Abstract
CRISPR/Cas9 is a versatile genome-editing technology that is widely used for studying the functionality of genetic elements, creating genetically modified organisms as well as preclinical research of genetic disorders. However, the high frequency of off-target activity (≥50%)—RGEN (RNA-guided endonuclease)-induced mutations at sites other than the intended on-target site—is one major concern, especially for therapeutic and clinical applications. Here, we review the basic mechanisms underlying off-target cutting in the CRISPR/Cas9 system, methods for detecting off-target mutations, and strategies for minimizing off-target cleavage. The improvement off-target specificity in the CRISPR/Cas9 system will provide solid genotype–phenotype correlations, and thus enable faithful interpretation of genome-editing data, which will certainly facilitate the basic and clinical application of this technology.
Keywords: CRISPR/Cas9, off-target
The CRISPR (clustered, regularly interspaced, short palindromic repeats)/Cas9 system, which is found in diverse bacterial and archaeal species, has been used successfully to edit eukaryotic genomes.1,2 It now also holds great promise in diverse fields such as animal disease modeling, material science, genetically modified plant technology, biofuel technology, gene therapy, and drug development. In addition, CRISPR/Cas9 technology has substantially accelerated the understanding of functional organization of the genome at the systems level and thus helps to establish solid causal links between genetic variations and biological phenotypes.3,4,5,6 However, off-target mutations observed at frequencies greater than the intended mutation, which may cause genomic instability and disrupt the functionality of otherwise normal genes, is still one major concern when applying CRISPR/Cas9 system to biomedical and clinical application.7,8,9,10,11,12
Mechanism of Off-Target Effects In Crispr/Cas9 System
The CRISPR/Cas9 system functions as the RNA-based adaptive immune system in bacteria and archaea.13 The type II CRISPR system, which includes CRISPR-associated nuclease 9 (Cas9), is derived from Streptococcus pyogenes. Native CRISPR system confers resistance to viruses by incorporating short repeats of the viral DNA into the bacterial genome. When a bacterial colony is infected the second time, transcripts of these repeats direct a nuclease to the complementary DNA from the invading virus and thus destroy the viral DNA.14,15 To enable its gene-targeting capacity in the eukaryotic cell, the CRISPR/Cas9 system can be reconstituted in mammalian cells using the following three minimal components: Cas9, a specificity-determining CRISPR RNA (crRNA), and an auxiliary trans-activating RNA (tracrRNA).1,2,8 The crRNA and tracrRNA duplexes can also be fused to generate a chimeric single-guide RNA (sgRNA). The first ~20 nucleotides of the sgRNA are complementary to the target DNA sequence, followed by a sequence called the protospacer adjacent motif (PAM), typically NGG.16,17,18
Although the targeting specificity of Cas9 is believed to be tightly controlled by the 20-nt guide sequence of the sgRNA and the presence of a PAM adjacent to the target sequence in the genome, potential off-target cleavage activity could still occur on DNA sequence with even three to five base pair mismatches in the PAM-distal part of the sgRNA-guiding sequence.2,7,8,9,10 Moreover, previous studies have demonstrated that different guide RNA structures can affect the cleavage of on-target and off-target sites.8 Crystal structure studies and single-molecule DNA curtain experiments suggest that while the PAM site is essential for the initiation of Cas9 binding, the seed sequence corresponding to 3′ end of the crRNA complementary recognition sequence, directly adjacent to PAM, is also critical for subsequent Cas9 binding, R-loop formation, and activation of nuclease activities in Cas9.19,20
sgRNA
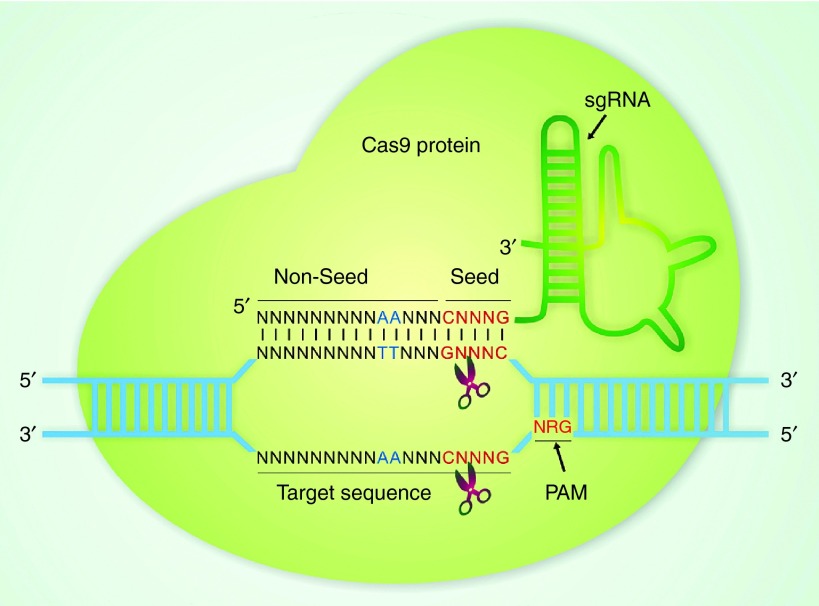
SgRNA comprises the seed sequence and nonseed sequence (Figure 1). Several initial studies have shown that the 10–12 base pairs adjacent to the PAM (3′ end of the guide RNA), called the “seed sequence”, determine Cas9 specificity and is generally more important than the rest of the guide RNA sequences.2,15 However, chromatin immunoprecipitation followed by sequencing (ChIP-seq) of DNA bound to catalytically dead Cas9 (dCas9) in murine embryonic stem cells demonstrates that only one to five base pairs of the immunoprecipitated DNA match to the guide region, suggesting that the one to five base pairs of guide region proximal to the PAM mark the true “seed region”.21 These results were also confirmed by Yu Zhang and his colleagues by ChIP-seq analysis of human genome after targeting with CRISPR/Cas922, and were consistent with the previous observations that base-pairing adjacent to the PAM is critical for DNA targeting.17,23 Yet the ChIP-seq assay for detecting off target sites of dCas9 only capture the PAM-proximal binding event but not cleavage event, leading to over-prediction of off-target sites. Pelletier and his colleagues reported that sequences distal to PAM, which possibly triggers a conformational change of Cas9, engage CRISPR/Cas9 DNA target cleavage, and thus using sgRNAs that places equal emphasis on seed sequence and PAM-distal target sequences will lead to lower off-target editing compared to ChIP-seq data only using seed regions.24 The results could be considered to be a supplement to what Sharp and colleagues have reported in their genome-wide binding maps of dCas9 study through ChIP-seq in mouse embryonic stem cells, in which they have revealed well-defined seed region for target binding and very abundant off-target binding sites, however, majority of which do not undergo substantial cleavage by Cas9.21
Figure 1.
Base preferences for CRISPR/Cas9 system. Cas9 protein is shown in orange. PAM and seed sequences are shown in red. Base preferences are shown in blue. Scissors indicate cleavage sites. The single-letter DNA code is used (N=A, T, C, G; R=G or A).
The seed sequence influences the specificity of Cas9-sgRNA binding through multiple potential mechanisms. The sequence of the seed region determines the frequency of a “seed + NGG” in the genome, and controls the effective concentration of the Cas9-sgRNA complex (Cas9 binding or sgRNA abundance and specificity).21,25 Meanwhile, U-rich seeds are likely to result in decreased sgRNA abundance and increased specificity since multiple U's in the sequence can induce termination of sgRNA transcription.21,25 In general, mismatches of the one to five base pairs at the 5′ end of sgRNAs are better tolerated than those at the 3′ end. Single and double mismatches are tolerated to various degrees depending on their position along the guide RNA-DNA interface.10,15,23 It has also been reported that sgRNAs with exceptionally low or high GC content tends to be less active.24,26 In a study of CRISPR/Cas9-mediated mutagenesis in Drosophila, Ren et al. observed positive correlation between mutagenesis efficiency and GC content of the most proximal region to the PAM sequence of the sgRNAs, based on their evidence that sgRNAs with at least four GCs in the sequence of the six base pairs that are most proximal to the PAM sequence have a heritable mutation (that means mutation generated by effective sgRNA of the CRISPR/Cas9 system from parents is able to pass on to the next generation) rate over 60%, suggesting that effective sgRNAs can be selected according to the GC content of the sequence proximal to PAM.27 When choosing an appropriate sgRNA sequence, guanine is strongly preferred, and cytosine is strongly unfavorable, as the first base of the seed sequence immediately adjacent to the PAM. Conversely, there is a preference for cytosine, but not guanine, at position 5 that is fifth base proximal to PAM. Adenine is favored in the middle of the sgRNA, and cytosine is disfavored at position 18.21,25,26,28 (Figure 1) These design principles are probably based upon the theory that guanine-rich sequences can fold into stable noncanonical structures called G-quadruplexes in vivo, which contributes to sgRNA stability.29 However, this base preference is largely depending on the target site in many cases,10 and the tail sequence of tracrRNA is also very critical for Cas9 activity in vivo.8
Pam
The sequence of the PAM also influences the activity of sgRNA (Figure 1). Initial results indicated that NGG (N is A, T, C, or G) is the canonical sequence for the PAM. However, recent studies have suggested that the type II CRISPR system can also use NRG (Figure 1) (where R is G or A) as PAM sequence, albeit with only one-fifth of the binding efficiency compared to NGG. Several studies reported that the NRG sequence is the predominant noncanonical PAM for CRISPR/Cas9-mediated DNA cleavage at the human EMX locus.8,23 The binding frequency of each base in the PAM sequence is different. The first nucleotide is the least conserved, with G in nearly 50% of binding sites, while the second position with G in >90% of the binding sites,8,30 suggesting that NRG is not the optimal PAM for the designing of CRISPR/Cas9 sequences. Therefore, the exact effect of NRG PAM sequence on DNA cleavage of Cas9 is largely unclear.31
When sgRNAs corresponding to a target DNA site are designed via common CRISPR/Cas9 design tools (Table 1), every sgRNA has its own PAM, typically NGG. However, in practice, alternatively NGG may be not exist corresponding to the effective sgRNA if we want to achieve precise insertion or precise point mutation in the genome, and thus NRG (R=G or A) sequence can be considered as alternative, albeit low cleavage efficiency.31 In the cases that both NGG and NRG(R=G or A) are not able to provide optimized design of sgRNAs, one may choose the other CRISPR/Cas9 system (Cas9 orthologues, Streptococcus thermophilus Cas9 and Staphylococcusaureus Cas9). When applying these Cas9 orthologues for gene editing, NGA, NAC (Figure 2) can be considered without causing higher off-target effects as compared to wild-type SpCas9.32
Table 1. Common CRISPR/Cas9 design tools.
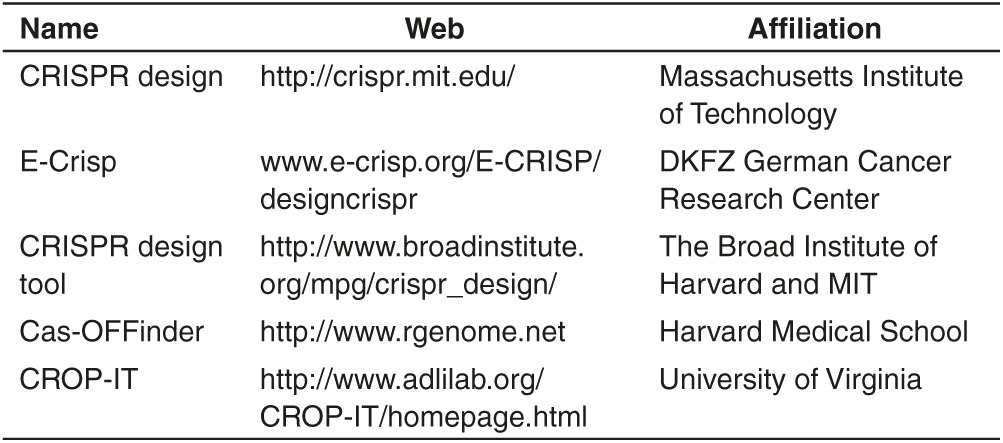
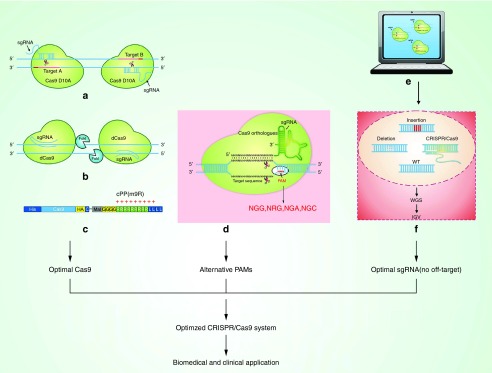
Figure 2.
Methods for optimal CRISPR/Cas9 system for biomedical and clinical application. (a) Double nicking by RNA-Guided CRISPR Cas9 for enhanced genome-editing specificity. Cas9 protein is shown in orange. Scissors indicate cleavage sites. (b) Fusion of catalytically inactive Cas9 to Fok I nuclease. (c) Delivery of Cas9 protein. cPP, cell-penetrating peptide. The single-letter codes for amino acids are used. C, cysteine; G, glycine; R, arginine; L, leucine. His, Histidine tag; HA, hemagglutinin tag; Mal, maleimide. (d) Alternative PAMs for CIRSPR/Cas9 system. (e) SgRNA design—CRISPR/Cas9 design tools. (f) Method of off-target detection-Degenome-seq. IGV, Integrative Genomics Viewer; WGS, whole-genome sequencing; WT, wild type.
Cas9 Protein and Other Factors
Direct delivery of purified Cas9 protein and sgRNA into cells has been reported to result in reduced off-target effects compared to the delivery of plasmid sequences encoding Cas9 and sgRNA, because Cas9-sgRNA ribonucleoprotein complexes cleave chromosomal DNA almost immediately after delivery and are degraded rapidly in cells.33,34
Off-target effects might be cell-type-specific and highly depending on the integrity of double-stranded breaks (DSBs) repair pathways of particular cell type.22 For example, nuclease off-target effects can occur in transformed human cell lines with dysregulated DSBs repair pathways, while whole-genome sequencing of healthy human pluripotent stem cell clones with relatively intact DSBs repairing capability have revealed very few off-target mutations attributable to the nucleases.35,36 Furthermore, it has been reported that methylation of DNA at CpG sites may impede the binding efficiency of Cas9 in cells, and small molecules that enhance CRISPR genome editing by promoting precise genome editing via homology-directed repair (e.g., L755507, a 73-adrenergic receptor agonist, and Brefeldin A, an inhibitor of intracellular protein transport from the ER to the Golgi apparatus) or sequence-specific gene knockout via nonhomologous end-joining (NHEJ) (e.g., azidothymidineorTrifluridine) in pluripotent stem cells are being studied.21,22,30,37 Moreover, toward the goal of achieving more efficient genome editing with CRISPR/Cas9, more explorative studies by employing epigenetic, DSBs repairing pathways modulators, such as small molecules or RNAi strategies that can further enhance or inhibit specific genome-editing pathways of Cas9 via homology-directed repair or NHEJ, need to be more extensively examined.
In summary, the seed sequence and the PAM, which are indispensable components of CRISPR/Cas9, need to be carefully designed. Moreover, the application of purified Cas9 protein as well as modulatory small molecules of DSBs pathways, which influence both on-target efficiency and off-target specificity, should be considered in individual application.
Methods of Off-Target Detection
Detecting off-target sites in a highly sensitive and comprehensive manner remains a key challenge in the field of gene editing.38 The T7 endonuclease I assay was initially used to detect off-target mutations, but this assay suffers poor sensitivity (it cannot detect off-target mutations that occur at frequencies <1%), and it is neither practical nor cost-effective for large-scale screening.39,40 Various advanced methods for off-targeting detecting including deep sequencing (measure off-target mutations at frequencies ranging from 0.01 to 0.1%11), web-based prediction tools, and ChIP-seq have been recently developed and widely adapted(Table 1).41,42,43 The Web-based algorithms have an innate limitation as the tools assume that off-target sequences are closely related to the on-target site, which may miss detrimental off-target sites with less sequence similarity. ChIP-seq has also been used to identify off-target binding sites for sgRNAs complexed with catalytically inactive Cas9 (dCas9). Encouragingly, majority of published works suggest that very few, if any, off-target cleavage sites were caused by active Cas9 nuclease.21,22,30,41,42,44
Genome-wide, unbiased identification of DSBs enabled by sequencing (GUIDE-seq) that has been reported recently, based on global capturing of DSBs introduced by RNA-guided endonucleases (RGEN), enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases.45 GUIDE-seq consists of a two-stage process. First, DSBs induced by RGEN in the genomes of living human cells are tagged by integration of a blunt, 34 bp double-stranded oligodeoxynucleotide at these breaks via an end-joining process consistent with NHEJ. Second, dsODN integration sites in genomic DNA are precisely mapped at the nucleotide level using unbiased amplification and next-generation sequencing.45 This method extends the detecting range of off-target mutagenesis frequencies to as low as 0.12%, far beyond the existing computational methods or ChIP-seq. Of note, some sgRNAs may induce extremely low frequency of, or perhaps no, undesired mutations (perhaps beyond the detecting limitation of GUIDE-seq technology). Another newly developed, high-throughput, genome-wide, translocation sequencing (HTGTS) method, which identifies translocations of yeast I-SceImeganuclease-generated “bait” DSBs at target sites, introduced into the genome of mouse cells to other “prey” cellular DSBs genome-wide. This HTGTS method not only reveals 10-folds more off-target sites caused by certain previously characterized nucleases, but also provides nucleotide-level resolution of junctions.46 This method further improves the detection of off-target sites through the identification of translocations between homologous chromosomes, an undesired collateral effect that has not been detected previously. The strategies outlined here can be used as part of a rigorous preclinical pathway for objectively assessing the potential off-target effects of any RGEN proposed for therapeutic use, thereby substantially improving the prospects for eventual translation of these reagents to the clinic.
Although in silico and in vitro methods are used to screen for potential off-target sites, they cannot precisely predict mutations that occur in vivo.7,8,10,11 Thus, efficient methods to detect where the off-target mutations occur in vivo need to be explored. Linear double-stranded integrase-defective lentiviral vectors (IDLV) preferentially incorporate into DNA DSBs by NHEJ, making it possible to tag nuclease-generated DSBs in vivo and has been used routinely to detect the off-target cleavage sites of zinc finger nucleases.47,48 A modified IDLV system was able to identify off-target cleavage sites with a frequency as low as 1% when CRISPR/Cas9 and transcription activator-like effector nucleases (TALEN) systems were used to target the Wiskott-Aldrich Syndrome (WAS) and tyrosine aminotransferase (TAT) genes. Therefore, this method can be an alternative tool to validate the target specificity of CRISPR/Cas9 and other gene-editing technologies in preclinical applications.49
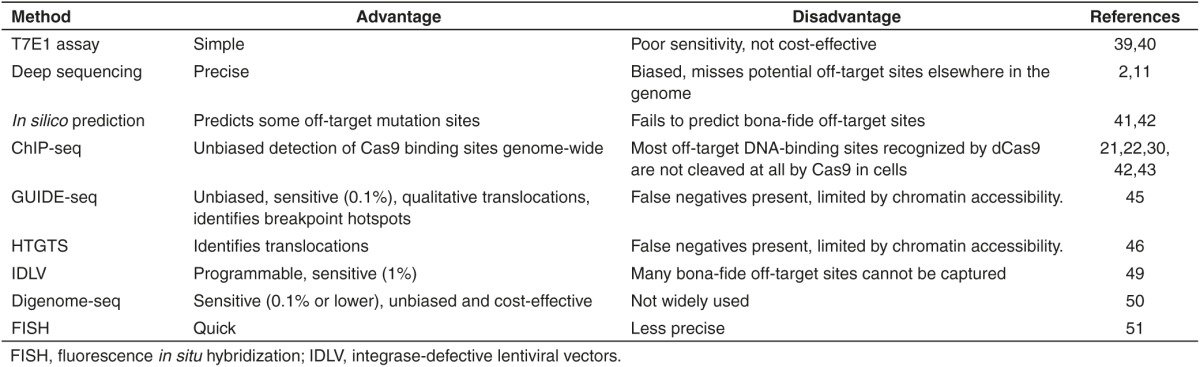
Although various approaches have been shown to reveal the frequency of off-target mutations by at least an order of magnitude at several sites, it still remains unknown whether these RGEN variants are free of off-target effects in the entire genome. Recently, Digenome-seq, in vitro Cas9-digested whole-genome sequencing, has emerged as a robust, sensitive, unbiased, and cost-effective method for profiling genome-wide off-target effects of programmable nucleases, including Cas9, in human cells.50 Digenome-seq also consists of a two-stage process. First, intact genomic DNA was isolated from transfected cells without RGEN and transfected cells with RGEN. Then DNA isolated from RGEN-transfected was digested in vitro with RGEN or not and RGEN transfected cells were digested in vitro with RGEN, both followed by whole-genome sequencing. Second, these digests can produce many DNA fragments with identical 5′ ends, which give rise to sequence reads that are vertically aligned at cleavage sites. In contrast, all other sequence reads would be aligned in a staggered manner. After mapping sequence reads to the reference genome, the authors used the Integrative Genomics Viewer (IGV) to observe patterns of sequence alignments at the on-target and the off-target sites.50 This method has demonstrated that Cas9 off-target effects can be avoided by replacing “promiscuous” sgRNA gX19 or GX19 sgRNAs with ggX20 sgRNAs (“g” and “G” represent a mismatched guanine and matched guanine, respectively), improving target specificity for gene and cell therapy applications.38 In addition, off-target effects of dCas9-FokI, paired Cas9 nickases, or other programmable nucleases could be monitored carefully via Digenome-seq to avoid unwanted mutations. Of course, the verification of animal experiments in vivo using Digenome-seq need to be further studied. Although different methods of off-target detection have their own characteristic advantages (Table 2), Digenome-seq is currently considered to be the gold standard for validating the specificity of next-generation genome-editing tools.
Table 2. Methods of off-target detection.
Off-target assessment can be time consuming and expensive, especially for simple in vitro projects. Yet some reports in the literature suggest that the frequency of off-target mutations is higher in in vitro cellular experiments compared to in vivo animal experiments. When performing in vitro cellular experiments, one can choose one of the unbiased methods for off-target detection as listed in Table 2, depending on the requirement of specific experiment, to reveal potential off-target sites, which may confound the interpretation of experimental results. The fluorescence in situ hybridization (FISH)-based methods for off-target identification, which is fast but less precise, can be also used as an alternative.51 Interestingly, when it comes to animal experiments, for many model species modified by CRIPSR/Cas9 including mouse,52,53 pig,54 or monkey,55,56 no off-target mutations, and no detectable damage at known off-target sites57 have been reported so far. There are a few possibilities that might explain the no reported off-target cleavage in animals. Firstly, it is possible to control the expression level of Cas9 protein by injection of Cas9 mRNA or protein, which not only acts much faster than plasmid transfection, but also degrades rapidly. Secondly, researchers tend to keep the founder animals with right on-target mutations, and do not maintain the animals without on-target mutations, leading to off-target mutations being diluted over generations in small animals, even though the founder (F0) might harbor possible off-target mutations.58 In addition, the off-target sites may be in a noncoding region,25 so, the likelihood of generating an off-target phenotype is small. Thirdly, the methods and experiments employed for detecting off-target cleavage in most of the published animal model studies were not performed in genome-wide, high resolution, and unbiased way, which may mask some low frequency off-target editing sites. Thus, much more comprehensive and stringent examination of off-target cleavage sites at the animal level should be performed for future studies.
Strategies for Minimizing Off-Target Effects
Various strategies have been reported to reduce RGEN off-target effects.50 First, the sgRNA sequence can be altered. Truncation of the 3′ end of sgRNA (derived from tracrRNA domain that interacts with Cas9), shortening the region complementary to the target site at the 5′ end of the sgRNA by as many as 3 nt (tru-gRNA) or addition of two guanine nucleotides to the 5′ end of the sgRNA (just before the 20-nt complementary region) improves target specificity, decreasing undesired mutagenesis at some off-target sites by 5,000-fold.9,11,59 Meanwhile, RGENs using these altered sgRNAs also have decreased on-target activity. Efforts toward modifications of sgRNA sequences to enhance the specificity of sgRNAs without compromising on-target efficiency have not provided consistent results.11,60 In addition, chromatin accessibility has been reported to be one major determinant of in vivo binding. As Zhang and Sharp reported recently, there are hundreds of thousands of “seed+NGG” sites in the genome, yet <1% are actually bound by dCas9, and most of the matches are in promoters, enhancers and genes.21 Therefore, when we design sgRNA, we should choose sgRNAs in promoters, enhancers, and genes as far as possible to improve the target cleavage efficiency. However, those sites cleaved by CRISPR/Cas9 system can be both on-target and off-targets sites and we need to balance it according to different experimental purposes.
Second, one potential strategy for minimizing off-target effects is to control the concentration of the Cas9-sgRNA complex by titrating the amount of Cas9 and sgRNA delivered. However, increasing specificity by reducing the amount of transfected DNA also leads to a reduction in on-target cleavage. Therefore, a balance between on-target cleavage efficiency and off-target effects has to be considered. Nonetheless, future optimization of both Cas9 and sgRNA design may improve Cas9 specificity without sacrificing cleavage efficiency.8,9,10,30
Third, the wild-type Cas9 nuclease can be replaced with D10 mutant nickase version of Cas9 and paired with two sgRNAs that each cleaves only one strand. Paired nicking strategy substantially reduces the off-target activity by 50–1,500-fold in cell lines and facilitates gene knockout in mouse zygotes without sacrificing on-target cleavage efficiency.61 This versatile strategy enables a wide variety of genome-editing applications that require high specificity. Additionally, the repair of two nicks generated by double nicking produces 5' overhangs, leading to the formation of indels much more frequently than 3' overhang.11,46 Comicroinjection of mouse embryos with wild-type Cas9 and sgRNAs induces on-target and off-target mutations that are transmissible to the offspring. Conversely, double nicking can be used in vivo to efficiently mutate single or multiple genes with minimal off-target mutations while preserving on-target cleavage efficiency. There is also an established framework for the optimal design of paired sgRNAs and computational tools for predicting potential off-target sites for sgRNA pairs.57 The optimal pair of Cas9 sites are in a tail-to-tail orientation, separated by −10 bp to +30 bp, with the sequence 5'-CCN(32–72)GG-3'. This method is applicable for editing the genome of any model organism and minimizes confounding problems of off-target mutations, thereby holds great application potential in clinical gene therapy.
Fourth, to further improve DNA cleavage specificity, fusions of catalytically inactive Cas9 with FokI nuclease domain (fCas9) have been generated, which edits target DNA sites with >140-fold higher specificity than wild-type Cas9 and at least fourfold than that of paired nickases at loci with highly similar off-target sites.62,63 This work provides the foundation for the further characterization and improvement of Cas9 specificity and cleavage activity in vitro and in vivo. Recently, tru-RFN with 19 bp half-site complementarity lengths that combines truncated guide RNAs with dimerization-dependent RNA-guided FokI-dCas9 nucleases decreases off-target cleavage by 40% relative to standard RFNs, providing a useful and further improved tool for high-precision genome-editing applications in human cells.64 We believe that the synergistic use of double nicking and fCas9 offers a promising route for mitigating the effects of off-target CRISPR/Cas9 activity, and may perhaps be a useful approach for exploring therapeutic applications.7 Recently, a strategy that improves the efficiency of precise genome editing with CRISPR-Cas9 via inhibition of nonhomologous end joining using the inhibitor Scr7 to target DNA ligase IV has also been reported, providing another opportunity to improve the target specificity.65 Moreover, a simple and effective way to codeliver chemically modified sgRNAs with Cas9 mRNA or protein has been recently described.66 It is an efficient RNA- or ribonucleoprotein-based delivery method for the CRISPR/Cas9 system, with lower cytotoxicity in primary cells than DNA plasmid-based systems, enabling the expansion and widespread application of this technology.
Perspectives On Improved Crispr/Cas9 In Gene-Editing Field
The growth of any new technology, including CRISPR/Cas9, demands progressive enhancement. In 2 years, research in CRISRP/Cas9 has made huge strides in the evolution of gene editing. RGENs are a promising new member in the growing family of programmable nucleases, which include ZFNs and TALENs, but have more severe off-target effects than other nucleases due to their inherent structure and mechanism.11,44 The optimization of various components of the CRISPR/Cas9 system enables us to reduce the off-target activity without sacrificing on-target cleavage efficiency. In addition, strategies for minimizing off-target mutations (Figure 2), such as double nickases Cas9, delivery of purified Cas9 protein and fusion of dCas9 to FokI nuclease, illuminate the path for mitigating off-target effects, which may eventually lead to therapeutic applications. In addition, new methods for detecting off-target mutations, in particular Digenome-seq, will enable the detection of rare and detrimental off-target sites before moving to gene or cell therapy.
Many new tools are available for researchers keen on exploiting the CRISPR/Cas9 system for various applications. An improved sgRNA screening library has been established by Feng Zhang's group for single- or paired-vector systems to deliver Cas9 and sgRNA.67 Moreover, CRISRP/Cas9 can also be modified to control gene transcription,68,69,70,71,72 and a novel split-Cas9 architecture for inducible genome editing and transcription modulation has also been reported.73 Donald Zack's report of successfully modifying endogenous genes using H1 promoter-expressed gRNAs, which can be used to target both AN19NGG and GN19NGG genomic sites, will increase the coverage of potential target sites, in particular relevant to human disease-specific loci.74 Recently, another research team engineered smaller-sized Cas9 orthologues (Streptococcus thermophilus Cas9 and Staphylococcus aureus Cas9) with altered PAM specificities, which exhibits improved specificity in human cells without sacrificing target recognition efficiency,32 thereby getting rid of the PAM recognition limitation. In view of numerous previous optimization and perhaps lots of ongoing work, the SpCas9 platform will likely revolutionize the gene-editing field. By combining all these available new methods, hopefully we will be able to eventually manipulate and further correct many (if not all) disease-related gene mutations in a cost-effectively, simple, reliable, efficient, and precise way (Figure 2). Recently, CRISPR/Cpf1, another new gene-editing tool in class 2 CRISPR-Cas system that has the different properties from CRISPR/Cas9 was reported, in which mature crRNAs that begins with 19 nt of the direct repeat followed by 23–25 nt of the spacer sequence (like sgRNA in CRISR/Cas9 system) and Cpf1 needed can efficiently cleave target DNA proceeded by a short T-rich PAM, in contrast to the G-rich PAM following the target DNA for Cas9 systems, and a staggered DNA double-stranded breaks with a 4 or 5-nt 5`overhang was generated by Cpf1 could provide an effective way to precisely introduce DNA into the genome via non-homology-directed repair mechanisms. So, it has the potential to substantially advance researchers ability to manipulate eukaryotic genomes.75 Furthermore, other gene-modifying technologies based on DNA methylation,76 optogenetics,77,78 or mutagens79,80,81 (base analog, base modifier, or intercalating dye) may potentially enhance gene-editing technology, providing they can be linked with sgRNA. No doubt, new generation of gene-editing tools beyond the already ZFNs, TALENs, CRISPR/Cas9, and CRISPR/Cpf1, may be on the way or await exploration in nature.
Acknowledgments
The authors thank Yang Zhou from Massachusetts Institute of Technology for their comments and help for improving our manuscript. This work was supported by grants from the Ministry of Science of China (2012CBA01302), the Science and Technology Department of Guangdong Province, China (2014B020225007), Program for New Century Excellent Talents in University (NCET-12–1078), the National Natural Science Foundation of China (31071279), and a Science and Technology Innovation Project Grant from the Department of Education of Guangdong Province (2013KJCX0029).
References
- Mali, P, Yang, L, Esvelt, KM, Aach, J, Guell, M, DiCarlo, JE et al. (2013). RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong, L, Ran, FA, Cox, D, Lin, S, Barretto, R, Habib, N et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, PD, Lander, ES and Zhang, F (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna, JA and Charpentier, E (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Cox, DB, Platt, RJ and Zhang, F (2015). Therapeutic genome editing: prospects and challenges. Nat Med 21: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C, Abalde-Atristain, L, He, C, Brodsky, BR, Braunstein, EM, Chaudhari, P et al. (2015). Efficient and allele-specific genome editing of disease loci in human iPSCs. Mol Ther 23: 570–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali, P, Aach, J, Stranges, PB, Esvelt, KM, Moosburner, M, Kosuri, S et al. (2013). CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, PD, Scott, DA, Weinstein, JA, Ran, FA, Konermann, S, Agarwala, V et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak, V, Lin, S, Guilinger, JP, Ma, E, Doudna, JA and Liu, DR (2013). High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31: 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y, Foden, JA, Khayter, C, Maeder, ML, Reyon, D, Joung, JK et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31: 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, SW, Kim, S, Kim, Y, Kweon, J, Kim, HS, Bae, S et al. (2014). Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan-Curay, J, O'Reilly, M, Kohn, DB, Cannon, PM, Bao, G, Bushman, FD et al. (2015). Genome editing technologies: defining a path to clinic. Mol Ther 23: 796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath, P and Barrangou, R (2010). CRISPR/Cas, the immune system of bacteria and archaea. Science 327: 167–170. [DOI] [PubMed] [Google Scholar]
- Gasiunas, G, Barrangou, R, Horvath, P and Siksnys, V (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109: E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M, Chylinski, K, Fonfara, I, Hauer, M, Doudna, JA and Charpentier, E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica, FJ, Díez-Villaseñor, C, García-Martínez, J and Almendros, C (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155(Pt 3): 733–740. [DOI] [PubMed] [Google Scholar]
- Sternberg, SH, Redding, S, Jinek, M, Greene, EC and Doudna, JA (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, C, Niewoehner, O, Duerst, A and Jinek, M (2014). Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu, H, Ran, FA, Hsu, PD, Konermann, S, Shehata, SI, Dohmae, N et al. (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156: 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M, Jiang, F, Taylor, DW, Sternberg, SH, Kaya, E, Ma, E et al. (2014). Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343: 1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X, Scott, DA, Kriz, AJ, Chiu, AC, Hsu, PD, Dadon, DB et al. (2014). Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 32: 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, J, Lu, G, Xie, Z, Lou, M, Luo, J, Guo, L et al. (2014). Genome-wide identification of CRISPR/Cas9 off-targets in human genome. Cell Res 24: 1009–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W, Bikard, D, Cox, D, Zhang, F and Marraffini, LA (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic, R, Miura, H, Malina, A, Robert, F, Ethier, S, Schmeing, TM et al. (2014). Protospacer adjacent motif (PAM)-distal sequences engage CRISPR Cas9 DNA target cleavage. PLoS One 9: e109213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T, Wei, JJ, Sabatini, DM and Lander, ES (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, JG, Hartenian, E, Graham, DB, Tothova, Z, Hegde, M, Smith, I et al. (2014). Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32: 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, X, Yang, Z, Xu, J, Sun, J, Mao, D, Hu, Y et al. (2014). Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep 9: 1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon, JA, Valen, E, Thyme, SB, Huang, P, Akhmetova, L, Ahkmetova, L et al. (2014). Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 9: e98186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos, MA, Vejnar, CE, Beaudoin, JD, Fernandez, JP, Mis, EK, Khokha, MK et al. (2015). CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods 12: 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu, C, Arslan, S, Singh, R, Thorpe, J and Adli, M (2014). Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol 32: 677–683. [DOI] [PubMed] [Google Scholar]
- Zhang, Y, Ge, X, Yang, F, Zhang, L, Zheng, J, Tan, X et al. (2014). Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci Rep 4: 5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver, BP, Prew, MS, Tsai, SQ, Topkar, VV, Nguyen, NT, Zheng, Z et al. (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523: 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S, Kim, D, Cho, SW, Kim, J and Kim, JS (2014). Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24: 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna, S, Kwaku Dad, AB, Beloor, J, Gopalappa, R, Lee, SK and Kim, H (2014). Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res 24: 1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C, Gore, A, Yan, W, Abalde-Atristain, L, Li, Z, He, C et al. (2014). Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 15: 12–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veres, A, Gosis, BS, Ding, Q, Collins, R, Ragavendran, A, Brand, H et al. (2014). Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 15: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C, Liu, Y, Ma, T, Liu, K, Xu, S, Zhang, Y et al. (2015). Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16: 142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, R, von Kalle, C and Schmidt, M (2015). Mapping the precision of genome editing. Nat Biotechnol 33: 150–152. [DOI] [PubMed] [Google Scholar]
- Cho, SW, Kim, S, Kim, JM and Kim, JS (2013). Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 230–232. [DOI] [PubMed] [Google Scholar]
- Kim, HJ, Lee, HJ, Kim, H, Cho, SW and Kim, JS (2009). Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res 19: 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, FA, Hsu, PD, Wright, J, Agarwala, V, Scott, DA and Zhang, F (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer, F, Kerr, G and Boutros, M (2014). E-CRISP: fast CRISPR target site identification. Nat Methods 11: 122–123. [DOI] [PubMed] [Google Scholar]
- Singh, R, Kuscu, C, Quinlan, A, Qi, Y and Adli, M (2015). Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res 43: e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel, A, Fine, EJ, Bao, G and Porteus, MH (2015). Quantifying on- and off-target genome editing. Trends Biotechnol 33: 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, SQ, Zheng, Z, Nguyen, NT, Liebers, M, Topkar, VV, Thapar, V et al. (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33: 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frock, RL, Hu, J, Meyers, RM, Ho, YJ, Kii, E and Alt, FW (2015). Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol 33: 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, R, Lombardo, A, Arens, A, Miller, JC, Genovese, P, Kaeppel, C et al. (2011). An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol 29: 816–823. [DOI] [PubMed] [Google Scholar]
- Osborn, MJ, Starker, CG, McElroy, AN, Webber, BR, Riddle, MJ, Xia, L et al. (2013). TALEN-based gene correction for epidermolysis bullosa. Mol Ther 21: 1151–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X, Wang, Y, Wu, X, Wang, J, Wang, Y, Qiu, Z et al. (2015). Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol 33: 175–178. [DOI] [PubMed] [Google Scholar]
- Kim, D, Bae, S, Park, J, Kim, E, Kim, S, Yu, HR et al. (2015). Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods 12: 237–43, 1 p following 243. [DOI] [PubMed] [Google Scholar]
- Paulis, M, Castelli, A, Lizier, M, Susani, L, Lucchini, F, Villa, A et al. (2015). A pre-screening FISH-based method to detect CRISPR/Cas9 off-targets in mouse embryonic stem cells. Sci Rep 5: 12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, V, Shen, B, Zhang, W, Hodgkins, A, Keane, T, Huang, X et al. (2015). Off-target mutations are rare in Cas9-modified mice. Nat Methods 12: 479. [DOI] [PubMed] [Google Scholar]
- Shen, B, Zhang, J, Wu, H, Wang, J, Ma, K, Li, Z et al. (2013). Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res 23: 720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai, T, Teng, F, Guo, R, Li, W and Zhou, Q (2014). One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res 24: 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, H, Feng, C, Teng, F, Yang, S, Hu, B, Niu, Y et al. (2015). One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Res 25: 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, Y, Shen, B, Cui, Y, Chen, Y, Wang, J, Wang, L et al. (2014). Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156: 836–843. [DOI] [PubMed] [Google Scholar]
- Shen, B, Zhang, W, Zhang, J, Zhou, J, Wang, J, Chen, L et al. (2014). Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods 11: 399–402. [DOI] [PubMed] [Google Scholar]
- Li, D, Qiu, Z, Shao, Y, Chen, Y, Guan, Y, Liu, M et al. (2013). Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 31: 681–683. [DOI] [PubMed] [Google Scholar]
- Fu, Y, Sander, JD, Reyon, D, Cascio, VM and Joung, JK (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusk, N (2015). Next-generation CRISPRs. Nat Methods 12: 36. [Google Scholar]
- Ran, FA, Hsu, PD, Lin, CY, Gootenberg, JS, Konermann, S, Trevino, AE et al. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger, JP, Thompson, DB and Liu, DR (2014). Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol 32: 577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, SQ, Wyvekens, N, Khayter, C, Foden, JA, Thapar, V, Reyon, D et al. (2014). Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 32: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyvekens Nicolas, TVV, Khayter Cyd, Joung J. Keith, and Tsai Shengdar Q (2015). Dimeric CRISPR RNA-Guided FokI-dCas9 Nucleases Directed by Truncated gRNAs for Highly Specific Genome Editing. Hum Gene Ther 26: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama, T, Dougan, SK, Truttmann, MC, Bilate, AM, Ingram, JR and Ploegh, HL (2015). Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33: 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel, A, Bak, RO, Clark, JT, Kennedy, AB, Ryan, DE, Roy, S, et al. (2015). Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 6, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana, NE, Shalem, O and Zhang, F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11: 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann, S, Brigham, MD, Trevino, AE, Joung, J, Abudayyeh, OO, Barcena, C et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, LA, Larson, MH, Morsut, L, Liu, Z, Brar, GA, Torres, SE et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder, ML, Linder, SJ, Cascio, VM, Fu, Y, Ho, QH and Joung, JK (2013). CRISPR RNA-guided activation of endogenous human genes. Nat Methods 10: 977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, LS, Larson, MH, Gilbert, LA, Doudna, JA, Weissman, JS, Arkin, AP et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, AW, Wang, H, Yang, H, Shi, L, Katz, Y, Theunissen, TW et al. (2013). Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res 23: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche, B, Volz, SE and Zhang, F (2015). A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol 33: 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan, V, Wahlin, K, Maruotti, J and Zack, DJ (2014). Expansion of the CRISPR-Cas9 genome targeting space through the use of H1 promoter-expressed guide RNAs. Nat Commun 5: 4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche, B, Gootenberg, JS, Abudayyeh, OO, Slaymaker, IM, Makarova, KS, Essletzbichler, P, et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, ZD and Meissner, A (2013). DNA methylation: roles in mammalian development. Nat Rev Genet 14: 204–220. [DOI] [PubMed] [Google Scholar]
- Deisseroth, K (2011). Optogenetics. Nat Methods 8: 26–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein, LR and Gersbach, CA (2015). A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 11: 198–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondo, Y (2008). Trends in large-scale mouse mutagenesis: from genetics to functional genomics. Nat Rev Genet 9: 803–810. [DOI] [PubMed] [Google Scholar]
- Lada, AG, Stepchenkova, EI, Waisertreiger, IS, Noskov, VN, Dhar, A, Eudy, JD et al. (2013). Genome-wide mutation avalanches induced in diploid yeast cells by a base analog or an APOBEC deaminase. PLoS Genet 9: e1003736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, W and Vinograd, J (1968). The interaction of closed circular DNA with intercalative dyes. I. The superhelix density of SV40 DNA in the presence and absence of dye. J Mol Biol 33: 141–171. [DOI] [PubMed] [Google Scholar]