Abstract
Objective
Normal adipose tissue growth and function is critical to maintaining metabolic homeostasis and its excess (e.g. obesity) or absence (e.g. lipodystrophy) is associated with severe metabolic disease. The goal of this study was to understand the mechanisms maintaining healthy adipose tissue growth and function.
Methods
Adipose tissue senses and responds to systemic changes in growth factor and nutrient availability; in cells mTORC1 regulates metabolism in response to growth factors and nutrients. Thus, mTORC1 is poised to be a critical intracellular regulator of adipocyte metabolism. Here, we investigate the role of mTORC1 in mature adipocytes by generating and characterizing mice in which the Adiponectin-Cre driver is used to delete floxed alleles of Raptor, which encodes an essential regulatory subunit of mTORC1.
Results
RaptorAdipoq-cre mice have normal white adipose tissue (WAT) mass for the first few weeks of life, but soon thereafter develop lipodystrophy associated with hepatomegaly, hepatic steatosis, and insulin intolerance. RaptorAdipoq-cre mice are also resistant to becoming obese when consuming a high fat diet (HFD). Resistance to obesity does not appear to be due to increased energy expenditure, but rather from failed adipose tissue expansion resulting in severe hepatomegaly associated with hyperphagia and defective dietary lipid absorption. Deleting Raptor in WAT also decreases C/EBPα expression and the expression of its downstream target adiponectin, providing one possible mechanism of mTORC1 function in WAT.
Conclusions
mTORC1 activity in mature adipocytes is essential for maintaining normal adipose tissue growth and its selective loss in mature adipocytes leads to a progressive lipodystrophy disorder and systemic metabolic disease that shares many of the hallmarks of human congenital generalized lipodystrophy.
Keywords: mTORC1, Raptor, Rapamycin, Lipodystrophy, Obesity, White Adipose Tissue (WAT)
Highlights
-
•
A new model of Raptor (mTORC1) loss in adipose tissue is described.
-
•
Young fat Raptor KO mice have normal WAT mass.
-
•
Fat Raptor KO mice progressively develop lipodystrophy and metabolic disease.
-
•
Fat Raptor KO mice resist obesity but develop severe hepatomegaly.
1. Introduction
White adipose tissue (WAT) functions both as the body's major energy storage site, and as a critical endocrine tissue, and interest in understanding its biology has intensified with the obesity epidemic. Obesity (defined as a BMI > 30) results from energy imbalance and can lead to ectopic lipid deposition in non-adipose tissues (e.g. the liver), type 2 diabetes, cardiovascular disease, and some cancers. Obesity now affects more than 1 in 3 adults in the United States, and between 8% and 25% of adults in countries of the European Union, making this a major international clinical problem of this era [1], [2], [3]. Lack of adipose tissue or lipodystrophy also associates with severe metabolic complications. For example, patients suffering from congenital generalized lipodystrophy (or Berardinelli-Seip Syndrome) also develop insulin resistance, hypertriglyceridemia, and fatty liver disease, which can lead to hepatomegaly and liver failure. Thus, normal adipose tissue growth and function is critical to maintaining metabolic homeostasis and understanding the mechanisms that promote healthy fat has broad clinical implications.
The mechanistic target of rapamycin complex 1 (mTORC1) integrates multiple upstream signals from nutrient availability to promote anabolic metabolism. For example, mTORC1 detects intracellular amino acid availability through multiple sensors that converge upon the Rag GTPases to control mTORC1 subcellular localization, and circulating glucose levels through the insulin-signaling pathway, which promotes mTORC1 activity through the TSC/Rheb pathway [4], [5], [6]. Thus, mTORC1 is poised to be a critical regulator of adipocyte function. To test this, we conditionally deleted the essential mTORC1 regulatory subunit Raptor in mature adipocytes with Adiponectin-Cre, which is reported to have greater efficiency and specificity for mature adipocytes than aP2-Cre [7], [8], [9], [10], [11]. We find that RaptorAdipoq-Cre mice have normal WAT mass for the first few weeks of life, but progressively develop a lipodystrophy disorder resembling human congenital generalized lipodystrophy including insulin intolerance and hepatic steatosis. These and several additional characteristics of the RaptorAdipoq-Cre mice differ significantly from those of mice in which Raptor was deleted with aP2-Cre [12]. Our results provide a new framework for understanding how mTORC1 signaling helps maintain healthy adipose tissue and could provide insight into human lipodystrophy disorders.
2. Materials & methods
2.1. Mice
Raptor floxed mice are described in [13] and were backcrossed 10 generations to C57BL/6 and crossed to mice expressing the Adiponectin-Cre driver or the Ucp1-Cre driver (generous gifts of Evan Rosen). Floxed Cre-negative mice were used as controls. Mice were kept on a daily 12 h light/dark cycle and fed a normal chow diet (Prolab® Isopro® RMH 3000) from Lab Diet ad libitum at 22 °C. All animal experiments were approved by the University of Massachusetts Medical school animal care and use committee.
2.2. Antibodies and reagents
PPARγ antibody is from Santa Cruz (sc-7196). UCP1 antibody is from AbCam (ab-10983). All other antibodies were purchased from Cell Signaling Technologies: ACC (3676), ACLY (4332), AKT (9727), P-AKT-S473 (4058), ATGL (2439), FASN (3180), HSL (4107), P-HSL-S660 (4126), ULK1 (8054), P-ULK1-S757 (6888), 4EBP1 (9644), P-4EBP1-S65 (9456), P-4EBP1-T37/46. All other reagents were from Sigma–Aldrich.
2.3. Diet & metabolic studies
At 12 weeks of age, male mice were placed on a 60% high fat diet (HFD) (D12492 Harlan Laboratories) for 8 weeks. Body weight was recorded weekly. The analysis of blood metabolites was performed by the Joslin Diabetes Center and MMPC at the University of Cincinnati. For glucose tolerance tests (GTT) mice were fasted overnight (16 h) and then administered 2 g/kg of body weight of glucose or sodium pyruvate by intraperitoneal (i.p.) injection. For insulin tolerance tests (ITT), mice were fasted for 6 h before i.p. administration of 0.75 unit/kg of body weight of insulin. Blood glucose concentrations were measured before and after the injection at indicated time points.
2.4. Tissue harvest and histology
Adipose tissue depots were carefully dissected to avoid contamination from surrounding tissue. Samples for RNA or protein were snap frozen in liquid nitrogen and stored at −80 °C until analysis. For histology, tissue pieces were fixed in 10% formalin. Embedding, sectioning, and Hematoxylin & Eosin (HE) staining was done by the UMass Medical School Morphology Core. For Oil Red O staining, liver samples were embedded in OCT before sectioning and staining. For cell size measurements a minimum of 9–12 images were taken used per mouse (n = 3 wild type and 3 conditional knockouts). Image J was used to measure cell size and the distribution of cell size as percentage of total counted cells was analyzed.
2.5. Western blots
Tissue samples or cells were lysed in a buffer containing 50 mM Hepes, pH 7.4, 40 mM NaCl, 2 mM EDTA, 1.5 mM NaVO4, 50 mM NaF, 10 mM sodium pyrophosphate, 10 mM sodium β-glycerophosphate and 1% Triton X-100. Tissues were homogenized using a TissueLyser (Qiagen) in the same lysis buffer supplemented with 0.1% SDS, 1% sodium deoxycholate. Equal amounts of total protein were loaded into acrylamide/bis-acrylamide gels and transferred to PVDF membranes for detection with the indicated antibodies. Membranes were incubated with primary antibodies in 5% milk/PBST or 5% BSA/PBST overnight. Membranes were then incubated for 1hr with HRP-conjugated secondary antibodies. Western blots were developed by enhanced chemiluminescence (PerkinElmer) and detected by X-ray films. To obtain UCP1 signaling from WAT lysates, films were exposed for varying durations, up to 12 hrs for the indicated “long exposure (LE)”.
2.6. Lipolysis
Excised pgWAT tissue was incubated in 12 well cell culture dishes in DMEM with or without isoproterenol at 10 μM for 4 h, respectively, before collecting medium to measure glycerol concentration using a commercial kit (Sigma). The glycerol level was normalized with mass of the excised tissue.
2.7. Fecal lipids extraction
Mice were housed individually for 48 h and 1 g of dried feces was collected from each mouse. Feces were powderized using a tissue grinder, and rehydrated in 5 ml of normal saline. A 2:1 chloroform:methanol solution was used to extract lipids from the suspension.
2.8. Body composition and metabolic cages
Mice were placed into metabolic cages for 3 days at 25 days into their HFD feeding. Lean mass and whole body fat mass were measured noninvasively using 1H-MRS (Echo Medical Systems, Houston TX), and a 3-day measurement of physical activity, energy expenditure, respiratory exchange ratio, and food intake were conducted using metabolic cages (TSE Systems, Bad Homburg, Germany) by the UMass Mouse Metabolic Phenotyping Center. Thereafter, mice were kept on HFD in normal housing until dissection.
2.9. Gene expression analysis
Cells or tissues were lysed with Qiazol (Invitrogen) and total RNA was isolated with the RNeasy kit (Invitrogen). Equal amounts of RNA were retro-transcribed to cDNA using a High capacity cDNA reverse transcription kit (#4368813, Applied Biosystems). Quantitative RT-PCR was performed in 10 μL reactions using a StepOnePlus real-time PCR machine from Applied Biosystems using SYBR Green PCR master mix (#4309156, Applied Biosystems) according to manufacturer instructions. Relative mRNA expression was determined by the ΔCt method, and Tbp (TATA sequence binding protein) expression was used as a normalization gene in all conventional RT-PCR experiments.
2.10. Statistics
Unless otherwise stated, values given are mean ± SEM. Two-way ANOVA was performed where indicated. For most experiments, unpaired two-tailed Student's t test was used to determine statistical significance among two groups (*p < 0.05; **p < 0.01; ***p < 0.001).
3. Results and discussion
3.1. Adipocyte Raptor KO mice have normal WAT mass early in life
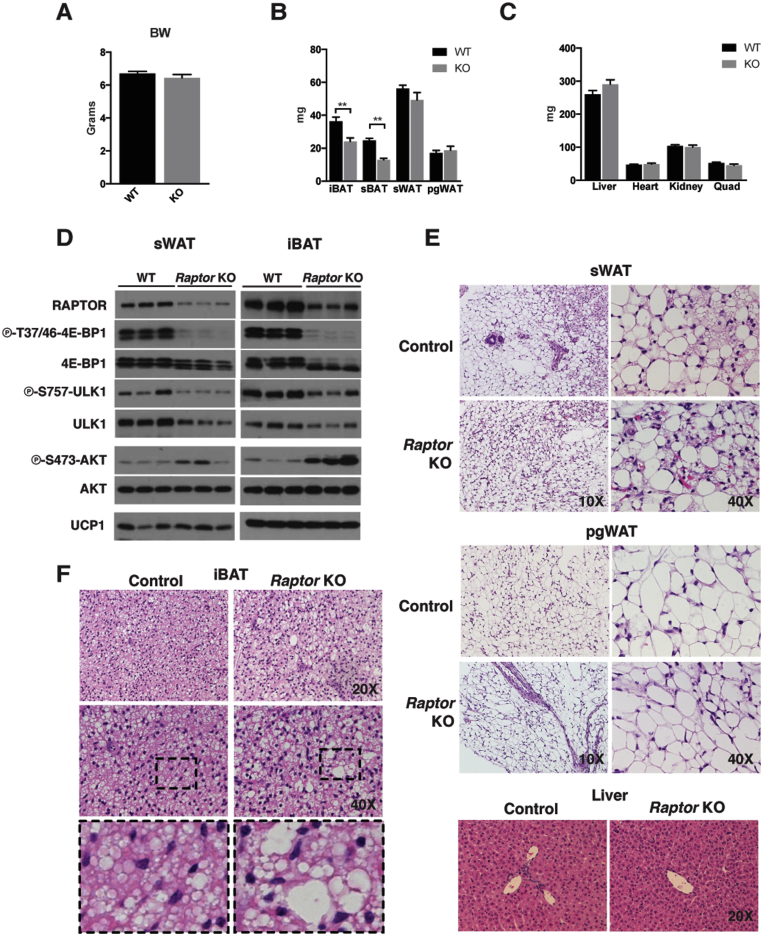
To investigate the role of mTORC1 in adipocytes, we generated Adiponectin-Cre;Raptorfl/fl mice (herein RaptorAdipoq-cre mice). RaptorAdipoq-cre mice are born at the expected Mendelian ratio and are grossly indistinguishable from their littermates at birth. At post-natal day 14 (P14), there is no difference in total body weight between control and RaptorAdipoq-cre mice (Figure 1A). The major subcutaneous and visceral perigonadal white adipose tissue depots (sWAT and pgWAT) are not significantly different in mass from controls at P14 (Figure 1B). Liver mass at P14 is also not significantly different though it is trending slightly larger in the KOs; the mass of other lean tissues such as the heart, kidney, and skeletal muscle is normal (Figure 1C). We confirmed Raptor loss by Western blots using whole sWAT and interscapular brown adipose tissue (iBAT) depots (Figure 1D); the residual signal reflects the non-adipocyte population (see Section 3.2). Inactivation of mTORC1 activity was confirmed by decreased phosphorylation of the mTORC1 substrates 4E-BP1 (on phospho-T37/46) and ULK1 (on phospho-S757), and by increased phosphorylation of AKT (on phospho-S473), which is caused by loss of the well-known mTORC1-mediated feedback inhibition of insulin/AKT signaling (Figure 1D) [4]. We also noted a slight decrease in total ULK1 protein in the KO tissues (Figure 1D).
Figure 1.
Adipocyte Raptor KO mice exhibit normal WAT mass and mild BAT defects early in life. (A) Body weight measurements of KO (n = 6) and wt (n = 6) mice at 14 days of age. (B) Fat tissue mass of KO and wt mice at 14 days of age. (C) Lean tissue mass of KO and wt mice at 14 days of age. (D) Western blots of whole tissue lysate from iBAT and sWAT depots for indicated proteins. (E,F) Representative H&E images of tissues at indicated magnification. (Data were analyzed by Student's t-test. Values expressed as mean + SEM. *p < 0.05; **p < 0.01; ***p < 0.001).
In post-natal development, the sWAT adipocytes exhibit characteristics of brown adipocytes including the presence of multi-locular lipid droplets and positive UCP1 expression (Figure 1D,E) [14], [15]. These adipocytes are called brite or beige adipocytes to distinguish them from classic brown adipocytes [16], and with age, they usually convert to or are replaced by more classic-looking unilocular white adipocytes. RaptorAdipoq-cre mice exhibit no defect in the post-natal formation of brite/beige adipocytes in the sWAT as determined by UCP1 protein expression (Figure 1D) and the appearance of multilocular adipocytes (Figure 1E). There is also no difference in the histological appearance of the pgWAT adipocytes or the liver at P14 (Figure 1E). Thus, Raptor is dispensable in white adipocytes for post-natal adipose tissue growth and normal adipocyte size, and for post-natal formation of brite/beige adipocytes in the sWAT.
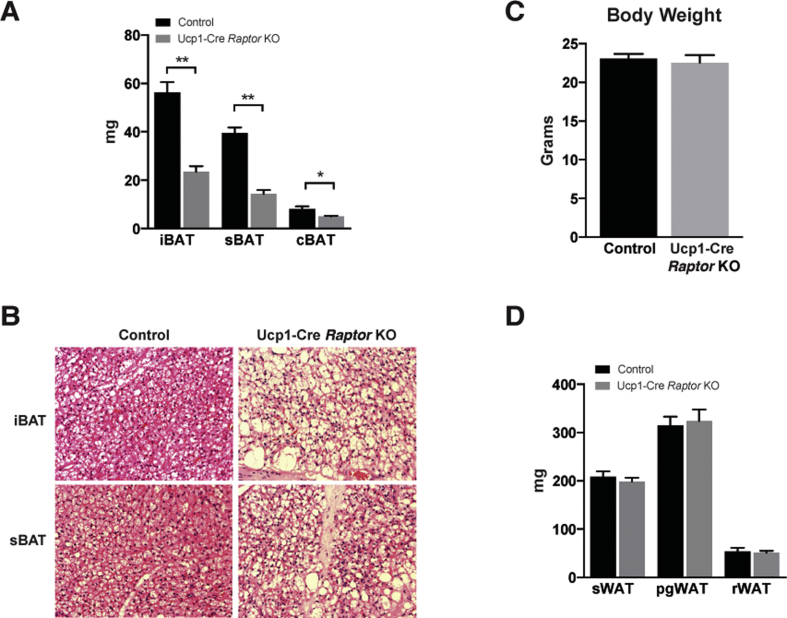
In contrast, the interscapular and subscapular brown adipose tissue depots (iBAT and sBAT) of RaptorAdipoq-cre mice are reduced in mass by 37% and 48% respectively at P14 (Figure 1B). We also noted an increase in the number of brown adipocytes containing large lipid droplets in the Raptor KO BAT (Figure 1F). To test whether this phenotype is tissue autonomous, we deleted Raptor with Ucp1-Cre. This also results in a significant size reduction of the BAT depots (Figure S1A) and the heterogeneous appearance of larger lipid droplets (Figure S1B). Total body mass and individual WAT depot mass was normal in RaptorUcp1-Cre mice (Figure S1C–D). Thus, while adipocyte Raptor is dispensable for post-natal WAT growth, it is required autonomously in brown adipocytes for normal post-natal BAT growth and lipid droplet size.
3.2. Adipocyte Raptor KO mice progressively develop lipodystrophy associated with hepatic steatosis and insulin intolerance
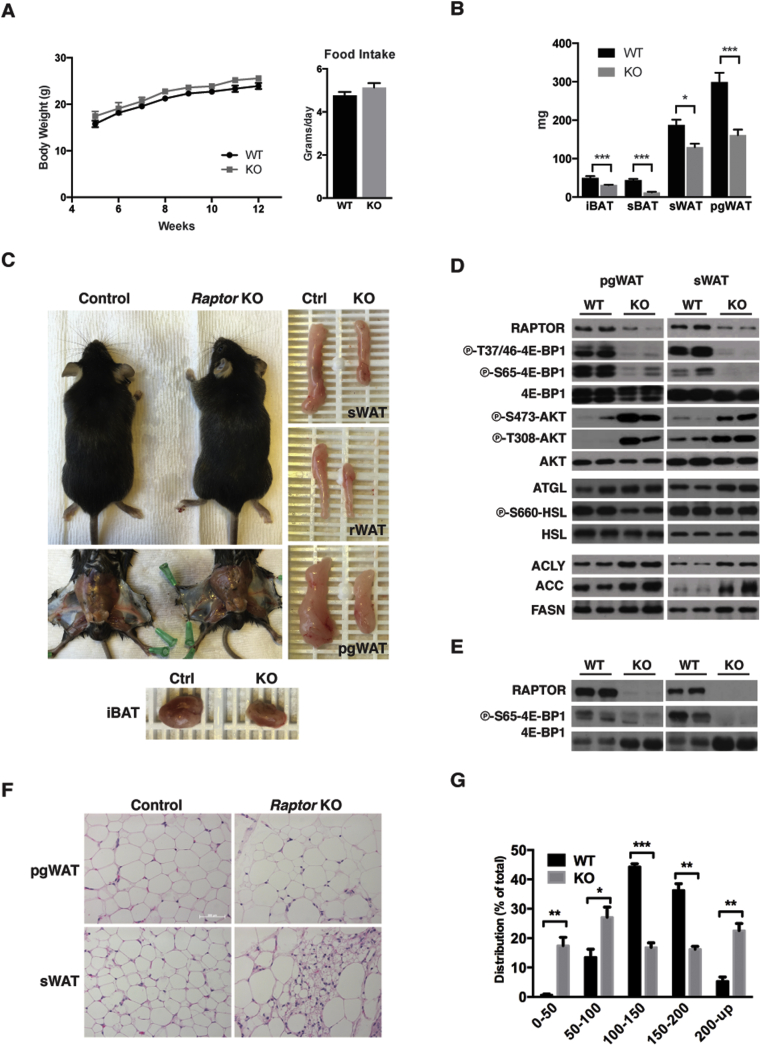
During the first 4 months of life, RaptorAdipoq-cre mice consuming a normal chow diet (NCD) tend to be slightly heavier than controls although this does not reach statistical significance at least by 12 weeks of age (p value = 0.066) (Figure 2A, left). RaptorAdipoq-cre mice also tend to eat slightly more food but again this was not statistically significant (p value = 0.25) (Figure 2A, right). However, despite their relatively normal body weight, RaptorAdipoq-cre mice fail to expand their white adipose tissues with age. For example, the KO sWAT, retroperitoneal WAT (rWAT) and pgWAT are 35%, 37%, and 54% smaller respectively in the KOs compared to age matched controls (Figure 2B,C); KO BAT tissues also remained small (Figure 2B,C). Thus, adipocyte Raptor KO mice progressively develop lipodystrophy with age.
Figure 2.
Adipocyte Raptor KO mice develop lipodystrophy with age. (A) Body weight growth chart of wt (n = 6) and KO (n = 6) mice up to 12 weeks. Food intake of adult KO and wt mice. (B) Fat tissue mass of KO and wt mice at ∼12 weeks of age. (C) Representative images of adult KO and wt mice, and indicated WAT and BAT depots. (D) Western blots from whole tissue lysate for indicated proteins. (E) Western blots from isolated mature adipocytes for indicated proteins. (F) Representative H&E images of WAT depots from KO and wt mice. (G) Distribution of adipocyte diameter (microns) in pgWAT depots from ∼12 week old mice on chow diet.
We also confirmed Raptor loss and mTORC1 inactivation in whole sWAT and pgWAT depots from the 12-week old mice (Figure 2D). The residual Raptor signal is from stromal vascular fraction (SVF) cells as Raptor is virtually undetectable in purified mature adipocytes (Figure 2E). Histological analysis of the white adipocytes from 12-week old RaptorAdipoq-cre mice revealed a heterogeneous size distribution pattern (Figure 2F). For example, pgWAT adipocytes in the RaptorAdipoq-cre mice show a bimodal distribution in which the relative number of small (<100 μM) and large (>200 μM) adipocytes is significantly increased compared to controls (Figure 2G). The mutant sWAT also contains two populations of adipocytes: a pool of large unilocular adipocytes and a pool of smaller adipocytes containing multiple lipid droplets (Figure 2F). Thus, with age the RaptorAdipoq-cre mice also cannot maintain normal adipocyte morphology.
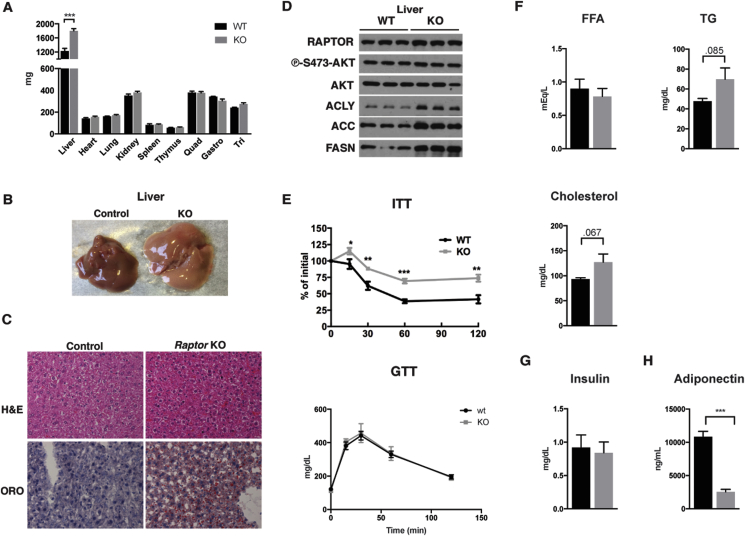
Strikingly, at 12-weeks of age there is also a 46% increase in liver mass in the RaptorAdipoq-cre mice compared to controls that is associated with a paler color suggesting hepatic steatosis (Figure 3A,B). The overgrowth of non-adipose tissues is largely restricted to the liver as the mass of other lean tissues including the heart and skeletal muscles is unchanged (Figure 3A). We confirmed hepatic steatosis by Oil Red O staining, which revealed a marked increase in lipid staining in the KO (Figure 3C). Consistently, expression of the major de novo lipogenesis (DNL) enzymes ATP-citrate lyase (ACLY), Acetyl-CoA Carboxylase (ACC) and Fatty Acid Synthase (FASN) are also increased in RaptorAdipoq-cre KO livers (Figure 3D). RaptorAdipoq-cre KO mice are also insulin intolerant exhibiting a 54% increase in AUC in insulin tolerance tests (Figure 3E, top); glucose tolerance is normal (Figure 3E, bottom). Serum chemistry analysis indicates normal circulating free fatty acids (FFAs) (p = 0.77), but a trending increase in circulating triacylglycerides (TAGs) (p = 0.0851) and cholesterol (p = 0.067) (Figure 3F). Insulin levels are not significantly different (Figure 3G). We conclude that the progressive lipodystrophy in RaptorAdipoq-cre mice is causing systemic metabolic disease including insulin intolerance, hyperlipidemia, and non-alcoholic fatty liver disease (NAFLD).
Figure 3.
The progressive lipodystrophy of adipocyte Raptor KO mice associates with hepatic steatosis and insulin intolerance. (A) Lean tissue mass of KO and wt mice at ∼12 weeks of age. (B) Representative images of liver from KO and wt mice at ∼12 weeks. (C) Representative H&E images of liver from KO and wt mice. (D) Western blots of liver lysate for indicated proteins. (E) ITT and GTT of adult KO and wt mice. (F) Serum chemistry levels of FFA, TG, and Cholesterol for adult wt and KO mice on chow diet. (G) Serum insulin levels for adult wt and KO mice. (H) Serum adiponectin levels for adult wt and KO mice. (Data were analyzed by Student's t-test. Values expressed as mean + SEM. *p < 0.05; **p < 0.01; ***p < 0.001).
3.3. Altered lipid metabolic pathways in adipocyte Raptor KO mice
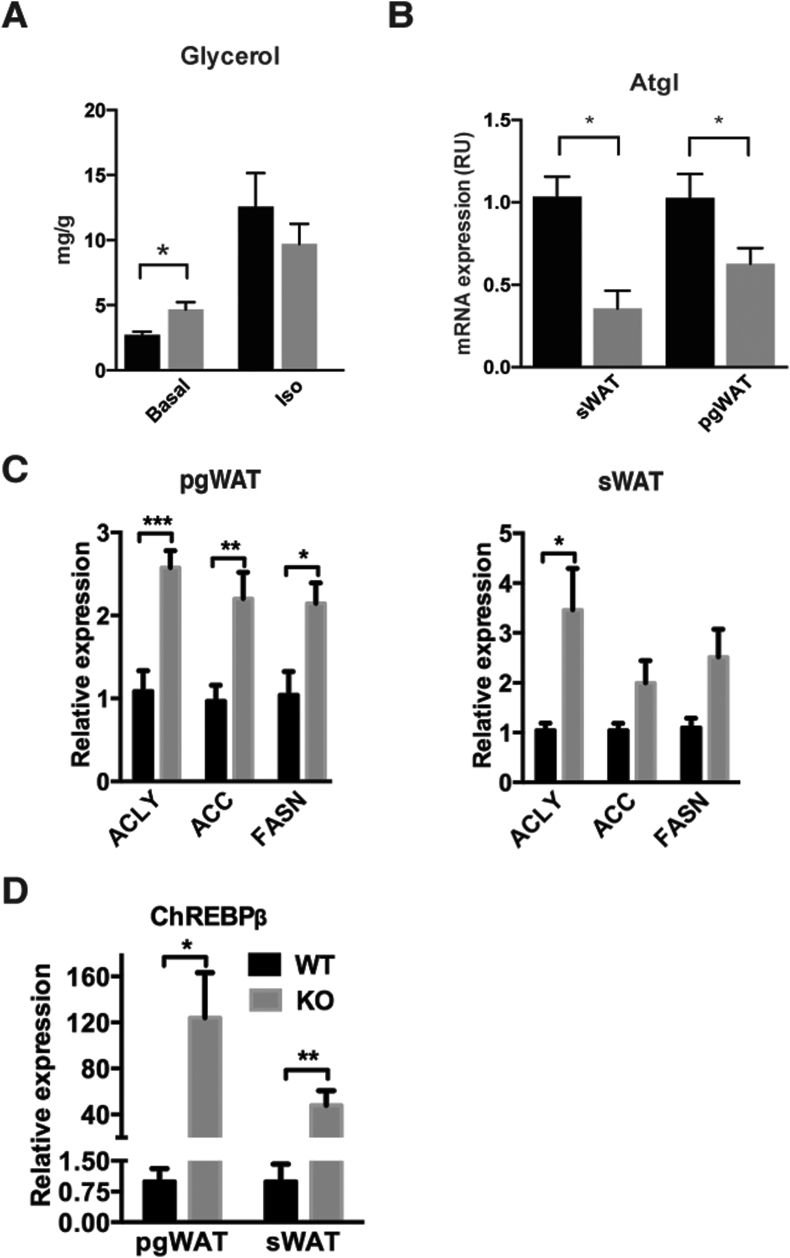
The mTORC1 inhibitor rapamycin can stimulate lipolysis in cultured adipocytes [17], [18]. Therefore, we examined the major regulators of lipolysis in Raptor-deficient adipose tissues. We did not observe a significant increase in hormone sensitive lipase (HSL) phosphorylation between control and KO in pgWAT at 12 weeks of age; in fact it was slightly lower (Figure 2D). We also did not detect increased adipose triglyceride lipase (ATGL) protein levels in the sWAT of RaptorAdipoq-cre mice; however, we could detect a slight increase in ATGL in the pgWAT (Figure 2D) that correlated with a modest increase in basal glycerol release from tissue explants (Figure 4A). However, Atgl mRNA was decreased in both WAT depots (Figure 4B). Additionally, we examined regulators of lipogenesis. In the pgWAT of RaptorAdipoq-cre mice we detected significant increases in Acly (2.5 fold), Acc (2.1 fold), and Fasn (2.1 fold) mRNA (Figure 4C left) and protein (Figure 2D). Similar increases occur in the sWAT although only the increase in Acly mRNA reaches significance even though ACLY, ACC and FASN proteins are all elevated in this depot (Figure 2, Figure 4C right). Consistently, expression of Chrebpβ, which encodes an isoform of ChREBP that potently induces transcription of the DNL genes [19], increases by 124-fold and 48-fold in pgWAT and sWAT respectively (Figure 4D). This is interesting because although DNL contributes only a fraction to the total lipid pool of adipocytes, increased Chrebpβ expression and DNL often correlate with insulin sensitivity [19] indicating this can be uncoupled in the RaptorAdipoq-cre mice. Moreover, while the mTORC1 substrate ULK1 is well known to control autophagy, in mature adipocytes it may have an additional role in lipogenesis downstream of mTORC1 as in vitro knockdown of Ulk1 in differentiated 3T3L1 adipocytes increases Acc and Fasn expression [20]. Thus, in vivo Raptor loss in mature adipocytes alters the normal regulation of lipid metabolic pathways, possibly with depot-dependent variances.
Figure 4.
Altered lipid metabolic pathways in the adipose tissues of adipocyte Raptor KO mice. (A) Concentration of glycerol from media of incubated excised pgWAT depots under specified conditions. (B) ATGL mRNA from whole tissue lysate. (C) Acly, Acc, and FasN mRNA expression in pgWAT and sWAT. (D) Chrebpβ mRNA expression in pgWAT and sWAT.
3.4. Raptor KO mice are resistant to diet-induced obesity but develop severe hepatic steatosis
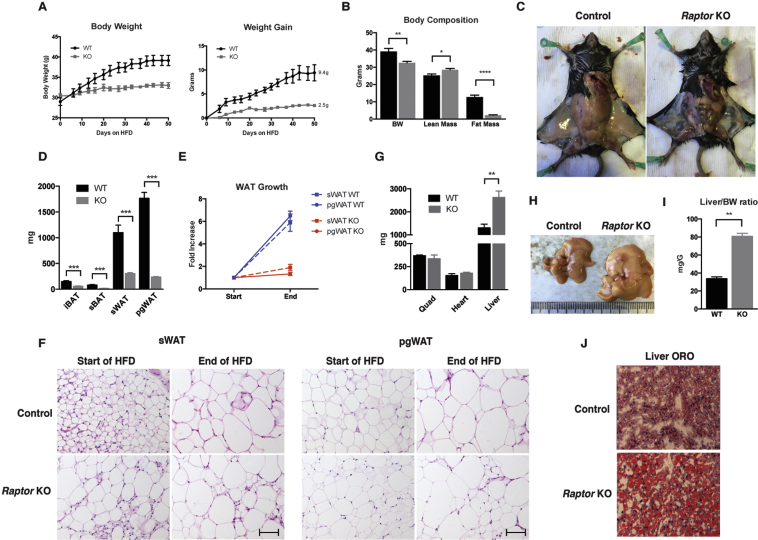
We next asked whether RaptorAdipoq-Cre mice could increase their fat mass if placed on an obesogenic high fat diet (HFD). Beginning at 12-weeks of age, RaptorAdipoq-Cre mice and their age matched controls were switched onto a HFD for 8-weeks. Controls gained on average 9.4 g of body weight while in contrast RaptorAdipoq-Cre mice gained only 2.5 g resulting in a 14% difference in total body mass between the two cohorts after 8 weeks (Figure 5A). Body composition analysis indicates the weight difference is due to a 5-fold decrease in fat mass in the mutants, which notably also have slightly higher lean mass (Figure 5B). The difference in fat mass is clearly evident upon dissection (Figure 5C) and individual tissue mass measurements (Figure 5D). Comparing the average mass of individual WAT depots at the start and end of HFD feeding shows that control sWAT increases by 5.9 fold and pgWAT by 6.5 fold while the mutant sWAT and pgWAT barely grow, increasing by only 1.9 fold and 1.3 fold, respectively (Figure 5E). Interestingly, comparing individual adipocyte size at the start and end of HFD feeding indicates that the control and KO adipocytes in both depots increase in size although the size heterogeneity in the KO tissue is still apparent (Figure 5F). Thus, despite the inability of whole Raptor KO depots to expand on HFD, Raptor KO adipocytes appear to retain some ability to grow by hypertrophy.
Figure 5.
Adipocyte Raptor KO mice are resistant to HFD obesity but suffer from more severe hepatic steatosis. (A) Body weight, and weight gain of mice over course of HFD. (B) Body weight and body composition after ∼4 weeks HFD. (C) Representative images of inguinal fat depots after HFD. (D) Adipose depot masses at the end of HFD course. (E) Fold increase in WAT depot mass after HFD feeding. (F) Representative H&E images of specified tissues before and after HFD. (G) Lean tissue mass after HFD. (H) Representative images of liver after HFD. (I) Liver/BW ratio of mice after HFD. (J) Representative ORO staining of liver samples after HFD. (Data were analyzed by Student's t-test. Values expressed as mean + SEM. *p < 0.05; **p < 0.01; ***p < 0.001).
Importantly, at the conclusion of HFD-feeding, the livers of the KO mice are now ∼80% larger than controls (as opposed to 46% larger at the start of feeding) resulting in a liver/body weight ratio that is now nearly twice of that of controls (Figure 5G,H,I). The livers of the KO mice also exhibit significantly more severe hepatic steatosis (Figure 5J). Muscle and heart mass remain unchanged between the cohorts (Figure 5G). Thus, the inability of adipose tissues to expand in HFD-fed RaptorAdipoq-Cre mice results in more redistribution of lipids to the liver.
3.5. Adipocyte Raptor KO mice have normal energy expenditure and a lipid absorption defect
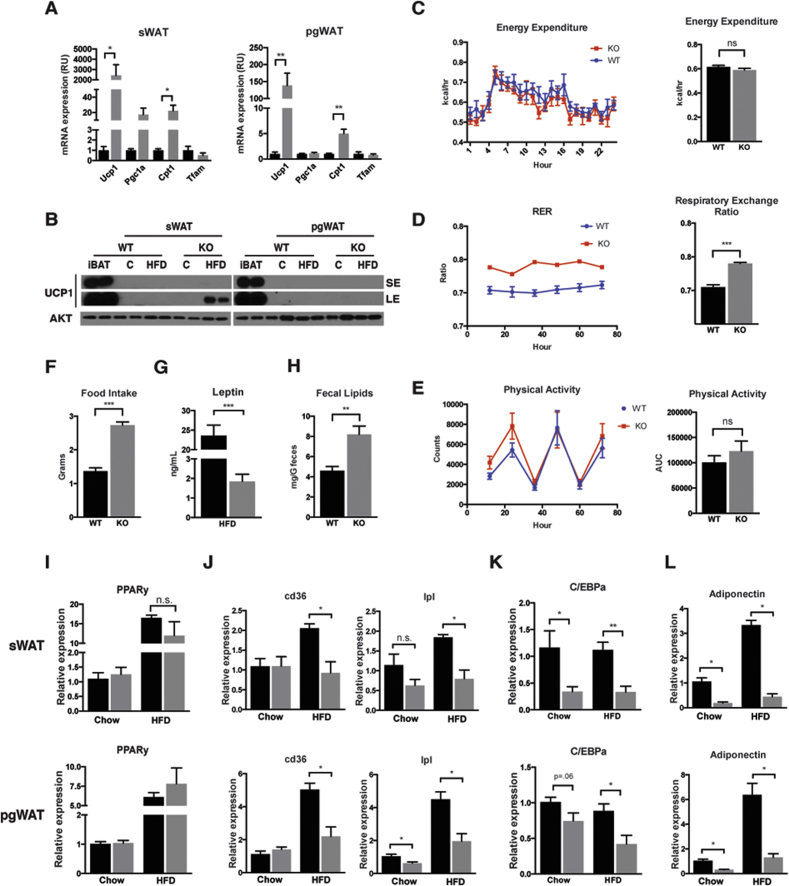
Next we explored possible mechanisms by which deleting Raptor in adipocytes restricts adipose tissue growth. One possibility is that Raptor increases energy expenditure (EE), which protects against obesity. We reasoned that the severe hepatic steatosis and hepatomegaly observed in both chow and HFD fed RaptorAdipo-Cre cohorts argues against this scenario. However, the concept was previously proposed in a study that used aP2-Cre to delete Raptor in adipose tissue [12]. This study observed normal food intake but resistance to HFD and improved insulin tolerance in RaptoraP2-Cre mice and this was attributed to increased in Ucp1 mRNA expression in pgWAT and elevated EE; hepatic steatosis and hepatomegaly was not observed in RaptoraP2-Cre mice. Indeed, we find that Ucp1 mRNA is increased in both the sWAT and pgWAT of RaptorAdipo-Cre mice consuming chow diet (Figure 6A). This correlates with elevated Pgc1α levels in sWAT, and elevated Cpt1 in both sWAT and pgWAT (Figure 6A). However, in chow fed mice UCP1 protein was barely detectable in either WAT depot (Figure 6B). Moreover, we could only detect a slight increase in UCP1 protein in the sWAT of HFD-fed RaptorAdipo-Cre mice after a long exposure (Figure 6B). This suggests that total UCP1 protein is only slightly elevated in the sWAT of RaptorAdipo-Cre mice.
Figure 6.
Energy utilization and adipocyte transcriptional regulation. (A) mRNA expression levels from whole tissue lysate from Chow fed mice for indicated genes. (B) Western blots of whole tissue lysate for indicated proteins from both chow and HFD mice, along with exposure times. SE is short exposure, LE is long exposure. (C) Mean energy expenditure per mouse over 24 hrs, with AUC. (D) Mean respiratory exchange ratio per mouse over 3 days, with AUC. (E) Mean activity per mouse over 3 days, with AUC. (F) Daily food intake per mouse after 4 weeks on HFD. (G) Serum leptin concentration after HFD. (H) Fecal lipid content after 4 weeks HFD. (I) PPARy mRNA expression levels in whole tissue lysate for respective mouse models. (J) mRNA expression levels of PPARy targets in whole tissue lysate for respective mouse models. (K) C/EBPa mRNA expression levels from whole tissue lysate for respective mouse models. (L) Adiponectin mRNA expression from whole tissue lysate for respective mouse models. (Data were analyzed by Student's t-test. Values expressed as mean + SEM. *p < 0.05; **p < 0.01; ***p < 0.001).
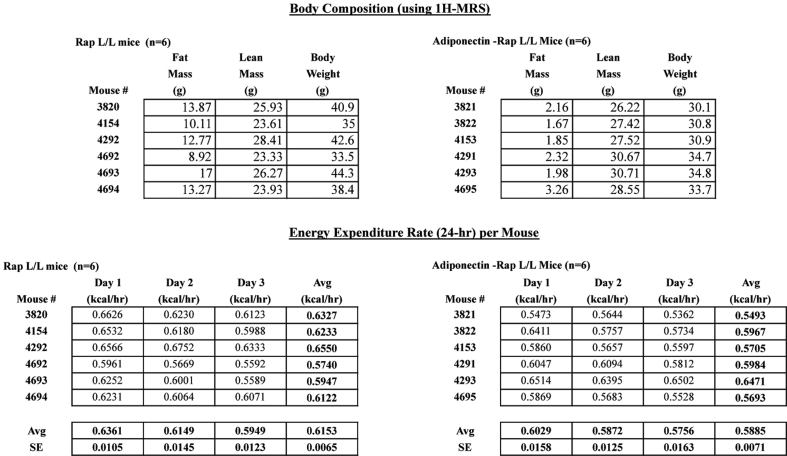
To better assess energy balance we also performed metabolic cage experiments. We found no difference in EE between HFD-fed RaptorAdipoq-cre mice and controls when expressing the data “per mouse” as recommend in [16] (Figure 6C). However, because it is often debated which method of reporting is correct, we provide the raw data in Table S1. We also found that the Respiratory Exchange Ratio (RER) is higher in RaptorAdipoq-cre mice indicating greater utilization of carbohydrates for fuel (Figure 6D). RaptorAdipoq-cre mice also exhibited normal physical activity (Figure 6E) yet interestingly, they are hyperphagic consuming nearly twice as much food as controls (Figure 6F). Hyperphagia is likely explained by the fact that HFD-fed RaptorAdipoq-cre mice have greatly reduced circulating leptin (Figure 6G). We also examined fecal lipids and found nearly twice the amount of lipid in the feces of RaptorAdipoq-cre mice (Figure 6H) indicating a lipid absorption defect. Thus, the mechanism causing resistance to obesity in RaptorAdipoq-cre mice cannot be explained by decreased food intake or increased EE.
Based on the increased EE reported in aP2-Cre;Raptor mice, it was suggested that adipose mTORC1 could be a potential target for anti-obesity drugs. However, our results indicate several important phenotypic differences compared to RaptoraP2-Cre mice (see detailed comparison in Table S2). These differences likely reflect recent data indicating that the aP2-Cre driver has limited efficiency for adipocytes and targets non-adipocytes such as endothelial cells [7], [8], [9], [10], [11]. It would be interesting to identify the exact target cell/mechanism responsible for the beneficial metabolic effects of deleting Raptor with aP2-Cre. Nevertheless, in light of our characterization of RaptorAdipoq-Cre mice, we would argue against prolonged targeting of mTORC1 in fat as a strategy to reduce obesity, though the effects of temporal or partial mTORC1 loss have not been thoroughly explored.
3.6. Decreased C/EBPα activity in Raptor-deficient WAT
We next asked whether adipocytes require mTORC1 to maintain expression of Pparγ, which encodes the master transcriptional regulator of adipocyte differentiation and function, and is a marker of adipocyte identity. We saw no significant changes in Pparγ mRNA expression in the sWAT or pgWAT from RaptorAdipoq-Cre mice eating chow or HFD (Figure 6I) suggesting Raptor is not required to maintain adipocyte identity per se. We also examined the expression of CD36 and Lpl, two Pparγ targets. In chow fed mice, Lpl tended to be lower while CD36 unaffected (Figure 6J); in the HFD group, both genes are reduced (Figure 6J). Thus, Pparγ activity, particularly during high fat feeding, may be altered in Raptor KO adipocytes.
In contrast to Pparγ, expression of the Pparγ co-regulator C/EBPα (the founding member of the CCAAT/enhancer-binding family) is greatly reduced in the sWAT of both chow and HFD-fed mice as well as in pgWAT, though to a lesser extent in this depot (Figure 6K). Interestingly, it was recently reported that C/EBPα in mature adipocytes is dispensable for terminal embryonic adipogenesis and adult adipocyte survival, but required for normal adipose tissue expansion during high fat diet feeding [21]. C/EBPα also promotes adiponectin expression [22], [23], [24]. Indeed, adiponectin mRNA expression in WAT (Figure 6L) and circulating Adiponectin levels (Figure 3H) are greatly reduced in RaptorAdipoq-Cre mice. Thus, decreased C/EBPα activity provides a possible mechanism to explain why only older RaptorAdipoq-Cre mice are resistant to adipose tissue expansion.
3.7. Comparison to human lipodystrophy
RaptorAdipoq-Cre mice bear phenotypic resemblance to human patients suffering from congenital generalized lipodystrophy or Berardinelli-Seip lipodystrophy [25]. These patients suffer from adipose tissue loss, and similar to the RaptorAdipoq-Cre model, exhibit insulin resistance, hepatic steatosis, hypertriglyceridemia, and increased RER. Nearly a quarter of the patients develop diabetes mellitus. Other clinical features can include ectopic lipid in muscle and hypertrophic cardiomyopathy. Berardinelli-Seip lipodystrophy is linked to mutations in AGPAT2 or BSCL2/seipin but the molecular basis of the disease is poorly understood. Interestingly, AGPAT2 and BSCL2 knockout models exhibit several phenotypic similarities to RaptorAdipoq-Cre mice [26], [27], [28]. Moreover, the mTORC1 substrate Lipin-1, a phosphatidic acid phosphatase mutated in fatty liver dystrophy (fld) mice, was recently shown to physically interact with Seipin and AGPAT2 [29], [30], [31]. Thus, RaptorAdipoq-Cre mice may be useful in understanding the mechanistic basis of human congenital lipodystrophy.
4. Conclusions
In this study we investigated the in vivo role of mTORC1 in mature adipocytes by deleting Raptor with Adiponectin-Cre. We conclude that mTORC1 activity in white adipocytes is dispensable for early post-natal WAT growth, but becomes essential for normal adipose tissue expansion with age. Moreover, adipocyte Raptor KO mice consuming a high fat diet are hyperphagic yet resistant to obesity. However, despite increased Ucp1 mRNA expression in WAT, these mice are not protected from obesity due to increased energy expenditure. Rather, they appear to have a defect in adipose tissue expansion, which redistributes lipids to the liver resulting in severe hepatomegaly and hepatic steatosis, and causes a dietary lipid absorption defect. It does not appear that mTORC1 activity is required in WAT to maintain Pparγ expression, but it may promote PPARγ activity towards certain targets. We also find that mTORC1 is required for normal C/ebpα and adiponectin expression providing a plausible link to the adipose tissue expansion defect in older mice. We found no obvious indication that adipocytes were dying in large numbers (i.e. crown-like structures) at least at the time-points examined. One hypothesis is that increased autophagy coupled with defective lipid metabolic pathways, perhaps in part through altered ULK1 and/or Lipin function, could cause defective triglyceride accumulation and/or adipose tissue wasting. Alternatively, mTORC1 in mature adipocytes may promote WAT expansion in older mice by regulating production of a pro-adipogenic paracrine signal. Interestingly, mTORC1 may have a unique function in neo-natal BAT development that requires further examination. Because mTORC1 has several known and putative substrates [32], [33] it will be important to continue delineating the expression and contributions of each downstream pathway in WAT maintenance in vivo using the most updated and precise genetic and metabolic strategies.
We also found that mTORC1 loss did not impair the post-natal BAT-like character of sWAT. Interestingly, it was recently reported that rapamycin blocks the “browning” of WAT in adult mice by inhibiting mTORC1 in mature adipocytes [34], [35] suggesting there may be differential requirements for mTORC1 in post-natal versus adult brite/beige adipocyte formation. Moreover, another study reported that increased mTORC1 signaling in BAT promotes a brown-to-white phenotypic switch, and in this model, rapamycin reversed the “whitening” of BAT [36]. Thus, the precise role of mTORC1 in BAT/WAT interconversions remains to be seen.
Notably, our conclusions are based on chronic (congenital) Raptor loss in adipocytes and thus the primary in vivo defect associated with losing mTORC1 activity cannot be determined by this approach. Moreover, it is clearly difficult to obtain the perfect Cre-driver for tissue-specific KO studies, thus future advances may lead to additional refinements in understanding mTORC1 signaling in fat. Nevertheless, RaptorAdipoq-Cre mice exhibit phenotypic similarities to human patients suffering from congenital generalized lipodystrophy suggesting this model may have significant clinical implications.
Acknowledgments
D.A.G. is supported by grants from the National Institutes of Health (R01DK094004 & R01CA196986) and a Leukemia and Lymphoma society Career Development Award. P.L.L. is supported by a predoctoral fellowship from the National Institute on Alcohol Abuse and Alcoholism (1F30AA024385-01). Y.T. is an American Cancer Society Postdoctoral Fellow. The metabolic cage studies were conducted by the UMass Mouse Pheonotyping Center (DK09300). We thank members of the Guertin lab for critical discussions.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.04.001.
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Fig. S1.
Ucp1-Cre driven Raptor KO reveal similar BAT phenotype as Adipoq-Cre driven mice. (A) BAT depot mass in adult Ucp1-Cre Raptor L/L KO and wt mice. (B) Representative H&E histology of adult wt and KO mice. (C) Body weight of adult mice. (D) WAT depot mass in adult mice.
References
- 1.Ogden C.L., Carroll M.D., Kit B.K., Flegal K.M. Prevalence of childhood and adult obesity in the United States, 2011–2012. JAMA. 2014;311:806–814. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.NIDDK, http://www.niddk.nih.gov/health-information/health-statistics/Pages/overweight-obesity-statistics.aspx.
- 3.Eurostat, http://ec.europa.eu/eurostat/en/web/products-press-releases/-/3-24112011-BP.
- 4.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolfson R.L., Chantranupong L., Saxton R.A., Shen K., Scaria S.M., Cantor J.R. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351:43–48. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chantranupong L., Scaria S.M., Saxton R.A., Gygi M.P., Shen K., Wyant G.A. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell. 2016 doi: 10.1016/j.cell.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eguchi J., Wang X., Yu S., Kershaw E.E., Chiu P.C., Dushay J. Transcriptional control of adipose lipid handling by IRF4. Cell Metabolism. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee K.Y., Russell S.J., Ussar S., Boucher J., Vernochet C., Mori M.A. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62:864–874. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F., Mullican S.E., DiSpirito J.R., Peed L.C., Lazar M.A. Lipoatrophy and severe metabolic disturbance in mice with fat-specific deletion of PPARgamma. Proceedings of the National Academy of Sciences U S A. 2013;110:18656–18661. doi: 10.1073/pnas.1314863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullican S.E., Tomaru T., Gaddis C.A., Peed L.C., Sundaram A., Lazar M.A. A novel adipose-specific gene deletion model demonstrates potential pitfalls of existing methods. Molecular Endocrinology. 2013;27:127–134. doi: 10.1210/me.2012-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeffery E., Berry R., Church C.D., Yu S., Shook B.A., Horsley V. Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte. 2014;3:206–211. doi: 10.4161/adip.29674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polak P., Cybulski N., Feige J.N., Auwerx J., Ruegg M.A., Hall M.N. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metabolism. 2008;8:399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Peterson T.R., Sengupta S.S., Harris T.E., Carmack A.E., Kang S.A., Balderas E. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue B., Rim J.S., Hogan J.C., Coulter A.A., Koza R.A., Kozak L.P. Genetic variability affects the development of brown adipocytes in white fat but not in interscapular brown fat. Journal of Lipid Research. 2007;48:41–51. doi: 10.1194/jlr.M600287-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez-Gurmaches J., Hung C.M., Sparks C.A., Tang Y., Li H., Guertin D.A. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metabolism. 2012;16:348–362. doi: 10.1016/j.cmet.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez-Gurmaches J., Hung C.M., Guertin D.A. Emerging complexities in adipocyte origins and identity. Trends in Cell Biology. 2016 doi: 10.1016/j.tcb.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakrabarti P., English T., Shi J., Smas C.M., Kandror K.V. Mammalian target of rapamycin complex 1 suppresses lipolysis, stimulates lipogenesis, and promotes fat storage. Diabetes. 2010;59:775–781. doi: 10.2337/db09-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soliman G.A., Acosta-Jaquez H.A., Fingar D.C. mTORC1 inhibition via rapamycin promotes triacylglycerol lipolysis and release of free fatty acids in 3T3-L1 adipocytes. Lipids. 2010;45:1089–1100. doi: 10.1007/s11745-010-3488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herman M.A., Peroni O.D., Villoria J., Schon M.R., Abumrad N.A., Bluher M. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 2012;484:333–338. doi: 10.1038/nature10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ro S.H., Jung C.H., Hahn W.S., Xu X., Kim Y.M., Yun Y.S. Distinct functions of Ulk1 and Ulk2 in the regulation of lipid metabolism in adipocytes. Autophagy. 2013;9:2103–2114. doi: 10.4161/auto.26563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q.A., Tao C., Jiang L., Shao M., Ye R., Zhu Y. Distinct regulatory mechanisms governing embryonic versus adult adipocyte maturation. Nature Cell Biology. 2015;17:1099–1111. doi: 10.1038/ncb3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park S.K., Oh S.Y., Lee M.Y., Yoon S., Kim K.S., Kim J.W. CCAAT/enhancer binding protein and nuclear factor-Y regulate adiponectin gene expression in adipose tissue. Diabetes. 2004;53:2757–2766. doi: 10.2337/diabetes.53.11.2757. [DOI] [PubMed] [Google Scholar]
- 23.Qiao L., Maclean P.S., Schaack J., Orlicky D.J., Darimont C., Pagliassotti M. C/EBPalpha regulates human adiponectin gene transcription through an intronic enhancer. Diabetes. 2005;54:1744–1754. doi: 10.2337/diabetes.54.6.1744. [DOI] [PubMed] [Google Scholar]
- 24.Qiao L., Schaack J., Shao J. Suppression of adiponectin gene expression by histone deacetylase inhibitor valproic acid. Endocrinology. 2006;147:865–874. doi: 10.1210/en.2005-1030. [DOI] [PubMed] [Google Scholar]
- 25.Patni N., Garg A. Congenital generalized lipodystrophies–new insights into metabolic dysfunction. Nature Reviews Endocrinology. 2015;11:522–534. doi: 10.1038/nrendo.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou H., Lei X., Benson T., Mintz J., Xu X., Harris R.B. Berardinelli-Seip congenital lipodystrophy 2 regulates adipocyte lipolysis, browning, and energy balance in adult animals. Journal of Lipid Research. 2015;56:1912–1925. doi: 10.1194/jlr.M060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cortes V.A., Curtis D.E., Sukumaran S., Shao X., Parameswara V., Rashid S. Molecular mechanisms of hepatic steatosis and insulin resistance in the AGPAT2-deficient mouse model of congenital generalized lipodystrophy. Cell Metabolism. 2009;9:165–176. doi: 10.1016/j.cmet.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L., Jiang Q., Wang X., Zhang Y., Lin R.C., Lam S.M. Adipose-specific knockout of SEIPIN/BSCL2 results in progressive lipodystrophy. Diabetes. 2014;63:2320–2331. doi: 10.2337/db13-0729. [DOI] [PubMed] [Google Scholar]
- 29.Talukder M.M., Sim M.F., O'Rahilly S., Edwardson J.M., Rochford J.J. Seipin oligomers can interact directly with AGPAT2 and lipin 1, physically scaffolding critical regulators of adipogenesis. Molecular Metabolism. 2015;4:199–209. doi: 10.1016/j.molmet.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huffman T.A., Mothe-Satney I., Lawrence J.C., Jr. Insulin-stimulated phosphorylation of lipin mediated by the mammalian target of rapamycin. Proceedings of the National Academy of Sciences U S A. 2002;99:1047–1052. doi: 10.1073/pnas.022634399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterfy M., Phan J., Xu P., Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nature Genetics. 2001;27:121–124. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- 32.Hsu P.P., Kang S.A., Rameseder J., Zhang Y., Ottina K.A., Lim D. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Y., Yoon S.O., Poulogiannis G., Yang Q., Ma X.M., Villen J. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tran C.M., Mukherjee S., Ye L., Frederick D.W., Kissig M., Davis J.G. Rapamycin blocks induction of the thermogenic program in white adipose tissue. Diabetes. 2016 doi: 10.2337/db15-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu D., Bordicchia M., Zhang C., Fang H., Wei W., Li J.L. Activation of mTORC1 is essential for beta-adrenergic stimulation of adipose browning. Journal of Clinical Investigation. 2016 doi: 10.1172/JCI83532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang X., Lan H., Tang H., Yuan F., Xu Y., Zhao J. Tuberous sclerosis complex 1-mechanistic target of rapamycin complex 1 signaling determines brown-to-white adipocyte phenotypic switch. Diabetes. 2015;64:519–528. doi: 10.2337/db14-0427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.