Abstract
Maize is one of the most important crops worldwide and is strongly dependent on arbuscular mycorrhiza (AM) fungi, organisms that form a mutualistic association with land plants. In maize, AM symbiosis enhances spike dry weight, spike length, spike circumference, and the dry weight and dimensions of the grain. Notwithstanding its ubiquitous nature, the detailed relationship between AM fungal colonization and plant development is not completely understood. To facilitate a better understanding of the effects of AM fungi on plants, the work reported here assessed the effects of a consortium of AM fungi on the kernel proteome of maize, cultivated in open-field conditions. To our knowledge, this is the first report of the modulation of a plant seed proteome following AM fungal inoculation in the field. Here, it was found that AM fungi modify the maize seed proteome by up-regulating enzymes involved in energetic metabolism, embryo development, nucleotide metabolism, seed storage and stress responses.
Mycorrhiza represent a widespread mutualistic association between most land plants, including agriculturally relevant species1, and arbuscular mycorrhizal (AM) fungi, a monophyletic group of soil microorganims belonging to the Glomeromycota phylum2. When the symbiosis is established, the fungus grows within the cells of the roots forming arbuscules, which is the main site of nutrient exchange between the fungus and the plant. Moreover, the fungus develops an extensive extraradical mycelium that enhances the absorption ability of the plant root system3. The success of AM symbiosis is mostly due to the benefits that both partners gain from this relationship. The fungal partner takes up both water and mineral nutrients, mainly phosphorus and nitrogen, from the soil, through its mycelium, and transfers these compounds via the symbiotic interface to the plant root cells4,5. In turn, the plant supplies the fungus with about 10–20% of the plant’s photosynthates. This symbiosis directly influences plant responses and plant physiology, both in the target organ (roots) and in shoots, and as recently demonstrated in fruits6,7,8,9. As a consequence of this plant-fungal relationship, the AM symbiosis enhanced yield and improved fruit quality (taste and vitamin concentration) in strawberry fruits6,7,8,9; modulated sugar and carotenoid concentrations in tomato fruits10; increased the accumulation of carotenoids, chlorophylls and tocopherol in green and red leaf lettuces11; improved the yield and quality of saffron (Crocus sativus L.)12; increased growth, flavour and yield in Allium sativum L. cultivated in field conditions13; impacted the phenolic content and antioxidant properties of artichoke leaves14; and modulated essential oil production in Artemisia annua L.15 and in Ocimum basilicum L.16,17. A large body of evidence has shown that the protein profile of Pteris vittata and Medicago truncatula root18,19,20, and P. vittata, Populus alba and Zea mays leaf21,22,23, are affected by AM symbiosis. The above mentioned studies found that the plant traits that were positively affected by AM fungi included photosynthesis, carbon fixation and energy production in leaves, and glycolysis in roots. Notwithstanding these results, there is very little data regarding the impact of AM fungi on the plant seed proteome.
Maize (Zea mays L.) is one of the most important crops worldwide. Its economic and nutritional value is mainly due to the high starch content that represent about 75% of mature seed weight24. Maize is strongly dependent on mycorrhizae25. For example, in maize, the AM symbiosis enhances spike dry weight, spike length, spike circumference, and the dry weight and dimensions of the grain26. To better understand the effect of AM fungi on maize, the present study was undertaken with the aim of assessing the effects of a consortium of AM fungi on the maize kernel proteome, cultivated in open-field conditions.
Results and Discussion
According to FAO, cereals are defined as a group of species generally, but not exclusively, belonging to the gramineous family (i.e. Poaceae) that produce dry seeds rich in starch. Of the cereals, the most commonly cultivated plant is maize; this is because of its multiple uses, as a food and feed, and as a source of raw materials for industrial applications, such as the production of bioplastics and biofuels. While it has been known for some time that maize is a mycorrhiza-dependent plant25, the effects of the interaction between AM fungi and seeds has not been exhaustively investigated. To the best of our knowledge, this is the first report describing the effects of AM fungal inoculation, in open field conditions, on seed protein composition using a proteomic approach. As reported previously26, maize plant roots are naturally colonized by autochthonous AM fungi. Perhaps not surprisingly, using field soil, the frequency and the intensity of the mycorrhizal colonization, as well as arbuscule abundance, were significantly higher in plants treated with an AM fungal inoculum than in control plants. For example, the mycorrhizal colonization degree (M%) in mycorrhiza inoculated plants (MIC) was 27.7 ± 4.6 while in control plants (CTRL) was 6.9 ± 0.9. Moreover, it has been demonstrated26 that AM fungal inoculation increased maize plant growth and grain yield. In particular, spikes produced by MIC plants were greater in both number and size than those produced by the CTRL plants. In addition, the number, the dry weight, the size and the morphology of kernels were also increased by mycorrhizal inocula.
In the present work, seeds from CTRL and MIC plants collected 20 days after flowering (DAF) and 60 DAF were used for biochemical and proteomic analyses. Maize seeds accumulate large amount of proteins beginning with the first phases of seed development (Table 1), however, the differences between the amounts of protein in four protein different classes in CTRL and MIC plants were not statistically significant (p > 0.05). This data is consistent with previous results27 in a study examining the early accumulation of proteins in developing kernels. We subsequently investigated the possible modulation of the relative amounts of the different seed protein classes as a consequence of the AM inoculation. The results are shown in Table 1. According to Osborne28, seed proteins may be classified into groups according to their solubility in a series of solvents as albumins (water), globulins (dilute aqueous salt solutions), prolamins (alcohol solutions) and glutelins (dilute alkali or acid). At 20 DAF, the albumin fraction represented the majority of the kernel proteins in both CTRL and MIC samples. At 60 DAF, when the seeds were fully mature, the albumin content was significantly reduced with more albumin fraction in CTRL than in MIC plants. The albumin fraction consits largely of metabolically active proteins and, thus, a greater amount of this kind of proteins was expected in the first sampling, when intensive seed filling occurs. Mature seeds contain a limited set of enzymes, with the majority of them necessary to sustain the ability of the seed to resume metabolic activities during germination. Globulins, which are deposited in the embryo and in the outer aleurone layer29, were accumulated in a larger amount in mature seeds than in the 20 DAF seeds; they were positively affected by AM treatment. The globulin fraction is a heterogeneous group that includes the 7S proteins (also called vicilins), which include the major maize storage protein globulin30, the 11S storage proteins (legumins), various kind of defence proteins and the lipid transfer protein (LTP), one of the main maize seed allergens31. Zeins are prolamins and are the main storage proteins in the starchy endosperm tissue32, accounting for about 45–50% of the total maize seed proteins33. Zeins are classified according to structural features as α-, β-, γ- and ω-zeins, the first of which is the most abundant and is encoded by at least four gene families34. It has been shown that the accumulation of zeins begins very early (15 DAF) and continues through most of seed development34. In these experiments, the presence of zeins reached about 32% in 20 DAF CTRL plant. Moreover, AM treatment boosted their relative amount up to 42%. In mature seeds, zeins accounted for about 45–47% of the total seed proteins, independent of the presence of the AM fungal symbiosis. Glutelins, together with zeins, are major storage proteins of the seed endosperm. They represent the second largest protein fraction in mature seeds and show sequence similarities to other cereal storage proteins, such as gliadins and glutenins35. Overall, these results indicate that AM treatment does not greatly influence the accumulation of the analysed protein fractions either at the beginning of the seed filling process or in mature seeds.
Table 1. Relative amounts (%) of maize seed proteins.
| 20 DAF |
60 DAF |
|||
|---|---|---|---|---|
| CTRL | MIC | CTRL | MIC | |
| Albumin | 34.7 ± 5.3 aA | 31.1 ± 6.2 bA | 15.1 ± 1.7 aB | 10.3 ± 2.4 bB |
| Globulin | 8.1 ± 4.4 aA | 5.3 ± 2.6 aA | 9.9 ± 2.2 aA | 16.8 ± 2.2 bB |
| Prolamins | 32.1 ± 6.1 aA | 41.9 ± 5.7 aA | 44.6 ± 3.1 aB | 47.4 ± 2.6 aA |
| Glutelins | 20.0 ± 5.4 aA | 21.6 ± 5.2 aA | 27.9 ± 3.4 aA | 25.2 ± 3.9 aA |
Seed proteins were classified in groups according to their solubility in a series of solvents including albumin (soluble in water), globulins (soluble in dilute aqueous salt solution), prolamins (soluble in alcohol solution) and glutelins (soluble in dilute alkali). Data are expressed as means ± standard error (Three biological samples were analysed twice in duplicate). ANOVA followed by Fisher’s probable least-squares difference test used a cut-off significance at p = 0.05. Different letters indicate significantly different values based on one-way ANOVA (P < 0.05). Small letters indicate comparison between treatments (CTRL vs MIC) at the same time (20 or 60 DAF); capital letters indicate comparison between different times (20 DAF vs 60 DAF) in the same treatment (CTRL or MIC).
The 2D maps of seed proteins, stained with Colloidal Coomassie, showed a mean of 750 reproducible spots (Figs 1(a,b) and 2(a,b)). Significant variations were detected for 141 spots, of which 131 were MS/MS identified (93%). Table 2 lists the information regarding modulated proteins: spot number, number of identified peptides, sequence coverage, optical density variation using colour code, ANOVA P-value, protein name and Blast results when present, theorical molecular weight and pI, accession number and reference organism, and the biological process in which the identified protein is believed to be involved. Supplementary Tables S1, S2 and S3 list optical density raw data (as well as the statistical differences and P values), MS/MS results and BLAST results, respectively.
Figure 1.
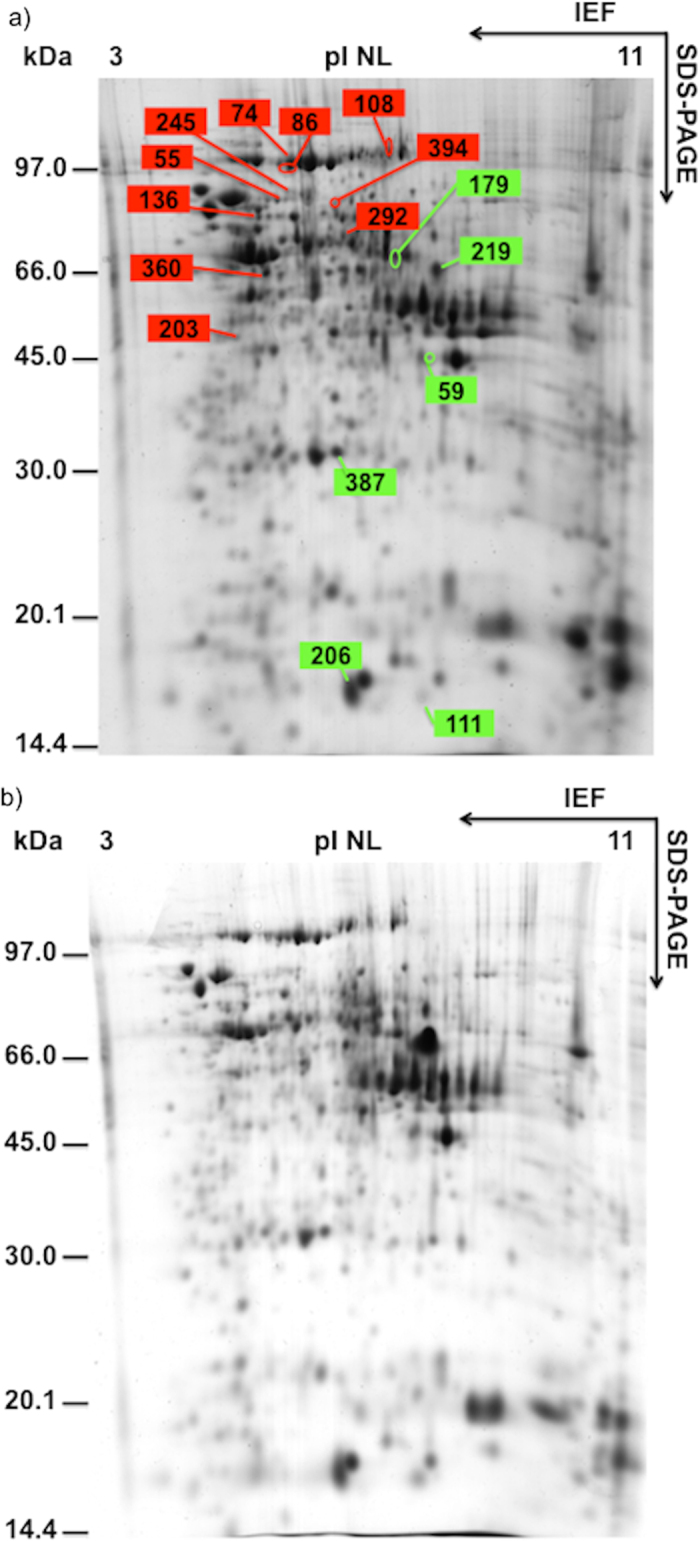
((a) (CTRL), (b) (MIC)). 2D maps of seed proteins extracted from seeds at 20 days after flowering (DAF), stained with Colloidal Coomassie. The assigned spots in the map were those modulated by AM symbiosis (green, up-regulated spots; red, down-regulated spots).
Figure 2.
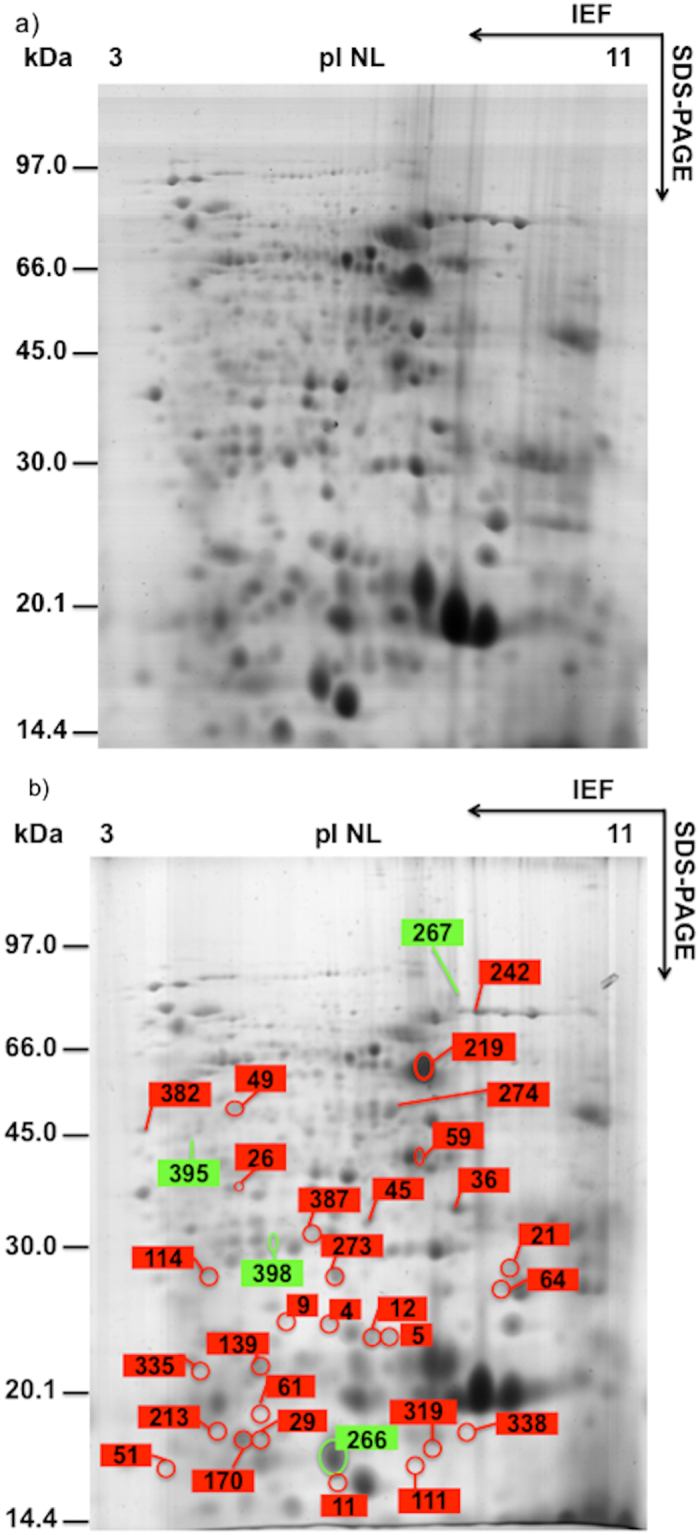
((a) (CTRL), (b) (MIC)). 2D maps of seed proteins extracted from seeds at 60 days after flowering (DAF), stained with Colloidal Coomassie. The assigned spots in the map were those modulated by AM symbiosis (green, up-regulated spots; red, down-regulated spots).
Table 2. Information regarding modulated proteins spot number, number of identified peptides, sequence coverage, optical density variation using colour code, ANOVA P-value, protein name and Blast results when present, theoretical molecular weight and pI (experimental pI data were not reported because the isoelectrofocusing was performed on non-linear IPG strips and image analysis software was not able to precisely calculate the pI), accession number and reference organism, biological process in which the identified protein was involved.
| Spot | N. Peptides | Seq. Coverage | Fungus effect at 20 DAF | Fungus effect at 60 DAF | Ripening effect on CTRL | Ripening effect on MIC | P-Value | Protein name/Blast result | Mr (kDa)/pI Theor | AC number/reference organism | Biological process |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 114 | 8 | 47% |  |
 |
 |
<0.0001 | Unknown/Adenine phosphoribosyl transferase | 19336/5.14 | gi|194701624/Zea mays | Adenine salvage | |
| 381 | 11 | 18% |  |
0.0357 | Acetolactate synthase 1 | 68887/6.69 | gi|75102649/Zea mays | Amino-acid biosynthesis | |||
| 219 | 7 | 22% |  |
 |
 |
 |
<0.0001 | ATP synthase beta chain | 45679/4.92 | gi|149798689/Eriosorus cheilanthoides | ATP synthesis coupled proton transport |
| 388 | 3 | 8% |  |
0.0344 | IAA-glu synthetase | 49679/5.75 | gi|162460991/Zea mays | Auxin conjugation | |||
| 24 | 5 | 7% |  |
 |
<0.0001 | Putative aconitate hydratase | 98021/5.67 | gi|75225211/Oryza sativa | Carbohydrate metabolism | ||
| 164 | 9 | 22% |  |
 |
<0.0001 | Phosphoglucomutase 2 | 63002/5.47 | gi|162459678/Zea mays | Carbohydrate metabolism | ||
| 274 | 12 | 34% |  |
 |
0.0135 | Sorbitol dehydrogenase | 39063/6.27 | gi|77378040/Zea mays | Carbohydrate metabolism | ||
| 380 | 13 | 37% |  |
 |
0.0018 | Fructokinase 2 | 35459/5.34 | gi|162460525/Zea mays | Carbohydrate metabolism | ||
| 54 | 11 | 22% |  |
 |
<0.0001 | Phosphoglucomutase 1 | 63058/5.46 | gi|162463106/Zea mays | Carbohydrate metabolism/glucose metabolism | ||
| 67 | 17 | 36% |  |
 |
<0.0001 | Phosphoglucomutase 2 | 63002/5.47 | gi|162459678/Zea mays | Carbohydrate metabolism/glucose metabolism | ||
| 245 | 3 | 8% |  |
 |
0.0272 | Phosphoglucomutase 2 | 63002/5.47 | gi|162459678/Zea mays | Carbohydrate metabolism/glucose metabolism | ||
| 112 | 12 | 53% |  |
 |
0.0002 | Unknown/Aldolase 1 | 38566/7.52 | gi|194690156/Zea mays | Carbohydrate metabolism/Glycolysis | ||
| 136 | 16 | 41% |  |
 |
0.0002 | Phosphoglycerate mutase | 60592/5.29 | gi|551288/Zea mays | Carbohydrate metabolism/Glycolysis | ||
| 145 | 20 | 62% |  |
 |
<0.0001 | 3-phosphoglycerate kinase | 42413/5.65 | gi|194707626/Zea mays | Carbohydrate metabolism/Glycolysis | ||
| 163 | 14 | 63% |  |
 |
0.0002 | Glyceraldehyde-3-phosphate dehydrogenase | 24930/8.44 | gi|293887/Zea mays | Carbohydrate metabolism/Glycolysis | ||
| 196 | 18 | 61% |  |
 |
0.0004 | Glyceroldehyde-3-phosphate dehydrogenase | 36428/6.61 | gi|162458671/Zea mays | Carbohydrate metabolism/Glycolysis | ||
| 205 | 17 | 64% |  |
0.0045 | Glyceroldehyde-3-phosphate dehydrogenase | 36519/6.41 | gi|162461501/Zea mays | Carbohydrate metabolism/Glycolysis | |||
| 249 | 4 | 7% |  |
 |
<0.0001 | Phosphoglycerate mutase | 60592/5.29 | gi|551288/Zea mays | Carbohydrate metabolism/Glycolysis | ||
| 251 | 10 | 49% |  |
 |
0.0003 | Glyceroldehyde-3-phosphate dehydrogenase | 36428/6.61 | gi|162458671/Zea mays | Carbohydrate metabolism/Glycolysis | ||
| 278 | 10 | 35% |  |
 |
0.0070 | Enolase1 | 48033/5.20 | gi|162458207/Zea mays | Carbohydrate metabolism/Glycolysis | ||
| 305 | 15 | 67% |  |
 |
0.0002 | Glyceraldehyde-3-phosphate dehydrogenase | 24930/8.44 | gi|293887/Zea mays | Carbohydrate metabolism/Glycolysis | ||
| 309 | 4 | 9% |  |
0.0112 | Unknown/Phosphofructokinase | 60980/5.96 | gi|194700662/Zea mays | Carbohydrate metabolism/Glycolysis | |||
| 311 | 17 | 51% |  |
 |
0.0013 | Unknown/Phosphoglycerate kinase | 42413/5.65 | gi|194707626/Zea mays | Carbohydrate metabolism/Glycolysis | ||
| 321 | 17 | 48% |  |
 |
0.0079 | Enolase1 | 48033/5.20 | gi|162458207/Zea mays | Carbohydrate metabolism/Glycolysis | ||
| 326 | 2 | 8% |  |
 |
<0.0001 | Enolase1 | 48033/5.20 | gi|162458207/Zea mays | Carbohydrate metabolism/Glycolysis | ||
| 368 | 10 | 31% |  |
 |
0.0100 | Enolase2 | 48132/5.70 | gi|162460735/Zea mays | Carbohydrate metabolism/Glycolysis | ||
| 36 | 7 | 42% |  |
 |
 |
<0.0001 | Prohibitin3 | 30580/7.00 | gi|162462359/Zea mays | Cell growth | |
| 45 | 4 | 22% |  |
 |
 |
<0.0001 | Prohibitin 2 | 30702/6.55 | gi|162462211/Zea mays | Cell growth | |
| 358 | 26 | 55% |  |
 |
0.0018 | Protein disulfide isomerase | 56838/5.01 | gi|162461063/Zea mays | Cell redox homeostasis | ||
| 29 | 4 | 40% |  |
 |
 |
<0.0001 | Actin depolymerizing factor | 15890/5.46 | gi|162459533/Zea mays | Cytoskeleton | |
| 170 | 5 | 40% |  |
 |
 |
<0.0001 | Actin depolymerizing factor | 15890/5.46 | gi|162459533/Zea mays | Cytoskeleton | |
| 252 | 11 | 51% |  |
0.0003 | Actin | 41699/5.24 | gi|53759189/Saccharum officinarum | Cytoskeleton | |||
| 340 | 7 | 21% |  |
 |
0.0033 | Hypothetical protein LOC100191561/Actin | 41699/5.24 | gi|212274479/Zea mays | Cytoskeleton | ||
| 398 | 7 | 40% |  |
0.0409 | Hypothetical protein LOC100193683/Proteasome subunit alpha type 2 | 25848/5.53 | gi|212720956/Zea mays | Defense response to bacterium | |||
| 382 | 2 | 8% |  |
 |
0.0145 | Unknown/Ankyrin repeat domain-containing protein 2 | 36227/4.50 | gi|194707992/Zea mays | Defense response to bacterium, incompatible interaction | ||
| 61 | 15 | 91% |  |
 |
 |
<0.0001 | Late embryogenesis abundant protein Lea14-A | 16078/8.05 | gi|195658529/Zea mays | Defense response to dessiccation | |
| 104 | 3 | 14% |  |
 |
0.0007 | Unknown/Dessication-related protein | 34010/4.82 | gi|194708240/Zea mays | Defense response to dessiccation | ||
| 1 | 3 | 16% |  |
 |
<0.0001 | Late embryogenesis abundant protein, group 3 | 18588/7.85 | gi|195605580/Zea mays | Embryo development ending in seed dormancy | ||
| 363 | 9 | 44% |  |
0.009 | Unknown/APx1 - Cytosolic Ascorbate Peroxidase | 27368/5.65 | gi|195654277/Zea mays | Embryo development ending in seed dormancy | |||
| 385 | 10 | 22% |  |
0.0314 | Unknown/Vacuolar ATP synthase catalytic subunit A | 68376/5.30 | gi|195658441/Zea mays | Embryo development ending in seed dormancy | |||
| 387 | 10 | 48% |  |
 |
 |
 |
0.0012 | Unknown/APx1 - Cytosolic Ascorbate Peroxidase | 27368/5.65 | gi|195654277/Zea mays | Embryo development ending in seed dormancy |
| 403 | 7 | 36% |  |
0.0189 | Unknown/APx2 - Cytosolic Ascorbate Peroxidase | 27211/5.28 | gi|194707280/Zea mays | Embryo development ending in seed dormancy | |||
| 328 | 4 | 8% |  |
0.0273 | Unknown/2-isopropylmalate synthase B | 67138/7.02 | gi|195604800/Zea mays | Glucosinolate biosynthesis process | |||
| 339 | 17 | 30% |  |
0.0463 | Unknown/2-isopropylmalate synthase B | 67138/7.02 | gi|195604800/Zea mays | Glucosinolate biosynthesis process | |||
| 185 | 2 | 3% |  |
 |
0.0003 | Putative aconitate hydratase | 98021/5.67 | gi|75225211/Oryza sativa | Glyoxylate and dicarboxylate metabolism | ||
| 186 | 3 | 6% |  |
 |
0.0009 | Putative aconitate hydratase | 98021/5.67 | gi|75225211/Oryza sativa | Glyoxylate and dicarboxylate metabolism | ||
| 147 | 5 | 16% |  |
 |
<0.0001 | Catalase isozyme 1 | 56841/7.40 | gi|115679/Zea mays | Hydrogen peroxide | ||
| 399 | 3 | 7% |  |
0.0451 | Non-photosynthetic NADP-malic enzyme | 70622/6.46 | gi|37147841/Zea mays | Malate metabolic process | |||
| 9 | 2 | 17% |  |
 |
 |
<0.0001 | Unknown/Splicing factor | 19898/11.53 | gi|194695412/Zea mays | Nucleic acid binding | |
| 266 | 2 | 10% |  |
 |
0.0093 | Glycine-rich RNA binding protein | 15908/5.22 | gi|20257707/Zea mays | Nucleic acid binding | ||
| 282 | 2 | 9% |  |
 |
0.0006 | Unknown/Plasminogen activator inhibitor 1 RNA-binding protein | 40439/5.72 | gi|194701098/Zea mays | Nucleic acid binding | ||
| 111 | 3 | 7% |  |
 |
 |
 |
0.0001 | Nucleoside diphosphate kinase 1 | 16835/6.30 | gi|50096951/Oryza sativa | Nucleotide metabolism |
| 319 | 4 | 19% |  |
 |
0.0144 | Nucleoside diphosphate kinase 1 | 16835/6.30 | gi|50096951/Oryza sativa | Nucleotide metabolism | ||
| 69 | 16 | 54% |  |
 |
<0.0001 | Unknown/Glucose and ribitol dehydrogenase homolog | 32924/5.78 | gi|194699516/Zea mays | Oxidation-reduction process | ||
| 334 | 6 | 29% |  |
0.0023 | Carbonyl reductase 1 | 32662/6.16 | gi|195650645/Zea mays | Oxidation-reduction process | |||
| 335 | 3 | 20% |  |
 |
 |
<0.0001 | Unknown/Peroxiredoxin | 17312/4.85 | gi|194698866/Zea mays | Oxidation-reduction process | |
| 43 | 17 | 27% |  |
 |
<0.0001 | C4-specific pyruvate orthophosphate dikinase | 102343/5.50 | gi|31322754/Miscanthus x giganteus | Photosynthesis | ||
| 52 | 34 | 51% |  |
 |
<0.0001 | Chain A, Pyruvate Phosphate Dikinase | 95132/5.27 | gi|62738111/Zea mays | Photosynthesis | ||
| 38 | 28 | 43% |  |
 |
<0.0001 | Pyruvate orthophosphate dikinase | 102444/5.71 | gi|168586/Zea mays | Photosynthesis | ||
| 53 | 12 | 22% |  |
 |
<0.0001 | Pyruvate orthophosphate dikinase | 102444/5.71 | gi|168586/Zea mays | Photosynthesis | ||
| 74 | 13 | 18% |  |
 |
 |
<0.0001 | Pyruvate orthophosphate dikinase | 102471/5.52 | gi|6274486/Saccharum officinarum | Photosynthesis | |
| 96 | 13 | 19% |  |
 |
0.0002 | Pyruvate orthophosphate dikinase | 102471/5.52 | gi|6274486/Saccharum officinarum | Photosynthesis | ||
| 107 | 17 | 21% |  |
 |
<0.0001 | Pyruvate orthophosphate dikinase | 102471/5.52 | gi|6274486/Saccharum officinarum | Photosynthesis | ||
| 207 | 2 | 10% |  |
0.0047 | QM protein | 24903/10.27 | gi|162458844/Zea mays | Photosynthesis | |||
| 276 | 5 | 10% |  |
 |
0.0014 | Pyruvate, orthophosphate dikinase | 102444/5.71 | gi|168586/Zea mays | Photosynthesis | ||
| 108 | 14 | 24% |  |
 |
0.0006 | Os02g0519900/Elongation factor 2 | 93961/5.85 | gi|115446385/Oryza sativa | Protein biosynthesis | ||
| 139 | 2 | 10% |  |
 |
 |
<0.0001 | Translation initiation factor 5A | 17486/5.61 | gi|162458009/Zea mays | Protein biosynthesis | |
| 174 | 13 | 35% |  |
 |
0.0003 | Unknown/Eukariotic translation initiation factor 3 subunit 7 | 64846/5.51 | gi|194704818/Zea mays | Protein biosynthesis | ||
| 178 | 12 | 23% |  |
 |
<0.0001 | Os02g0519900/Elongation factor 2 | 93961/5.85 | gi|115446385/Oryza sativa | Protein biosynthesis | ||
| 360 | 12 | 38% |  |
 |
0.0041 | Translational initiation factor eIF-4A | 46952/5.37 | gi|162458395/Zea mays | Protein biosynthesis | ||
| 286 | 5 | 27% |  |
0.0055 | Unknown/Proteasome subunit alpha type 5 | 25961/4.76 | gi|195635461/Zea mays | Protein catabolic process | |||
| 66 | 5 | 39% |  |
 |
<0.0001 | Unknown/Putative chaperonin 21 precursor | 25739/8.49 | gi|194688414/Zea mays | Protein folding | ||
| 265 | 10 | 27% |  |
 |
0.0005 | Os02g0102900 /RuBisCO large subunit-binding protein | 63759/5.77 | gi|115443643/Oryza sativa | Protein folding | ||
| 284 | 16 | 33% |  |
0.0145 | Unknown/T-complex protein 1 subunit alpha | 59158/5.78 | gi|195636596/Zea mays | Protein folding | |||
| 287 | 11 | 22% |  |
 |
0.0211 | Os06g0114000/Chaperonin 60 Beta | 64046/5.60 | gi|115466004/Oryza sativa | Protein folding | ||
| 318 | 9 | 48% |  |
 |
0.0003 | Peptidyl-prolyl cis-trans isomerase | 18337/8.91 | gi|118104/Zea mays | Protein folding | ||
| 86 | 13 | 22% |  |
 |
0.0029 | OSJNBa0039C07.4/ATP dependent Clp protease ATP-binding subunit | 98436/5.79 | gi|38347158/Oryza sativa | Protein metabolic process | ||
| 148 | 7 | 13% |  |
 |
<0.0001 | Os12g0230100/ATP dependent Clp protease | 101954/6.62 | gi|115487910/Oryza sativa | Protein metabolic process | ||
| 338 | 3 | 34% |  |
 |
0.0100 | Unknown/NADH ubiquinone oxidoreductase B22-like subunit | 13346/8.01 | gi|195605254/Zea mays | Respiratory chain | ||
| 214 | 4 | 15% |  |
 |
<0.0001 | Glyoxalase I | 32336/5.59 | gi|162461576/Zea mays | Response to salt stress | ||
| 292 | 13 | 25% |  |
 |
0.0416 | Unknown/Ketol-acid reductoisomerase | 62963/6.31 | gi|194693902/Zea mays | Response to salt stress | ||
| 337 | 11 | 27% |  |
0.0084 | Alanine aminotransferase 2 | 53000/6.23 | gi|195625602/Zea mays | Response to salt stress | |||
| 357 | 7 | 18% |  |
0.0293 | Adenosine kinase | 36009/5.23 | gi|4582787/Zea mays | Response to salt stress | |||
| 12 | 6 | 41% |  |
 |
 |
<0.0001 | Unknown/Transcription factor homolog | 17757/6.62 | gi|194695608/Zea mays | Response to salt stress | |
| 134 | 13 | 19% |  |
 |
<0.0001 | Putative aconitate hydratase 1 | 106913/6.63 | gi|92429669/Sorghum bicolor | Response to salt stress | ||
| 189 | 3 | 27% |  |
 |
0.0034 | Unknown/Mitochondrial F0 ATP synthase D chain | 19915/5.19 | gi|194701816/Zea mays | Response to salt stress | ||
| 192 | 10 | 20% |  |
0.0014 | Methionine synthase protein | 83736/5.93 | gi|18483235/Sorghum bicolor | Response to salt stress | |||
| 273 | 7 | 44% |  |
 |
0.0003 | Unknown/Superoxide dismutase 3 | 25571/7.11 | gi|194689068/Zea mays | Response to salt stress | ||
| 395 | 8 | 29% |  |
 |
0.019 | Hypothetical protein LOC100191638/Salt tolerance protein | 35252/4.92 | gi|212274681/Zea mays | Response to salt stress | ||
| 15 | 3 | 6% |  |
 |
<0.0001 | Vicilin-like embryo storage protein | 66122/6.23 | gi|22284/Zea mays | Seed storage | ||
| 21 | 4 | 19% |  |
 |
0.0004 | Zein-alpha 19D1 precursor | 26616/9.21 | gi|162458484/Zea mays | Seed storage | ||
| 26 | 3 | 6% |  |
 |
 |
<0.0001 | Vicilin-like embryo storage protein | 66122/6.23 | gi|22284/Zea mays | Seed storage | |
| 44 | 9 | 20% |  |
 |
<0.0001 | Vicilin-like embryo storage protein | 66122/6.23 | gi|22284/Zea mays | Seed storage | ||
| 51 | 3 | 6% |  |
 |
<0.0001 | Vicilin-like embryo storage protein | 66122/6.23 | gi|22284/Zea mays | Seed storage | ||
| 188 | 4 | 14% |  |
 |
0.0031 | Vicilin-like embryo storage protein | 66122/6.23 | gi|22284/Zea mays | Seed storage | ||
| 242 | 7 | 19% |  |
 |
0.0010 | Vicilin-like embryo storage protein | 66122/6.23 | gi|22284/Zea mays | Seed storage | ||
| 261 | 1 | 10% |  |
0.0229 | Zein protein precursor | 19448/8.05 | gi|168664/Zea mays | Seed storage | |||
| 354 | 4 | 6% |  |
 |
0.0187 | Vicilin-like embryo storage protein | 66122/6.23 | gi|22284/Zea mays | Seed storage | ||
| 64 | 1 | 23% |  |
 |
0.0003 | z1B alpha zein protein | 16047/8.00 | gi|157780962/Zea mays | Seed storage (nutrient reservoir activity) | ||
| 179 | 10 | 34% |  |
 |
0.0135 | Legumin 1 | 52798/6.20 | gi|162460908/Zea mays | Seed storage (nutrient reservoir activity) | ||
| 50 | 11 | 51% |  |
 |
<0.0001 | Unknown/Chitinase | 29193/8.44 | gi|194702870/Zea mays | Somatic embryogenesis/fruit development | ||
| 210 | 11 | 22% |  |
0.0232 | Granule-bound starch synthase precursor | 66567/6.59 | gi|33321047/Zea mays | Starch metabolic process | |||
| 342 | 10 | 22% |  |
 |
0.0036 | Granule-bound starch synthase precursor | 66567/6.59 | gi|33321047/Zea mays | Starch metabolic process | ||
| 83 | 10 | 38% |  |
 |
0.0002 | Unknown/Stress responsive protein | 38371/6.30 | gi|194707628/Zea mays | Stress response | ||
| 4 | 6 | 30% |  |
 |
 |
<0.0001 | 22.0 kDa class IV heat shock protein precursor | 22872/6.01 | gi|195644560/Zea mays | Stress response | |
| 13 | 3 | 26% |  |
 |
<0.0001 | Heat shock protein 17.2 | 17152/5.54 | gi|162459222/Zea mays | Stress response | ||
| 49 | 4 | 16% |  |
 |
0.0018 | Activator of 90 kDa heat shock protein ATPase | 38577/5.33 | gi|195651993/Zea mays | Stress response | ||
| 55 | 10 | 21% |  |
 |
 |
<0.0001 | Heat shock 70 kDa protein | 72704/5.62 | gi|195649437/Zea mays | Stress response | |
| 59 | 14 | 54% |  |
 |
 |
 |
<0.0001 | Unknown/Stress responsive protein | 38371/6.30 | gi|194707628/Zea mays | Stress response |
| 87 | 2 | 3% |  |
0.0059 | Putative heat shock protein | 82531/5.43 | gi|37718900/Oryza sativa | Stress response | |||
| 149 | 15 | 26% |  |
 |
0.0027 | Heat-shock protein 101 | 101069/5.85 | gi|162458166/Zea mays | Stress response | ||
| 154 | 10 | 21% |  |
0.0036 | Unknown/NADH ubiquinone oxidoreductase 75 kDa subunit | 80628/6.10 | gi|194688928/Zea mays | Stress response | |||
| 172 | 8 | 31% |  |
 |
<0.0001 | Lactoylglutathione lyase | 35140/6.62 | gi|195639070/Zea mays | Stress response | ||
| 197 | 8 | 42% |  |
 |
<0.0001 | Unknown/Stress responsive protein | 37857/6.70 | gi|194703432/Zea mays | Stress response | ||
| 213 | 4 | 42% |  |
0.0002 | Hypothetical protein Z477F24.14/Lactoylglutathione lyase | 15597/4.94 | gi|48374986/Zea mays | Stress response | |||
| 272 | 6 | 41% |  |
0.0012 | Hypothetical protein LOC100191552/Heat shock protein 17.9 | 17869/6.86 | gi|212276212/Zea mays | Stress response | |||
| 312 | 11 | 41% |  |
 |
<0.0001 | Unknown/Stress responsive protein | 37857/6.70 | gi|194703432/Zea mays | Stress response | ||
| 356 | 18 | 29% |  |
0.0149 | Unknown/Stromal 70 kDa heat shock-related protein | 74625/5.08 | gi|195657157/Zea mays | Stress response | |||
| 376 | 15 | 21% |  |
 |
0.0134 | Unknown/Heat shock 70 kDa protein | 72704/5.62 | gi|195649437/Zea mays | Stress response | ||
| 352 | 7 | 19% |  |
 |
<0.0001 | Unknown/Putative aminotransferase | 49566/6.55 | gi|195634861/Zea mays | Systemic acquired resistance | ||
| 336 | 4 | 32% |  |
 |
0.0030 | Unknown/40S ribosomal protein S7 | 22198/9.76 | gi|195605060/Zea mays | Translation | ||
| 268 | 6 | 32% |  |
 |
<0.0001 | Malate dehydrogenase 5 | 35567/5.77 | gi|162464321/Zea mays | Tricarboxylic acid cycle | ||
| 394 | 5 | 12% |  |
 |
0.0150 | Succinate dehydrogenase flavoprotein subunit | 67941/6.08 | gi|195647178/Zea mays | Tricarboxylic acid cycle | ||
| 5 | 3 | 8% |  |
 |
 |
<0.0001 | Fasciclin-like arabinogalactan protein 8 precursor | 44699/6.56 | gi|195607426/Zea mays | Unknown | |
| 78 | 8 | 49% |  |
 |
0.0013 | Unknown/Carbonyl reductase | 20867/6.84 | gi|194701990/Zea mays | Unknown | ||
| 206 | 5 | 53% |  |
 |
0.0009 | Chain A, Bifunctional Hageman Factor AMYLASE INHIBITOR FROM MAIZE | 13570/6.51 | gi|157830250/Zea mays | Unknown | ||
| 329 | 2 | 12% |  |
 |
0.0066 | Unknown/DREPP4 protein | 22630/4.89 | gi|194690236/Zea mays | Unknown | ||
| 343 | 5 | 16% |  |
0.0098 | Unknown/Seed protein | 26645/7.11 | gi|195626982/Zea mays | Unknown | |||
| 362 | 3 | 13% |  |
 |
<0.0001 | Unknown/Malonyl-CoA:ACP transacylase 1-3 | 38593/5.64 | gi|195638470/Zea mays | Unknown | ||
| 392 | 11 | 24% |  |
 |
0.0400 | Unknown/UDP-glucose pyrophosphorylase | 52056/5.30 | gi|194688950/Zea mays | Unknown | ||
| 17 | 7 | 38% |  |
0.0013 | Rab28 | 27693/4.90 | gi|22460/Zea mays | Unknown | |||
| 6 |  |
0.0141 | ND | ||||||||
| 11 |  |
 |
 |
<0.0001 | ND | ||||||
| 40 |  |
 |
<0.0001 | ND | |||||||
| 175 |  |
 |
0.0003 | ND | |||||||
| 200 |  |
 |
0.0045 | ND | |||||||
| 203 |  |
 |
<0.0001 | ND | |||||||
| 257 |  |
 |
0.0001 | ND | |||||||
| 262 |  |
 |
<0.0001 | ND | |||||||
| 267 |  |
 |
0.0071 | ND | |||||||
| 389 |  |
 |
0.0034 | ND |
Despite the fact that the root is the organ that is colonised by AM fungi, the physiology of the entire plant is affected by the symbiosis, with interaction with the fungus having been reported to modulate photosynthesis, leaf hydration, reproduction and fruit quality in both maize and other plant species6,26,36,37. This modulation of the plant physiology changes according to the different stages of a plant’s development and which plant organs are being analyzed. As shown in Table 2, the maize seed proteome of mycorrhizal plants differed either at the beginning of seed development (20 DAF) or at the end of maturation (60 DAF).
Effects during seed development
A detailed examination of the results revealed that at 20 DAF the AM symbiosis induced the up-regulation of enzymes involved in energetic metabolism, the latter stages of embryo development, nucleotide metabolism, seed storage and stress responses. AM fungi enhances primary metabolism by up-regulating ATP synthase (spot 219); this protein is a key enzyme whose expression is linked to respiratory and photosynthetic phosphorylation, both of which are major processes in the energetic metabolism of above-ground plant tissues. The up-regulation of the cytosolic ascorbate peroxidase (spot 387), a major enzyme involved in detoxification of hydrogen peroxide, was also induced by AM fungi; its expression may be linked with embryo development. Thus, Méchin et al.24 reported that this enzyme is modulated in maize seeds 14 days after pollination24, whereas Finnie et al.38 showed that a cytosolic form of this enzyme was only detectable in an early developmental stage of barley seeds.
The overexpression of nucleoside diphosphate kinase 1 (spot 111) may lead to reduced constitutive reactive oxygen species (ROS) levels and enhaced tolerance to multiple environmental stress39. The expression of nucleoside diphosphate kinase has been reported to increase in response to drought and salinity, thus it is expected to accumulate in the late phases of embryogenesis. This enzyme also plays significant roles in hormone responses, heat stress and, in general, growth and development39. An increase of ROS could induce the observed increase of heat shock 70 kDa protein (spot 55) expression.
The accumulation of Legumin 1 (spot 179), a storage protein found in maize seeds, is a confirmation of the data of globulin quantification and is linked with the seed storage process. The AM symbiosis induced down regulation of three starch granule-associated proteins, namely phosphoglucomutase 2 (spot 245), phosphoglycerate mutase (spot 136), and a pyruvate ortophosphate dikinase (spot 74) as well as seven proteins involved in cellular metabolic processes, an elongation factor 2 (spot 108), a translational initiation factor eIF-4A (spot 360), an ATP-dependent Clp protease ATP-binding subunit (spot 86), a ketol-acid reductoisomerase (spot 292), a stress responsive protein (spot 59) and succinate dehydrogenase flavoprotein subunit (spot 394). In order to use their stored carbon reserves, plants must be able to degrade their starch granules to oligosaccharides and monosaccharides. In particular, as previously reported, phosphoglucomutase 2 converts glucose 1-phosphate to glucose 6-phosphate facilitating the use of this compound in glycolysis40. The orthophosphate dikinase partly controls the composition of the storage protein fractions and the starch-protein balance24. The classical role of orthophosphate dikinase in both C3 and C4 plants involves catalyzing the reversible reaction of pyruvate, ATP and phosphate to phosphoenol-pyruvate, AMP and diphosphate. In rice, the expression of ortophosphate dikinase was found to be highest at 5–15 days after pollination; after that time this enzyme was likely rapidly degraded or inactivated through phosphorylation41. This pool of inactivated orthophosphate dikinase was also present in mature seeds, suggesting a role in developmental processes during seed germination39,42.
Effects at seed maturation
At 60 DAF, the presence of AM fungi induced the modulation of 33 maize seed proteins, 4 up-regulated and 29 down-regulated. The degradation of the reserves (starch and storage proteins) and of some functional proteins could provide enough energy and amino acids for seed germination and for embryo development43. This could explain the up-regulation of proteasome proteins (spot 398) and the strong down regulation of different enzymes in AM-treated plants, such as adenine phosphoribosyl transferase (spot 114), ATP synthase beta chain (spot 219), sorbitol dehydrogenase (spot 274), prohibitin 3 and 2 (spots 36 and 45, respectively), two actin depolymerizing factor (spots 29 and 70), ankyrin repeat domain-containing protein 2 (spot 382), and late embryogenesis abundant protein Leal 4-A (spot 61).
Maize seeds acquire the ability to germinate during the stage of maturation drying44. The decreased water content plays an important role for the seeds to acquire the ability to germinate and for protection against fungal infection. Germination is a potentially stressful process and the reactivation of metabolism may provide an important source of ROS44. This can explain the increase in the abundance of proteins linked to the ROS response and AM symbiosis an overexpression of the same proteins such as salt tolerance protein (395) and down-regulation of APx-1 cytosolic ascorbate peroxidase (387), splicing factor (9), two spots belonging to nucleoside diphosphate kinase 1 (spots 111 and 319) that can lead to decreased constitutive reactive oxygen species (ROS) levels and enhanced tolerance to multiple environmental stress39, a peroxiredoxin (335), superoxide dismutase3 (273), a 22 kDa heat shock protein (4), the activator of a 90 kDa heat shock protein ATPase (49), and a stress responsive protein (59). The down regulation, in AM-treated seeds, of different isoforms of storage proteins such as Zein-alpha 19D1 precursor (21), vicilin-like embryo storage protein (spots 26, 51 and 242), z1B alpha zein protein (64) and lactoylglutathione lyase (213) could be linked with the seed protein turnover induced by embryo maturation.
Ripening effect on maize seed proteome
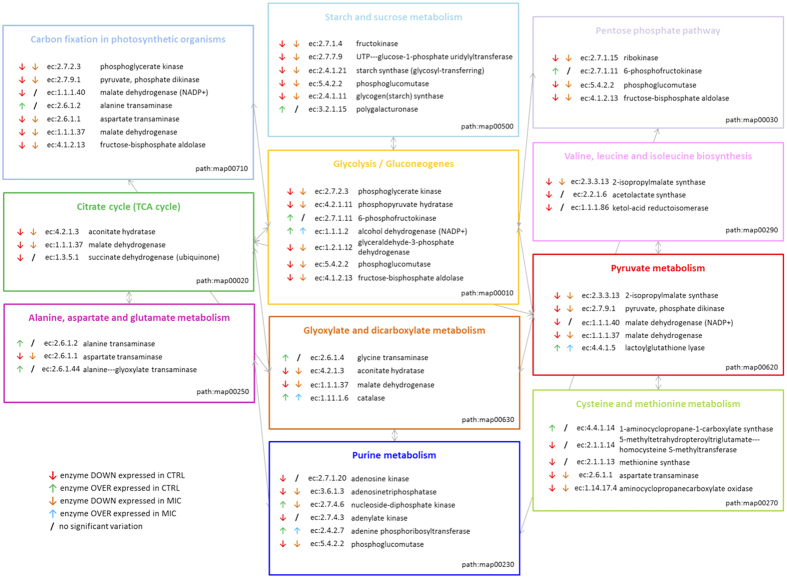
Maize is an excellent model for research on cereal seed development because of the relatively large size of both its embryo and endosperm. Despite the importance of seed maturation information for agricultural purpose, there is scant data available in literature regarding the effects of root AM fungal inoculation on seed maturation. Kegg maps (Fig. 3 and Table S4) summarizes the main biochemical pathways involved in the maize seed proteome modification during ripening, i.e. carbon fixation; starch and sucrose metabolism; the pentose phosphate pathway; the citrate cycle; glycolysis/gluconeogenesis; valine, leucine and isoleucine biosynthesis; alanine, aspartate and glutamate metabolism; glyoxylate and dicarboxylate metabolism; pyruvate metabolism; purine metabolism; and cysteine and methionine metabolism. Both in control and in mycorrhizal plants, seed maturation induced the same proteome evolution with the exception of: malate dehydrogenase, succinate dehydrogenase, adenosine kinase, adenylate kinase, acetolactate synthase, ketol-acid reductoisomerase, homocysteine S-methyltransferase and methionine synthase (down-regulated in CTRL and not modified in MIC); alanine transaminase, alanine-glyoxylate transaminase, polygalacturonase, 6-phosphofructokinase, glycine transaminase, 1-aminocyclopropane-1-carboxylate synthase (up-regulated in CTRL and not modified in MIC); nucleoside diphosphate kinase (up-regulated in CTRL and down-regulated in MIC); and zein-protein precursor (up-regulated in MIC and not modified in CTRL). These changes in protein abundance could be linked with the higher content of starch in the seeds of plants treated with mycorrhizal fungi.
Figure 3. Kegg maps summarizing the main biochemical pathways involved in proteome modification during ripening: carbon fixation, starch and sucrose metabolism, pentose phosphate pathway, citrate cycle, glycolysis/gluconeogenesis, valine, leucine and isoleucine biosynthesis, alanine, aspartate and glutamate metabolism, glyoxylate and dicarboxylate metabolism, pyruvate metabolism, purine metabolism, cysteine and methionine metabolism.
Our results are in agreement with those of Huang et al.45, who reported, on the basis of the metabolic and functional features of maize embryos, the identification of proteins classified into 7 major categories belonging to 3 functional groups: protein metabolism (26%), stress response (21%) and carbohydrate and energy metabolism (17%). At maturity, the maize seed accumulates large amounts of starch and storage proteins45. However, proteins involved in stress response (24%) were often up-regulated during seed maturation45.
A large body of literature describes the effects of AM fungi on the physiology of whole plants6,7,16,17, with a particular focus on fruit composition. The work presented here is a first step in filling the gap in the knowledge of the effect of AM fungi on seed composition. In the work described here, it was demonstrated that AM fungi strongly modify the seed proteome, particularly up-regulating enzymes involved in energy metabolism, embryo development, nucleotide metabolism, seed storage and stress responses.
Finally, this work underlines the importance of using soil microorganisms as inocula in field production to sustainably improve crop quality.
Materials and Methods
Experimental Field, Plant Growth And Seed Sampling
The experiment was conducted as described in Berta et al.26. In accordance with standard agricultural practices, field soil was fertilized with potassium sulfate (400 Kg/ha) and 18/46 N/P (350 Kg/ha). Corn seeds (Zea mays var. Ostiglia) were sown on 14th March 2013 in double rows. Three double lines (200 plants each) were treated with AM inoculum. An uninoculated double row was selected ramdomly as a control. The AM inoculum (Mybasol s.r.l., Alessandria, Italy), consisted of root fragments, spores, and hyphae of Rhizophagus intraradices, Glomus aggregatum, Glomus viscosum, Claroideoglomus etunicatum, and Claroideoglomus claroideum produced on sorghum, containing about 85,000 infective propagules l−1, was applied every 6 cm using a drip irrigation tube. During the growth period, diseases and insects were adequately controlled. Caryopsis harvest started from the 26th of July, 20 DAF and ended on the 4th of September, 60 DAF. During each sampling date, tillers of three kernels of control (CTRL) and mycorrhizal (MIC) plants were open, 25 g of grains from half of each ear, were collected, immediately frozen in liquid nitrogen, and stored at −80 °C.
Five, randomly selected, roots per treatment were used to evaluate frequency (F%), mycorrhizal degree (M%) and arbuscule abundance (A%)46.
Selective extraction of different protein classes
Ten grams of seeds were ground in a mortar using liquid nitrogen and extracted twice with milliQ water containing a protease inhibitor cocktail (Sigma-Aldrich), in the ratio 1:10 (p/v), at 4 °C for 2 hours. The slurries were centrifuged at 10,000 × g for 15 min. The two supernatants (albumin fraction) were pooled and stored at −20 °C, whereas the pellet was extracted twice with Tris-HCl 50 mM, pH 8.0, containing 0.3 M NaCl. The slurries were centrifuged at 10,000 ×g for 15 min. The supernatants (globulin fraction) were pooled and stored at −20 °C. The insoluble pellet was extracted twice with 70% ethanol containing 0.2% 2-mercaptoethanol. After stirring for 3 hours at 4 °C, the suspension was centrifuged at 10,000 × g for 30 min at 4 °C. The supernatants (prolamin fraction), were pooled and dried with a Rotavapor device. The insoluble pellet was then resuspended in 0.1 M NaOH to extract the glutelin fraction at 4 °C for 2 hours.
Three biological replicates were analysed in triplicate. Protein concentrations were determined according to Bradford47.
Seed water content was determined by placing one gram of ground seeds at 110 °C and then in a jar containing silica gel. Samples were analyzed twice in duplicate.
Proteomic analysis
Proteins were extracted according to Bona et al.48. The pellet was resuspended in 1 ml of solubilization buffer containing 7 M urea, 2M thiourea, 4% CHAPS, 100 mM DTT, 1% IPG buffer (3–11 NL) and quantified by the method of Bradford47. Aliquots of 700 μg of protein extracts were mixed with a rehydration buffer (8 M urea, 4% w/v CHAPS, 18 mM DTT, 0.5% 3–11 IPG Buffer), focused at 60 kVh at 20 °C on precast 13 cm NL pH 3–11 strips in an IPG-Phor unit (GE Healthcare Bio-Sciences) and separated on 12% gels at 10 °C under constant amperage (30 mA per gel) with a Protean Plus Dodeca gel (BioRad). At least ten replicates were run, two analytical replicates per five biological replicates.
Gels were stained according to Candiano et al.49, and then digitized in a GS 710 densitometer (Bio-Rad). The gel images were analyzed using SameSpot (Progenesis v. 2006) (build 3419. 12870). Differential expression analysis was performed: i) comparing the quantity of matched spots in the CTRL at 20 DAF versus MIC plants at 20 DAF (to evaluate the effect of AM fungus addition at the beginning of maturation); ii) comparing the quantity of matched spots in the CTRL at 60 DAF versus MIC plants at 60 DAF (to evaluate the effect of AM fungus addition at the end of the maturation period); iii) comparing the quantity of matched spots in the CTRL at 20 DAF versus CTRL plants at 60 DAF (to evaluate protein changes during maturation); iv) comparing the quantity of matched spots in the MIC plants at 20 DAF versus MIC plants at 60 DAF (to evaluate protein changes during maturation in AM plants). The software created a quantitative table with all normalized optical spot densities that allowed us to perform an analysis of variance to detect statistical differences between the quantitation of the same spot in all replicates.
Protein identification by nano-LC-Q-TOF MS/MS
For MS analysis, spots of interest were cut from the gel and destained overnight with a solution of 25 mM ammonium bicarbonate and 50% acetonitrile. The proteins were digested with trypsin (Roche, Segrate, Milano, Italy) in-gel digested as described by Hellmann et al.50. All nano-HPLC-MS/MS experiments were performed on a Q-TOF mass spectrometer Q-Star XL (AB Sciex, Concord, Ontario, Canada) controlled by the Analyst QS 1.1 software (AB Sciex) connected to an Ultimate 3000 nano-HPLC system. The peptide pellets were resuspended in 10 μl of solvent A (95% v/v water, 5% v/v acetonitrile, 0.1%v/v formic acid). Five microliters of each sample were loaded onto the precolumn, 300 μm i.d. × 5 mm, C18 PepMap, 5 μm beads, 100 Å, (LC-Packings) and washed for 5 min using a flow rate of 40 μl min−1 solvent A. The peptides were subsequently eluted at 300 nl min−1 from the precolumn over an analytical column, 15 cm × 75 μm, C18 PepMap100, 3 μm beads, 100 Å (LCPackings) using a 35 min gradient from 5 to 60% solvent B (5% v/v water, 95% v/v acetonitrile, 0.1% v/v formic acid) delivered at 300 μl min−1. The analytical column was connected with a 15 μm inner diameter Silica Tip (Pico Tip) nanospray emitter (New Objective, Woburn, MA). The spray voltage (set between 1800 and 2000 V) was applied to the emitter through a stainless steel union and tuned to get the best signal intensity using a standard BSA tryptic digest before every sample’s batch submission. The QStar-XL was operated in information-dependent acquisition (IDA) mode. Mass spectra were acquired from 400 to 1800 m/z. The two most intense ions with charge states between 1 and 4 in each survey scan were selected for the MS/MS experiment. MS/MS data were acquired from 60 to 1800 m/z. Each acquisition cycle was comprised of a 1 s MS and a 3 s MS/MS. The MS to MS/MS switch threshold was set to 15 counts per second (c.p.s.). All precursor ions subjected to MS/MS in the previous cycle were automatically excluded for 60 s using a 3 amu.
Homology-driven proteomics
Mascot Distiller (Matrix Science, London, UK) was used to create peak lists from MS and MS/MS raw data. Mascot Server (Matrix Science) was used for database searching versus NCBInr. The last check for proteins homology assignments was made versus NCBInr 20151214 (78002046 sequences; 28422168805 residues). Carbamidomethylation of cysteine residues, oxidation of methionine, deamidation of asparagine and glutamine were set as possible variable modifications and trypsin was selected as the protease. One missed cleavage site was allowed, and the peptide MS and MS/MS tolerance was set respectively to 100 ppm and 0.2 Da. Positive identifications were assigned with a minimum of two unique peptides with at least one peptide having a significant ion score (underlined in red in Table S2 in the supporting information). Considering the scarce number of corn sequences in the databases, if we obtained an automatic hit without a significant score, sequence tags were manually interpreted from the ESI-MS/MS spectra to confirm the hypothetical assignment. We also accepted hits identified by at least one peptide with a significant ion score according to the MASCOT MS/MS ion search algorithm as being confident assignments. When a protein has only one spectrum with a significant Mascot score, but in the results there are more spectra with lower scores, they were manually inspected and if they had a pattern compatible with the theoretical peptide, they were considered for homology searching. The sequence obtained from the manually reconstructed peptide was submitted to MS homology and if the first positive hit was the same protein or a homologue sequence of the one automatically recognized, the peptide was inserted in the table as assigned to the protein. This approach allow the use of partial “de novo” sequences that can be more fitting to the sequences in the database51.
Blast2GO data analysis
To perform the Blast2GO analysis (http://www.blast2go.com/b2ghome) we downloaded the protein FASTA sequences from http://www.ncbi.nlm.nih.gov using the GI code ID.
Data analysis was performed with Blast2GO standard parameters.
The EC annotations, obtained by mapping from equivalent GO annotations, were visualized reconstructing the structure of the Gene Ontology relationships and ECs on KEGG maps (http://www.genome.jp/kegg). In KEGG maps were displayed the enzymatic functions of sequences in the context of the metabolic pathways in which they participate.
Statistical analysis
Data were analyzed by a one-way ANOVA followed by Fisher’s test with cut-off significance at p = 0.05 using Stat View 4.5 (Abacus Concepts) software.
Additional Information
How to cite this article: Bona, E. et al. Arbuscular mycorrhizal symbiosis affects the grain proteome of Zea mays: a field study. Sci. Rep. 6, 26439; doi: 10.1038/srep26439 (2016).
Supplementary Material
Acknowledgments
This work was supported by Dipartimento di Scienze e Innovazione Tecnologica, Università del Piemonte Orientale and by Mossi and Ghisolfi s.r.l. The authors wish to thank the farm Mossi and Ghisolfi S.r.l. and Dr. Alessandro Ausano for his precious help throughout the experiments; Dr. Giorgia Novello for the help during paper revisions; Prof. Bernard Glick for the English Language revision.
Footnotes
Author Contributions B.E. organized the sampling, performed protein extractions, two-DE analysis, image analysis, protein digestion, MS data elaboration and wrote the paper; S.A. performed the extraction and the quantification of the different protein categories and wrote the paper; M.F. performed MS analysis and cooperated to MS data elaboration; B.L. performed BLAST 2GO analysis; C.A., G.D. and C.P. performed biological experiments in field; N.M. cooperated in statistical data analysis and in the manuscript revision; G.E. cooperated in the paper writing; C.M. cooperated in data elaboration and manuscript revision; G.B. coordinated biological experiments, data analysis and paper writing. All authors revised the manuscript.
References
- Smith S. E. & Read D. Mycorrhizal symbiosis 3rd edn (eds Smith S. E., Read D. ) The symbionts forming arbuscular mycorrhizas, 13–41 (Academic press, New York, 2008). [Google Scholar]
- Schlussler A., Schwarzott D. & Walker C. A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol. Res. 105, 1413–1421 (2001). [Google Scholar]
- Sawers R. J. H., Gutjahr C. & Paszkowski U. Cereal mycorrhiza: an ancient symbiosis in modern agriculture. Trends Plant Sci. 13, 93–7 (2008). [DOI] [PubMed] [Google Scholar]
- Javot H., Pumplin N. & Harrison M. J. Phosphate in the arbuscular mycorrhizal symbiosis: transport properties and regulatory roles. Plant. Cell Environ. 30, 310–22 (2007). [DOI] [PubMed] [Google Scholar]
- Bonfante P. & Genre A. Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat Commun 1, 1–11 (2010). [DOI] [PubMed] [Google Scholar]
- Lingua G. et al. Arbuscular mycorrhizal fungi and plant growth-promoting pseudomonads increases anthocyanin concentration in strawberry fruits (Fragaria x ananassa var. Selva) in conditions of reduced fertilization. Int. J. Mol. Sci. 14, 16207–16225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bona E. et al. AM fungi and PGP pseudomonads increase flowering, fruit production, and vitamin content in strawberry grown at low nitrogen and phosphorus levels. Mycorrhiza 25, 181–193 (2015). [DOI] [PubMed] [Google Scholar]
- Castellanos-Morales V., Villegas-Moreno J., Vierheilig H. & Cárdenas-Navarro R. Nitrogen availability drives the effect of Glomus intraradices on the growth of strawberry (Fragaria x ananassa Duch.) plants. J. Sci. Food Agric. 92, 2260–2264 (2012). [DOI] [PubMed]
- Castellanos-Morales V. et al. Root colonisation by the arbuscular mycorrhizal fungus Glomus intraradices alters the quality of strawberry fruits (Fragaria × ananassa Duch.) at different nitrogen levels. J. Sci. Food Agric. 90, 1774–1782 (2010). [DOI] [PubMed] [Google Scholar]
- Copetta A., Bardi L., Bertolone E. & Berta G. Fruit production and quality of tomato plants (Solanum lycopersicum L.) are affected by green compost and arbuscular mycorrhizal fungi. Plant Biosyst. 145, 106–115 (2011). [Google Scholar]
- Baslam M., Esteban R., García-Plazaola J. I. & Goicoechea N. Effectiveness of arbuscular mycorrhizal fungi (AMF) for inducing the accumulation of major carotenoids, chlorophylls and tocopherol in green and red leaf lettuces. Appl. Microbiol. Biotechnol. 97, 3119–3128 (2013). [DOI] [PubMed] [Google Scholar]
- Aimo S. et al. Use of arbuscular mycorrhizal fungi and beneficial soil bacteria to improve yield and quality of saffron (Crocus sativus L.). ISHS Acta Hortic. 850, 159–162 (2010). [Google Scholar]
- Borde M., Dudhane M. & Jite P. K. Role of bioinoculant (AM Fungi) increasing in growth, flavor content and yield in Allium sativum L. under field condition. Not. Bot. Horti Agrobot. Cluj-Napoca 37, 124–128 (2009). [Google Scholar]
- Ceccarelli N. et al. Mycorrhizal colonization impacts on phenolic content and antioxidant properties of artichoke leaves and flower heads two years after field transplant. Plant Soil 335, 311–323 (2010). [Google Scholar]
- Chaudhary V. & Kapoor R. & Bhatnagar, a. K. Effectiveness of two arbuscular mycorrhizal fungi on concentrations of essential oil and artemisinin in three accessions of Artemisia annua L. Appl. Soil Ecol. 40, 174–181 (2008). [Google Scholar]
- Copetta A., Lingua G. & Berta G. Effects of three AM fungi on growth, distribution of glandular hairs, and essential oil production in Ocimum basilicum L. var. Genovese. Mycorrhiza 16, 485–94 (2006). [DOI] [PubMed] [Google Scholar]
- Copetta A., Lingua G., Bardi L., Masoero G. & Berta G. Influence of arbuscular mycorrhizal fungi on growth and essential oil composition in Ocimum basilicum var. Genovese. Caryologia 60, 106–110 (2007). [Google Scholar]
- Bona E. et al. Proteomic analysis as a tool for investigating arsenic stress in Pteris vittata roots colonized or not by arbuscular mycorrhizal symbiosis. J. Proteomics 74, 1338–50 (2011). [DOI] [PubMed] [Google Scholar]
- Aloui A. et al. On the mechanisms of cadmium stress alleviation in Medicago truncatula by arbuscular mycorrhizal symbiosis: a root proteomic study. Proteomics 9, 420–33 (2009). [DOI] [PubMed] [Google Scholar]
- Valot B., Negroni L., Zivy M., Gianinazzi S. & Dumas-Gaudot E. A mass spectrometric approach to identify arbuscular mycorrhiza-related proteins in root plasma membrane fractions. Proteomics 6 Suppl 1, S145–55 (2006). [DOI] [PubMed] [Google Scholar]
- Bona E. et al. Proteomic analysis of Pteris vittata fronds: two arbuscular mycorrhizal fungi differentially modulate protein expression under arsenic contamination. Proteomics 10, 3811–3834 (2010). [DOI] [PubMed] [Google Scholar]
- Lingua G. et al. Effects of heavy metals and arbuscular mycorrhiza on the leaf proteome of a selected poplar clone: a time course analysis. PLoS One 7, e38662 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requejo R. & Tena M. Maize response to acute arsenic toxicity as revealed by proteome analysis of plant shoots. Proteomics 6 Suppl 1, S156–62 (2006). [DOI] [PubMed] [Google Scholar]
- Méchin V., Thévenot C., Le Guilloux M., Prioul J.-L. & Damerval C. Developmental analysis of maize endosperm proteome suggests a pivotal role for pyruvate orthophosphate dikinase. Plant Physiol. 143, 1203–19 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann M. et al. Mycorrhizal phosphate uptake pathway in maize: vital for growth and cob development on nutrient poor agricultural and greenhouse soils. Front. Plant Sci. 4, 1–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berta G. et al. Maize development and grain quality are differentially affected by mycorrhizal fungi and a growth-promoting pseudomonad in the field. Mycorrhiza 24, 161–70 (2014). [DOI] [PubMed] [Google Scholar]
- Cully D. et al. Endosperm protein synthesis and L-[35S] methionine incorporation in maize kernels cultured in vitro. Plant Physiol. 74, 389–394 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne T. B. Monographs on Biochemistry 2nd edn (eds Longmans, Green and Co) The vegetable proteins XIII-154 (London, 1924).
- Shewry P. & Halford N. Cereal seed storage proteins: structure, properties and role in grain utilization. J. Exp. Bot. 53, 947–958 (2002). [DOI] [PubMed] [Google Scholar]
- Kriz A. L. Seed proteins (eds Shewry P. R. & Casey R. ) 7S Globulins of cereals 477–498 (Kluwer Academic Publishers, Dordrecht, 1999). [Google Scholar]
- Egger M., Hauser M., Mari A., Ferreira F. & Gadermaier G. The role of lipid transfer proteins in allergic diseases. Curr. Allergy Asthma Rep 10, 326–335 (2010). [DOI] [PubMed] [Google Scholar]
- Woo Y., Hu D., Larkins B. & Jung R. Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell 13, 2297–2317 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soave C., Tardani L., Di Fonzo N. & Salamini F. Zein level in maize endosperm depends on a protein under control of the opaque-2 and opaque-6 loci. Cell 27, 403–410 (1981). [DOI] [PubMed] [Google Scholar]
- Feng L. et al. Expressional profiling study revealed unique expressional patterns and dramatic expressional divergence of maize α-zein super gene family. Plant Mol. Biol. 69, 649–659 (2009). [DOI] [PubMed] [Google Scholar]
- Prat S., Cortadas J., Puigdomènech P. & Palau J. Nucleic acid (cDNA) and amino acid sequences of the maize endosperm protein glutelin-2. Nucleic Acids Res. 13, 1493–1504 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. & Harrier L. A. Expression studies of plant genes differentially expressed in leaf and root tissues of tomato colonised by the arbuscular mycorrhizal fungus Glomus mosseae. Plant Mol. Biol. 51, 619–29 (2003). [DOI] [PubMed] [Google Scholar]
- Liu J. et al. Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J 50, 529–544 (2007). [DOI] [PubMed] [Google Scholar]
- Finnie C., Melchior S., Roepstorff P. & Svensson B. Proteome analysis of grain filling and seed maturation in barley. Plant Physiol 129, 1308–1319 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salekdeh G. H. & Komatsu S. Crop proteomics: aim at sustainable agriculture of tomorrow. Proteomics 7, 2976–96 (2007). [DOI] [PubMed] [Google Scholar]
- Koziol A. G., Marquez B. K., Huebsch M. P., Smith J. C. & Altosaar I. The starch granule associated proteomes of commercially purified starch reference materials from rice and maize. J. Proteomics 75, 993–1003 (2012). [DOI] [PubMed] [Google Scholar]
- Chastain C. & Challet R. Regulation of pyruvate, orthophosphate dikinase by ADP-/Pi-dependent reversible phosphorilation in C3 and C4 plants. Plant Physiol. Biochem. 41, 523–532 (2003). [Google Scholar]
- Chastain C., Heck J., Colquhaun T., Voge D. & Gu X. Posttranslational regulation by pyruvate, orthophosphate dikinase in developing rice (Oryza sativa) seeds. Planta 224, 924–934 (2006). [DOI] [PubMed] [Google Scholar]
- Yang P., Li X., Wang X., Chen H., Chen F. & Shen S. Proteomic analysis of rice (Oryza sativa) seeds during germination. Proteomics 7, 3358–3368 (2007). [DOI] [PubMed] [Google Scholar]
- Wang W.-Q. et al. Proteomic comparison between maturation drying and prematurely imposed drying of Zea mays seeds reveals a potential role of maturation drying in preparing proteins for seed germination, seedling vigor, and pathogen resistance. J. Proteome Res. 13, 606–26 (2014). [DOI] [PubMed] [Google Scholar]
- Huang H., Møller I. M. & Song S.-Q. Proteomics of desiccation tolerance during development and germination of maize embryos. J. Proteomics 75, 1247–62 (2012). [DOI] [PubMed] [Google Scholar]
- Trouvelot A., Kough J. & Gianinazzi-Pearson V. Mycorrhizae: physiology and genetics (eds Gianninazzi-Pearson, V. & Gianinazzi, S.) Mesure du taux de mycorhization VA d’un système radiculaire. Recherche de méthodes d’estimation ayant une signification fonctionnelle, 217–221 (INRA, Paris, 1986).
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- Bona E., Marsano F., Cavaletto M. & Berta G. Copper stress in Cannabis sativa roots: morphological and proteomic analysis. Proteomics 60, 96–101 (2007). [DOI] [PubMed] [Google Scholar]
- Candiano G. et al. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327–1333 (2004). [DOI] [PubMed] [Google Scholar]
- Hellman U., Wernstedt C., Gonez J. & Heldin C. Improvement of an ‘In-Gel’ digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem. 224, 451–455 (1995). [DOI] [PubMed] [Google Scholar]
- Shevchenko A. et al. Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal. Chem. 73, 1917–1926 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.