Abstract
The facultative anaerobic Gram-negative species Escherichia albertii has been isolated from human faeces in gastrointestinal infection and from a range of wild bird species. Here we report the first case of a febrile infection associated with E. albertii bacteraemia in a 76-year-old woman with gastric dysplasia.
Keywords: Bacteraemia, bacterial identification methods, Escherichia, facultative anaerobes, gut microbiota
Escherichia coli is the single commonest Gram-negative species isolated from bacteraemic patients in most hospital centres. Other Escherichia species are much less common and are possibly overlooked unless detailed identification procedures are carried out in all cases of presumptive Escherichia species bacteraemia. Escherichia albertii was first described in 2003 in association with gastrointestinal infection in Bangladeshi children [1]. The species was subsequently identified in fatal infections of wild birds in several continents [2] but has yet to be identified in a bacteraemic human infection.
We describe a case of E. albertii infection in a 76-year-old woman. The patient had multiple comorbidities: a recent pelvic fracture from which she was convalescing, a dysplastic polyp of the gastric fundus with carcinoma-in-situ, hypothyroidism due to a previous thyroidectomy for papillary carcinoma, epilepsy and hypertension. She was admitted from residential care with a febrile illness of undetermined cause, an oral temperature peaking at 38.7°C, tachycardia at 139 beats per minute and a respiratory rate of 26 breaths per minute. Blood cultures collected during febrile episodes over a 24-hour period resulted in isolation of an oxidase-negative, Gram-negative bacillus from two sets of aerobic and anaerobic bottles inoculated on separate occasions. MALDI-TOF (matrix-assisted laser desorption/ionization time-of-flight mass spectrometry) analysis with direct extraction from blood cultures resulted in presumptive identification of Escherichia coli (score > 2.0; Biotyper, Bruker Daltonik, Germany). Targeted identification of blood culture isolates identified E. coli (score > 2.0, no alternative species listed). The patient was treated initially with piperacillin/tazobactam intravenously for 72 hours and then after defervescence with oral ciprofloxacin 500 mg twice daily. There were no clinical features of gastrointestinal infection. A midstream urine specimen contained >100 × 106 leucocytes per litre and a few bacteria on centrifuged deposit, but no significant bacterial growth. She defervesced rapidly and made an uneventful clinical recovery.
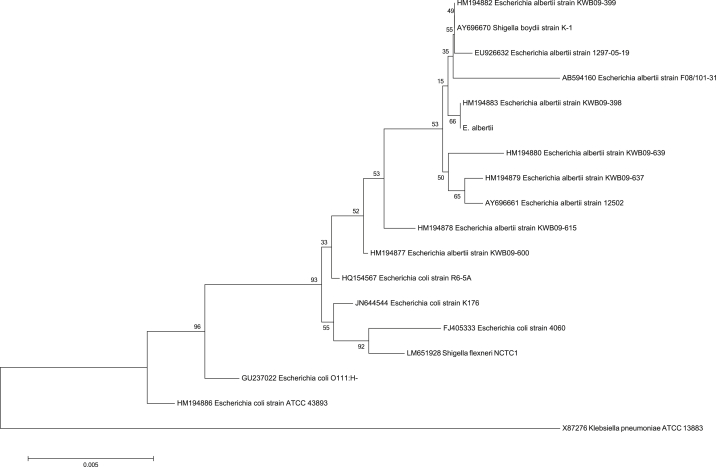
The possibility of an alternative aetiology was only considered when a molecular method of identifying common causes of bacteraemia (film array; BioFire, bioMérieux, France) produced a discrepant result: positive for Enterobacteriaceae but negative for E. coli. All isolates were oxidase-negative, Gram-negative bacilli on 5% horse blood agar and initially non-lactose-fermenting colonies on MacConkey agar. We identified them by substrate utilization (API 20E, bioMérieux), resulting in a low discrimination E. coli identification (results of tests against were urease positive, rhamnose and melibiose negative). Glucose, lactose and ONPG (O-nitrophenyl-β-d-galactopyranoside) tests in the panels were consistently positive. An isolate extract was prepared for 16S-based identification, which was consistent with E. albertii (Fig. 1). Though the patient had no history of gastrointestinal infection symptoms such as diarrhoea, we sought evidence of Shiga toxin [3] by PCR assay and tissue culture cytotoxin assay. Both were negative. The patient had no recent contact with birds, caged or otherwise, young children or other family members with active diarrhoea; no recent history of travel outside the Perth metropolitan area; and no investigations in the preceding months for diarrhoea or other clinical features of gastrointestinal infection. No bird die-off had been reported in the state during the previous 6 months.
Fig. 1.
Neighbour-joining tree depicting 16S rRNA gene sequence of STRAIN PERTH and related taxa. GenBank accession numbers are shown in brackets.
The aetiology of this infection would have been missed had it not been for the discrepancy we recorded between the MALDI-TOF results and the film array identification of the blood culture isolates because the urinary leucocytosis was interpreted as presumptive evidence of urosepsis, and the initial identification of E. coli isolated from repeated blood cultures was taken as further evidence of urosepsis. The phenotypic features of E. albertii are sufficiently variable to render classic phenotypic identification methods unreliable [4], and even the use of substrate utilization leant towards a low-discrimination E. coli result for all blood culture isolates. Although the erroneous identification of E. coli did not result in inappropriate clinical management, the opportunity for early public health intervention against a cluster of gastrointestinal infection cases might have been missed if the strain had been Shiga toxin positive [5], [6]. It is of particular note the woman had no epidemiologic connection with sick birds or children, as documented in early reports of E. albertii infection [1], [2]. E. albertii has recently been recognized as an early speciation event close to the separation of E. coli and Shigella species and has complex genomic diversity [7]. The addition of clinically significant E. albertii strains to culture collections used for identification databases will assist in timely recognition of this emerging pathogen and will help clarify whether this species is responsible for a wider spectrum of human disease than originally thought.
Conflict of interest
None declared.
References
- 1.Huys G., Cnockaert M., Janda J.M., Swings J. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Int J Syst Evol Microbiol. 2003;53:807–810. doi: 10.1099/ijs.0.02475-0. [DOI] [PubMed] [Google Scholar]
- 2.Ooka T., Seto K., Kawano K. Clinical significance of Escherichia albertii. Emerg Infect Dis. 2012;18:488–492. doi: 10.3201/eid1803.111401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandal L.T., Tunsjø H.S., Ranheim T.E., Løbersli I., Lange H., Wester A.L. Shiga toxin 2a in Escherichia albertii. J Clin Microbiol. 2015;53:1454–1455. doi: 10.1128/JCM.03378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abbott S.L., O’Connor J., Robin T., Zimmer B.L., Janda J.M. Biochemical properties of a newly described Escherichia species, Escherichia albertii. J Clin Microbiol. 2003;41:4852–4854. doi: 10.1128/JCM.41.10.4852-4854.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ooka T., Tokuoka E., Furukawa M. Human gastroenteritis outbreak associated with Escherichia albertii, Japan. Emerg Infect Dis. 2013;19:144–146. doi: 10.3201/eid1901.120646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asoshima N., Matsuda M., Shigemura K. Identification of Escherichia albertii as a causative agent of a food-borne outbreak occurred in 2003. Jpn J Infect Dis. 2014;67:139–140. doi: 10.7883/yoken.67.139. [DOI] [PubMed] [Google Scholar]
- 7.Retchless A.C., Lawrence J.G. Phylogenetic incongruence arising from fragmented speciation in enteric bacteria. Proc Natl Acad Sci U S A. 2010;107:11453–11458. doi: 10.1073/pnas.1001291107. [DOI] [PMC free article] [PubMed] [Google Scholar]