serial dilutions of peptide or antibiotic in 10 mM phosphate buffer for 3 h at 37°C. Dilutions were plated in triplicate on tryptic soy agar with 1% cysteine for 24 h; colonies were counted to determine survival (n = 3). This experiment was performed three independent times. Bacterial survival was calculated by a ratio of the number of colonies on each experimental plate to the average number of colonies on the control plates lacking peptide or antibiotic application. The EC50 was determined using GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA, USA) to plot the percent survival versus log of peptide or antibiotic concentration (log μg/mL) and fitting data to a standard sigmoidal dose–response curve as previously described (Blower et al., 2015).
Waxworm Infection
Galleria mellonella larvae (Vander horst Wholesale, St. Mary’s, OH, USA, 16 per group) were infected following previous reports (Aperis et al., 2007; Dean et al., 2011; McKenney et al., 2012). G. mellonella were infected by injecting 10 μL of bacteria into the hemocoel via a right proleg and incubated at 37°C. Each larva received bacterial concentrations of 1 × 106, 5 × 105, 1 × 105, 5 × 104, 1 × 104, 5 × 103, 1 × 103, 5 × 102, or 1 × 102 CFU/mL with 16 larvae per group. Waxworms were examined once a day for death. Bacterial concentrations were verified via retrospective plating and counting of CFUs.
Murine Infection
BALB/c mice (Harlan, Frederick, MD, USA, five per group) were infected intranasally with 20 μL of the following concentrations of bacteria: 1 × 106, 5 × 105, 1 × 105, 5 × 104, 1 × 104, 5 × 103, or 1 × 103 CFU/20 μL. Mice were examined twice a day for signs of illness or death. Bacterial concentrations were verified via retrospective plating and counting of CFUs. Animal experiments were approved by and conducted in compliance with regulations of the Institutional Animal Care and Use Committee (Protocol # 0236) of George Mason University. All experiments were carried out in accordance with the National Research Council’s Guide for the Care and Use of Laboratory Animals (2011) and the Public Health Service Policy on Humane Care and Use of Laboratory Animals (2002).
Statistical Analysis
Antimicrobial EC50 assays were performed in triplicate with n = 3 for each experiment, and representative experiments are shown. Standard deviations of the mean of each set are represented on each graph as error bars. Additionally the confidence interval (95%) is provided for EC50 determinations to demonstrate statistical overlap of data. Student’s t-test was performed and p values of p < 0.05 was considered statistically different.
The survival curves were performed with an n = 16 for G. mellonella and an n = 5 for BALB/c mice and were analyzed using the Mantel–Cox test, which is used to test the null hypothesis that survival curves are not different between groups. This test does not assume a normal distribution, allows for censored data, and is based off of the chi-squared test, which allows for a minimum of five samples.
Results
During pulmonary tularemia infections, bacteria colonize the alveolar macrophages, the lungs, and the liver (Hall et al., 2007; Faron et al., 2015). F. philomiragia was evaluated to see if it infected cell lines representative of these systems in vitro: murine macrophage cells, J774A.1, human Type II alveolar epithelial cells, A549, and human hepatocyte-like cells, HepG2. These cell lines have been previously shown by us and others to be susceptible to infection by F. tularensis subsp. novicida and LVS (Qin and Mann, 2006; Han et al., 2008; Amer et al., 2010; Bradburne et al., 2013).
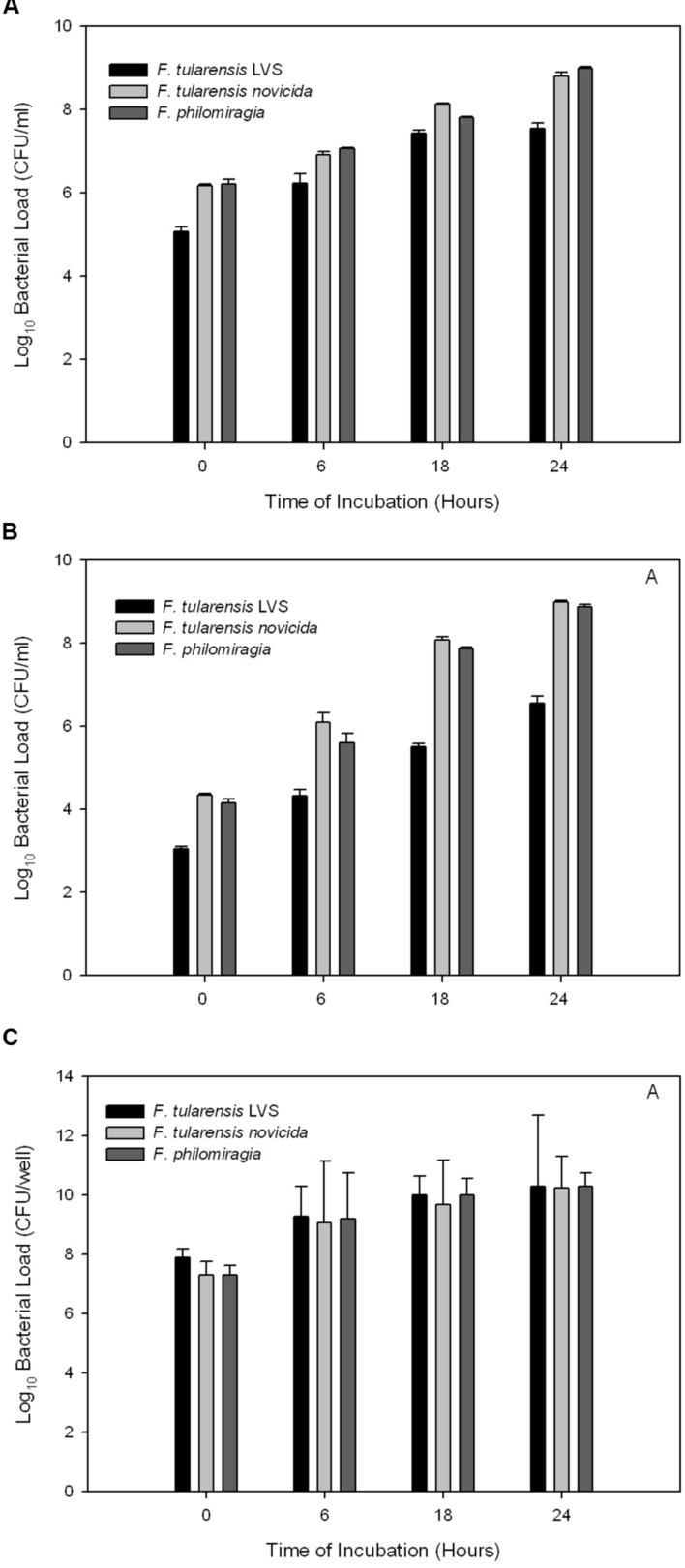
Francisella tularensis readily infects macrophages and proliferates within these cells (Anthony et al., 1991; Golovliov et al., 1997; Lai et al., 2001; Bolger et al., 2005). It is characteristic for Francisella replication to occur with little cytotoxicity until the cell becomes overburdened (at about 48 h post infection) and will experience cell death. F. philomiragia was found to be able to infect and proliferate in murine macrophages at 24 h to higher levels than what was seen for F. tularensis subsp. novicida and subsp. LVS (Figure 1A, p < 0.05). These results with the in vitro macrophage model suggest that F. philomiragia may be capable of infecting mammalian macrophages in vivo. Furthermore, these results suggest that F. philomiragia may be able to infect the alveolar macrophages in the lungs of near-drowning victims, which results in the clinical disease resembling tularemia that can afflict these patients.
FIGURE 1.
Bacterial replication following infection of cultured mammalian cells. Francisella philomiragia, F. tularensis subsp. novicida, and F. tularensis subsp. LVS infection of (A) murine macrophage J774A.1 cells, (B) human lung epithelial A549 cells, and (C) human hepatocytes HepG2 cells.
Human alveolar epithelial cells are known to be infected by Francisella both in vitro and in vivo (Hall et al., 2007; Faron et al., 2015). Here, experiments utilizing A549 cells showed that F. philomiragia infects this cell type to a lesser extent than F. tularensis subsp. novicida but more than subsp. LVS (Figure 1B, p < 0.05). The infection of this cell type suggests another potential mechanism by which the near-drowning infections in human could occur by this organism due to the direct inoculation of the lung.
Francisella infection of and proliferation in hepatocytes has been observed in human tularemia patients and animal models (Conlan and North, 1992; Lamps et al., 2004; Rasmussen et al., 2006; Ray et al., 2010). The fully virulent F. tularensis subsp. SchuS4 replicates well in cultured HepG2 cells (Qin and Mann, 2006). In these experiments, F. tularensis subsp. LVS infected human hepatocyte-like cells well, and replicated faster than F. tularensis subsp. novicida and F. philomiragia. However, by 24 h post infection, there were no differences between the bacterial burdens of the three Francisella species in HepG2 cells (Figure 1C, p > 0.05). This suggests that the F. philomiragia infections could potentially lead to liver damage, consistent with a tularemia infection.
Francisella tularensis is said to be a “stealth” pathogen, promoting its intracellular survival by not causing high cytotoxicity, among other mechanisms (Sjostedt, 2006; Jones et al., 2014). It was previously shown that infections of A549 cells by F. tularensis subsp. LVS (at 500 MOI) for 24 h did not cause significant cytotoxicity, although CFU increased significantly (Han et al., 2008; Bradburne et al., 2013). This high MOI of 500 is standard for Francisella infection protocols, as it is not taken up into non-phagocytic cells readily (Lai et al., 2001; Lai and Sjostedt, 2003; Telepnev et al., 2003). These studies were expanded to all three strains of Francisella investigated here (F. tularensis subsp. LVS, F. tularensis subsp. novicida, and F. philomiragia) and J774A.1, A549, and HepG2 cells.
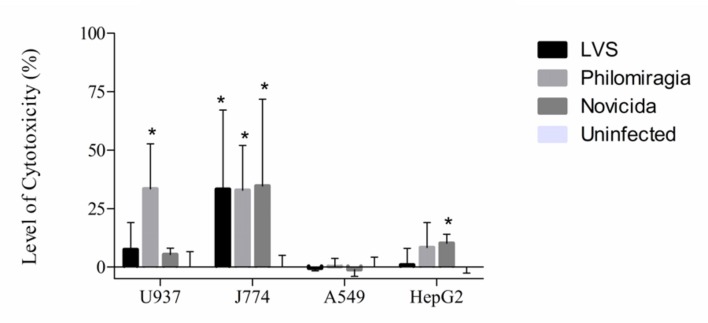
This study confirmed that F. tularensis subsp. LVS is not significantly cytotoxic toward A549 cells and, furthermore, it was found that F. tularensis subsp. novicida and F. philomiragia also displayed little cytotoxicity in this cell line at 24 h (Figure 2). HepG2s were also minimally affected by cytotoxic effects of F. tularensis subsp. LVS and displayed only 10 and 8% cytotoxicity from F. tularensis subsp. novicida and F. philomiragia infections at 24 h, respectively. The murine macrophage cell line, J774A.1, demonstrated greater susceptibility to the cytotoxic effects of Francisella, with all strains demonstrating about 33% cytotoxicity at 24 h (p < 0.05) consistent with previous reports (Lai et al., 2001; Lai and Sjostedt, 2003). However, additional cytotoxicity studies showed that F. philomiragia is highly cytotoxic to the human macrophage-like cell line, U937, (33%, p < 0.05) while F. tularensis subsp. LVS and subsp. novicida showed only 7 and 5% cytotoxicity, respectively. The differences between the human and murine macrophage cell lines are not yet understood in regard to Francisella infections. Previously, differences in Francisella intracellular replication have been noted between rat and murine macrophages (Anthony et al., 1991), however, other causes for the cytotoxicity differences other than species of origin are possible. These findings are consistent with the intracellular replication lifestyle of other Francisella species (Sjostedt, 2006).
FIGURE 2.
Cytotoxicity of Francisella strains to cultured mammalian cells. Cytotoxicity of F. philomiragia to cultured mammalian cells, compared to F. tularensis subsp. LVS and F. tularensis subsp. novicida.
Here, F. philomiragia was found to infect and replicate in the same cell types and have the same general level of cytotoxicity in those cell types as F. tularensis subsp. LVS and subsp. novicida, with the exception of the U937 cells (p < 0.05). The susceptibility of F. philomiragia to two antimicrobial peptides, LL-37, a human cathelicidin, and mCRAMP, a murine cathelicidin, known to be expressed by host cells and have killing activity against F. tularensis subsp. novicida and subsp. LVS was examined (Amer et al., 2010). This is important because host defense against Francisella infection relies not only on antibody production, but also on the response of the innate immune system (Allen, 1962; Metzger et al., 2007; Kirimanjeswara et al., 2008).
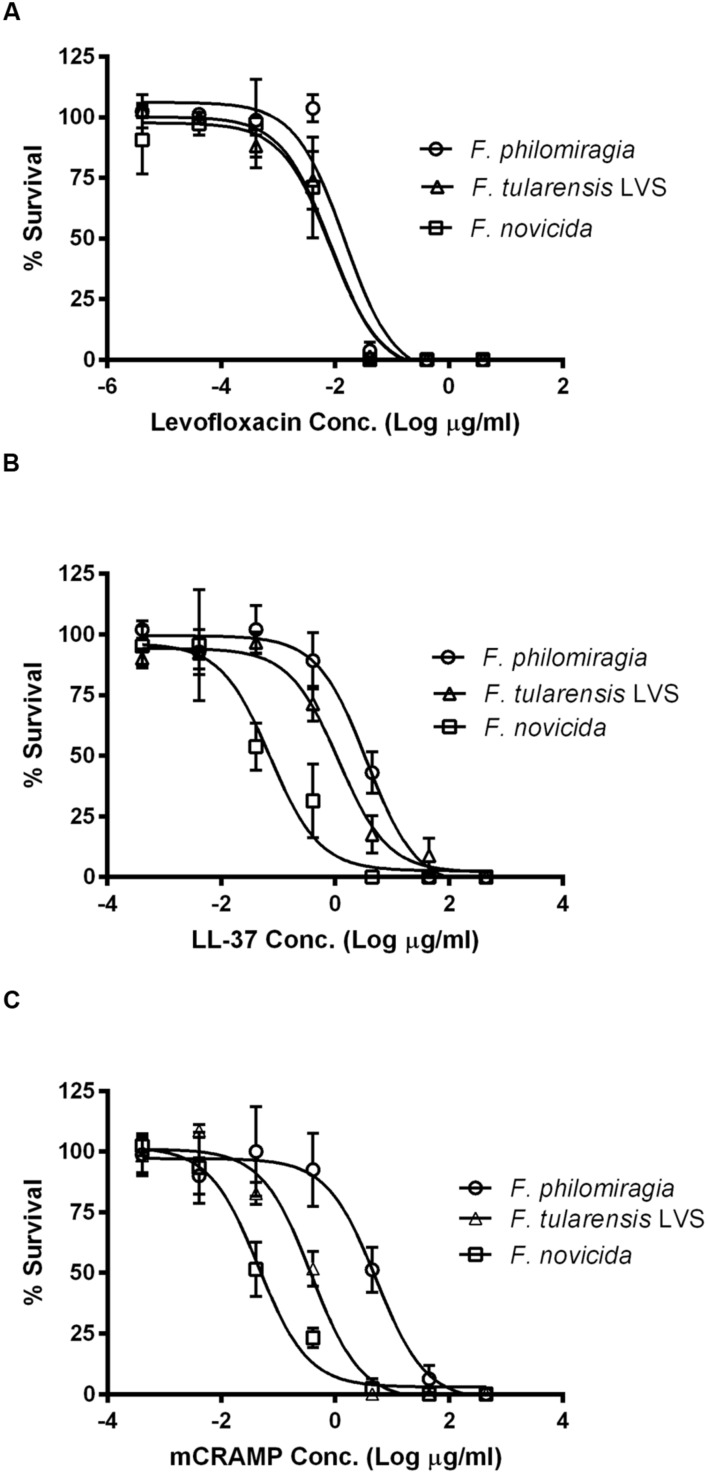
Francisella species, including F. philomiragia, are known to be highly susceptible to levofloxacin under MIC conditions (Nelson et al., 2010; Georgi et al., 2012), thus it was used as a control for the EC50 antimicrobial assays. The EC50 for levofloxacin of F. philomiragia is 0.0146 μg/mL (14.6 ng/mL; Figure 3A) while the F. tularensis subsp. LVS EC50 is 0.00827 μg/mL (8.27 ng/mL) and the F. tularensis subsp. novicida EC50 is 0.00843 μg/mL (8.43 ng/mL; Table 1). These values are statistically the same within the 95% confidence intervals (p > 0.05) and are consistent with the MICs previously reported (Georgi et al., 2012).
FIGURE 3.
Antimicrobial (EC50) assays for Francisella strains. F. philomiragia, F. tularensis subsp. novicida, and F. tularensis subsp. LVS susceptibility against (A) levofloxacin and the cathelicidins (B) LL-37 and (C) mCRAMP.
Table 1.
Summary of Antimicrobial (EC50) assays for Francisella strains.
| Francisella tularensis subsp. LVS | Francisella tularensis subsp. novicida | Francisella philomiragia | ||
|---|---|---|---|---|
| Levofloxacin | EC50 (μg/ml) | 0.00827 | 0.00843 | 0.0146 |
| 95% CI | (0.00524–0.0131) | (0.00413–0.0172) | (0.00696–0.0305) | |
| mCRAMP | EC50 (μg/ml) | 0.381 | 0.0453 | 5.27 |
| 95% CI | (0.239–0.607) | (0.0284–0.0723) | (2.93–9.46) | |
| LL-37 | EC50 (μg/ml) | 1.15 | 0.0724 | 3.61 |
| 95% CI | (0.604–2.18) | (0.0331–0.158) | (2.36–5.53) |
EC50 values plus 95% confidence intervals for F. philomiragia, F. tularensis subsp. novicida, and F. tularensis subsp. LVS susceptibility against levofloxacin and the cathelicidins LL-37 and mCRAMP.
The sensitivity of F. philomiragia to cationic antimicrobial peptides has not been well studied. This organism is highly resistant to colistin and polymyxin B, which are cationic cyclic peptide antibiotics (Petersen et al., 2009; Stephens et al., 2016). It was previously demonstrated that expression of the human cathelicidin LL-37 in A549 cells is strongly induced by F. tularensis subsp. novicida infection (Amer et al., 2010). This is of interest as Francisella bacteria replicate directly in the cytosol of the infected cells (Wehrly et al., 2009), and thus the bacteria may be able to be killed by expression of these innate immunity peptides by the afflicted cell.
The antimicrobial peptides, LL-37 and mCRAMP, were tested for their killing activity against F. philomiragia in 10 mM phosphate buffer, pH 7.2. The EC50 of LL-37 against F. philomiragia was determined to be 3.61 μg/mL (Figure 3B). In contrast to Francisella sensitivity to LL-37 in other species (EC50 of 1.15 and 0.0724 μg/mL in F. tularensis subsp. LVS and novicida; Amer et al., 2010; Flick-Smith et al., 2013), F. philomiragia is more resistant to this human cathelicidin peptide (p < 0.05). The EC50 of LL-37 against F. tularensis subsp. novicida found here is statistically similar to previously reported values due to overlapping 95% confidence interval values (Amer et al., 2010). Some small difference could also be due to the “host-adapted phenotype” growth conditions used to grow the bacteria for this study (BHI pH 6.8) compared to growth in Tryptic Soy Broth with Cysteine (TSB-C) media that was used previously (Amer et al., 2010). Growth in BHI (pH 6.8) is known to alter the surface carbohydrate and gene expression in a way that mimics the “host-adapted” phenotype of Francisella (Zarrella et al., 2011). In conclusion, F. philomiragia is more resistant to LL-37 than other Francisella species that were verified here (F. tularensis subsp. LVS EC50 = 1.15 (threefold), p = 0.011, and F. tularensis subsp. novicida EC50 = 0.0724 μg/mL (50-fold), p = 0.0011; Amer et al., 2010; Flick-Smith et al., 2013).
Francisella susceptibility to mCRAMP has not been previously reported. The EC50 of mCRAMP against F. philomiragia was determined to be 5.27 μg/mL (Figure 3C). No previous reports of Francisella susceptibility to mCRAMP were found; however, for this murine cathelicidin peptide, F. philomiragia is significantly more resistant (p < 0.05) than F. tularensis subsp. novicida (EC50 = 0.0453 μg/mL) or F. tularensis subsp. LVS (EC50 = 0.381 μg/mL). Here it was found that F. philomiragia is 14–116-fold more resistant to mCRAMP (EC50 = 5.27 μg/mL) than F. tularensis subsp. LVS (0.381 μg/mL, p = 0.007) and subsp. novicida (0.0453 μg/mL, p = 0.0051).
This increased resistance of F. philomiragia to cationic antimicrobial peptides could be due to differences in the LPS (Siddaramappa et al., 2012) or other surface properties of F. philomiragia compared to F. tularensis subsp. novicida or LVS perhaps due to differential expression of high-molecular weight carbohydrates in the “host-adapted” phenotype (Zarrella et al., 2011).
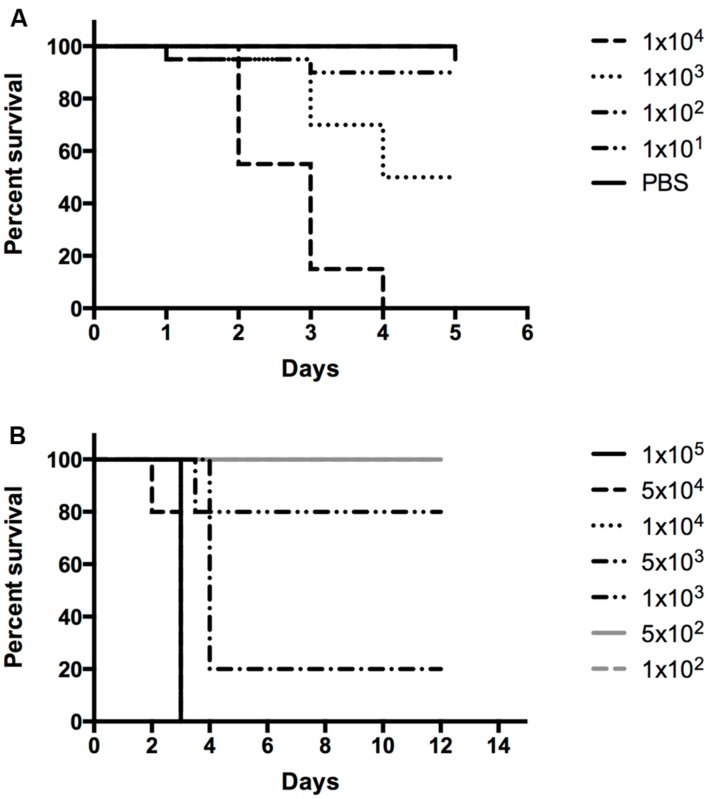
Galleria mellonella has been demonstrated to be a useful model for Francisella infection (Aperis et al., 2007; Ahmad et al., 2010; McKenney et al., 2012; Sprynski et al., 2014), thus a survival curve evaluating the mortality of G. mellonella during F. philomiragia infection was examined. As shown in Figure 4A, F. philomiragia injection is lethal to G. mellonella in a manner similar to F. tularensis subsp. LVS (Aperis et al., 2007; Ahmad et al., 2010). The LD50 of F. philomiragia in G. mellonella is ~1.8 × 103 CFU/mL or ~18 CFU with an inoculation volume of 10 μL. For comparison, the LD50 of F. tularensis subsp. novicida in G. mellonella is ~1.2 × 102 CFU/mL or ~1 CFU due to an inoculation volume of 10 μL (McKenney et al., 2012). The mean and median times to death for F. philomiragia are 2.79 and 3 days, respectively. Since F. philomiragia is able to infect G. mellonella similarly to other laboratory strains of Francisella, infection of BALB/c mice by the intranasal route of infection was tested for this organism.
FIGURE 4.
In vivo survival assays. F. philomiragia survival curve in (A) G. mellonella and (B) BALB/c mice infected by the pulmonary route.
BALB/c mice are a common experimental model for Francisella infections and they are susceptible to Francisella infection by the pulmonary route (intranasal or aerosol), among other routes; no studies have been reported for F. philomiragia infections of insect models, mice, rats, or marmosets (Chen et al., 2003; Conlan et al., 2003; Cowley and Elkins, 2011). To conform to these standards and expand the in vivo results obtained with G. mellonella, a survival curve evaluating the lethality of F. philomiragia in BALB/c mice when delivered via intranasal administration (mimicking near-drowning experiences) was examined. As shown in Figure 4B, intranasal F. philomiragia is lethal to BALB/c mice with an approximate LD50 of 3.45 × 103 CFU. This is very comparable to the intranasal F. tularensis subsp. LVS LD50 (1 × 103 CFU) in the same species of mice but is higher than the 100 CFU intranasal LD50 of F. tularensis subsp. novicida (Aperis et al., 2007).
Discussion
Multiple reports of severe pneumonic infections of humans following near-drowning experiences (Hollis et al., 1989; Wenger et al., 1989) suggested that direct or large inoculation of F. philomiragia into the lung by this method is sufficient to allow for infection of normal, healthy human lungs, potentially via infection of the alveolar macrophages and/or lung epithelial cells. However, this organism is not generally regarded as a human pathogen and its ability to infect mammalian cells is generally uncharacterized. In addition, the highly related organism, F. noatunensis, was found to be unable to infect laboratory mice (Mikalsen et al., 2009). Thus, F. philomiragia was compared to F. tularensis subsp. novicida and subsp. LVS regarding its ability to infect human and murine cells was further studied.
Francisella philomiragia was shown to be capable of infecting a murine macrophage cell line, J774A.1, which are commonly used for Francisella studies, with statistically higher levels (p < 0.05) than more commonly studied strains of Francisella (Hegedus et al., 2008; Pechous et al., 2008; Ahmad et al., 2010). Similarly, F. philomiragia was also found to infect a human Type II alveolar epithelial cell line, A549, at statistically higher levels than F. tularensis subsp. LVS. This is the first demonstration of F. philomiragia infecting Type II alveolar epithelial cells and is a significant contribution to the understanding of the potential interactions of F. philomiragia within the human lung. These findings suggest a potential mechanism by which near-drowning in brackish water known to contain F. philomiragia (Ottem et al., 2007) could potentially lead to infection through interaction of the bacteria with Type II alveolar epithelial cells of the lung and/or alveolar macrophages (Gentry et al., 2007; Hall et al., 2007; Craven et al., 2008; Faron et al., 2015). Furthermore, these results suggest that aerosol exposure to F. philomiragia could potentially lead to pulmonary infections in humans if inhaled via an aerosol. Given the wide distribution of F. philomiragia, in particular its known presence in various bodies of water within the United States, this potential route of infection should be further investigated.
In addition, it was demonstrated that F. philomiragia infects HepG2 cells, a human hepatocyte-like cell line. This finding suggests that F. philomiragia may be able to replicate in the liver in infected near-drowning victims. The liver is one of the main organs infected by F. tularensis strains and liver failure following overwhelming organ infection is thought to be the primary cause of death in mice suffering from tularemia (Conlan and North, 1992). Patients suffering from F. philomiragia pneumonia should be closely observed for sequelae similar to those found in tularemia infections caused by F. tularensis species.
Cytotoxicity data after 24 h of infection show that F. philomiragia is similar to F. tularensis subsp. novicida and subsp. LVS in most of the studied cell lines. Little cytotoxicity was seen in A549 cells (~0%, similar to other species) and HepG2 cells (8%, more than subsp. LVS but similar to subsp. novicida), and moderate cytotoxicity in J774A.1 cells (32%, similar to other species). However, the U937 human macrophage-like cell line only showed high cytotoxicity (33%) from F. philomiragia and not the other Francisella strains studied. This observation will be the subject of future investigation to understand the difference in U937 susceptibility.
Susceptibility testing using the antimicrobial peptides LL-37, a human cathelicidin, and mCRAMP, a murine cathelicidin, showed that these peptides were highly active in vitro against F. philomiragia. Despite being active in killing the bacteria in vitro, this antimicrobial peptide host defense mechanism is clearly insufficient to control F. philomiragia infections in infected cells or in vivo.
Francisella philomiragia was found to be lethal for both in vivo models tested: G. mellonella and BALB/c mice. G. mellonella has been demonstrated to be a useful in vivo model for Francisella infection (Aperis et al., 2007; Ahmad et al., 2010; McKenney et al., 2012; Sprynski et al., 2014), thus the survival of G. mellonella during F. philomiragia infection was examined. In G. mellonella, F. philomiragia was shown to be fatal in concentrations similar to F. tularensis subsp. LVS, with an LD50 of 18 CFU. This similarity supports the ability of G. mellonella to be used as an effective model for Francisella infection but also suggests that F. philomiragia is capable of infecting a range of hosts similar to other Francisella strains.
Francisella philomiragia is not generally regarded as a pathogen of humans or animals but is considered an environmental species of the genus (Anda et al., 2001; Tarnvik et al., 2004; Verhoeven et al., 2010). In some cases, F. philomiragia infections in near-drowning victims individuals are observed (Wenger et al., 1989; Ender and Dolan, 1997). An intranasal infection of mice by F. philomiragia was used to mimic lung exposure seen in drowning victims and test the susceptibility BALB/c mice to this organism. F. philomiragia was shown to be fatal in BALB/c mice by intranasal-delivered inoculum concentrations similar to F. tularensis subsp. LVS, with an LD50 of 3.45 × 103 CFU; however, this is significantly higher than the 10 CFU LD50 seen with F. tularensis subsp. novicida. Thus, contrary to the result for F. noatunensis (Mikalsen et al., 2009), F. philomiragia is able to infect laboratory mice. These results call for further studies to determine the full host range of F. philomiragia.
Conclusion
These studies show that F. philomiragia results in similar in vitro and in vivo infections to the F. tularensis subspecies novicida and LVS for the evaluated strains. It was demonstrated for the first time that there is potential for significant and robust F. philomiragia infection in macrophages, lung, and liver cells. F. philomiragia infection of human alveolar epithelial cells and macrophages suggests a mechanism for infection in the lungs of near-drowning patients. The high level of F. philomiragia intracellular replication in all three cell types suggests that F. philomiragia follows an infection course similar to tularemia caused by F. tularensis subspecies. It was previously demonstrated that infections of G. mellonella and pulmonary infections of BALB/c mice were fatal with similar LD50s to F. tularensis subsp. LVS. The results of these in vitro and in vivo experiments confirm earlier suggestions that F. philomiragia may be an emerging opportunistic human pathogen (Mailman and Schmidt, 2005; Sjodin et al., 2012) and that cellular and animal models of Francisella infection could also be used to study F. philomiragia. It would be of interest to evaluate all the available F. philomiragia strains for their ability to infect the various tissue culture and murine models.
It was found that F. philomiragia is comparable to the other Biosafety level 2 strains of Francisella in many respects but unusual in its effect on human U937 cells. This finding will open some interesting new avenues of research regarding pathogenesis and virulence of F. philomiragia. In addition, this work also positions F. philomiragia as another important organism in the field of Francisella research, especially for researchers interested in questions of microbial ecology or environmental persistence of members of the genus Francisella.
Author Contributions
All authors listed have made substantial, direct and intellectual contribution to the work, and approved it for publication. MLV conceived the study; MLV and CNP wrote the manuscript; CNP, SLP, RJB, SA, and MM contributed experimental data and contributed to the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge general technical support from Ms. SuHua Han, Albert O. Nwabueze, GMU, helpful discussions from Allison P. Belsches Jablonski, Lynchburg College, and the donation of U937 cells from Dr. R. Hakami, GMU. This work was partially supported by funded from DTRA (HDTRA1-11-1-0054) to MvH, the U.S. Department of Homeland Security under Cooperative Agreement Number DHS 2010-ST-061-AG0002 (CP), and Virginia’s Commonwealth Health Research Board (APBJ/MvH). Animal studies were performed with support from the GMU Foundation. The authors gratefully acknowledge the Open Access Publishing Fund of George Mason University to support the publication of this article.
References
- Ahmad S., Hunter L., Qin A., Mann B. J., van Hoek M. L. (2010). Azithromycin effectiveness against intracellular infections of Francisella. BMC Microbiol. 10:123 10.1186/1471-2180-10-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen W. P. (1962). Immunity against tularemia: passive protection of mice by transfer of immune tissues. J. Exp. Med. 115 411–420. 10.1084/jem.115.2.411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amer L. S., Bishop B. M., van Hoek M. L. (2010). Antimicrobial and antibiofilm activity of cathelicidins and short, synthetic peptides against Francisella. Biochem. Biophys. Res. Commun. 396 246–251. 10.1016/j.bbrc.2010.04.073 [DOI] [PubMed] [Google Scholar]
- Anda P., Segura del Pozo J., Diaz Garcia J. M., Escudero R., Garcia Pena F. J., Lopez Velasco M. C., et al. (2001). Waterborne outbreak of tularemia associated with crayfish fishing. Emerg. Infect. Dis. 7(3 Suppl.), 575–582. 10.3201/eid0703.010340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony L. D., Burke R. D., Nano F. E. (1991). Growth of Francisella spp. in rodent macrophages. Infect. Immun. 59 3291–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aperis G., Fuchs B. B., Anderson C. A., Warner J. E., Calderwood S. B., Mylonakis E. (2007). Galleria mellonella as a model host to study infection by the Francisella tularensis live vaccine strain. Microbes Infect. 9 729–734. 10.1016/j.micinf.2007.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrada Z. L., Telford S. R., III (2010). Diversity of Francisella species in environmental samples from Martha’s Vineyard. Massachusetts. Microb. Ecol. 59 277–283. 10.1007/s00248-009-9568-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower R. J., Barksdale S. M., van Hoek M. L. (2015). Snake cathelicidin NA-CATH and smaller helical antimicrobial peptides are effective against Burkholderia thailandensis. PLoS Negl. Trop Dis. 9:e0003862 10.1371/journal.pntd.0003862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger C. E., Forestal C. A., Italo J. K., Benach J. L., Furie M. B. (2005). The live vaccine strain of Francisella tularensis replicates in human and murine macrophages but induces only the human cells to secrete proinflammatory cytokines. J. Leukoc. Biol. 77 893–897. 10.1189/jlb.1104637 [DOI] [PubMed] [Google Scholar]
- Bradburne C. E., Verhoeven A. B., Manyam G. C., Chaudhry S. A., Chang E. L., Thach D. C., et al. (2013). Temporal transcriptional response during infection of type II alveolar epithelial cells with Francisella tularensis live vaccine strain (LVS) supports a general host suppression and bacterial uptake by macropinocytosis. J. Biol. Chem. 288 10780–10791. 10.1074/jbc.M112.362178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Shen H., Webb A., KuoLee R., Conlan J. W. (2003). Tularemia in BALB/c and C57BL/6 mice vaccinated with Francisella tularensis LVS and challenged intradermally, or by aerosol with virulent isolates of the pathogen: protection varies depending on pathogen virulence, route of exposure, and host genetic background. Vaccine 21 3690–3700. [DOI] [PubMed] [Google Scholar]
- Conlan J. W., Chen W., Shen H., Webb A., KuoLee R. (2003). Experimental tularemia in mice challenged by aerosol or intradermally with virulent strains of Francisella tularensis: bacteriologic and histopathologic studies. Microb. Pathog. 34 239–248. 10.1016/S0882-4010(03)00046-9 [DOI] [PubMed] [Google Scholar]
- Conlan J. W., North R. J. (1992). Early pathogenesis of infection in the liver with the facultative intracellular bacteria Listeria monocytogenes, Francisella tularensis, and Salmonella typhimurium involves lysis of infected hepatocytes by leukocytes. Infect. Immun. 60 5164–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cora M. C., Neel J. A., Tarigo J., Post K., Barnes J. (2010). Francisella philomiragia septicemia in a dog. J. Vet. Intern. Med. 24 969–972. 10.1111/j.1939-1676.2010.0545.x [DOI] [PubMed] [Google Scholar]
- Cowley S. C., Elkins K. L. (2011). Immunity to Francisella. Front. Microbiol. 2:26 10.3389/fmicb.2011.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven R. R., Hall J. D., Fuller J. R., Taft-Benz S., Kawula T. H. (2008). Francisella tularensis invasion of lung epithelial cells. Infect. Immun. 76 2833–2842. 10.1128/IAI.00043-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S., Mohapatra N. P., Schlesinger L. S., Gunn J. S. (2010). Regulation of francisella tularensis virulence. Front. Microbiol. 1:144 10.3389/fmicb.2010.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport K. W., Daligault H. E., Minogue T. D., Bishop-Lilly K. A., Broomall S. M., Bruce D. C., et al. (2014). Whole-genome sequences of nine francisella isolates. Genome Announc 2:5 10.1128/genomeA.00941-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean S. N., Bishop B. M., van Hoek M. L. (2011). Susceptibility of Pseudomonas aeruginosa Biofilm to Alpha-Helical Peptides: D-enantiomer of LL-37. Front. Microbiol. 2:128 10.3389/fmicb.2011.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender P. T., Dolan M. J. (1997). Pneumonia associated with near-drowning. Clin. Infect. Dis. 25 896–907. 10.1086/515532 [DOI] [PubMed] [Google Scholar]
- Faron M., Fletcher J. R., Rasmussen J. A., Apicella M. A., Jones B. (2015). Interactions of Francisella tularensis with alveolar type II epithelial cells and the murine respiratory epithelium. PLoS ONE 10:e0127458 10.1371/journal.pone.0127458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick-Smith H. C., Fox M. A., Hamblin K. A., Richards M. I., Jenner D. C., Laws T. R., et al. (2013). Assessment of antimicrobial peptide LL-37 as a post-exposure therapy to protect against respiratory tularemia in mice. Peptides 43 96–101. 10.1016/j.peptides.2013.02.024 [DOI] [PubMed] [Google Scholar]
- Gentry M., Taormina J., Pyles R. B., Yeager L., Kirtley M., Popov V. L., et al. (2007). Role of primary human alveolar epithelial cells in host defense against Francisella tularensis infection. Infect. Immun. 75 3969–3978. 10.1128/IAI.00157-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi E., Schacht E., Scholz H. C., Splettstoesser W. D. (2012). Standardized broth microdilution antimicrobial susceptibility testing of Francisella tularensis subsp. holarctica strains from Europe and rare Francisella species. J. Antimicrob. Chemother. 67 2429–2433. 10.1093/jac/dks238 [DOI] [PubMed] [Google Scholar]
- Golovliov I., Ericsson M., Sandstrom G., Tarnvik A., Sjostedt A. (1997). Identification of proteins of Francisella tularensis induced during growth in macrophages and cloning of the gene encoding a prominently induced 23-kilodalton protein. Infect. Immun. 65 2183–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. D., Craven R. R., Fuller J. R., Pickles R. J., Kawula T. H. (2007). Francisella tularensis replicates within alveolar type II epithelial cells in vitro and in vivo following inhalation. Infect. Immun. 75 1034–1039. 10.1128/IAI.01254-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Bishop B. M., van Hoek M. L. (2008). Antimicrobial activity of human beta-defensins and induction by Francisella. Biochem. Biophys. Res. Commun. 371 670–674. 10.1016/j.bbrc.2008.04.092 [DOI] [PubMed] [Google Scholar]
- Hegedus C. M., Skibola C. F., Warner M., Skibola D. R., Alexander D., Lim S., et al. (2008). Decreased urinary beta-defensin-1 expression as a biomarker of response to arsenic. Toxicol. Sci. 106 74–82. 10.1093/toxsci/kfn104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis D. G., Weaver R. E., Steigerwalt A. G., Wenger J. D., Moss C. W., Brenner D. J. (1989). Francisella philomiragia comb. nov. (formerly Yersinia philomiragia) and Francisella tularensis biogroup novicida (formerly Francisella novicida) associated with human disease. J. Clin. Microbiol. 27 1601–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. L., Daligault H. E., Davenport K. W., Coyne S. R., Frey K. G., Koroleva G. I., et al. (2015). Genome sequencing of 18 francisella strains to aid in assay development and testing. Genome Announc 3 e00147 10.1128/genomeA.00147-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. D., Faron M., Rasmussen J. A., Fletcher J. R. (2014). Uncovering the components of the Francisella tularensis virulence stealth strategy. Front. Cell Infect. Microbiol. 4:32 10.3389/fcimb.2014.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay W., Petersen B. O., Duus J. O., Perry M. B., Vinogradov E. (2006). Characterization of the lipopolysaccharide and beta-glucan of the fish pathogen Francisella victoria. FEBS J. 273 3002–3013. 10.1111/j.1742-4658.2006.05311.x [DOI] [PubMed] [Google Scholar]
- Kirimanjeswara G. S., Olmos S., Bakshi C. S., Metzger D. W. (2008). Humoral and cell-mediated immunity to the intracellular pathogen Francisella tularensis. Immunol. Rev. 225 244–255. 10.1111/j.1600-065X.2008.00689.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitmann L., Terriou L., Launay D., Caspar Y., Courcol R., Maurin M., et al. (2015). Disseminated infection caused by Francisella philomiragia. France, 2014. Emerg. Infect. Dis. 21 2260–2261. 10.3201/eid2112.150615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X. H., Golovliov I., Sjostedt A. (2001). Francisella tularensis induces cytopathogenicity and apoptosis in murine macrophages via a mechanism that requires intracellular bacterial multiplication. Infect. Immun. 69 4691–4694. 10.1128/IAI.69.7.4691-4694.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X. H., Sjostedt A. (2003). Delineation of the molecular mechanisms of Francisella tularensis-induced apoptosis in murine macrophages. Infect. Immun. 71 4642–4646. 10.1128/IAI.71.8.4642-4646.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamps L. W., Havens J. M., Sjostedt A., Page D. L., Scott M. A. (2004). Histologic and molecular diagnosis of tularemia: a potential bioterrorism agent endemic to North America. Mod. Pathol. 17 489–495. 10.1038/modpathol.3800087 [DOI] [PubMed] [Google Scholar]
- Le Pihive E., Blaha D., Chenavas S., Thibault F., Vidal D., Valade E. (2009). Description of two new plasmids isolated from Francisella philomiragia strains and construction of shuttle vectors for the study of Francisella tularensis. Plasmid 62 147–157. 10.1016/j.plasmid.2009.07.001 [DOI] [PubMed] [Google Scholar]
- Mailman T. L., Schmidt M. H. (2005). Francisella philomiragia adenitis and pulmonary nodules in a child with chronic granulomatous disease. Can. J. Infect. Dis. Med. Microbiol. 16 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauel M. J., Soto E., Moralis J. A., Hawke J. (2007). A piscirickettsiosis-like syndrome in cultured Nile tilapia in Latin America with Francisella spp. as the pathogenic agent. J. Aquat. Anim. Health 19 27–34. 10.1577/H06-025.1 [DOI] [PubMed] [Google Scholar]
- McKenney E. S., Sargent M., Khan H., Uh E., Jackson E. R., San Jose G., et al. (2012). Lipophilic prodrugs of FR900098 are antimicrobial against Francisella novicida in vivo and in vitro and show GlpT independent efficacy. PLoS ONE 7:e38167 10.1371/journal.pone.0038167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D. W., Bakshi C. S., Kirimanjeswara G. (2007). Mucosal immunopathogenesis of Francisella tularensis. Ann. N. Y. Acad. Sci. 1105 266–283. 10.1196/annals.1409.007 [DOI] [PubMed] [Google Scholar]
- Mikalsen J., Colquhoun D. J. (2009). Francisella asiatica sp. nov. isolated from farmed tilapia (Oreochromis sp.) and elevation of Francisella philomiragia subsp. noatunensis to species rank as Francisella noatunensis comb. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 10.1099/ijs.0.002139-0 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Mikalsen J., Olsen A. B., Rudra H., Moldal T., Lund H., Djonne B., et al. (2009). Virulence and pathogenicity of Francisella philomiragia subsp. noatunensis for Atlantic cod, Gadus morhua L., and laboratory mice. J. Fish Dis. 32 377–381. 10.1111/j.1365-2761.2008.00987.x [DOI] [PubMed] [Google Scholar]
- Nano F. E., Schmerk C. (2007). The Francisella pathogenicity island. Ann. N. Y. Acad. Sci. 1105 122–137. 10.1196/annals.1409.000 [DOI] [PubMed] [Google Scholar]
- Nelson M., Lever M. S., Dean R. E., Pearce P. C., Stevens D. J., Simpson A. J. (2010). Bioavailability and efficacy of levofloxacin against Francisella tularensis in the common marmoset (Callithrix jacchus). Antimicrob. Agents Chemother. 54 3922–3926. 10.1128/AAC.00390-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostland V. E., Stannard J. A., Creek J. J., Hedrick R. P., Ferguson H. W., Carlberg J. M., et al. (2006). Aquatic Francisella-like bacterium associated with mortality of intensively cultured hybrid striped bass Morone chrysops x M. saxatilis. Dis. Aquat. Organ. 72 135–145. 10.3354/dao072135 [DOI] [PubMed] [Google Scholar]
- Ottem K. F., Nylund A., Karlsbakk E., Friis-Moller A., Krossoy B., Knappskog D. (2007). New species in the genus Francisella (Gammaproteobacteria; Francisellaceae); Francisella piscicida sp. nov. isolated from cod (Gadus morhua). Arch. Microbiol. 188 547–550. 10.1007/s00203-007-0274-1 [DOI] [PubMed] [Google Scholar]
- Pechous R. D., McCarthy T. R., Mohapatra N. P., Soni S., Penoske R. M., Salzman N. H., et al. (2008). A Francisella tularensis Schu S4 purine auxotroph is highly attenuated in mice but offers limited protection against homologous intranasal challenge. PLoS ONE 3:e2487 10.1371/journal.pone.0002487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J. M., Carlson J., Yockey B., Pillai S., Kuske C., Garbalena G., et al. (2009). Direct isolation of Francisella spp. from environmental samples. Lett. Appl. Microbiol. 48 663–667. 10.1111/j.1472-765X.2009.02589.x [DOI] [PubMed] [Google Scholar]
- Qin A., Mann B. J. (2006). Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 6:69 10.1186/1471-2180-6-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen J. W., Cello J., Gil H., Forestal C. A., Furie M. B., Thanassi D. G., et al. (2006). Mac-1+ cells are the predominant subset in the early hepatic lesions of mice infected with Francisella tularensis. Infect. Immun. 74 6590–6598. 10.1128/IAI.00868-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray H. J., Chu P., Wu T. H., Lyons C. R., Murthy A. K., Guentzel M. N., et al. (2010). The Fischer 344 rat reflects human susceptibility to francisella pulmonary challenge and provides a new platform for virulence and protection studies. PLoS ONE 5:e9952 10.1371/journal.pone.0009952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relich R. F., Humphries R. M., Mattison H. R., Miles J. E., Simpson E. R., Corbett I. J., et al. (2015). Francisella philomiragia Bacteremia in a patient with acute respiratory insufficiency and acute-on-chronic kidney disease: a case report and review of the literature. J. Clin. Microbiol. 53 3947–3950. 10.1128/JCM.01762-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddaramappa S., Challacombe J. F., Petersen J. M., Pillai S., Kuske C. R. (2012). Genetic diversity within the genus Francisella as revealed by comparative analyses of the genomes of two North American isolates from environmental sources. BMC Genomics 13:422 10.1186/1471-2164-13-422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjodin A., Svensson K., Ohrman C., Ahlinder J., Lindgren P., Duodu S., et al. (2012). Genome characterisation of the genus Francisella reveals insight into similar evolutionary paths in pathogens of mammals and fish. BMC Genomics 13:268 10.1186/1471-2164-13-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostedt A. (2006). Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microbes Infect. 8 561–567. 10.1016/j.micinf.2005.08.001 [DOI] [PubMed] [Google Scholar]
- Sjostedt A. (2007). Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann. N. Y. Acad. Sci. 1105 1–29. 10.1196/annals.1409.009 [DOI] [PubMed] [Google Scholar]
- Sprynski N., Valade E., Neulat-Ripoll F. (2014). Galleria mellonella as an infection model for select agents. Methods Mol. Biol. 1197 3–9. 10.1007/978-1-4939-1261-2_1 [DOI] [PubMed] [Google Scholar]
- Stephens M. D., Hubble V. B., Ernst R. K., van Hoek M. L., Melander R. J., Cavanagh J., et al. (2016). Potentiation of Francisella resistance to conventional antibiotics through small molecule adjuvants. Med. Chem. Commun. Adv. 7 128–131. 10.1039/C5MD00353A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnvik A., Priebe H. S., Grunow R. (2004). Tularaemia in Europe: an epidemiological overview. Scand. J. Infect. Dis. 36 350–355. 10.1080/00365540410020442 [DOI] [PubMed] [Google Scholar]
- Telepnev M., Golovliov I., Grundstrom T., Tarnvik A., Sjostedt A. (2003). Francisella tularensis inhibits Toll-like receptor-mediated activation of intracellular signalling and secretion of TNF-alpha and IL-1 from murine macrophages. Cell Microbiol. 5 41–51. 10.1046/j.1462-5822.2003.00251.x [DOI] [PubMed] [Google Scholar]
- Verhoeven A. B., Durham-Colleran M. W., Pierson T., Boswell W. T., Van Hoek M. L. (2010). Francisella philomiragia biofilm formation and interaction with the aquatic protist Acanthamoeba castellanii. Biol. Bull. 219 178–188. [DOI] [PubMed] [Google Scholar]
- Wehrly T. D., Chong A., Virtaneva K., Sturdevant D. E., Child R., Edwards J. A., et al. (2009). Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol. 11 1128–1150. 10.1111/j.1462-5822.2009.01316.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger J. D., Hollis D. G., Weaver R. E., Baker C. N., Brown G. R., Brenner D. J., et al. (1989). Infection caused by Francisella philomiragia (formerly Yersinia philomiragia). A newly recognized human pathogen. Ann. Intern. Med. 110 888–892. 10.7326/0003-4819-110-11-888 [DOI] [PubMed] [Google Scholar]
- Whipp M. J., Davis J. M., Lum G., de Boer J., Zhou Y., Bearden S. W., et al. (2003). Characterization of a novicida-like subspecies of Francisella tularensis isolated in Australia. J. Med. Microbiol. 52(Pt 9), 839–842. 10.1099/jmm.0.05245-0 [DOI] [PubMed] [Google Scholar]
- Whitehouse C. A., Kesterson K. E., Duncan D. D., Eshoo M. W., Wolcott M. (2012). Identification and characterization of Francisella species from natural warm springs in Utah, USA. Lett. Appl. Microbiol. 54 313–324. 10.1111/j.1472-765X.2012.03214.x [DOI] [PubMed] [Google Scholar]
- Zarrella T. M., Singh A., Bitsaktsis C., Rahman T., Sahay B., Feustel P. J., et al. (2011). Host-adaptation of Francisella tularensis alters the bacterium’s surface-carbohydrates to hinder effectors of innate and adaptive immunity. PLoS ONE 6:e22335 10.1371/journal.pone.0022335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeytun A., Malfatti S. A., Vergez L. M., Shin M., Garcia E., Chain P. S. (2012). Complete genome sequence of Francisella philomiragia ATCC 25017. J. Bacteriol. 194:3266 10.1128/JB.00413-12 [DOI] [PMC free article] [PubMed] [Google Scholar]