Abstract
The acute hepatic porphyrias are caused by inherited enzymatic deficiencies in the heme biosynthesis pathway. Induction of the first enzyme 5-aminolevulinic acid synthase 1 (ALAS1) by triggers such as fasting or drug exposure can lead to accumulation of neurotoxic heme intermediates that cause disease symptoms. We have demonstrated that hepatic ALAS1 silencing using siRNA in a lipid nanoparticle effectively prevents and treats induced attacks in a mouse model of acute intermittent porphyria. Herein, we report the development of ALN-AS1, an investigational GalNAc-conjugated RNAi therapeutic targeting ALAS1. One challenge in advancing ALN-AS1 to patients is the inability to detect liver ALAS1 mRNA in the absence of liver biopsies. We here describe a less invasive circulating extracellular RNA detection assay to monitor RNAi drug activity in serum and urine. A striking correlation in ALAS1 mRNA was observed across liver, serum, and urine in both rodents and nonhuman primates (NHPs) following treatment with ALN-AS1. Moreover, in donor-matched human urine and serum, we demonstrate a notable correspondence in ALAS1 levels, minimal interday assay variability, low interpatient variability from serial sample collections, and the ability to distinguish between healthy volunteers and porphyria patients with induced ALAS1 levels. The collective data highlight the potential utility of this assay in the clinical development of ALN-AS1, and in broadening our understanding of acute hepatic porphyrias disease pathophysiology.
Keywords: RNAi, GalNAc-conjugated siRNA, exosome, circulating RNA, heme biosynthetic disorder
Introduction
The acute hepatic porphyrias (AHP) are inherited metabolic disorders caused by deficiencies of specific enzymes in the heme biosynthetic pathway.1 Of the four AHPs, the most common is acute intermittent porphyria (AIP), an autosomal dominant disease due to the half-normal activity of porphobilinogen deaminase (PBGD). These diseases are characterized by life-threatening acute neurovisceral attacks that involve the autonomic, peripheral, and central nervous systems and typically manifest with severe abdominal pain, tachycardia, hypertension, motor weakness, and psychiatric symptoms.1 Acute attacks are triggered by a number of factors, including cytochrome p450-inducing drugs, dieting, and hormonal fluctuations, all of which result in the increased expression of hepatic 5-aminolevulinic acid synthase (ALAS1), the first and rate-limiting enzyme of the heme biosynthetic pathway.2,3,4,5,6,7,8 When ALAS1 activity is induced, the inherited enzyme deficiency becomes limiting and results in the increased hepatic production of the upstream neurotoxic porphyrin precursors, 5-aminolevulinic acid (ALA), and porphobilinogen (PBG), which accumulate in the plasma and urine during attacks.9,10,11
In recent years, it has become clearer that the symptoms of acute attacks are mainly attributed to the accumulation of the neurotoxic metabolites, ALA and/or PBG, rather than heme deficiency in neuronal tissues.12 In particular, the observations that (i) nonporphyric liver transplant recipients who receive livers from symptomatic AIP donors (i.e., domino liver transplantations) subsequently develop acute porphyric attacks and (ii) that AIP patients with recurrent porphyric attacks who undergo orthotopic liver transplant have normalization of ALA and PBG levels as well as attack cessation13,14,15,16 strongly support that the pathogenic mechanism is related to the increased production of porphryin precursors in the liver. Therefore, inhibition of ALAS1 overexpression and reduction in ALA and PBG levels specifically within the liver should both prevent and treat the debilitating attacks of AIP. While the current standard of care, administration of blood-derived heme, normalizes elevated ALA and PBG levels through negative feedback inhibition of ALAS1, its slow onset of effect, negative side effects such as iron overload and phlebitis, requirement for administration through a central vein, and the potential for tachyphylaxis with repeat usage, highlight the need for a more effective therapeutic approach.17,18,19,20,21
RNA interference (RNAi) is a naturally occurring biological process by which small interfering RNAs (siRNA) selectively and robustly silence target mRNA, effectively suppressing inhibition of the corresponding disease-causing protein.22,23 The ability to leverage RNAi to selectively suppress disease-causing genes has become a highly promising, clinically validated therapeutic approach for the treatment of a host of genetic, metabolic and infectious diseases.24,25 The recent advances in lipid nanoparticle (LNP) and N-acetylgalactosamine (GalNAc)-siRNA-conjugate platforms have allowed for efficient and liver-specific delivery of siRNA, thus enabling development of RNAi-based therapeutics for liver-expressed disease targets.26,27,28,29,30 Given that the liver is the primary site of pathology in the AHPs, we are developing an RNAi therapeutic targeting liver ALAS1, called ALN-AS1, as a potential novel therapy for AHP patients with recurrent attacks. A marked upregulation of ALAS1 mRNA and enzyme activity in livers from patients that have undergone liver transplant provides clinical evidence that ALAS1 expression contributes to the active disease state.31 Conversely, the standard of care for AHP attacks, heme, is thought to impart a therapeutic effect through inhibition of ALAS1 level and activity.17,18,19,32
We have previously demonstrated successful RNAi-mediated silencing of hepatic ALAS1 as measured by plasma and urinary heme intermediate levels in preclinical models using an LNP-formulated ALAS1-targeting siRNA.32 Given that ALAS1 is not a secreted protein, determining the extent and kinetics of ALAS1 mRNA reduction due to RNAi-mediated silencing is difficult without performing serial liver biopsies, an invasive procedure that is not readily applicable to clinical studies. Hepatocytes secrete high levels of mRNAs in exosomes that can be collected and purified from serum or urine, which reflect levels of mRNA expression from liver.33 Exosomes have been shown to be important for cell to cell communication and are being used widely in the cancer diagnostics and biomarker fields.34 We have previously demonstrated the ability to detect mRNAs encoding tissue-specific gene transcripts, as well as their RNA-mediated reduction with siRNA administration in vivo, in biological fluids including serum and cerebrospinal fluid using a circulating extracellular RNA detection (cERD) method.35 In an effort to extend this approach to monitor ALAS1 reduction with ALN-AS1 treatment, we developed a target-specific, cERD method to quantify hepatic ALAS1 mRNA levels using serum, and further optimized this method to use urine. We demonstrate herein a striking correlation between serum, urine, and liver ALAS1 transcript levels across a number of preclinical species including mice, rats, and non-human primates (NHP) following subcutaneous dosing with a GalNAc-siRNA conjugate targeting ALAS1 (ALN-AS1). Moreover, we utilized this method in donor-matched human serum and urine samples obtained from healthy volunteers and patients to assess if there were differences in the “basal” ALAS1 transcript levels between these two groups.
Results
Detection of urine ALAS1 mRNA levels representative of liver levels in WT rats
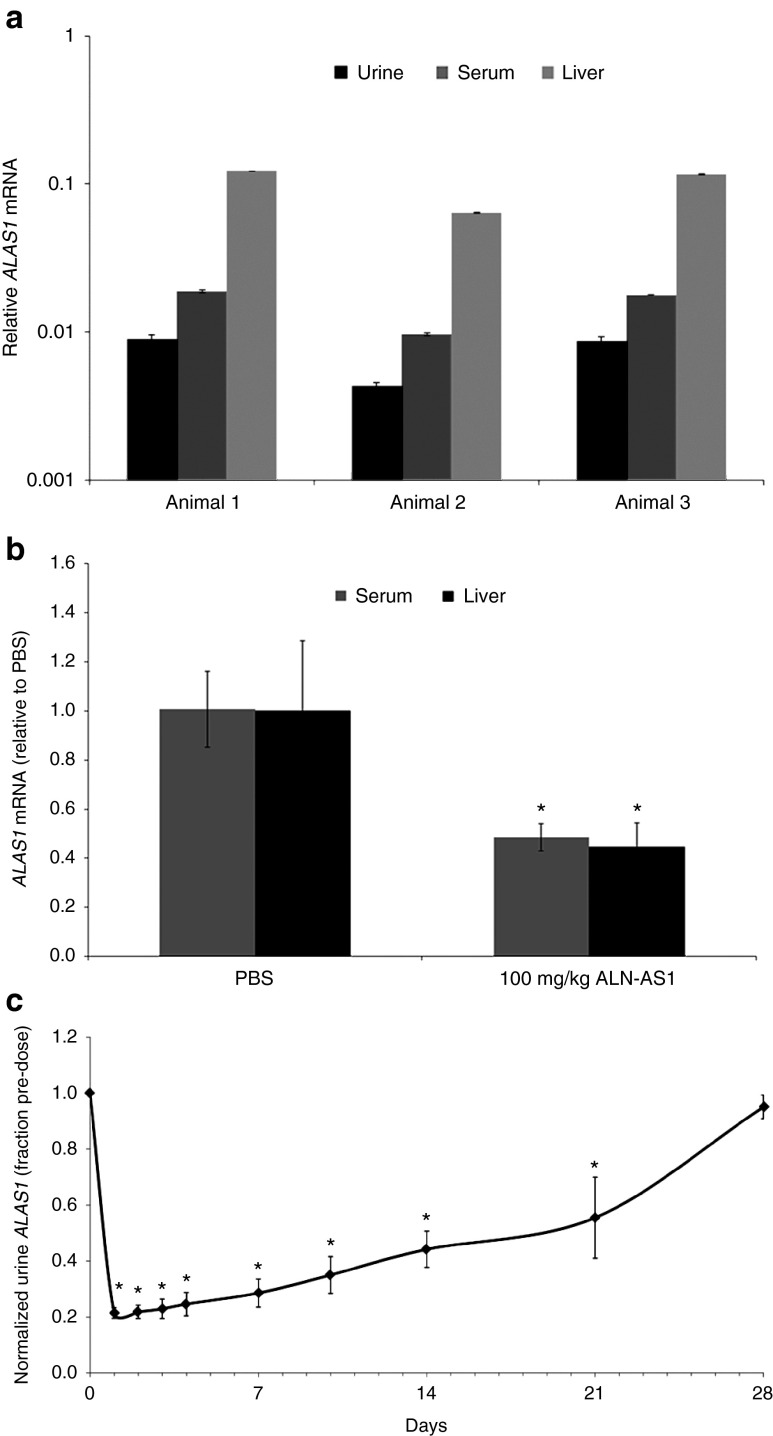
To determine whether baseline hepatic ALAS1 mRNA expression could be detected in biological fluids, filtered serum, and urine from naive Sprague Dawley (SD) rats (n = 3) were subjected to high-speed centrifugation, and total RNA was isolated from the resulting pellets as previously described.35 Total RNA was extracted from livers harvested from the same animals. As shown in Figure 1a, mRNA presumably corresponding to the liver expressed ALAS1 could be detected in both the urine and serum by reverse-transcription quantitative PCR (RT-qPCR). Although the absolute expression levels relative to GAPDH varied across matrices with the highest expression in liver, followed by serum then urine, a correlation was observed in the relative expression of ALAS1 for urine and serum samples across the group of naive animals.
Figure 1.
Detection of ALAS1 RNA from rodent serum or urine and correlation with RNAi-mediated gene silencing in liver. (a) Relative levels of ALAS1, as measured by quantitative polymerase chain reaction in RNA isolated from filtered, centrifuged rat serum. Values are relative to GAPDH levels; error bars, SD in experimental replicates. (b) Silencing of ALAS1 mRNA in rat liver and serum 72 hours after subcutaneous administration of 10 mg/kg ALN-AS1 or phosphate-buffered saline (PBS) control (vehicle). Levels of ALAS1 were normalized to GAPDH levels. ALAS1 liver and serum mRNA levels from individual animals were normalized to those from a group average of three PBS-treated livers and serum, respectively. Each graphical data point represents the percent remaining ALAS1 mRNA relative to the group average PBS-treated ALAS1 mRNA ± SD of the group. *Significance was determined by Student's t-test (P < 0.05). (c) Time course of ALAS1 mRNA silencing normalized to GAPDH monitored in urine following subcutaneous administration of 10 mg/kg ALN-AS1. ALAS1 transcript from individual animals was normalized to their respective individual predose ALAS1 level at each timepoint. Each graphical data point represents the fraction remaining ALAS1 relative to predose for the group average of four animal samples assayed in technical triplicates ± the standard deviation of the group. *Significance was determined by Student's t-test (P < 0.001).
To evaluate whether RNAi-mediated silencing of hepatic ALAS1 results in a concomitant reduction in circulating serum ALAS1 mRNA levels, SD rats (n = 3 per group) were administered a single 10 mg/kg subcutaneous dose of ALN-AS1, while control animals received phosphate-buffered saline (PBS) vehicle. As illustrated in Figure 1b, treatment with ALN-AS1 resulted in a 56 ± 9% reduction of ALAS1 mRNA in the liver and a 52 ± 5% reduction in the serum at 72 hours postadministration. These data confirmed the ability to detect comparable siRNA-mediated silencing of the liver-expressed target gene via circulating serum transcript levels. A similar level of urine ALAS1 mRNA reduction was observed when rats were treated under similar experimental conditions (data not shown).
To determine if cERD could be used to monitor the kinetics of ALAS1 transcript reduction, serial urine samples were collected from rats (n = 4) prior to administration and at sparse time points following a single dose of ALN-AS1 at 10 mg/kg, out to 28 days post-treatment. As shown in Figure 1c, a maximum reduction of 80% ALAS1 mRNA occurs as early as 24 hours postdose and is maintained for approximately 3 additional days. Normalization of ALAS1 mRNA levels after ALN-AS1 treatment to predose control samples from the same individual animal helped reduce the significant interanimal variability that exists in ALAS1 transcript levels, thus allowing detection of a more substantial degree of knockdown than observed when normalizing to a separate PBS control group. Recovery to within 80–90% of baseline levels occurs between 21–28 days postdose.
Detection of urine ALAS1 mRNA changes in a rat AIP model
We next determined if changes in hepatic ALAS1 transcript levels could be detected by cERD in rodent models of AIP. A rat AIP model was developed by administering to wild-type female SD rats a single dose of a PBGD siRNA that decreased hepatic PBGD mRNA to ~20% of normal levels (data not shown). When given four serial injections of phenobarbital (PB), a prototypic inducer of hepatic ALAS1 expression, the PBGD siRNA-treated rats accumulated high levels of ALA and PBG in their urine, thereby mimicking the biochemical abnormalities of an acute attack (data not shown).
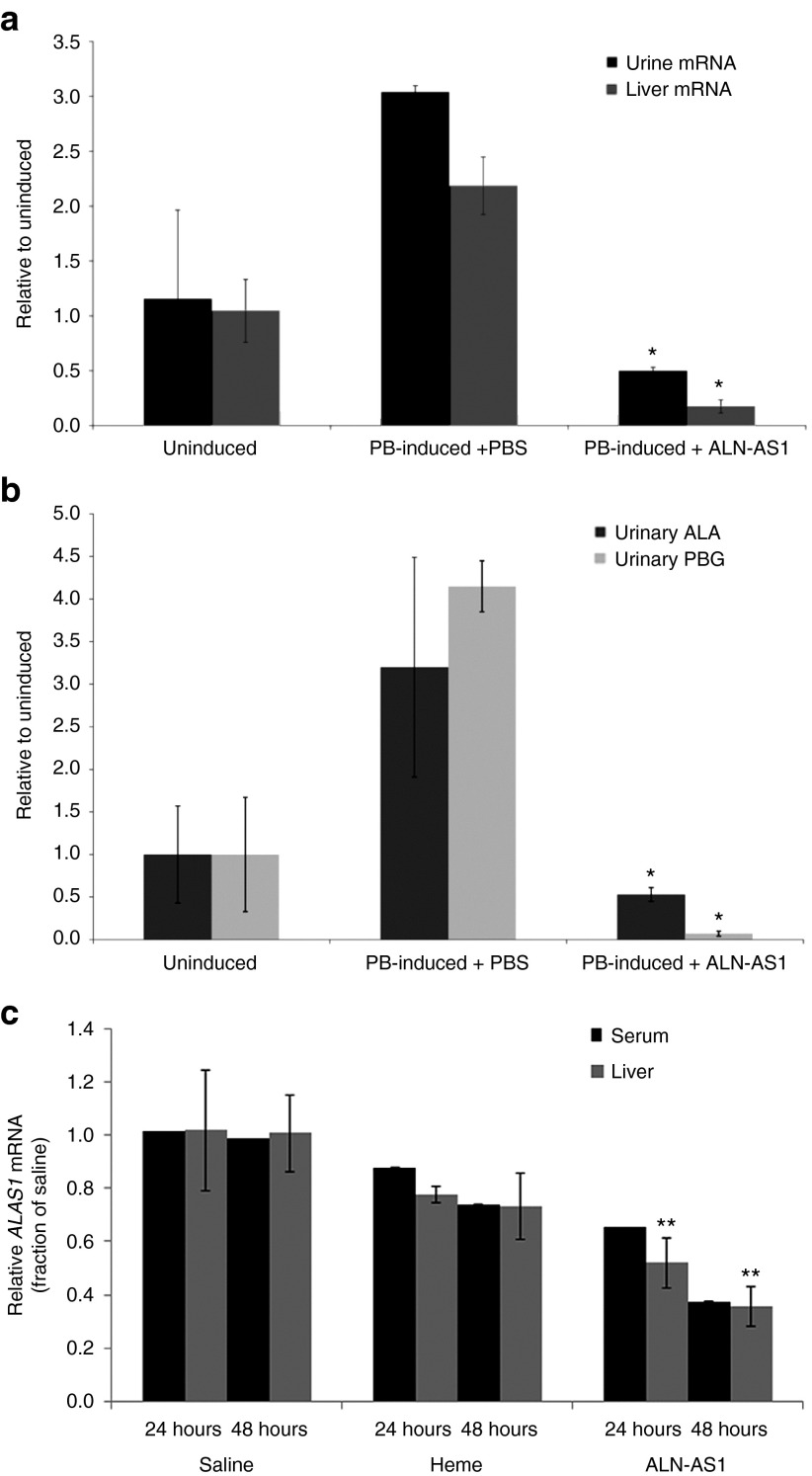
To evaluate if ALN-AS1 could prevent PB-induced hepatic ALAS1 expression in this model and if changes in hepatic ALAS1 expression could be detected in the urine, ALN-AS1 or PBS was administered prophylactically, twice weekly for 3 weeks at a dose of 3 mg/kg. The PBGD siRNA was administered 6 days prior to the final ALN-AS1 dose and was followed by the PB induction phase. As shown in Figure 2a, animals (n = 3 per group) treated with PBS exhibited a 2.5-fold increase in hepatic ALAS1 mRNA over mean baseline levels 24 hours after the final PB dose. In contrast, animals treated with ALN-AS1 maintained baseline ALAS1 liver transcript levels after PB induction. A correlation with liver ALAS1 mRNA was observed at the level of urine, with approximately a threefold induction in ALAS1 mRNA observed in the induced, but untreated animals, and preservation of baseline transcript levels in the prophylactic ALN-AS1 treatment group. Moreover, these results were consistent with the levels of urinary ALA and PBG, with over three- and fourfold inductions, respectively, over mean baseline levels in PBS control animals at 24 hours postfinal PB injection. In contrast, animals treated with prophylactic ALN-AS1 exhibited porphyrin precursor levels comparable to those observed in uninduced animals (Figure 2b). Thus, prophylactic injection of ALN-AS1 completely prevented both the PB-induced upregulation of hepatic ALAS1 expression and the accumulation of urinary ALA and PBG. The ALAS1 mRNA changes in the liver closely correlated with levels of ALAS1 mRNA measured in urine in this induced rat AIP model (Figure 2a).
Figure 2.
Silencing of liver ALAS1 mRNA upon prophylactic and acute administration of ALN-AS1 quantified by circulating mRNA levels in rodent models of acute intermittent porphyria. Wild-type rats were administered ALN-AS1 or phosphate-buffered saline (PBS) prophylactically, twice weekly for 3 weeks. Six days prior to the final dose, all animals were i.v. administered Porphobilinogen deaminase siRNA. This was followed by a challenge with 4-daily phenobarbital (PB) injections to induce ALAS1 levels in all animals, excluding the “Uninduced” cohort. Liver and urine samples were collected 24 hours after the final PB injection. (a) ALAS1 mRNA levels in liver and urine and (b) urinary ALA and porphobilinogen concentrations were measured. Data are presented as mean ± SD (n = 3). *Significance was determined by Student's t-test (P < 0.05). (c) Acute intermittent porphyria mice were biochemically induced with four daily doses of 3,5-diethoxycarbonyl-1,4-dihydro-2,4,6-trimethylpyridine and PB and treated with a single subcutaneous injection of ALN-AS1 (20 mg/kg), saline, or intravenous heme (4.0 mg/kg) (n = 7–10 per group) ~4 hours after the third PB dose. The heme treatment group was administered a second dose of heme 24 hours after the first. Terminal blood collections were made from cohorts sacrificed at 24 and 48 hours post-treatment (n = 7–10/time point), at which times livers were harvested from a subset of mice (n = 4). Serum and liver transcript levels were determined and are presented as mean ± SD. **Significance for ALN-AS1-treated liver samples relative to Heme-treated animals was determined by Student's t-test (P < 0.05).
Detection of urine ALAS1 mRNA changes in a mouse AIP model after ALN-AS1 or heme treatment
To investigate the ability to detect changes in hepatic ALAS1 transcript levels by cERD with acute treatment with ALN-AS1, a well-characterized mouse AIP model was utilized. These mice have two hypomorphic PBGD alleles and about 30% of normal PBGD enzyme activity in their livers.32 A biochemical attack was induced by administering four daily doses of PB and 3,5-diethoxycarbonyl-1,4-dihydro-2,4,6-trimethylpyridine (DDC), an inhibitor of ferrochelatase, the last enzyme in the heme biosynthetic pathway. This combination was previously shown to achieve markedly higher plasma and urinary ALA and PBG concentrations compared to PB alone.32 Following the third PB/DDC dose, the mice were administered a single subcutaneous injection of 20 mg/kg ALN-AS1 or PBS to evaluate the ability of ALN-AS1 to reduce ALAS1 liver transcript levels. A high dose of ALN-AS1 was used to more rapidly decrease ALAS1. As expected, pooled serum samples (n = 7–10 per group, per time point) taken at 24 and 48 hours post-PBS administration show no decrease in ALAS1 transcript, which is mirrored by liver transcript levels in a subset of mice (n = 4) (Figure 2c). In contrast, animals treated with ALN-AS1 exhibited a 40% lowering of liver transcript levels at 24 hours, with further reduction to 65% at 48 hours, relative to PBS-treated animals. These liver ALAS1 data correlate well with circulating serum ALAS1 mRNA levels measured by cERD (Figure 2c).
To directly compare the effectiveness of ALN-AS1 versus heme to reduce the elevated levels of ALAS1 mRNA in the AIP mouse model, an additional cohort of PB/DDC-induced mice (n = 7–10 per group, per time point) was administered either a single IV dose (24-hour sacrifice group) or two IV doses (48-hour sacrifice group) of 4 mg/kg heme. As shown in Figure 2c, a more rapid reduction in serum ALAS1 mRNA was observed with subcutaneously administered ALN-AS1 versus IV administered heme treatment (40 versus 15% AS1 suppression at 24 hours, respectively). This separation between the two treatments was even more pronounced at 48 hours, with 65 versus 25% reduction detected in the serum for ALN-AS1 and heme, respectively. Similar to the PBS and ALN-AS1-treated animals, liver mRNA levels from the heme treatment group were well-correlated with transcript levels in serum at both time points. Collectively, these data suggest that RNAi-mediated ALAS knockdown by ALN-AS1 could work as fast, or faster, than heme in reducing elevated ALA and PBG levels. Moreover, they demonstrate that cERD accurately assayed both the induced expression of hepatic ALAS1 mRNA as well as the kinetics of reduction that occurred with administration of either heme or ALN-AS1.
Monitoring urine or serum ALAS1 mRNA in NHPs after ALN-AS1 dosing
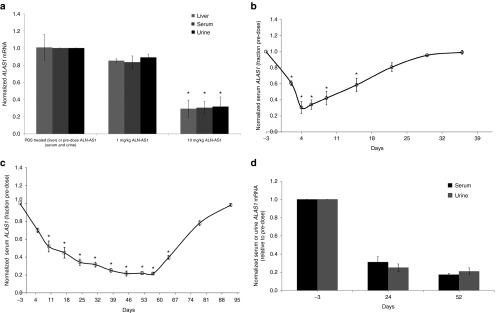
To understand whether this cERD method could be applied to monitor ALAS1 mRNA changes in NHPs, an ALAS1 silencing study was conducted in the cynomolgus monkey (Macaca fascicularis). Animal-matched serum, urine, and liver biopsies were analyzed for ALAS1 transcript levels at select time points (n = 3 per time point). Serum and urine collected prior to dosing served as intra-animal baseline controls, while PBS-treated animals served as normalizing controls for liver biopsy samples. As shown in Figure 3a, treatment with a single subcutaneous injection of 1 or 10 mg/kg ALN-AS1 resulted in reduction in liver, serum, and urine ALAS1 transcript levels, of approximately 20 and 75% respectively, relative to predose serum and urine for circulating RNA levels, or PBS-treated animals for liver transcript levels. Kinetics of ALAS1 mRNA silencing in NHP was also monitored following administration of ALN-AS1using sequential serum and urine collections taken at various time points postsingle or multidose administration (n = 3 per time point). As shown in Figure 3b, animals administered a single 10 mg/kg dose of ALN-AS1 achieved rapid and robust serum ALAS1 suppression, with a nadir of 75% suppression observed by day 4. Full recovery to baseline levels was demonstrated by 28 days postdose. A similar level of ALAS1 suppression was achieved upon administering 8 weekly doses at 5 mg/kg (n = 3 per timepoint). As shown in Figure 3c, a maximum of 80% serum ALAS1 mRNA reduction occurs at 30 days postfirst dose at 5 mg/kg, a level of ALAS1 suppression which is maintained throughout the remainder of the dosing phase, and out to 7 days postfinal dose administration on day 53. Recovery to within 90–95% of baseline levels occurs by 42 days postfinal dose. As shown in Figure 3d, animal-matched urine samples taken at day 24 and day 52 illustrate a clear correlation between circulating ALAS1 transcript in both serum and urine over time, with 75 and 80% mRNA suppression at days 24 and 52, respectively.
Figure 3.
Evaluation of ALAS1 mRNA silencing in circulation and liver in nonhuman primates treated with ALN-AS1. (a) Silencing of liver, serum, and urine ALAS1 mRNA normalized to GAPDH in cynomolgus monkey (M. fascicularis) 96 hours after subcutaneous administration of indicated doses of ALN-AS1. ALAS1 liver mRNA levels from individual animals were normalized to those from a group average of three PBS-treated liver biopsies. Each graphical data point represents the percent remaining ALAS1 mRNA relative to the group average PBS-treated ALAS1 mRNA ± SD of the group. ALAS1 serum and urine transcript from individual animals was normalized to their respective individual predose ALAS1 level. Each graphical data point represents the fraction remaining ALAS1 relative to predose for the group average of three animal samples assayed in technical triplicates ± the standard deviation of the group. *Significance relative to PBS-treated samples for liver transcript and relative to pre-ALN-AS1 treatment for serum and urine was determined by Student's t-test (P < 0.05). (b) Time course of ALAS1 mRNA silencing in cynomolgus monkey serum following a single subcutaneous administration of ALN-AS1 at 10 mg/kg. ALAS1 serum transcript from individual animals was normalized to their respective individual predose ALAS1 level at each timepoint. Each graphical data point represents the fraction remaining ALAS1 relative to predose for the group average of three animal samples assayed in technical triplicates ± the standard deviation of the group. (c) Time course of ALAS1 silencing in cynomolgus monkey serum following subcutaneous administration of 5 mg/kg once weekly for 8 weeks of ALN-AS1 and (d) comparison between serum and urine ALAS1 levels following subcutaneous administration of 5 mg/kg once weekly for 8 weeks of ALN-AS1. Levels of ALAS1 were normalized to those of GAPDH. ALAS1 serum and urine transcript from individual animals was normalized to their respective individual predose ALAS1 level at each timepoint. Each graphical data point represents the fraction remaining ALAS1 relative to pre-dose for the group average of three animal samples assayed in technical triplicates ± the standard deviation of the group. *Significance relative to pre-ALN-AS1 treatment for serum and urine was determined by Student's t-test (P < 0.05).
Validation of cERD method in healthy volunteer and AIP patient samples
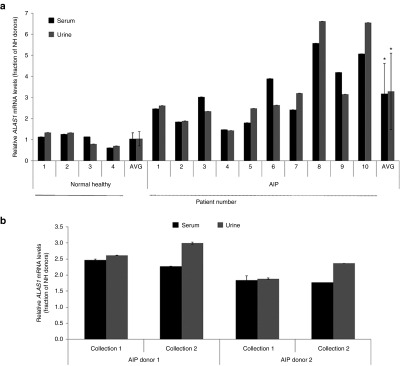
In an effort to understand whether circulating serum and urine ALAS1 mRNA could be detected in human samples, cERD analysis was completed on filtered, donor-matched serum and urine samples from four normal healthy (NH) donors. As shown in Figure 4a, similar levels of ALAS1 mRNA could be detected in both serum and urine of each donor-matched sample. We then assessed ALAS1 transcript levels in urine and serum samples collected from a cohort of ten individuals with biochemically and/or DNA-confirmed AIP, to investigate if there was a detectable upregulation of ALAS1 transcript levels relative to samples from normal healthy donors, as would be predicted. It is important to note that these serum and urine samples were collected from AIP donors at a time when they were not having active symptoms to better understand the basal ALAS1 mRNA levels of these donors, versus the levels that would be observed during a crisis. As illustrated in Figure 4a, samples from AIP donors had generally higher levels of ALAS1 mRNA compared to those of NH samples, with a high degree of correlation between circulating serum and urine levels. On average, the ALAS1 mRNA levels in the urine and serum samples of AIP donors were elevated approximately threefold over those of NH donors. Notably, samples collected from two AIP donors on two separate dates, spaced approximately 3 weeks apart, demonstrated the ability to consistently monitor ALAS1 transcript levels over time for a given individual (Figure 4b).
Figure 4.
Assessment of ALAS1 transcript levels in normal and acute intermittent porphyria (AIP) human donor-matched serum and urine samples. (a) ALAS1 mRNA levels in filtered AIP donor-matched serum and urine (n = 10) relative to those from normal healthy volunteers (n = 4). Values are relative to GAPDH levels; error bars, SD in experimental replicates. *Significance was determined by Student's t-test (P < 0.05). (b) Comparison of ALAS1 mRNA levels in AIP donor-matched, circulating serum and urine collected at two different time points, 3 weeks apart (n = 2). Values are relative to GAPDH levels; error bars, SD in experimental replicates.
Discussion
In this report, we demonstrate that the cERD assay provides an accurate and noninvasive way to monitor changes in liver ALAS1 mRNA with ALN-AS1 treatment in several preclinical species. Accurate detection of tissue RNA status from circulating RNA is particularly advantageous in the case of ALAS1 since it is not a secreted protein, and thus serial liver biopsies would be required to determine the kinetics of target downregulation and recovery after treatment with ALN-AS1. Importantly, the cERD assay was also able to detect increased basal ALAS1 mRNA levels in urine and serum samples from AIP patients compared to healthy volunteers, suggesting that it may also have utility in the clinic by providing more extensive data on ALAS1 changes in AHP patients, with and without therapeutic interventions.
The cERD assay was able to accurately characterize the kinetics of ALAS1 transcript silencing in rodents following treatment with ALN-AS1. Whereas this type of characterization would typically require terminal liver harvest from a designated cohort of animals at each specified time point, we were instead able to monitor the pharmacodynamic response using serial urine collections from a single group of animals over time for these studies. Importantly, the cERD assay demonstrated consistent results regardless of whether serum or urine was examined. We optimized the urine cERD assay by employing a lyophilization procedure to concentrate the urine. This may reduce the need for serial blood collections from individual animals, which is invasive and subject to volume restrictions. Changes in ALAS1 transcript were also successfully monitored in a rat model of induced AIP, which more closely represents a clinically relevant state. Prophylactic injection of ALN-AS1 completely prevented the phenobarbital (PB)-induced upregulation of hepatic ALAS1 expression monitored in urine, as well as the accumulation of plasma and urinary ALA and PBG metabolites. The cERD method also detected more rapid and robust lowering in ALAS1 mRNA with ALN-AS1, as compared to a clinical dosing regimen of heme in a mouse model of AIP. Although the administered dose of 20 mg/kg ALN-AS1 is relatively high, the results provide important proof of concept that the onset of effect for ALN-AS1 may be rapid enough to enable treatment of an established acute attack. Additional dose titration studies in the rodent disease models have shown that weekly dosing of approximately 1–3 mg/kg are sufficient to completely prevent ALA/PBG induction (data not shown) suggesting that it should also be possible to titrate lower doses for acute treatment.
NHPs have been shown to be the most predictive species for the pharmacodynamic responses of RNAi therapeutics in humans, however, obtaining serial liver biopsies to evaluate the activity of ALN-AS1 is invasive and impractical.29,30 The cERD method detected a rapid nadir for ALAS1 suppression around day 4 following a single ALN-AS1 dose, and a sustained knockdown of the ALAS1 target following weekly dosing with ALN-AS1. It is possible that mild, chronic suppression of ALAS1 with weekly, or potentially less frequent dosing, may prevent the induction of acute attacks in severe patients who suffer from multiple recurrent attacks. Notably, transcript recovery back to baseline occurred within a month following discontinuation of dosing, and importantly, no evidence of overshooting or a rebound effect was observed.
To determine the clinical utility of the cERD assay, we demonstrated the ability to detect the ALAS1 transcript in human donor-matched serum and urine samples. A strong correlation between the urine and serum mRNA levels was observed. In addition, we observed limited variability in samples from the same patient collected on different days, which is important if the assay is to be used to monitor transcript changes after ALN-AS1 dosing. Baseline ALAS1 levels were threefold higher in the serum or urine samples obtained from AIP patients compared to normal healthy (NH) donors. These increased values are very consistent with recent findings of ALAS1 mRNA levels directly monitored in an explanted liver from an AIP patient.31 Furthermore, a review of the deidentified medical records for each of the AIP donors suggested that patients with more disease activity tended to have higher ALAS1 mRNA upregulation (Supplementary Table S1). These data are also consistent with the prior literature demonstrating the central role of ALAS1 upregulation in the pathophysiology of AIP attacks, and strengthens the therapeutic hypothesis for an RNAi therapeutic targeting ALAS1.13,14,15,16,31
While much is known about ALAS1 regulation from in vitro work in hepatocytes, and from in vivo animal models of AIP, little is known about how ALAS1 mRNA changes in AIP patients over the course of their disease. The ability to noninvasively detect serum and urine ALAS1 mRNA in human samples using the cERD assay may facilitate an enhanced understanding of the pathogenesis of AHP, as well as aid in the clinical development of ALN-AS1, by providing more complete data on ALAS1 mRNA changes in the liver at baseline, and following interventions like ALN-AS1. As part of an ongoing natural history study, urine, and serum samples are being collected from AIP patients with multiple recurrent attacks. These samples are being analyzed by cERD to further characterize the kinetics of ALAS1 change throughout the course of their disease (Trial Search (Internet). EXPLORE c.2014- (cited 2015) https://clinicaltrials.gov/ct2/show/NCT02240784?term=porphyria+natural+history&rank=1). Since ALAS1 is also upregulated in patients with other hepatic porphyrias such as variegate porphyria and hereditary coproporphyria, it is possible that ALN-AS1 may impact those diseases as well. The cERD assay described herein can help establish ideal ALN-AS1 dose levels and regimens through monitoring ALAS1 suppression. The collective data highlight the potential utility of this assay to monitor liver ALAS1 gene modulation, broaden our understanding of AIP disease pathophysiology, and aid in the clinical development of novel therapeutics such as ALN-AS1, for the treatment of the acute hepatic porphyrias.
Materials and methods
Nonhuman primate studies. All NHP studies were conducted at Charles River Laboratories in experimentally naive male Chinese cynomolgus monkeys (Macaca fascicularis) between 2 to 4 years of age and 2 to 4 kg in body weight. Animals were housed in accordance with the USDA Animal Welfare Act (9 Code of Federal Regulations, parts 1, 2, and 3) and as described in the Guide for the Care and Use of Laboratory Animals. Subcutaneous ALN-AS1 was administered at a dose volume of 2 ml/kg as a series of subcutaneous bolus doses, divided into two or three dose sites of ≥1.2 ml and ≤2 ml per injection site, into the scapular and mid-dorsal regions. Where applicable, PBS served as the control article and was administered at a dose volume equal to the respective test article volume. Serum, urine, and/or liver biopsies were collected at specified time points for downstream pharmacodynamic analyses.
Rodent AIP model studies
Rat AIP model. Wild-type (Sprague Dawley) female rats were injected subcutaneously with either 3 mg/kg ALN-AS1 or PBS biweekly for 3 weeks on days 0, 3, 7, 10, 14, and 17. On day 11, all groups were treated intravenously with a Porphobilinogen deaminase-targeting (PBGD) siRNA in an AF-011 lipid nanoparticle to decrease the levels of PBGD in liver to mimic the inherited enzyme deficiency in patients with AIP. Following PBGD knockdown for 72 hours, rats were administered four daily intraperitoneal doses of Phenobarbital (PB) (75 mg/kg) on days 14, 15, 16, and 17 to induce hepatic ALAS1 levels. Overnight urines were collected day18, the morning after the fourth dose of PB, to measure ALA and PBG and ALAS1 mRNA levels. Animals were then sacrificed on day 18, and serum and liver were harvested to measure ALAS1 transcript.
Mouse AIP model. 32,36 The previously described T1/T2 AIP mice were bred and housed in a barrier facility at the Icahn School of Medicine at Mount Sinai. Animal procedures were reviewed and approved by the Icahn School of Medicine at Mount Sinai Institutional Animal Care and Use Committee. Combined PB/DDC inductions were performed in male AIP mice (40–120 days old) as previously described.32 DDC (20 mg/kg/day) and PB (90, 100, 110, and 90 mg/kg/day) were administered on days 0, 1, 2, and 3, and mice were treated with either a single dose of subcutaneous ALN-AS1 (20 mg/kg), saline, or intravenous heme (4.0 mg/kg) on day 2, ~4 hours after the third PB/DDC dose. The heme-treated mice received a second heme infusion (4.0 mg/kg) 24 hours after the first, unless they were sacrificed at this specific time point. Terminal blood collections were performed at the indicated times, after deeply anesthetizing the mice with ketamine/xylazine. A subset of mice was subsequently perfused with saline, and their livers were harvested.
DDC was resuspended in corn oil and administered by oral gavage, while PB was administered intraperitoneally. Heme (Panhematin) was reconstituted in a 25% human albumin solution immediately before use, in amber tubes, and administered via tail vein injections. The siRNA and heme formulations were diluted accordingly so that the mice consistently received a volume of 0.01 ml/g weight.
Urine ALA and PBG measurements. Urine ALA and PBG were measured using a commercial kit from BIORAD (Hercules, CA) as per manufacturer's instructions. ALA standards and samples were run as per the manufacturer's instructions with minor variation. PBG standards (Sigma Aldrich, St. Louis, MO) were diluted serially from 500 mg/l. Urinary creatinine levels were measured using the Olympus AU400 Chemistry Analyzer (Olympus, Center Valley, PA).
Serum/urine samples and liver biopsy collections. Terminal rodent blood samples were collected by cardiac puncture, while nonterminal NHP or human blood samples were collected from a peripheral vessel into tubes containing no additive (serum separator tubes). Blood samples were allowed to clot at room temperature for at least 30 minutes and then centrifuged within 1 hour of collection. Serum was harvested (~3 ml rodent serum, 4 ml NHP serum, and ~10 ml human serum) and pooled for the mouse studies (n = 7–10). Samples were snap frozen on dry ice and stored at −70 °C until determination of serum ALAS1 levels.
All rodent and NHP urine samples were collected overnight on ice at the specified time points. First morning, urine samples were collected from normal healthy volunteers and AIP patients. Samples were frozen as a single aliquot in a 50 ml conical tube, and stored at −70 °C until determination of urine ALAS1 levels (~15–20 ml rodent urine, ~30 ml of NHP and human urine).
Livers were harvested from terminal rodent studies, while nonterminal NHP liver biopsies were collected as per the contract facilities' protocols at the specified time points. The liver tissues were flash frozen in liquid nitrogen and stored in a freezer at −70 °C until determination of ALAS1 levels.
Liver AS1 transcript measurement methods. The liver samples were homogenized and lysed in 700 µl Qiazol (Qiagen, Valencia, CA) using a TissueLyser processor for 2 minutes at 50 Hz, as per general recommendations from the manufacturer (Qiagen). One hundred and forty microliters of chloroform were thoroughly mixed with each sample, and lysates were cleared by centrifugation at full speed for 15 minutes in a table top centrifuge. Total RNA was isolated from the supernatant using the RNeasy Plus Micro kit (QIAGEN# 74034) according to the manufacturer's protocol. RNA concentrations were determined using a NanoDrop Spectrophotometer and 500 ng was used for cDNA synthesis (High Capacity cDNA synthesis kit, ABI #4374967). Singleplex qPCR was performed using 50 ng of cDNA and the Roche LightCycler480 Probes Master Mix in the Roche 480 LightCycler Real Time PCR System (Roche, Branchburg, NJ). Commercially available ALAS1 (catalog # Rn00577936_M1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (4352338E) TaqMan primer/probes from Life Technologies (Grand Island, NY) were used for analyses of rat samples, while a rhesus ALAS1 Taqman primer probe (Rh00963540_m1, Life Technologies) and a custom designed Cynomolgus monkey-specific GAPDH TaqMan primer/probe (Table 1) were used for NHP analyses. Relative ALAS1 transcript levels were determined by the comparative Ct method, using GAPDH as an internal control. mRNA levels from treated biopsies were expressed as the percent remaining ALAS1 mRNA relative to those from a group of PBS-treated animals (group average ± SD). ALAS1 Ct values from naive matrices across species ranged from 20 to 22 for liver transcript levels.
Table 1. Custom designed Taqman primer and probe sequences for detection of cynomolgus GAPDH.

Lyophilization of urine samples for circulating extracellular RNA extraction. 0.2-micron filtered urine samples were lyophilized over a 2- to 3-day period using a Labconco 12-L FreeZone Freeze Dry Console (Kansas City, MO). Lyophilized samples were resuspended in 8 ml of Qiagen nuclease-free water (Valencia, CA) and subjected to differential centrifugation. Samples were first centrifuged at 3,000×g for 10 minutes. The resulting supernatants were centrifuged at 17,000×g for 20 minutes. Exosomes were isolated from the supernatants of the second centrifugation as outlined in the “AS1 Circulating Extracellular RNA Detection” method listed below.
AS1 circulating extracellular RNA detection methods. 0.2-micron filtered serum samples were thawed on ice and centrifuged at 3,000×g for 10 minutes, and lyophilized urine samples prepared as previously described. Lithium chloride (Life Technologies) was added to all serum and urine samples at a final concentration of 1M. Following 1 hour incubation on ice, the samples were ultracentrifuged at 200,000×g for 120 minutes (Beckman Coulter, Brea, CA), and supernatants were decanted. One milliter of trizol (Life Technologies) was added to the pellets, and the samples were vortexed for 60 seconds. Samples were transferred to 1.5 ml microcentrifuge tubes (Life Technologies), and 0.2 ml chloroform (Sigma Aldrich) was added to each sample and mixed thoroughly by inversion. Samples were centrifuged at 16,000×g at 4 °C for 20 minutes. The upper aqueous phase was transferred to a fresh 1.5 ml ultracentrifuge tube and precipitated with an equal volume of isopropanol, 1 μl of GenElute Linear Polyacrylamide (Sigma Aldrich), and 1/10th volume of 3 M sodium acetate, pH 5.5 or less. Samples were centrifuged at 16,000×g at 4 °C for 10 minutes. The resulting RNA pellet was washed twice with ice cold 70% ethanol, air-dried, and resuspended in 20 μl RNase-free water (Life Technologies). 10 μl was used for cDNA synthesis (High Capacity cDNA synthesis kit, Life Technologies). 2 μl of cDNA was used for singleplex qPCR reactions, which were performed as described above. Commercially available TaqMan primer/probes from Life Technologies were used to quantitate human ALAS1 (catalogue#: HS00963534-M1) and GAPDH (4352934E) transcripts. The comparative Ct method was used to quantify fold change relative to an untreated control calibrator. The ALAS1 mRNA levels were normalized to those of GAPDH. ALAS1 Ct values from naive matrices across species ranged from 28 to 30 for serum mRNA and 30 to 32 for urine mRNA levels.
Human serum and urine sample acquisition. All deidentified samples from human donors were obtained through third-party contractors according to facility-established protocols. Donor-matched serums (10 ml) and urines (30 ml) from Normal Healthy donors were acquired from Bioreclamation IVT (Baltimore, MD). The same donor-matched matrices and deidentified medical records were obtained from confirmed AIP patients through Sanguine Biosciences (Valencia, CA).
SUPPLEMENTARY MATERIAL Table S1. Correlation of serum ALAS1 transcript levels in AIP donors with de-identified medical record information.
Acknowledgments
We thank the American Porphyria Foundation and Sanguine Biosciences for their assistance in acquiring donor-matched serum and urine samples, as well as facilitating the transfer of deidentified medical records from confirmed acute intermittent porphyria patients. This work was supported in part by a National Institutes of Health (NIH) Career Development Award (K01 DK087971) to M.Y. A.C., A.L., T.R., M.M., S.K, D.F., S.M., K.C., A.S., M.M., R.M., K.F., A.S., and W.Q. are employees and shareholders of Alnylam Pharmaceuticals. M.Y. and R.J.D. are consultants to Alnylam Pharmaceuticals.
Supplementary Material
Correlation of serum ALAS1 transcript levels in AIP donors with de-identified medical record information.
References
- Anderson, KE, Sassa, S, Bishop, DF, Desnick, RJ (2001). Disorders of heme biosynthesis: X linked sideroblastic anemia and the porphyrias. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds.). The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill, New York. pp. 2961–3062. [Google Scholar]
- Granick, S (1963). Induction of the synthesis of delta-aminolevulinic acid synthetase in liver parenchyma cells in culture by chemical that induce acute porphyria. J Biol Chem 238: 2247–2249. [PubMed] [Google Scholar]
- de Matteis, F (1967). Disturbances of liver porphyrin metabolism caused by drugs. Pharmacol Rev 19: 523–557. [PubMed] [Google Scholar]
- Sassa, S, Bradlow, HL and Kappas, A (1979). Steroid induction of delta-aminolevulinic acid synthase and porphyrins in liver. Structure-activity studies and the permissive effects of hormones on the induction process. J Biol Chem 254: 10011–10020. [PubMed] [Google Scholar]
- Miller, LK and Kappas, A (1974). The effect of progesterone on activities of delta-aminolevulinic acid synthetase and delta-aminolevulinic acid dehydratase in estrogen-primed avian oviduct. Gen Comp Endocrinol 22: 238–244. [DOI] [PubMed] [Google Scholar]
- Handschin, C, Lin, J, Rhee, J, Peyer, AK, Chin, S, Wu, PH et al. (2005). Nutritional regulation of hepatic heme biosynthesis and porphyria through PGC-1alpha. Cell 122: 505–515. [DOI] [PubMed] [Google Scholar]
- Roveri, G, Nascimbeni, F, Rocchi, E and Ventura, P (2014). Drugs and acute porphyrias: reasons for a hazardous relationship. Postgrad Med 126: 108–120. [DOI] [PubMed] [Google Scholar]
- Bishop, DF and Desnick, RJ (1982). Assays of the heme biosynthetic enzymes. Preface. Enzyme 28: 91–93. [PubMed] [Google Scholar]
- Lathrop, JT and Timko, MP (1993). Regulation by heme of mitochondrial protein transport through a conserved amino acid motif. Science 259: 522–525. [DOI] [PubMed] [Google Scholar]
- Yamamoto, M, Kure, S, Engel, JD and Hiraga, K (1988). Structure, turnover, and heme-mediated suppression of the level of mRNA encoding rat liver delta-aminolevulinate synthase. J Biol Chem 263: 15973–15979. [PubMed] [Google Scholar]
- Drew, PD and Ades, IZ (1989). Regulation of the stability of chicken embryo liver delta-aminolevulinate synthase mRNA by hemin. Biochem Biophys Res Commun 162: 102–107. [DOI] [PubMed] [Google Scholar]
- Bissell, DM, Lai, JC, Meister, RK, Blanc, PD (2015). Role of delta-aminolevulinic acid in the symptoms of acute porphyria. Am J Med 128: 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowman, JK, Gunson, BK, Bramhall, S, Badminton, MN and Newsome, PN (2011). Liver transplantation from donors with acute intermittent porphyria. Ann Intern Med 154: 571–572. [DOI] [PubMed] [Google Scholar]
- Seth, AK, Badminton, MN, Mirza, D, Russell, S and Elias, E (2007). Liver transplantation for porphyria: who, when, and how? Liver Transpl 13: 1219–1227. [DOI] [PubMed] [Google Scholar]
- Soonawalla, ZF, Orug, T, Badminton, MN, Elder, GH, Rhodes, JM, Bramhall, SR et al. (2004). Liver transplantation as a cure for acute intermittent porphyria. Lancet 363: 705–706. [DOI] [PubMed] [Google Scholar]
- Stojeba, N, Meyer, C, Jeanpierre, C, Perrot, F, Hirth, C, Pottecher, T et al. (2004). Recovery from a variegate porphyria by a liver transplantation. Liver Transpl 10: 935–938. [DOI] [PubMed] [Google Scholar]
- Anderson, KE, Bloomer, JR, Bonkovsky, HL, Kushner, JP, Pierach, CA, Pimstone, NR et al. (2005). Recommendations for the diagnosis and treatment of the acute porphyrias. Ann Intern Med 142: 439–450. [DOI] [PubMed] [Google Scholar]
- Mustajoki, P, Tenhunen, R, Tokola, O and Gothoni, G (1986). Haem arginate in the treatment of acute hepatic porphyrias. Br Med J (Clin Res Ed) 293: 538–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkowsky, HL, Tschudy, DP, Collins, A, Doherty, J, Bossenmaier, I, Cardinal, R et al. (1971). Repression of the overproduction of porphyrin precursors in acute intermittent porphyria by intravenous infusions of hematin. Proc Natl Acad Sci USA 68: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustajoki, P and Nordmann, Y (1993). Early administration of heme arginate for acute porphyric attacks. Arch Intern Med 153: 2004–2008. [PubMed] [Google Scholar]
- Goetsch, CA and Bissell, DM (1986). Instability of hematin used in the treatment of acute hepatic porphyria. N Engl J Med 315: 235–238. [DOI] [PubMed] [Google Scholar]
- Elbashir, SM, Harborth, J, Lendeckel, W, Yalcin, A, Weber, K and Tuschl, T (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498. [DOI] [PubMed] [Google Scholar]
- Fire, A, Xu, S, Montgomery, MK, Kostas, SA, Driver, SE and Mello, CC (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Sehgal, A, Vaishnaw, A and Fitzgerald, K (2013). Liver as a target for oligonucleotide therapeutics. J Hepatol 59: 1354–1359. [DOI] [PubMed] [Google Scholar]
- Vaishnaw, AK, Gollob, J, Gamba-Vitalo, C, Hutabarat, R, Sah, D, Meyers, R et al. (2010). A status report on RNAi therapeutics. Silence 1: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc, A, Querbes, W, De, S, Qin, J, Frank-Kamenetsky, M, Jayaprakash, KN et al. (2010). Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther 18: 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman, M, Ansell, SM, Mui, BL, Tam, YK, Chen, J, Du, X et al. (2012). Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem Int Ed Engl 51: 8529–8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, JK, Willoughby, JL, Chan, A, Charisse, K, Alam, MR, Wang, Q et al. (2014). Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc 136: 16958–16961. [DOI] [PubMed] [Google Scholar]
- Coelho, T, Adams, D, Silva, A, Lozeron, P, Hawkins, PN, Mant, T et al. (2013). Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med 369: 819–829. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, K, Frank-Kamenetsky, M, Shulga-Morskaya, S, Liebow, A, Bettencourt, BR, Sutherland, JE et al. (2014). Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: a randomised, single-blind, placebo-controlled, phase 1 trial. Lancet 383: 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda, M, Erwin, A, Liu, LU, Balwani, M, Chen, B, Kadirvel, S et al. (2015) Liver transplantation for acute intermittent porphyria: biochemical and pathologic studies of the explanted liver. Mol Med 21: 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda, M, Gan, L, Chen, B, Kadirvel, S, Yu, C, Phillips, JD et al. (2014). RNAi-mediated silencing of hepatic Alas1 effectively prevents and treats the induced acute attacks in acute intermittent porphyria mice. Proc Natl Acad Sci USA 111: 7777–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, DD and Gercel-Taylor, C (2013). The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Front Genet 4: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salido-Guadarrama, I, Romero-Cordoba, S, Peralta-Zaragoza, O, Hidalgo-Miranda, A and Rodríguez-Dorantes, M (2014). MicroRNAs transported by exosomes in body fluids as mediators of intercellular communication in cancer. Onco Targets Ther 7: 1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal, A, Chen, Q, Gibbings, D, Sah, DW and Bumcrot, D (2014). Tissue-specific gene silencing monitored in circulating RNA. RNA 20: 143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg, RL, Porcher, C, Grandchamp, B, Ledermann, B, Bürki, K, Brandner, S et al. (1996). Porphobilinogen deaminase deficiency in mice causes a neuropathy resembling that of human hepatic porphyria. Nat Genet 12: 195–199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation of serum ALAS1 transcript levels in AIP donors with de-identified medical record information.