Abstract
Strain GD1T gen. nov., sp. nov., is the type strain of the newly proposed genus and species Drancourtella massiliensis, belonging to the Clostridiales order. This strain, isolated from the stool of a healthy person, is a Gram-positive rod, oxygen intolerant and nonmotile, with spore-forming activity. The features of this organism and its genome sequence are described. The draft genome is 3 057 334 bp long with 45.24% G + C content; it contains 2861 protein-coding genes and 64 RNA genes.
Keywords: Anaerobe, culturomics, Drancourtella massiliensisgen. nov. et sp. nov., gut microbiota, taxonogenomics
Introduction
The gut microbiota is an important part of the Human Microbiome Project. Indeed, 53% of cultivated bacterial species from human are isolated from the gut [1], and 80% of gut phylotypes found using the metagenomic method, which are mainly anaerobic strains [2], are not yet cultivated [3]. Those phylotypes belong especially to Firmicutes and Proteobacteria. The culturomics project consists in the use of many different culture methods to cultivate the most bacterial species possible from a single plurimicrobial sample—notably the use of multiple anaerobic conditions [4]. With the aim of cultivating the most anaerobic species possible from human stool, we used culturomics methods to study the fresh stool of a healthy French person.
The new genus we propose here is close to Ruminococcus torques, according to 16S rRNA phylogenetic analysis, part of Clostridiales order in Firmicutes phylum, which was separated to a different clade according to 16S rRNA analysis [5]. Cluster XIV notably contains Ruminococcus, Coprococcus and Blautia genera. Ruminococcus was first described from rumen fluid. Human infections caused by these bacteria have been reported [6], [7], [8], [9].
We propose the novel genus and species Drancourtella massiliensis for a strain isolated from the fresh stool of a healthy French person, which is phylogenetically close to Ruminococcus torques. Type strain is GD1T (= CSUR P1506 = DSM 100357).
Material and Methods
Ethics approval and sample collection
After receiving signed informed consent, approved by the Institut Fédératif de Recherche 48 (Faculty of Medicine, Marseille, France) under agreement 09-022, a stool specimen was collected at La Timone Hospital Marseille (France) in January 2015. The specimen was from a healthy French man (body mass index 23.2 kg/m2), 28 years old, with no current treatment, especially no antibiotics.
Isolation and growth conditions of strain
The stool specimen was incubated at 37°C into an anaerobic blood bottle Bactec Lytic/10 Anaerobic/F (Becton Dickinson, Le Pont de Claix, France) supplemented with 5% sheep's blood, after a thermal shock of 20 minutes at 80°C. Then dilution cultures were performed, and characterization of growth conditions was tested as previously described [10]. Finally, sporulation, different pH levels and NaCl concentrations were tested in the agar plate under the best culture conditions [11].
Strain identification
Identification of colonies was performed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) Microflex spectrometer (Bruker Daltonics, Leipzig, Germany) as previously described, and the spectrum was compared with our database (which includes the Bruker database and our own collection) [10], [12]. In case of nonidentification, i.e. if the spectrum did not find a match in the database (score <1.7), we proceeded to spectrum verification. If the spectrum was without background noise, and after exclusion of culture contamination, 16S rRNA was sequenced as previously described [10]. In case of a sequence similarity value lower than 96%, the species is considered to be a new genera without performing DNA-DNA hybridization, as suggested by Stackebrandt and Ebers [13].
Morphologic and biochemical characterization
Morphologic characterization was first performed by observation of Gram staining and motility of the fresh sample. Negative staining was then performed using bacteria fixed with 2.5% glutaraldehyde, deposited on carbon formvar film and then incubated for 1 second on ammonium molybdate 1%, dried on blotting paper and finally observed using a TECNAI G20 transmission electron microscope (FEI, Limeil-Brevannes, France) at an operating voltage of 200 keV. Biochemical features, such as oxidase, catalase, API 50CH, 20A and ZYM strips (bioMérieux, Marcy l'Étoile, France), were investigated according to the manufacturer's instructions. Cellular fatty acids were analysed from two samples prepared with approximately 10 mg of bacterial biomass each collected from several culture plates. Fatty acid methyl esters were prepared as described [14] and gas chromatography mass spectrometry (GC/MS) analyses were carried out as described previously [15].
Antibiotic susceptibility
Antibiotic susceptibility was tested with the diffusion method according to the CASFM/EUCAST 2015 recommendations for fastidious anaerobes (http://www.sfm-microbiologie.org/UserFiles/files/casfm/CASFM_EUCAST_V1_2015.pdf) using a suspension of 1 McFarland on Wilkins Chalgren agar (Sigma using a bacterial dilution with a turbidity equivalent to the McFarland 1.0 standard. Wilkins Chalgren agar (Sigma, Aldrich, Steinheim, Germany) supplemented with 5% sheep blood was used for the experiment.. Incubations were done in anaerobic conditions at 37°C, and reading was done at 48 hours using the Sirscan system (i2a, Montpellier, France) and eye controlled.
Genome sequencing, annotation and comparison
DNA extraction, in view of whole-genome sequencing, consisted of growing the species on Columbia agar supplemented with 5% sheep's blood (bioMérieux) at 37°C in an anaerobic atmosphere. Bacteria grown on three petri dishes were collected and resuspended in 4 × 100 μL Tris-EDTA (TE) buffer. Then 200 μL of this suspension was diluted in 1 mL TE buffer for lysis treatment that included a 30-minute incubation with 2.5 μg/μL lysozyme at 37°C, followed by an overnight incubation with 20 μg/μL proteinase K at 37°C [16]. As previously described [10], [15], [17], our genomic platform uses a protocol including proteinase K incubated overnight in order to digest contaminating proteins. Extracted DNA was then purified using three successive phenol–chloroform extractions and ethanol precipitations at −20°C overnight. After centrifugation, the DNA was resuspended in 160 μL TE buffer. The whole genome was then sequenced using the MiSeq Technology (Illumina, San Diego, CA, USA) with the mate-pair strategy as previously described [17]. Open reading frames (ORFs) were predicted using Prodigal [18] with default parameters, but the predicted ORFs were excluded if they were spanning a sequencing gap region (contain N). The predicted bacterial protein sequences were searched against the Clusters of Orthologous Groups (COGs) database using BLASTP (E value 1e-03, coverage 0.7 and identity percentage 30%). If no hit was found, a search was performed against the NR database using BLASTP with an E value of 1e-03, coverage 0.7 and identity percentage of 30%, If the sequence lengths were smaller than 80 aa, we used an E value of 1e-05. The tRNAScanSE tool [19] was used to find tRNA genes, whereas ribosomal RNAs were found by using RNAmmer [20]. Lipoprotein signal peptides and the number of transmembrane helices were predicted using Phobius [21]. ORFans, sequences that did not blast in the BLASTP program to a known sequence, have been defined by sequences with an E value smaller than 1e-3 in case of a sequence length higher than 80 aa, and an E value smaller than 1e-5 in case of a sequence length smaller than 80 aa. Such parameter thresholds have already been used in previous works to define ORFans [22]. For each selected genome, the complete genome sequence, proteome genome sequence and Orfeome genome sequence were retrieved from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/news/08-26-2014-new-genomes-FTP-live/). All proteomes were analysed with proteinOrtho [23]. Then for each couple of genomes a similarity score was computed. This score is the mean value of nucleotide similarity between all couples of orthologues between the two genomes studied (AGIOS) [24]. An annotation of the entire proteome was performed to define the distribution of the functional classes of predicted genes according to the COGs of proteins (using the same method as for the genome annotation). Annotation and comparison processes were performed in the Multi-Agent software system DAGOBAH [25], which include Figenix libraries [26] that provide pipeline analysis.
Results
Classification and features
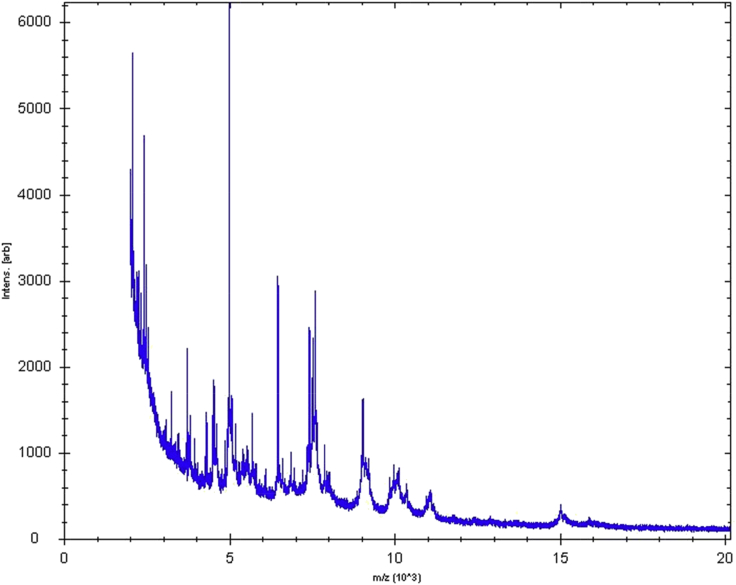
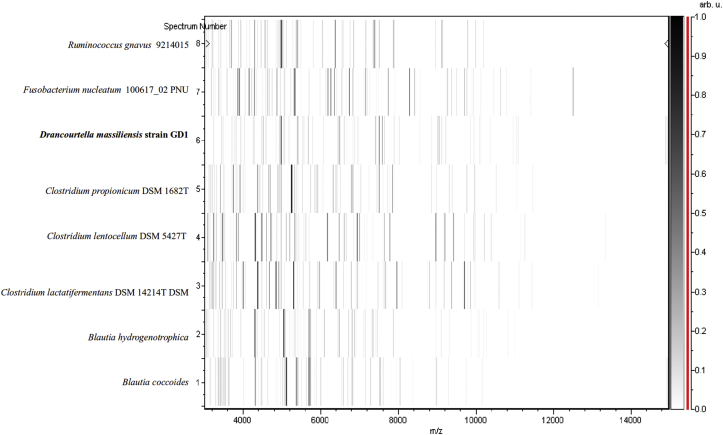
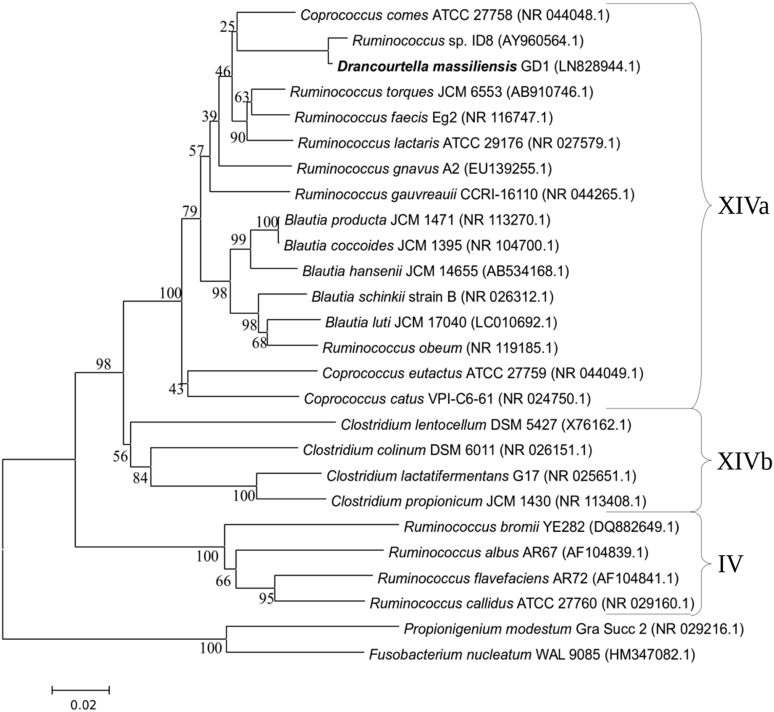
Type strain GD1T was first isolated after 10 days' anaerobic incubation of the stool sample in the presence of sheep's blood, after thermal shock, and then cultivated on Columbia agar supplemented with 5% sheep's blood under anaerobic conditions. MALDI-TOF spectrum of GD1T did not match anything in our database or Brucker's database, even though there was no background noise (Fig. 1, Fig. 2). 16S sequencing of the subunit of rRNA shows complete sequence of 1476 pb, with nucleotide BLAST results indicating Ruminococcus torques JCM6553 as the most closely cultured species, at 93.8% (Fig. 3). This result allowed us to define a new genus according to the thresholds delimited by Stackebrandt and Ebers [13]. The GD1T 16S rRNA accession number from the EBI Sequence Database is LN828944. A gel view was performed in order to observe the spectra differences of Drancourtella massiliensis with other close bacteria (Fig. 2).
Fig. 1.
Reference mass spectrum from Drancourtella massiliensis strain GD1T. Spectra from 12 individual colonies were compared and reference spectrum generated.
Fig. 2.
Gel view comparing Drancourtella massiliensis to other phylogenetically close species. Gel view displays raw spectra of loaded spectrum files arranged in pseudo-gel-like look. x-axis records m/z value. Left y-axis displays running spectrum number originating from subsequent spectra loading. Peak intensity is expressed by greyscale scheme code. The colour bar and right axis indicate the intensity each MALDI-TOF MS peak is displayed with and peak intensity in arbitrary units. Displayed species are indicated at left.
Fig. 3.
Phylogenetic tree highlighting position of Drancourtella massiliensis gen. nov., sp. nov., strain GD1T relative to other phylogenetically close type strains. Clusters are made according to Collins et al.[5]. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained with Kimura two-parameter model using neighbour-joining method with 1000 bootstrap replicates within MEGA6. Scale bar = 1% nucleotide sequence divergence.
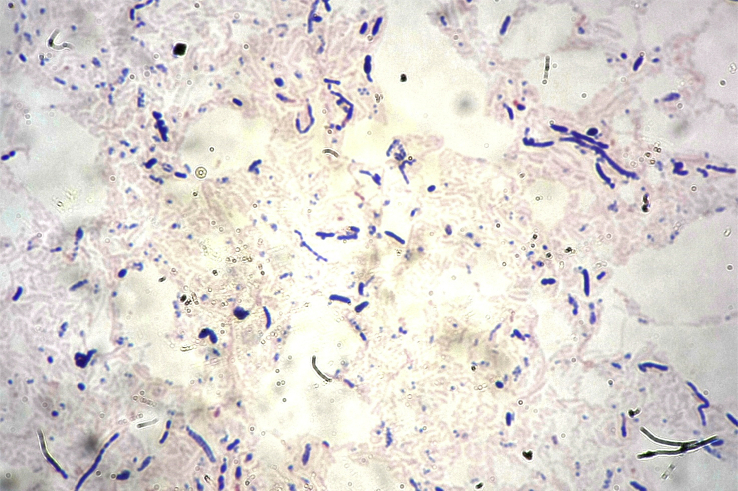
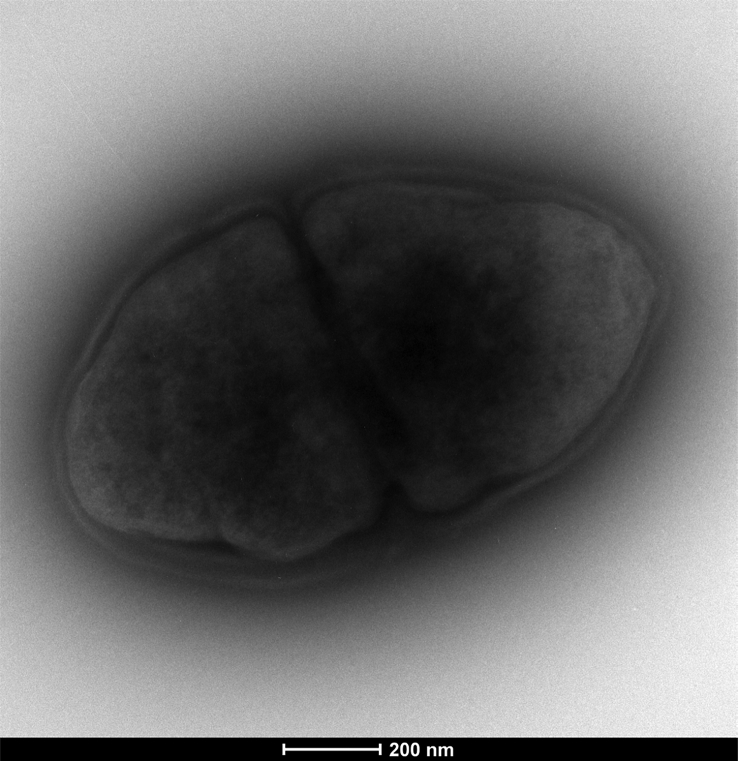
Tested culture conditions have identified optimal growth at 37°C after 48 hours under anaerobic conditions, but a little growth appeared in microaerophilic conditions, suggesting a relative oxygen tolerance. The pH range for growth is 6.5 to 7.0, and NaCl concentration needs to be lower than 10 g/L. The GD1T strain appears to be approximately 2 mm in size, homogeneous, translucent and smooth, nonhemolytic, nonmotile, with spore-forming activity colonies. Gram colouration was positive, and bacteria demonstrated a bacilli aspect under the microscope (Fig. 4). Electronic microscopy revealed small rods about 500 nm in size (Fig. 5). Classification and principal phenotypic features are listed in Table 1.
Fig. 4.
Gram staining of Drancourtella massiliensis strain GD1T.
Fig. 5.
Transmission electron microscopy of Drancourtella massiliensis strain GD1T using TECNAI G20 (FEI) at operating voltage of 200 keV. Scale bar = 200 nm.
Table 1.
Classification and general features of Drancourtella massiliensis strain GD1T
| Property | Term |
|---|---|
| Current classification | Domain: Bacteria |
| Phylum: Firmicutes | |
| Class: Clostridia | |
| Order: Clostridiales | |
| Family: Ruminococcaceae | |
| Genus: Drancourtella | |
| Species: Drancourtella massiliensis | |
| Type strain: GD1T | |
| Gram stain | Positive |
| Cell shape | Rod |
| Motility | Nonmotile |
| Sporulation | Sporulating |
| Temperature range | Mesophilic |
| Optimum temperature | 37°C |
Catalase and oxidase production reactions were negatives. Using an API 50CH strip, positive reactions were observed for d-ribose, methyl-αd-glucopyranoside and d-turanose and for potassium 5-ketogluconate. Negative reactions were observed for all others. Using an API 20A strip, all reactions were negative. Using an API ZYM strip, reactions were positive for leucine arylamidase, valine arylamidase, cysteine arylamidase, naphthol-AS-BI-phosphohydrolase, β-galactosidase and N-acetyl-β-glucosaminidase and were negative for others. The differences of characteristics compared with other representatives of the family Ruminococcaceae are detailed in Table 2. The major fatty acids detected (16:0 and 14:0) are saturated species. The GC/MS results indicated lower amounts of unsaturated acids and other saturated compounds (Table 3).
Table 2.
Differential characteristics of Drancourtella massiliensis strain GD1T, Ruminococcus faecis Eg2T, Ruminococcus torques ATCC27756 and Ruminococcus lactaris ATCC29176
| Property | D. massiliensis | R. faecis | R. torques | R. lactaris |
|---|---|---|---|---|
| Cell diameter (μm) | 0.5 | 1.0 | NA | NA |
| Oxygen requirement | Anaerobic | Anaerobic | Anaerobic | Anaerobic |
| Gram stain | + | + | + | + |
| Salt requirement | <10 g/L | NA | NA | NA |
| Motility | − | − | NA | NA |
| Endospore formation | + | − | − | − |
| Indole | − | − | + | − |
| Production of: | ||||
| Alkaline phosphatase | − | + | NA | NA |
| Catalase | − | + | NA | NA |
| Oxidase | − | − | NA | NA |
| Nitrate reductase | − | − | + | − |
| Urease | − | − | NA | NA |
| β-Galactosidase | − | − | − | + |
| N-acetyl-glucosamine | − | − | + | + |
| Acid from: | ||||
| l-Arabinose | − | − | + | − |
| Ribose | + | NA | NA | NA |
| Mannose | − | − | + | − |
| Mannitol | − | − | NA | NA |
| Saccharose | − | − | + | − |
| d-Glucose | − | + | − | − |
| d-Fructose | − | NA | NA | NA |
| d-Maltose | − | + | + | − |
| d-Lactose | − | + | + | − |
| Habitat | Human gut | Human gut | Human gut | Human gut |
+, positive result; −, negative result; v, variable result; w, weakly positive result; NA, data not available.
Table 3.
Total cellular fatty acid composition
| Fatty acids | IUPAC name | Mean relative %a |
|---|---|---|
| 16:0 | Hexadecanoic acid | 39.5 ± 4.1 |
| 14:0 | Tetradecanoic acid | 21.4 ± 9.6 |
| 18:1n9 | 9-Octadecenoic acid | 8.2 ± 5.2 |
| 16:1n7 | 9-Hexadecenoic acid | 6.3 ± 0.9 |
| 18:0 | Octadecanoic acid | 6.2 ± 1.6 |
| 18:1n7 | 11-Octadecenoic acid | 3.8 ± 0.5 |
| 13:0 | Tridecanoic acid | 3.3 ± 1.8 |
| 18:2n6 | 9,12-Octadecadienoic acid | 3.2 ± 0.8 |
| 14:1n5 | 9-Tetradecenoic acid | 2.7 ± 0.4 |
| 12:0 | Dodecanoic acid | 1.4 ± 0.8 |
| 15:0 | Pentadecanoic acid | 1.4 ± 0.8 |
| 5:0 anteiso | 2-methyl-butanoic acid | TR |
| 15:0 anteiso | 12-methyl-tetradecanoic acid | TR |
| 15:1n5 | 10-Pentadecenoic acid | TR |
| C14:03OH | 3-hydroxy-Tridecanoic acid | TR |
| 17:0 | Heptadecanoic acid | TR |
TR, trace amounts (<1%).
Mean peak area percentage calculated from analysis of FAMEs in three sample preparations ± standard deviation (n = 3).
Concerning susceptibility on β-lactam, the GD1T strain was susceptible to amoxicillin, ticarcillin, cefepime, vancomycin, teicoplanin and linezolid but resistant to ceftriaxone and ceftazidime. Concerning aminoglycoside antibiotics, cells were susceptible to gentamicin and resistant to tobramycin and amikacin. GD1T strain was resistant to sulfamethoxazole, aztreonam, ciprofloxacin and ofloxacin, and susceptible to imipenem, fosfomycin, clindamycin, metronidazole, rifampicin and colistin.
Genomic characterization and comparison
The genome is 3 057 334 bp long with 45.24% G + C content (Table 4). It is composed of seven scaffolds (composed of seven contigs). On the 2925 predicted genes, 2861 were protein-coding genes, and 64 were RNAs (two genes are 5S rRNA, two genes are 16S rRNA, three genes are 23S rRNA and 57 genes are tRNA genes). A total of 1969 genes (68.82%) were assigned as putative function (by COGs or by NR BLAST). Seventy-eight genes (2.73%) were identified as ORFans. The remaining genes (703 genes, 24.57%) were annotated as hypothetical proteins. Table 5 summarizes the distribution of genes into COGs functional categories. The genome sequence has been deposited in GenBank under accession number CVPG00000000.
Table 4.
Nucleotide content and gene count levels of genome
| Attribute | Genome (total) |
|
|---|---|---|
| Value | % of totala | |
| Size (bp) | 3 057 334 | 100 |
| G + C content (bp) | 1 383 137 | 45.24 |
| Coding region (bp) | 2 772 730 | 90.7 |
| Total genes | 2925 | 100 |
| RNA genes | 64 | 2.18 |
| Protein-coding genes | 2861 | 97.81 |
| Genes with function prediction | 1969 | 67.32 |
| Genes assigned to COGs | 1763 | 61.62 |
| Genes with peptide signals | 245 | 8.56 |
| Genes with transmembrane helices | 664 | 23.2 |
| Genes with Pfam domains | 2693 | 92 |
COGs, Clusters of Orthologous Groups database.
Total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome.
Table 5.
Number of genes associated with 25 general COGs functional categories
| Code | Value | % of totala | Description |
|---|---|---|---|
| J | 152 | 5.31 | Translation |
| A | 0 | 0 | RNA processing and modification |
| K | 177 | 6.18 | Transcription |
| L | 115 | 4.02 | Replication, recombination and repair |
| B | 0 | 0 | Chromatin structure and dynamics |
| D | 23 | 0.8 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 71 | 2.48 | Defense mechanisms |
| T | 56 | 1.96 | Signal transduction mechanisms |
| M | 95 | 3.32 | Cell wall/membrane biogenesis |
| N | 3 | 0.10 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 0 | 0 | Extracellular structures |
| U | 22 | 0.77 | Intracellular trafficking and secretion |
| O | 59 | 2.06 | Posttranslational modification, protein turnover, chaperones |
| C | 98 | 3.43 | Energy production and conversion |
| G | 202 | 7.06 | Carbohydrate transport and metabolism |
| E | 227 | 7.93 | Amino acid transport and metabolism |
| F | 62 | 2.16 | Nucleotide transport and metabolism |
| H | 67 | 2.34 | Coenzyme transport and metabolism |
| I | 44 | 1.53 | Lipid transport and metabolism |
| P | 104 | 3.64 | Inorganic ion transport and metabolism |
| Q | 28 | 0.98 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 237 | 8.28 | General function prediction only |
| S | 111 | 3.88 | Function unknown |
| — | 1098 | 38.38 | Not in COGs |
COGs, Clusters of Orthologous Groups database.
Total is based on total number of protein-coding genes in annotated genome.
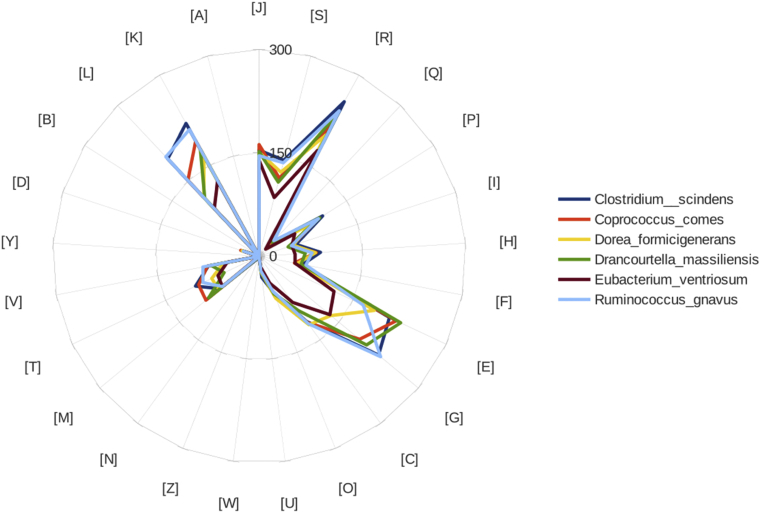
The draft genome sequence of Drancourtella massiliensis (3.05 Mb) is smaller than those of Ruminococcus gnavus, Clostridium scindens, Coprococcus comes and Dorea formicigenerans (3.72, 3.62, 3.24 and 3.19 Mb respectively) but larger than those of Eubacterium ventriosum (2.87 Mb). The G + C content of Drancourtella massiliensis is smaller than those of Clostridium scindens (45.24 and 46.35% respectively), but larger than those of Ruminococcus gnavus, Coprococcus comes, Dorea formicigenerans and Eubacterium ventriosum (42.52, 42.49, 40.97 and 34.92% respectively). The gene content of Drancourtella massiliensis is smaller than those of Ruminococcus gnavus, Clostridium scindens, Coprococcus comes and Dorea formicigenerans (2861, 3762, 3995, 3913 and 3277 respectively) but larger than those of Eubacterium ventriosum (2802). Fig. 6, Fig. 7 demonstrate that the distribution of genes into COGs categories was similar in all compared genomes. To evaluate the genomic similarity among the studied strains, we determined two parameters, digital DNA-DNA hybridization (dDDH), which exhibits a high correlation with DDH (Table 6) [27], [28], and AGIOS (Table 7) [24], which was designed to be independent from DDH. Drancourtella massiliensis shared 1235, 922, 1082, 1202 and 1266 orthologous proteins with Dorea formicigenerans, Eubacterium ventriosum, Coprococcus comes, Ruminococcus gnavus and Clostridium scindens respectively (Table 7).
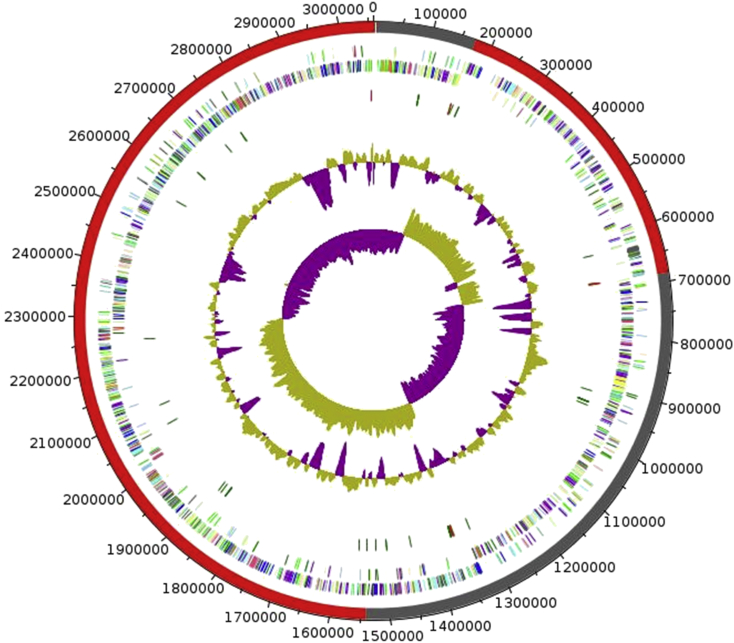
Fig. 6.
Graphical circular map of chromosome. From outside to center: Genes on forward strand coloured by COGs categories (only genes assigned to COGs), genes on reverse strand coloured by COGs categories (only genes assigned to COGs), RNA genes (tRNAs green, rRNAs red), GC content and GC skew.
Fig. 7.
Distribution of functional classes of predicted genes according to Clusters of Orthologous Groups (COGs) database of proteins.
Table 6.
Pairwise comparison of Drancourtella massiliensis (upper right) with eight other species using GGDC, formula 2 (DDH estimates based on identities/HSP length)a
| Dorea formicigenerans | Eubacterium ventriosum | Coprococcus comes | Ruminococcus gnavus | Clostridium scindens | Drancourtella massiliensis | |
|---|---|---|---|---|---|---|
| D. formicigenerans | 100% ± 00 | 32.8% ± 2.56 | 39.4% ± 2.70 | 25.6% ± 2.59 | 22.8% ± 2.62 | 22.9% ± 2.56 |
| E. ventriosum | 100% ± 00 | 38.9% ± 2.56 | 32.2% ± 2.55 | 28% ± 2.54 | 25.5% ± 2.53 | |
| C. comes | 100% ± 00 | 23.1% ± 2.58 | 22.5% ± 2.56 | 21.5% ± 2.56 | ||
| R. gnavus | 100% ± 00 | 25.7% ± 2.58 | 22.2% ± 2.56 | |||
| C. scindens | 100% ± 00 | 22.2% ± 2.57 | ||||
| D. massiliensis | 100% ± 00 |
DDH, DNA-DNA hybridization; GGDC, Genome-to-Genome Distance Calculator; HSP, high-scoring segment pairs.
Confidence intervals indicate inherent uncertainty in estimating DDH values from intergenomic distances based on models derived from empirical test data sets (which are always limited in size). These results are in accordance with 16S rRNA (Fig. 3) and phylogenomic analyses as well as GGDC results.
Table 7.
Numbers of orthologous protein shared between genomes (upper right) and average percentage similarity of nucleotides corresponding to orthologous protein shared between genomes (lower left) and numbers of proteins per genome (bold)
| Dorea formicigenerans | Eubacterium ventriosum | Coprococcus comes | Ruminococcus gnavus | Clostridium scindens | Drancourtella massiliensis | |
|---|---|---|---|---|---|---|
| D. formicigenerans | 3277 | 987 | 1194 | 1234 | 1337 | 1235 |
| E. ventriosum | 66.46 | 2802 | 870 | 954 | 946 | 922 |
| C. comes | 71.29 | 65.98 | 3913 | 1075 | 1144 | 1082 |
| R. gnavus | 69.73 | 65.47 | 70.00 | 3762 | 1269 | 1202 |
| C. scindens | 70.87 | 63.52 | 68.76 | 69.15 | 3995 | 1266 |
| D. massiliensis | 68.07 | 63.73 | 68.96 | 69.31 | 68.68 | 2861 |
Conclusion
Anaerobic conditions of culturomics have permitted the culture of the first strain of the new genus Drancourtella. Taxogenomics studies confirmed this species to be Drancourtella massiliensis gen. nov., sp. nov.
Taxonomic and nomenclatural proposals
Description of Drancourtella gen. nov.
The Drancourtella (dran.cour.tel'la, N.L. gen. fem.) name was chosen in honor of the French microbiologist Michel Drancourt (Université de la Méditerranée, Marseille, France).
Gram-positive bacilli. Spore forming and nonmotile. Drancourtella massiliensis is anaerobic. Oxidase and catalase negative. Indole negative, nitrate reductase negative. Habitat is the human gut. Type species is Drancourtella massiliensis.
Description of Drancourtella massiliensis gen. nov., sp. nov.
Drancourtella massiliensis (ma.si.li.en'sis, L. fem. adj. Massiliensis, from Latin Massilia, the city where the bacteria was isolated, Marseille).
D. massiliensis strain GD1T (= CSUR P1506 = DSM 100357) presented as translucent colonies 2 mm in diameter, isolated from the human stool of a healthy French person. Bacteria appeared as Gram-positive bacilli 0.5 μm long. Metabolism was strictly anaerobic, and growth was optimal at 37°C. D. massiliensis was spore forming and nonmotile. Oxidase and catalase were negative. Positive reactions were observed for d-ribose, methyl-αd-glucopyranoside, d-turanose and for potassium 5-ketogluconate, leucine arylamidase, valine arylamidase, cysteine arylamidase, naphthol-AS-BI-phosphohydrolase, β-galactosidase and N-acetyl-β-glucosaminidase. Resistance for ceftriaxone, ceftazidime, tobramycin, amikacin, sulfamethoxazole, aztreonam, ciprofloxacin and ofloxacin was observed.
This strain exhibited a G + C content of 45.24%. Its 16S rRNA sequence was deposited in GenBank under accession number LN828944, and the whole genome shotgun sequence was deposited in GenBank under accession number CVPG00000000. The type strain GD1T (= CSUR P1506 = DSM 100357) was isolated from the fecal flora of a healthy patient in France.
Acknowledgements
The authors thank the Xegen Company (http://www.xegen.fr/) for automating the genomic annotation process. This study was funded by the Fondation Méditerranée Infection. We thank K. Griffiths for English-language review and C. Andrieu for administrative assistance.
Conflict of Interest
None declared.
References
- 1.Hugon P., Dufour J.C., Colson P., Fournier P.E., Sallah K., Raoult D. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect Dis. 2015;15:1211–1219. doi: 10.1016/S1473-3099(15)00293-5. [DOI] [PubMed] [Google Scholar]
- 2.Faith J.J., Guruge J.L., Charbonneau M., Subramanian S., Seedorf H., Goodman A.L. The long-term stability of the human gut microbiota. Science. 2013;341(6141):1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins M.D., Lawson P.A., Willems A., Cordoba J.J., Fernandez-Garayzabal J., Garcia P. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 6.Hansen S.G.K., Skov M.N., Justesen U.S. Two cases of Ruminococcus gnavus bacteremia associated with diverticulitis. J Clin Microbiol. 2013;51:1334–1336. doi: 10.1128/JCM.03382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livaoğlu M., Yilmaz G., Kerimoğlu S., Aydin K., Karacal N. Necrotizing fasciitis with ruminococcus. J Med Microbiol. 2008;57(Pt 2):246–248. doi: 10.1099/jmm.0.47453-0. [DOI] [PubMed] [Google Scholar]
- 8.Sucu N., Köksal I., Yilmaz G., Aydin K., Caylan R., Aktoz Boz G. [Liver abscess and infective endocarditis cases caused by Ruminococcus productus] Mikrobiyoloji Bül. 2006;40:389–395. [PubMed] [Google Scholar]
- 9.Wang L., Christophersen C.T., Sorich M.J., Gerber J.P., Angley M.T., Conlon M.A. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol Autism. 2013;4:42. doi: 10.1186/2040-2392-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagier J.C., Elkarkouri K., Rivet R., Couderc C., Raoult D., Fournier P.E. Non contiguous-finished genome sequence and description of Senegalemassilia anaerobia gen. nov., sp. nov. Stand Genomic Sci. 2013;7:343–356. doi: 10.4056/sigs.3246665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aghnatios R., Cayrou C., Garibal M., Robert C., Azza S., Raoult D. Draft genome of Gemmata massiliana sp. nov, a water-borne Planctomycetes species exhibiting two variants. Stand Genomic Sci. 2015;10:120. doi: 10.1186/s40793-015-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seng P., Abat C., Rolain J.M., Colson P., Lagier J.C., Gouriet F. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51:2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 14.Myron Sasser. Bacterial identification by gas chromatographic analysis of fatty acids methyl esters (GC-FAME). MIDI 2006; (Technical Note 101).
- 15.Dione N., Sankar S.A., Lagier J.C., Khelaifia S., Michele C., Armstrong N. Genome sequence and description of Anaerosalibacter massiliensis sp. nov. New Microbes New Infect. 2016;10:66–76. doi: 10.1016/j.nmni.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sengüven B., Baris E., Oygur T., Berktas M. Comparison of methods for the extraction of DNA from formalin-fixed, paraffin-embedded archival tissues. Int J Med Sci. 2014;11:494–499. doi: 10.7150/ijms.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagier J.C., Bibi F., Ramasamy D., Azhar E.I., Robert C., Yasir M. Non contiguous-finished genome sequence and description of Clostridium jeddahense sp. nov. Stand Genomic Sci. 2014;9:1003–1019. doi: 10.4056/sigs.5571026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagesen K., Hallin P., Rødland E.A., Staerfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 22.Daubin V., Ochman H. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 2004;14:1036–1042. doi: 10.1101/gr.2231904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lechner M., Findeiss S., Steiner L., Marz M., Stadler P.F., Prohaska S.J. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabhan R., Rossi M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64(Pt 2):384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 25.Gouret P., Paganini J., Dainat J., Louati D., Darbo E., Pontarotti P. Evolutionary biology—concepts, biodiversity, macroevolution and genome evolution. Springer; Berlin: 2011. Integration of evolutionary biology concepts for functional annotation and automation of complex research in evolution: the multi-agent software system DAGOBAH; pp. 71–87. [Google Scholar]
- 26.Gouret P., Vitiello V., Balandraud N., Gilles A., Pontarotti P., Danchin E.G.J. FIGENIX: intelligent automation of genomic annotation: expertise integration in a new software platform. BMC Bioinformatics. 2005;6:198. doi: 10.1186/1471-2105-6-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auch A.F., von Jan M., Klenk H.P., Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2:117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meier-Kolthoff J.P., Auch A.F., Klenk H.P., Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]