Abstract
This research undertook the systematic analysis of the Klebsiella sp. D5A genome and identification of genes that contribute to plant growth-promoting (PGP) traits, especially genes related to salt tolerance and wide pH adaptability. The genome sequence of isolate D5A was obtained using an Illumina HiSeq 2000 sequencing system with average coverages of 174.7× and 200.1× using the paired-end and mate-pair sequencing, respectively. Predicted and annotated gene sequences were analyzed for similarity with the Kyoto Encyclopedia of Genes and Genomes (KEGG) enzyme database followed by assignment of each gene into the KEGG pathway charts. The results show that the Klebsiella sp. D5A genome has a total of 5,540,009 bp with 57.15% G + C content. PGP conferring genes such as indole-3-acetic acid (IAA) biosynthesis, phosphate solubilization, siderophore production, acetoin and 2,3-butanediol synthesis, and N2 fixation were determined. Moreover, genes putatively responsible for resistance to high salinity including glycine-betaine synthesis, trehalose synthesis and a number of osmoregulation receptors and transport systems were also observed in the D5A genome together with numerous genes that contribute to pH homeostasis. These genes reveal the genetic adaptation of D5A to versatile environmental conditions and the effectiveness of the isolate to serve as a plant growth stimulator.
Bacteria that efficiently colonize the rhizosphere and stimulate plant growth through direct or indirect mechanisms are referred to as plant growth promoting rhizobacteria (PGPR)1. In addition to possessing general plant growth promoting properties such as production of indole-3-acetic acid (IAA), siderophores2, 1-amino-cyclopropane-1-carboxylate (ACC) deaminase, hydrogen cyanate (HCN), nitrogenase3 and phosphate solubilization4, some PGPR also possess more environment specific plant growth promoting (PGP) traits such as heavy metal detoxifying activity5, salinity tolerance6, and biological control of phytopathogens and insects7.
PGPR have become of interest as inoculants for phytoremediation because of their diverse plant growth promoting capabilities8. Systematic analysis of whole genome data and the identification of genes that contribute to the beneficial activity of PGPR will aid our understanding of the molecular mechanisms of many bacterial species and also help in the development of PGPR assisted phytoremediation technology. Next generation sequencing technologies (NGS) have enabled whole genome sequencing of bacteria and other organisms9,10,11. NGS have recently been employed to study the genomes of several PGPR such as Pseudomonas sp.12, Bacillus sp.13, and Paenibacillus polymyxa14.
In 2013 a plant growth promoting bacterium, Klebsiella sp. D5A, was isolated from the rhizosphere soil of tall fescue (Testuca arundinacea L.) growing in a soil contaminated with oil and the PGP traits and environmental tolerance of this strain were quantified. The bacterium produced IAA, solubilized phosphate, synthesized sideropheres, and also had a strong ability to adapt to a high saline-alkaline environment and wide range of soil pH (4–10)15. Pot experiments indicate that inoculation with this isolate promoted the growth of host plants in a petroleum-contaminated saline-alkaline soil and enhanced phytoremediation efficiency. Klebsiella sp. is likely to be an effective PGPR due to some members being endophytic nitrogen-fixing bacteria16,17,18,19,20. Some Klebsiella genomes have now been reported18,20,21,22 but most investigations have focused on their pathogenic or N-fixation genes. By contrast, the genes that contribute to the beneficial activity of PGPR and especially salt tolerance and pH adaptability remain to be studied in detail.
Here, we report the complete genome sequence of Klebsiella sp. D5A to help reveal the complex biological mechanisms of D5A as a PGPR. The genomic analysis in this study also includes comparison of the PGP traits to four closely related and representative PGPR strains that have been studied previously, namely K. variicola 34218 (originally misclassified as K. pneumoniae and then clustered with K. variicola by Garza-Ramos et al.)23, K. variicola At-2219,20 and K. variicola DX12020, K. pneumoniae MGH7857818. The genome analysis will provide a fundamental basis for future studies towards fully understanding the functions of this organism. Furthermore, comparisons among the completely sequenced Klebsiella genomes will help to offer new insights into evolutionary changes in Klebsiella spp. and highlight the genes that may contribute to their plant growth-promoting properties.
Results
General genome features of Klebsiella sp. D5A
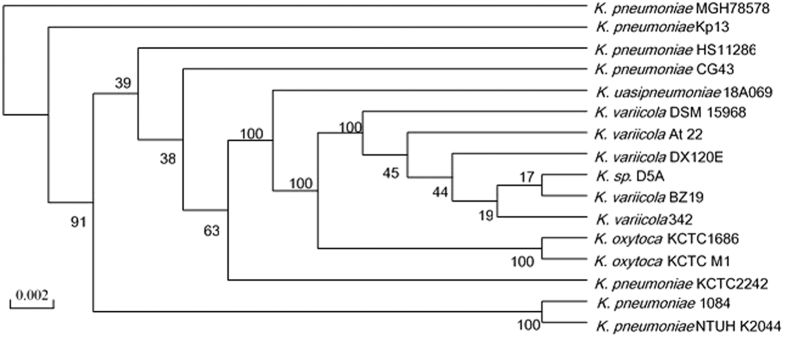
The genome of Klebsiella sp. D5A has a total of 5,540,009 bp with an average G + C content of 57.15% (Table 1). The genome contains 4999 predicted CDSs (coding sequence) with an average length of 944 bp. Among these CDSs, 4339 (86.8%) genes were classified into COG (Clusters of Orthologous Groups of proteins) families composed of 21 categories (Table 2). Coding regions cover 85.2% of the whole genome. Biological roles were assigned to 4129 (82.6%) genes of the predicted CDS based on similarity searches with the String and Nr database. The remaining 870 (17.4%) coding sequences were classified as proteins with unknown or hypothetical function. A total of four rRNAs comprising two 5S rRNAs, a single 16S rRNA, and a single 23S rRNA togther with 73 tRNA genes representing 37 amino acids were identified in the D5A genome. We established a phylogenetic tree of 16 completely sequenced Klebsiella based on 45 conserved genes. The tree shows that Klebsiella sp. D5A is most closely relate to K. variicola (Fig. 1).
Table 1. General features of the Klebsiella sp. D5A genome.
| Feature | Chromosome |
|---|---|
| Size (bp) | 5,540,009 |
| G + C content (%) | 57.15 |
| Number of CDSs | 4999 |
| Gene density | 0.902 genes per kb |
| Average CDS length (bp) | 944 |
| tRNA | 73 |
| rRNA | 4 |
| Number of genes with assigned function | 4129 (82.6%) |
| Number of genes without assigned function | 870 (17.4%) |
Table 2. COG functional categories of Klebsiella sp. D5A.
| Type | Functional categories | COG |
|---|---|---|
| Information storage and processing | [A] RNA processing and modification | 1 |
| [B] Chromatin structure and dynamics | 1 | |
| [J] Translation, ribosomal structure and biogenesis | 179 | |
| [K] Transcription | 413 | |
| [L] Replication, recombination and repair | 164 | |
| Cellular processes and signaling | [D] Cell cycle control, cell division, chromosome partitioning | 37 |
| [M] Cell wall/membrane/envelope biogenesis | 242 | |
| [N] Cell motility | 29 | |
| [O] Posttranslational modification, protein turnover, chaperones | 153 | |
| [T] Signal transduction mechanisms | 141 | |
| [U] Intracellular trafficking, secretion, and vesicular transport | 78 | |
| [V] Defense mechanisms | 50 | |
| Metabolism | [C] Energy production and conversion | 303 |
| [E] Amino acid transport and metabolism | 500 | |
| [F] Nucleotide transport and metabolism | 94 | |
| [G] Carbohydrate transport and metabolism | 508 | |
| [H] Coenzyme transport and metabolism | 155 | |
| [I] Lipid transport and metabolism | 129 | |
| [P] Inorganic ion transport and metabolism | 362 | |
| [Q] Secondary metabolites biosynthesis, transport and catabolism | 106 | |
| Poorly characterized | [R] General function prediction only | 297 |
| [S] Function unknown | 397 | |
| Total | 4339 |
Figure 1. Phylogenetic tree of 16 different Klebsiella species based on aligned concatenated sequences of 45 orthologous genes using the bootstrap method.
Numbers on nodes represent percentages of individual trees containing that relationship. The scale bar corresponds to the number of substitutions per site.
Genes related to plant growth promotion traits of Klebsiella sp. D5A
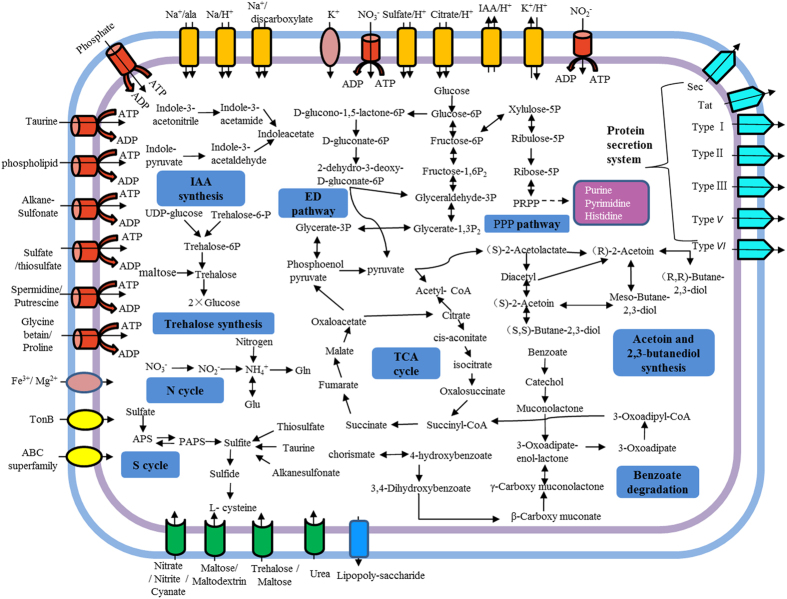
We identified genes in the D5A genome attributable to the production of IAA, solubilization of phosphate, synthesis of sideropheres, acetoin and 2,3-butanediol, suppression of pathogenic fungi, resistance to oxidative stress, and ability to break down toxic compounds and other abiotic stresses (Table S1). Our previous study shows that Klebsiella sp. D5A actively produces 112 mg L−1 IAA (Table 3) and IAA production is unaffected by pH values between 4 and 1015. Here, two proposed IAA biosynthesis pathways, indole-3-acetonitrile (IAN) and indole-3-pyruvate (IPyA) pathways are identified in the genome of D5A (Fig. 2) and four genes might be involved (Table S1). In the IAN pathway IAN can first be converted to indole-3-acetamide (IAM) by nitrile hydratase and then IAM is converted to IAA by amidase. In the IPyA pathway indole-3-pyruvate (IPyA) is converted to indole-3-acetaldehyde (IAAld) by indolepyruvate decarboxylase and then to IAA by aldehyde dehydrogenase. A search of four sequenced Klebsiella genomes (Table S2) for D5A-like IAA pathway associated genes reveals the presence of three orthologous genes in K. variicola 342, K. variicola At-22 and K. variicola DX120 compared to D5A. They lack a gene coding for aldehyde dehydrogenase. All of these genes responsible for IAA synthesis were absent from K. pneumoniae MGH78578.
Table 3. Biological and plant growth promotion properties and environmental tolerance of Klebsiella sp. D5A15.
| S. No. | Attribute | D5A |
|---|---|---|
| 1. | pH tolerance level | 3.5–10.5 |
| 2. | Optimum pH for growth | 4.0–10.0 |
| 3. | NaCl tolerance | up to 12% |
| 4 | Na2CO3 tolerance | 25 mM |
| 5. | IAA production | 112 mg L−1 |
| 6. | Phosphate solubilization | 131 mg L−1 |
| 7. | Siderophore production (A/Ar) | + |
| 8. | Growth on N-free agar medium | Growth observed |
Figure 2. Schematic overview of metabolic pathways and transport systems of Klebsiella sp. D5A.
The depicted pathways were predicted based on the genomic data of Klebsiella sp. D5A analyzed by Glimmer. Individual pathways are denoted by single headed arrows and reversible pathways are denoted by double-headed arrows.
Gluconic acid (GA) is recognized as one of the major organic acids in most bacteria responsible for the solubilization of mineral phosphates. The synthesis of GA is catalyzed by glucose dehydrogenase (GDH) and its co-factor pyrrolo-quinolone quinine (PQQ)24,25. As Table 3 shows, D5A can solubilize 131 mg L−1 phosphates. Accordingly, the D5A genome possesses genes encoding GDH activity and carries the redox co-factor pqq genes including pqqBCDEF while lacking the gene pqqA. Toyama and Lidstrom26 have reported that the enzyme encoded by pqqA is not essential for biosynthesis of PQQ in Methylobacterium. In addition, inorganic phosphate uptake in D5A may be promoted by one low-affinity phosphate transport system, PitA, and two high-affinity phosphate transport systems, PstBACS and PhnCDE2E1 (Table S1).
D5A carries genes coding for the synthesis of siderophores, such as entABECFD which are responsible for the conversion of chorismic acid to enterobactin and for the transport of this siderophore (entS) which is found ubiquitously among enterobacteria (Table S1). However, the genes involved in pyoverdine synthesis, the main class of siderophore, are absent from D5A. Furthermore, bacteria may also heterologously adopt siderophores produced by other organism via various siderophore receptors27. Klebsiella sp. D5A encoded 12 copies of genes for siderophore receptors including five TonB-dependent outer-membrane receptors, three putative ferric enterobactin receptors (fepA), a catecholate siderophore receptor (fiu), a ferric aerobactin receptor, a ferrioxamine receptor (foxA), a ferrichrome outer membrane receptor, and the ferric uptake regulator (fur) (Table S1). Furthermore, 43 additional ORFs (open reading frames) that encode iron transport such as the ferrous iron transporter which synthesizes proteins involved in Fe2+ capture and the sitABCD system involved in the transport of divalent cations such as Mn2+ and Fe2+ 22 were also determined in the D5A genome (Table S1). These indicate that although strain D5A cannot synthesis numerous sideropheres, it can heterologously obtain siderophores produced by other soil bacteria.
In addition to the above PGP traits, two growth-promoting volatile organic compounds (VOCs), acetoin and 2,3-butanediol, were reported to promote plant growth by stimulating root formation28 and increasing systemic disease resistance29 and drought tolerance30 in some other very efficient PGPR. Genes encoding enzymes including acetolactate synthase, acetolactate decarboxylase, and acetoin reductase (Table S1) which are involved in acetoin and 2,3-butanediol synthesis, were detected in the genome of D5A and the synthetic pathway is shown in Fig. 2. First, two pyruvate molecules are condensed into acetolactate catalyzed by acetolactate synthase, and then acetolactate is converted to acetoin by acetolactate decarboxylase and finally acetoin is converted to 2,3-butanediol catalyzed by acetoin reductase. When the genomes of the other four Klebsiella isolates were examined for acetion and 2,3-butanediol synthesis, partial or incomplete coding genes were observed while the synthesis of the two VOCs was not influenced.
It has been reported that PGPR may produce compounds such as phenazine and 4-hydroxybenzoate which function as antibiotics and suppress plant pathogenic microbes12,31. UbiC, involved in 4-hydroxybenzoate synthesis, and phzF, involved in phenazine synthesis, were identified in the D5A genome. Moreover, a homologue of the gene coding for chitinase enzyme was identified here which can dissolve the cell walls of pathogenic fungal and insect pests31,32. In addition to these we found the genes gabD and gabT which are responsible for the production of pest/disease inhibiting γ-aminobutyric acid (GABA)31 in the genome. A search for these genes in four Klebsiella genomes found that they were present in the four bacteria except that genes coding for chitinase enzyme were absent from K. variicola 342. This suggests that the synthesis of the three antimicrobial compounds is a widespread pathway in Klebsiella spp.
Furthermore, some other PGPR fitness conferring genes were also detected in the D5A genome. For example, the genes speA, speB and speE which encode for, respectively, arginine decarboxylase, agmatinase, and spermidine synthase may lead to the transformation of amino acids to PGP substances33. Moreover, the D5A genome encodes numerous proteins to protect the cell from oxidative stress: eight peroxidases, three catalases, four superoxide dismutases, three hydroperoxide reductases and 13 glutathione S-transferases (Table S1).
Production of cold-shock and heat-shock proteins by microoganisms can help their survival in harsh environments. The D5A genome carries the heat-shock protein genes dnaJ, dnaK, groEL, groES, htpG, and htpX and the cold-shock protein gene cspA (Table S1). Moreover, the gene coding for the enzyme rhodanese was found in the D5A genome and this is responsible for the detoxification of cyanide in organisms (Table S1). Cyanide is a potent cytotoxin which is produced by the hydrolysis of plant cyanogenic glycosides and may inhibit the cytochrome oxidases in the mitochondrial electron transport chain34.
Nitrogen fixation
Klebsiella sp. D5A is able to grow on nitrogen-free medium (Table 3) and this indicates that the strain is able to fix atmospheric nitrogen. Nitrogenase is the enzyme central to nitrogen fixation and it consists of Fe-protein encoded by nifH and MoFe-protein encoded by nifDK. Full assembly of the nitrogenase complex needs the products of at least twelve nif genes, especially for the processing of catalytic stability and nitrogenase metalloclusters (nifMZ, nifUS, and nifW) and for synthesis of a particular molybdenum cofactor (FeMo-co)35. The D5A genome contains all the above nif genes together with the NifA and NifL genes which are the positive/negative regulatory proteins for nif genes36. The rnfABCDEG operon, which encodes a membrane-bound protein complex related to electron transport to nitrogenase37, is also found in the D5A genome (Table S3). In contrast, comparative genomic analysis shows that key genes associated with nitrogen fixation including nitrogenase are absent from Klebsiella sp. MGH78578. It is therefore presumed that MGH78578 cannot fix nitrogen.
Genes putatively involved in salt tolerance
As Table 3 shows, D5A can grow well in 0–12% NaCl. Analysis of the genome reveals that strain D5A has a number of genes related to salt tolerance. For example, the key genes betA and betB for glycine-betaine synthesis that respectively encode choline dehydrogenase and betaine aldehyde dehydrogenase were found in D5A. They are considered to be the most effective genes responsible for salt tolerance38. In addition, trehalose can act as an osmoprotectant under environmental stresses such as high salt or drought, low temperature or osmotic stress in many organisms12. Trehalose accumulates in transgenic rice and enhances plant abiotic stress tolerance39. So far five trehalose biosynthetic pathways have been found in bacteria including treS, otsA/otsB, treP, treT and treY/treZ40. Here, two trehalose biosynthesis pathways, treS and otsA/otsB, were identified in the D5A genome. In the treS pathway maltose is converted to trehalose by trehalose synthase (treS). In the otsA/otsB pathway both glucose-6-phosphate and UDP-glucose can synthesize trehalose-6-phosphate catalyzed by trehalose-6-phosphate synthase (otsA) activity. Trehalose-6-phosphate is then formed from trehalose catalyzed by trehalose-6-phosphate phosphatase (otsB) activity. Eventually, trehalose may be hydrolyzed by trehalase (treA, treF) with the generation of two glucose molecules (Fig. 2, Table S1). This pathway has been recognized as a universal pathway present in microorganisms and it may further contribute to survival under harsh environmental conditions12.
Moreover, a number of osmoregulation receptors and transport systems were determined in the D5A genome. These genes can encode up to 24 two component systems (TCSs), among which 16 TCSs can be functionally assigned based on the KEGG database (Table S4). Of those 16 assigned TCSs, seven belong to the OmpR family, five to the NarL family, one to the LytT family, one to the NtrC family and two to the CitB family. The eight remaining TCS genes are annotated as sensor histidine kinase (Table S4). For example, the KdpD/KdpE TCSs that belong to the OmpR family have genes responsible for testing hyperosmotic stress and regulating the expression of genes in cell wall synthesis and for the accumulation of compatible solutes41. They may also activate the expression of the kdp operon which encodes the high-affinity K+ uptake system (Kdp) in response to high salt stress42. In addition, genes encoding transport systems such as K+ transport systems for K+ accumulation43 and Na+/H+ antiporters (nha) for importing H+ and pumping out Na+ 44 have also been found to resist hyperosmotic stress in the genome of D5A (Table S5).
Response of D5A to pH and genes putatively involved in wide pH adaptation
D5A was isolated from a saline-alkali oilfield which had a pH of 9.7 and it was adapted to a wide range of pH conditions as its growth was unaffected by pH values between 4 and 10. By contrast, common neutralophilic bacteria grow over a narrow range of external pH values (5.5–9.0) and maintain a near-neutral cytoplasmic pH that lies within a pH range of 7.5–7.745,46. Hence, they are able to acidify or alkalize the cytoplasm relative to the external environment to meet pH challenges by direct active uptake or efflux of protons. For instance, under conditions of acid challenge the gene kdpABC in the D5A genome that encodes the high-affinity K+ transport system can be activated by kdpD/kdpE. This may cause an inside-positive membrane potential by activating K+ influx and thus avert the ingress of protons47. Cytoplasmic pH has also been reported to be maintained by the metabolism of proton buffer molecules such as phosphate uptake (pstSCAB) and the expression of amino acid decarboxylase like arginine (speA), aspartate (panD) and lysine (cadA) decarboxylation, all of which were found in the D5A genome (Table S6). Moreover, some genes responsible for acid resistance occur in acid mine drainage environments as reported by Guazzaroni et al.48 including the nucleic acid-binding proteins of Hu (hupA, hupB), ClpXP (clpX, clpP) proteins, and Dps (dps), and the transcriptional repressor LexA (lexA) have also been found in the D5A genome. The genes encoding urease (ureABC) and urease accessory protein (ureEFGDH) that increases ammonia production are present in D5A and have an acid resistance ability49,50. In addition, several protective proteins including GroEL, DnaK, and HdeB chaperone that might be induced under acid stress48 were found in the D5A genome (Table S6).
For pH homeostasis under alkaline stress, inward transport of protons is an important adaptation mechanism that usually involves the activation of key cation/proton antiporters. A total of eight cation/proton antiporters exist in the D5A genome, including Na+/H+ antiporters, K+/H+ antiporters, Ca2+/H+ antiporters and some combinations of these cytoplasmic cations that exchange cations for external H+ moving inward (Table S6). A multidrug transporter gene (mdfA) was identified here which was earlier described as activating the outflow of numerous drug substrates in exchange for H+ and also catalyzing K+/H+ antiporter and Na+/H+ antiporter activity. This might support bacterial growth at pH > 9. Furthermore, it is well known that the genes encoding F1F0-ATP synthase which imports protons during ATP synthase activity will be induced under alkaline conditions. The D5A genome has an atp operon with the expected eight ATP synthase protein-encoding genes and an atpI gene whose protein product contributes to the stability and assembly of the synthase. Identification of the cls gene in D5A that encodes membrane cardiolipin in alkaliphiles will support oxidative phosphorylation by facilitating rapid proton transport along the membrane surface. In addition, expression of genes coding for periplasmic proteins such as OmpA, MalE and OmpX porins in the D5A genome likely leads to metabolic modes that are adapted to high pH. Some amino acid catabolism enzymes such as serine deaminase (sdaA) may also function at high pH (Table S6).
Degradation of aromatic compounds
D5A was isolated from an oil-polluted soil and can therefore potentially be adopted in oilfield bioremediation. Aromatic compounds have been recognized as the most recalcitrant and abundant pollutants in oilfields. Numerous genes associated with aromatic compounds have been determined in the D5A genome. For example the genes involved in 3-hydroxyphenylpropionate (3-HPP) catabolism including the mhpRABCDFET operon were identified here (Table S7). In addition, the D5A genome also possesses a complete β-ketoadipate pathway through the protocatechuate and catechol routes for further degradation of the ring cleavage products to TCA cycle intermediates (Fig. 2) which are derived from 4-hydroxybenzoate and benzoate separately. More than 19 genes involved in the protocatechuate (pca genes) and catechol (cat genes) branches of the β-ketoadipate catabolism pathway were identified in D5A (Table S7). Protocatechuate is recognized as one of the key intermediates during the catabolism of various aromatic compounds51. This pathway is considered to be one of the key routes for the degradation of aromatic compounds and is similar to those found in other Klebsiella genomes. This indicates that D5A has a broad potential for the degradation of aromatic compounds.
Central metabolic pathways
A schematic summary of the metabolic patterns in Klebsiella sp. D5A is shown in Fig. 2. The genome of Klebsiella sp. D5A shows that it carries genes consistent with the ability to survive in the soil environment and in plant rhizospheres. The genome also contains a complete carbohydrate metabolism pathway including glycolysis/gluconeogenesis, the tricarboxylic acid (TCA) cycle, pyruvate metabolism, and the pentose phosphate (PPP) and Entner-Doudoroff (ED) pathways.
Sulfur metabolism in D5A includes mineralization of organic sulfonates and assimilation of inorganic sulfate (Fig. 2). A total of 17 ORFs encoding inorganic thiosulfate or sulfate transporter and convert-related genes were present in the D5A genome (Table S8). Alkyl/aryl-sulfonates are considered to be key components of the sulfur present in agricultural soils34. In the genome of D5A, extracellular alkanesulfonates are first transported into the cell by aliphatic sulfonate ABC transport (ssuABC) and then catalyzed by alkanesulfonate monooxygenase (ssuD) and an NADPH-dependent FMN reductase (ssuE). Moreover, the D5A genome has four genes encoding taurine transporter-related proteins, tauABCD (Table S8). In addition, a transcriptional regulator coding gene cysB which can mediate global sulfur regulation in many gram-negative bacteria was identified in the D5A genome. This activates the transcription of cysteine synthesis genes under conditions of sulfur limitation52. Recently, hydrogen sulfide (H2S) produced by PGPR has been reported to increase seed germination and promote the growth of the plant roots that they colonize. The genes cysCIJN that are responsible for H2S biosynthesis as reported by Dooley et al.53 have been found in the D5A genome.
Secretion systems
D5A has seven potential protein secretion systems including Types I, II, III, V and VI, Tat (twin arginine translocation), and Sec (general secretory pathway) (Table S9). The Sec and Tat systems are the two widespread systems for transport across the cytoplasmic membrane. D5A has both of the secretion systems. In contrast to strain D5A, the other four Klebsiella genomes all lack the genes encoded for Types III and V but only K. variicola 342 possess the genes encoded for Type IV.
Discussion
In this study we report the whole genome sequencing and analysis of Klebsiella sp. D5A isolated from the rhizosphere soil of tall fescue growing in an oil-contaminated soil. The genome data of this strain support and extend various laboratory observations that have been reported in our previous study. Liu et al.15 found that Klebsiella sp. D5A exerts beneficial effects on plant growth as it may promote tall fescue germination rate, root length and shoot height. Consistent with the PGP properties that have been referred in our earlier study, we found genes attributable to IAA production, phosphate solubilization and siderophore synthesis. In addition, other PGP trait coding genes including acetoin and 2,3-butanediol synthesis, putrescine and spermidine synthesis and some other PGPR fitness conferring genes have also been found in the D5A genome. Genes with similar functions in other PGPR have been previously been reported from other studies12,34,54. In addition to their direct plant growth promoting abilities PGPR also support plant growth indirectly by suppressing pathogens55. In the D5A genome we have identified many genes that are well known to be responsible for the production of antimicrobial compounds such as 4-hydroxybenzoate, phenazine, chitinase and GABA. The genome also encodes enzymes including catalases, peroxidases, glutathione transferases and superoxide dismutases, all of which are responsible for resisting oxidative stresses in plants. Genome sequence analysis also shows that D5A has many genes corresponding to Sec, Tat and Types I, II, III, V, and VI (Table S9). Some earlier studies have reported that the presence of Type I–VI and Sec secretion systems in the rhizobacteria Variovorax paradoxus and Pseudomonas fluorescens may function in promoting plant growth12,34,56 and also help in their rhizosphere colonization57.
A fundamental role of the Klebsiella genome is its capacity to fix nitrogen19,20,58. 20 proposed nif-specific genes which are related to nitrogenase were all found in the D5A genome. In addition, sulfur is recognized as an essential nutrient for plant growth and is closely connected with stress tolerance in plants59. Generally, plants acquire sulfur from soils and rely on the mobilization of this sulfur for assimilation by plants as determined by the soil microbial community60. In the D5A genome we have found genes related to H2S synthesis and they may be an important sulfur source for plant growth31. We have also identified genes in the D5A genome that are involved in the degradation of aromatic compounds, suggesting a function in the catabolism of some organic compounds. The comparative genomics analysis of some PGP traits with four other representative Klebsiella PGPR strains reveals some conserved genes among the different Klebsiella species, such as IAA, solubilization of phosphates, scetoin and 2,3-butanediol synthesis and production of antimicrobial compounds, except for some strain-specific genes differentiated in each strain. This provides clues as to the characteristics common to Klebsiella PGPR.
Liu et al.15 found that strain D5A was capable of surviving and growing well under a wide range of NaCl concentrations which may help it establish well under harsh environmental conditions. The genes involved in betaine, trehalose, acetoin and 2,3-butanediol biosynthesis were detected in the D5A genome and these have been implicated in the survival of some microorganisms under saline or osmotic stress conditions38. In addition, many genes involved in osmosensing and regulation genes that belong to TCSs, as well as Na+/H+ antiporters (nha) for importing H+ and pumping out Na+ 44 were present in the D5A genome. Furthermore, the existence of K+ transporters may help the K+ influx that is regulated by KdpD/KdpE TCS and the possible accumulation of K+ inside the cells may be activated to resist osmotic pressure61. The coefficient of TCSs and related transporters might alter membrane permeability and activate the expression of osmoresponsive genes62. In addition, the presence of genes that regulate the production of cold-shock and heat-shock proteins and osmoregulants in the D5A genome may also help in adaptation to harsh environments for survival.
Strain D5A was isolated from a highly alkaline soil (pH 9.7) and previous experiments have indicated that it can adapt to a wide range of pH values as its growth was unaffected by pH levels between 4 and 1015. It is clear that adaptation to a wide range of environmental pH requires a robust internal pH homeostatic system for bacteria, thus maintaining a near-neutral cytoplasmic pH that is suitable for structural integrity and optimum functionality of the cytoplasmic proteins45,63. Numerous adaptive strategies are deployed for pH homeostasis under acid/alkaline pressures including variation in transport and metabolic patterns. The primary strategy for bacterial pH homeostasis is using the transporters that directly regulate the uptake or outflow of protons. For example, when D5A was exposed to acidic conditions the kdpABC genes in D5A that encode the ABC high-affinity potassium transport system were regulated by kdpD/kdpE and generated an inside-positive membrane potential through active influx of K+ to partially deflect the inward flow of protons whereas the expression of key cation/proton transporters that extrude cytoplasmic Na+, K+ and Ca2+ in exchange for H+ would be active and function under alkaline conditions. Among these cation/proton transporters, Na+/H+ antiporters (nhaA) are recognized as having the predominant role in alkaline pH homeostasis as the stoichiometry for nhaA is 2H+/1Na+. However, nhaA is a model of pH-regulated antiporter for pH homeostasis and is unable to support the growth of Escherichia coli at pH > 9 while mdfA can do so46. The gene mdfA that encodes multidrug translocase was found in the D5A genome. The expression of F1F0-ATP synthase that carries protons into the cell during ATP synthesis will decrease under acidic conditions and the hydrolysis of F1F0-ATPase that facilitates ATP-dependent H+ outflow will be upregulated64.
A secondary strategy for pH homeostasis is the remodeling of metabolic patterns that regulate proton generation or consumption by metabolic enzymes45. In the D5A genome, genes encoding amino acid decarboxylases such as arginine (speA), aspartate (panD) and lysine (cadA) decarboxylation would help the strain survive by acid tolerance by consuming protons and thus lead to decreased concentrations of free protons in the cytoplasm64. In contrast, challenges by alkaline conditions will lead to activation of amino acid deaminases such as serine deaminase (sdaA) determined in the D5A genome65. Deaminases can provide an acid-generating mechanism that is adaptive to alkaline conditions as decarboxylases promote alkalization that is adaptive to acid enviroments46. Moreover, genes encoding ClpXP protease, urease, the transcriptional repressor LexA and nucleic acid-binding proteins such as an RNA-binding protein, HU and Dps, that are found in the D5A genome (Table S6) have been reported to be able to expand the capability of bacteria to survive under severe acid stress48. The existence of these genes carried by the D5A genome provides a fundamental function for pH homeostasis, while the detailed elucidation of how they function or contribute to pH, cation or osmotic homeostasis requires further transcription studies.
Conclusions
The Klebsiella sp. D5A genome has a total of 5,540,009 bp with a 57.15% G+C content. Whole-genome comparison with 15 other completely sequenced Klebsiella strains reveals that D5A belongs to the K. variicola group. As confirmed from previous reports and supported by the genome analysis in the present study, Klebsiella sp. D5A is demonstrated to act as a PGPR as the genes potentially involved in plant growth promotion such as indole-3-acetic acid (IAA) biosynthesis, phosphate solubilization, siderophore production, acetoin and 2,3-butanediol synthesis, N2 fixation, chitinase, phenazine, 4-hydroxybenzoate, and H2S synthesis, and some other PGPR conferring genes were found. Comparative genomic analysis of four other representative PGPR with D5A reveals some conserved regions indicating common PGP characteristics among these Klebsiella PGPR. The Klebsiella sp. D5A genome also contains sets of catabolic genes involved in the degradation of aromatic compounds. Moreover, genes that contribute to the high salinity resistance of D5A were also observed including glycine-betaine synthesis, trehalose synthesis and a number of osmoregulation receptors and transport system coding genes. In addition, the strain grew well in the pH range 4–10 with the putative genes responsible for pH homeostasis but detailed elucidation of how these genes are regulated and function requires further transcription studies.
Materials and Methods
PGPR strain
Klebsiella sp. D5A is a previously studied gram-negative PGPR isolated from the roots of tall fescue. The 16S rRNA gene sequence of D5A exhibits the highest (99.5%) sequence similarity to Klebsiella variicola (HQ259961). Details of the plant growth promotion properties (IAA production, phosphate solubilization, siderophore production) and environmental tolerance (pH, NaCl, Na2CO3 tolerance level) of Klebsiella sp. D5A are shown in Table 3 15. Additionally, the nitrogen fixation property was tested according to Wu et al.66. Given the beneficial attributes of D5A, we chose to characterize them further at the genomic level.
Bacterial growth and DNA extraction
A single colony of Klebsiella sp. D5A grown on ADF agar medium67 was inoculated into 100 mL of LB medium (10 g tryptone, 5 g yeast extract and 10 g NaCl per liter) and shaken at 150 rpm at 30 °C for 48 h. Bacterial cells were collected by centrifugation and the genomic DNA was extracted with a Fast DNA SPIN kit (MP Biomedicals, Solon, OH) according to the manufacturer’s instructions and the DNA was checked on agarose gel.
Genome sequencing and annotation
The genome sequence of Klebsiella sp. D5A was determined by Shanghai Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China) using the HiSeq 2000 sequencing platform (Illumina Inc., San Diego, CA). The prepared DNA samples were used to construct the pair-end and mate-pair sequencing libraries. The average fragment sizes for the pair-end and mate-pair libraries were 300 and 3,000 bp, respectively. The read length was 101 bp. Low quality sequence data were cut and then the reads were assembled using the SOAPdenove v2.04 program (including GapCloser v1.12) (http://soap.genomics.org.cn/)68. Glimmer 3.0 (www.cbcb.umd.edu/software/glimmer) software was used to predict genes and Barrnap 0.4.2 and tRNA scan - SE v1.3.1 software to forecast the rRNA and tRNA of the genome. The protein sequences encoded by genes were blasted with each function database (KEGG, Nr, COG, String and GO).
Gene network/pathway analysis
Predicted and annotated gene sequences were analyzed for similarity with the KEGG enzyme database followed by assignment of each gene into the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway chart. Based on individual analysis results of the KEGG pathways, integrated biochemical pathway maps were constructed which demonstrated characteristic physiological features in the metabolism of D5A. The existence of a certain pathway was then determined and integrated when component genes within the corresponding pathway had been completely identified.
Phylogenetic analysis
The 16S rRNA gene sequences of Klebsiella sp. D5A were aligned with those of the publicly available Klebsiella genome sequences using the MAFFT (Multiple Alignment using Fast Fourier Transform) program based on 45 single copies of the homologous genes that were universally distributed in 16 analyzed Klebsiella genomes. The phylogenetic tree was constructed based on aligned concatenated sequences of these 45 genes using the bootstrap method available in RAxML (Randomized Axelerated Maximum Likelihood) which is a popular program for phylogenetic analysis of large datasets under maximum likelihood69.
Data submission
The whole genome shotgun project of Klebsiella sp. D5A has been deposited at DDBJ/EMBL/GenBank under the accession LOAR00000000. The version described in this paper is version LOAR01000000. This strain has also been deposited in the CGMCC (China General Microbiological Culture Collection Center) under the accession number NO. 7248.
Additional Information
How to cite this article: Liu, W. et al. Whole genome analysis of halotolerant and alkalotolerant plant growth-promoting rhizobacterium Klebsiella sp. D5A. Sci. Rep. 6, 26710; doi: 10.1038/srep26710 (2016).
Supplementary Material
Acknowledgments
We thank the Key Research Program of the Chinese Academy of Sciences (Grant No. KFZD-SW-303), the National Natural Science Foundation of China (41001182, 41201313) and the Key Projects in the National Science and Technology Pillar Program (2015BAD05B04) for financial support.
Footnotes
The authors declare no competing financial interests.
Author Contributions W.X.L. and Q.L.W. contributed equally to this work: analyzed data and wrote the manuscript. J.Y.H., C.T., Y.M.L. and P.C. provided ideas in data analysis and writing.
References
- Ahemad M. & Kibretm M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J King Saud Univ Sci 26, 1–20 (2014). [Google Scholar]
- Jahanian A., Chaichi M. R., Rezaei K., Rezayazdi K. & Khavazi K. The effect of plant growth promoting rhizobacteria (PGPR) on germination and primary growth of artichoke (Cynara scolymus). Int J Agric Crop Sci 4, 923–929 (2012). [Google Scholar]
- Glick B. R. Plant growth-promoting bacteria: mechanisms and applications. Scientifica doi: 10.6064/2012/963401 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahemad M. & Khan M. S. Evaluation of plant-growth-promoting activities of rhizobacterium Pseudomonas putida under herbicide stress. Ann Microbiol 62, 1531–1540 (2012). [Google Scholar]
- Ma Y., Rajkumar M., Luo Y. M. & Freitas H. Inoculation of endophytic bacteria on host and non-host plants-effects on plant growth and Ni uptake. J Hazard Mater 195, 230–237 (2011). [DOI] [PubMed] [Google Scholar]
- Tank N. & Saraf M. Salinity-resistant plant growth promoting rhizobacteria ameliorates sodium chloride stress on tomato plants. J Plant Interact 5, 51–58 (2010). [Google Scholar]
- Hynes R. K., Leung G. C. Y., Hirkala D. L. M. & Nelson L. M. Isolation, selection, and characterization of beneficial rhizobacteria from pea, lentil, and chickpea grown in western Canada. Can J Microbial 54, 248–258 (2008). [DOI] [PubMed] [Google Scholar]
- Liu W. X. et al. Rhizobacteria (Pseudomonas sp. SB) assist phytoremediation of oily-sludge-contaminated soil by tall fescue (Testuca arundinacea L.). Plant Soil 371, 533–542 (2013). [Google Scholar]
- Lai Q. & Shao Z. Genome sequence of an alkane-degrading bacterium, Alcanivorax pacificus type strain W11-5, isolated from deep sea sediment. J Bacteriol 194, 6936–6936 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu T. T. et al. Whole genome analysis of a community-associated methicillin-resistant Staphylococcus aureus ST59 isolate from a case of human sepsis and severe pneumonia in China. PloS One 9, e89235 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S. C. Next-generation sequencing transforms today’s biology. Nat Methods 5, 16–18 (2008). [DOI] [PubMed] [Google Scholar]
- Duan J., Jiang W., Cheng Z., Heikkila J. J. & Glick B. R. The complete genome sequence of the plant growth-promoting bacterium Pseudomonas sp. UW4. Plos One 8, 462–469 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. Y. et al. Genome sequence of the plant growth-promoting rhizobacterium Bacillus sp. strain JS. J Bacteriol 194, 3760–3761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. et al. Complete genome sequence of Paenibacillus polymyxa SQR-21, a plant growth-promoting rhizobacterium with antifungal activity and rhizosphere colonization ability. Genome Announce 2, e00281–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. X., Hou J. Y., Wang Q. L., Ding L. L. & Luo Y. M. Isolation and characterization of plant growth-promoting rhizobacteria and their effects on phytoremediation of petroleum-contaminated saline-alkali soil. Chemosphere 117, 303–308 (2014). [DOI] [PubMed] [Google Scholar]
- Bao G. et al. Genome sequence of Klebsiella oxytoca M5al, a promising strain for nitrogen fixation and chemical production. Genome Announce 1, e00074–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Lin L., Zhang Y., Sun L. & An Q. Genome sequence of Klebsiella oxytoca SA2, an endophytic nitrogen-fixing bacterium isolated from the pioneer grass Psammochloa villosa. Genome Announce 1, e00601–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts D. E. et al. Complete Genome Sequence of the N2-Fixing Broad Host Range Endophyte Klebsiella pneumoniae 342 and Virulence Predictions Verified in Mice. Plos Genet 4, e1000141 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto-Tomás A. A. et al. Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 326, 1120–1123 (2009). [DOI] [PubMed] [Google Scholar]
- Wei C. Y. et al. Endophytic nitrogen-fixing Klebsiella variicola strain DX120E promotes sugarcane growth. Biol Fertil Soils 50, 657–666 (2014). [Google Scholar]
- Andrade B. G. N., de-Veiga Ramos N., Marin M. F. A., Fonseca E. L. & Vicente A. C. P. The genome of a clinical Klebsiella variicola strain reveals virulence-associated traits and a pl9-like plasmid. FEMS Microbiol Lett 360, 13–16 (2014). [DOI] [PubMed] [Google Scholar]
- Ramos P. I. P. et al. Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms. BMC Genomics 15, 54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Ramos U. et al. Development of a Multiplex-PCR probe system for the proper identification of Klebsiella variicola. BMC microbiol 15, 64 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A. H. Future trends in research on microbial phosphate solubilization: one hundred years of insolubility. Dev Plant Soil Sci 102, 91–96 (2007). [Google Scholar]
- Wagh J., Shah S., Bhandari P., Archana G. & Kumar G. N. Heterologous expression of pyrroloquinoline quinone (pqq) gene cluster confers mineral phosphate solubilization ability to Herbaspirillum seropedicae Z67. Appl microbiol biotechnol 98, 5117–5129 (2014). [DOI] [PubMed] [Google Scholar]
- Toyama H. & Lidstrom M. E. pqqA is not required for biosynthesis of pyrroloquinoline quinone in Methylobacterium extorquens AM1. Microbiology 144, 183–191 (1998). [DOI] [PubMed] [Google Scholar]
- Yan Y. L. et al. Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc Natl Acad Sci 105, 7564–7569 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu C. M., et al. Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci USA 100, 4927–4932 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. M. et al. 2R, 3R-butanediol, a bacterial volatile produced by Pseudomonas chlororaphis O6, is involved in induction of systemic tolerance to drought in Arabidopsis thaliana. Mol Plant-Microbe Interact 21, 1067–1075 (2008). [DOI] [PubMed] [Google Scholar]
- Han S. H. et al. GacS-dependent production of 2R, 3R-butanediol by Pseudomonas chlororaphis O6 is a major determinant for eliciting systemic resistance against Erwinia carotovora but not against Pseudomonas syringae pv. tabaci in tobacco. Mol Plant-Microbe Interact 19, 924–930 (2006). [DOI] [PubMed] [Google Scholar]
- Gupta A. et al. Whole genome sequencing and analysis of plant growth promoting bacteria isolated from the rhizosphere of plantation crops coconut, cocoa and arecanut. Plos One 9, e104259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper J. E. et al. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. Plos Genet 8, 656–662 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassán F. et al. Cadaverine production by Azospirillum brasilense and its possible role in plant growth promotion and osmotic stress mitigation. Eur J Soil Biol 45, 12–19 (2009). [Google Scholar]
- Han J. I. et al. Complete genome sequence of the metabolically versatile plant growth-promoting endophyte Variovorax paradoxus S110. J Bacteriol 193, 1183–1190 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klipp W., Masepohl B., Gallon J. R. & Newton W. E. Genetics and Regulation of Nitrogen Fixation in Free-Living Bacteria, Vol. 2 (ed. Klipp W. et al.) (Springer Science & Business Media, 2004). [Google Scholar]
- Xie Z. H. et al. Interaction between NifL and NifA in the nitrogen-fixing Pseudomonas stutzeri A1501. Microbiology 152, 3535–3542 (2006). [DOI] [PubMed] [Google Scholar]
- Jeong H. S. & Jouanneau Y. Enhanced nitrogenase activity in strains of Rhodobacter capsulatus that overexpress the rnf genes. J Bacteriol 182, 1208–1214 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Wang Z. F. & Xu A. K. Advancement in the research of salt-alkali tolerance genes in alkaligrass. Acta prataculturae sinica 19, 260–266 (2010). [Google Scholar]
- Garg A. K. et al. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA 99, 15893–15903 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M. J., Primavesi L. F., Jhurreea D. & Zhang Y. Trehalose metabolism and signaling. Annu Rev Plant Biol 59, 417–441 (2008). [DOI] [PubMed] [Google Scholar]
- Moker N. et al. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol Microbiol 54, 420–438 (2004). [DOI] [PubMed] [Google Scholar]
- Heermann R. & Jung K. The complexity of the ‘simple’ two-component system KdpD/KdpE in Escherichia coli. FEMS Microbiol Lett 304, 97–106 (2010). [DOI] [PubMed] [Google Scholar]
- Epstein W. The roles and regulation of potassium in bacteria. Prog Nucleic Acid Res Mol Biol 75, 293–320 (2003). [DOI] [PubMed] [Google Scholar]
- Hunte C. et al. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435, 1197–1202 (2005). [DOI] [PubMed] [Google Scholar]
- Krulwich T. A., Sachs G. & Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9, 330–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padan E., Bibi E., Ito M. & Krulwich T. A. Alkaline pH homeostasis in bacteria: new insights. BBA-Biomembranes 1717, 67–88 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker-Austin C. & Dopson M. Life in acid: pH homeostasis in acidophiles. Trends Microbiol 15, 165–171 (2007). [DOI] [PubMed] [Google Scholar]
- Guazzaroni M. E., Morgante V., Mirete S. & González‐Pastor J. E. Novel acid resistance genes from the metagenome of the Tinto River, an extremely acidic environment. Environ microbiol 15, 1088–1102 (2013). [DOI] [PubMed] [Google Scholar]
- Booth I. R., Cash P. & O’byrne C. Sensing and adapting to acid stress. Antonie Van Leeuwenhoek 81, 33–42 (2002). [DOI] [PubMed] [Google Scholar]
- Ferrero R. L., Cussac V., Courcoux P. & Courcoux P. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J bacteriol 174, 4212–4217 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez J. I., Miñambres B., García J. L. & Díaz E. Genomic insights in the metabolism of aromatic compounds in Pseudomonas// In Pseudomonas (ed. Jiménez J. I. et al.) 425–462 (Springer US, 2004). [Google Scholar]
- Hryniewicz M. M. & Kredich N. M. The cysP promoter of Salmonella typhimurium: characterization of two binding sites for CysB protein, studies of in vivo transcription initiation, and demonstration of the anti-inducer effects of thiosulfate. J Bacteriol 173, 5876–5886 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley F. D., Nair S. P. & Ward P. D. Increased growth and germination success in plants following hydrogen sulfide administration. PLoS One 8, e62048 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez H., Fraga R., Gonzalez T. & Bashan Y. Genetics of phosphate solubilization and its potential applications for improving plant growth-promoting bacteria. Plant Soil 287, 15–21 (2006). [Google Scholar]
- Lugtenberg B. & Kamilova F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63, 541–556 (2009). [DOI] [PubMed] [Google Scholar]
- Preston G. M., Bertrand N. & Rainey P. B. Type III secretion in plant growth promoting Pseudomonas fluorescens SBW25. Mol Microbiol 41, 999–1014 (2001). [DOI] [PubMed] [Google Scholar]
- Barret M. et al. Characterization of the SPI-1 and Rsp type three secretion systems in Pseudomonas fluorescens F113. Environ Microbiol Rep 5, 377–386 (2013). [DOI] [PubMed] [Google Scholar]
- Iniguez A. L., Dong Y. & Triplett E. W. Nitrogen fixation in wheat provided by Klebsiella pneumoniae 342. Mol Plant Microbe Interact 17, 1078–1085 (2004). [DOI] [PubMed] [Google Scholar]
- Gill S. S. & Tuteja N. Cadmium stress tolerance in crop plants: probing the role of sulfur. Plant Signaling Behav 6, 215–222 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M. A., Fellows E. & Schmalenberger A. Rhizobacteria and plant sulfur supply. Adv Appl Microbiol 62, 235–268 (2007). [DOI] [PubMed] [Google Scholar]
- Nie Y. et al. The genome of the moderate halophile Amycolicicoccus subflavus DQS3-9A1T reveals four alkane hydroxylation systems and provides some clues on the genetic basis for its adaptation to a petroleum environment. Plos One 8, e70986 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. M. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol Mol Biol Rev 63, 230–262 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalva M. A., Tilburn J., Bignell E. & Arst H. N. Ambient pH gene regulation in fungi: making connections. Trends Microbiol 16, 291–300 (2008). [DOI] [PubMed] [Google Scholar]
- Richard H. & Foster J. W. Escherichia coli glutamate-and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriolo 186, 6032–6041 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale E. F. & Epps H. M. R. The effect of the pH of the medium during growth on the enzymic activities of bacteria (Escherichia coli and Micrococcus lysodeikticus) and the biological significance of the changes produced. Biochem J 36, 600 (1942). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Yue H., Lu J. & Li C. Characterization of rhizobacterial strain Rs-2 with ACC deaminase activity and its performance in promoting cotton growth under salinity stress. World J Microb Biot 28, 2383–2393 (2012). [DOI] [PubMed] [Google Scholar]
- Penrose D. M. & Glick B. R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant 118, 10–15 (2003). [DOI] [PubMed] [Google Scholar]
- Li R. Q. et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20, 265–272(2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.