Among both prospective parents and providers of medical care, genetic and social concerns peak during the perinatal period. Advances in genomics and assisted reproductive technology have created new opportunities to detect genetic disorders and susceptibilities at multiple times during perinatal care and thus are relevant to these concerns. Emerging therapies for single-gene disorders may reshape these discussions.
Practitioners working with persons wishing to be parents are encouraged to inquire about their genetic backgrounds and family histories, to counsel them about tests for disease-carrier status that are based on known population-specific risks,1 and to refer them, when appropriate, to specialists in high-risk pregnancy and genetics. Nonetheless, there are major differences across the world in the adoption and implementation of genetic education and screening practices by providers, women and their partners, and health payment systems.2,3 Such differences are to be expected because access to health care, along with the availability of genetic counseling and testing, varies.
Even in the best-case scenario, patients, practitioners, and policymakers face complicated choices when selecting which genomic techniques to use broadly or individually in assessing risk and in determining how laboratory findings should inform decision making as the options for genetic testing expand.4 For example, it is not always possible to predict a priori the severity of a clinical condition on the basis of a genotype. A laboratory result may be flawless, but the identified genetic variation may not be known to cause a disease (i.e., it is a variant of uncertain significance). Or the discovered mutation or variant in a known disease gene may not reliably correlate with phenotype because of the influence of modifiers, which can be genetic, epigenetic, or environmental.
Preconception Genetic Screening and Testing
Genetic risk, especially of known genetic conditions in the family or a previous pregnancy, should ideally be assessed before conception or the establishment of a pregnancy in the context of assisted reproductive technology. Genetic screening is offered for a particular condition (or group of conditions) in individuals, groups, or populations. A family history of the condition is not required for genetic screening. Genetic testing is generally carried out when there is suspicion that an individual is at increased risk because of family history or because of a positive result on a bio-chemical screening test.
The American Congress of Obstetricians and Gynecologists (ACOG) recommends that women be offered information about genetic risk, including the risk of carrying mutant alleles that cause cystic fibrosis, hemoglobinopathies, and diseases typically affecting those of Eastern European Jewish ancestry.1,5-10 The American College of Medical Genetics (ACMG) recommends a more extended screening panel for those of Eastern European Jewish ancestry and the offering of carrier testing for spinal muscular atrophy to all couples, regardless of race or ethnic background.11-16 Identifying carriers of autosomal recessive or X-linked conditions before conception allows more informed decisions about reproductive options.
Different methods are used for screening, depending on whether chromosomes, proteins, related products of a gene (e.g., RNA), or nuclear or mitochondrial DNA are examined. Contemporary carrier screening involves tests for the most common mutations and for specific diseases in specific populations. Recent advances in DNA sequencing and bioinformatics have led to an approach for identifying carriers of known mutations that cause more than 400 recessive genetic diseases.17 However, this approach may miss some mutations and thus not identify some carriers.
In the case of carrier screening for Tay–Sachs disease (hexosaminidase deficiency, which is most prevalent in persons of Eastern European Jewish ancestry), the hexosaminidase enzyme assay remains the primary method of screening because it has greater sensitivity than targeted DNA mutation analysis. (Screening for the three most common hexosaminidase gene mutations detects 92 to 94% of carriers.18) However, there are now genetic tests that use the less sensitive targeted-mutation strategy for Tay–Sachs disease and that simultaneously test for the presence of mutations causing other genetic conditions for which this population is at increased risk, thus trading higher sensitivity for Tay–Sachs carrier status for a broader range of disease detection. Consequently, clinicians who are recommending such screening should have knowledge of current professional society guidelines, provide informed consent about the sensitivity and specificity of tests, and be able to make an appropriate referral for complex results.
Preimplantation Genetic Screening
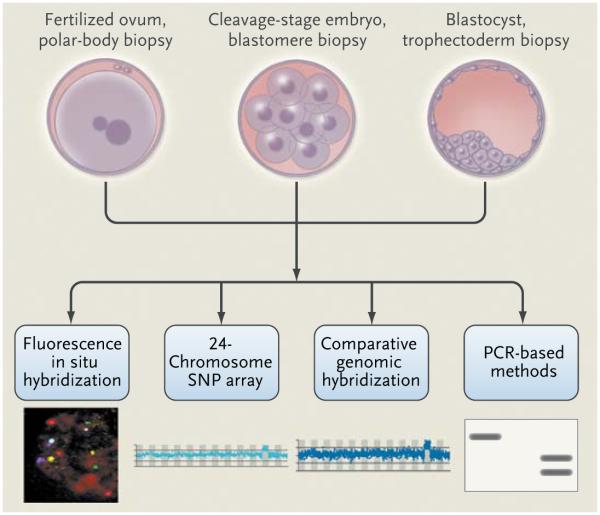
Preimplantation genetic screening involves the selection of embryos before transfer into the uterus to increase the success of assisted reproduction (Fig. 1, and interactive graphic, available with the full text of this article at NEJM.org). Genetic analysis is carried out on one or two blastomeres that are microsurgically removed from the embryo on day 3 of culture. Results are quickly obtained, so the selected embryos can be transferred on day 5 or frozen for future transfer. Fluorescence in situ hybridization (FISH), involving the use of fluorescently labeled DNA probes to paint fetal DNA in interphase nuclei, is usually used to detect chromosomal abnormalities (see the Glossary). Pre-implantation genetic screening has been applied in cases of advanced maternal age, repeated implantation failure, and idiopathic recurrent pregnancy loss and in order to improve pregnancy rates in single-embryo transfers.19-21
Figure 1. Methods for Preimplantation Genetic Screening and Diagnosis.
Material is obtained by microsurgical removal of polar bodies, blastomeres, or trophectoderm. The biopsy procedures generally do not affect the viability of the fertilized eggs or embryos. Depending on the specimen and analytic procedure, results can be obtained quickly and the selected embryos transferred without the need for cryopreservation while the genetic analysis is completed. Maternal genetic contributions (polar bodies, with evaluation of both bodies to determine the genetic status of the egg) and the genetic status of the embryo (blastomere and trophectoderm biopsy) can be analyzed by several different methods that provide varying amounts of information about sex chromosomes, chromosome copy number, and structural changes. These methods include fluorescence in situ hybridization (FISH), 24-chromosome single-nucleotide-polymorphism (SNP) arrays, and array comparative genomic hybridization. FISH methods detect a limited number of different chromosomes. Array comparative genomic hybridization and SNP arrays detect chromosome copy number as well as copy-number variations. In addition, SNP arrays can identify clinically significant uniparental disomy, consanguinity, and balanced translocations. Specific gene mutations may be identified with the use of methods based on polymerase-chain-reaction (PCR) assay. For mendelian disorders, a diagnosis can be obtained in 80% or more of samples. An inability to obtain a diagnosis is usually due to contamination or amplification failure. When a diagnosis is obtained for single-gene defects, it is highly accurate.
Because there is a high level of chromosomal mosaicism in the cleavage stages of embryonic development, which can confound the interpretation of findings or demand follow-up analysis, and because contemporary FISH methods do not capture the full complement of chromosome material, the extent to which preimplantation genetic screening is useful in improving pregnancy rates and outcomes is debated. Consequently, such genetic screening that is based on current FISH technology is not recommended for the indications noted above (i.e., advanced maternal age, repeated implantation failure, and idiopathic recurrent pregnancy loss and in order to improve pregnancy rates in single-embryo transfers).22 Analysis of polar bodies may yield improved pregnancy outcomes by detecting maternal genetic abnormalities in eggs, including meiotic errors that result in aneuploidy. Newer array-based methods, including 24-chromosome single-nucleotide-polymorphism (SNP) arrays (virtual karyotyping), will probably replace FISH because they provide more genetic information.23 This technology may increase the clinical use of preimplantation genetic screening.
Preimplantation Genetic Diagnosis
Preimplantation genetic diagnosis, which was introduced in 1990, allows for the selection of disease-free embryos for transfer into the uterus.24 Genetic analysis is usually carried out as described for preimplantation genetic screening. FISH is used to detect sex chromosomes and specific chromosomal abnormalities, or polymerase chain reaction (PCR) is used to amplify DNA for molecular diagnosis. The first births after the preimplantation genetic diagnosis of structural chromosomal abnormalities with the use of comparative genomic hybridization and microarray analyses were recently reported.25 Detection of mitochondrial DNA mutations is also possible, providing that they are prevalent in the mitochondrial pool.26
The first and second polar bodies can be analyzed to determine the presence of maternal genetic contributions (i.e., X-linked diseases and autosomal dominant diseases), including carrier states for Duchenne’s muscular dystrophy, incontinentia pigmenti, and neurofibromatosis type 2.27
The major monogenic dominant, recessive, and sex-linked diseases for which preimplantation genetic diagnosis has been used are listed in Table 1. With current methods, such diagnosis of mendelian disorders is highly accurate, with a misdiagnosis rate of less than 1%.28 Misdiagnosis has been attributed to laboratory error, including transfer of the wrong embryo, contamination by extra-embryonic material, allele dropout (when one of the alleles is not amplified on PCR), use of the wrong probes or primer sets, and chromosomal mosaicism.
Table 1.
Monogenic Diseases That Are Frequently Identified by Preimplantation Genetic Diagnosis.
| Disease | Genes |
|---|---|
| Dominant | |
| Huntington’s disease | HTT |
| Myotonic dystrophy | Type 1, DMPK; type 2, ZNF9 (CNBP) |
| Charcot–Marie–Tooth disease | Type 1A, PMP22 |
|
| |
| Recessive | |
| β-Thalassemia | HBB |
| Cystic fibrosis | CFTR |
| Spinal muscular atrophy (Werdnig–Hoffman disease) |
SMN1 |
| Sickle cell disease | HBB |
|
| |
| Sex-linked | |
| Fragile X syndrome | FMR1 |
| Duchenne’s muscular dystrophy | DMD |
| Hemophilia | Type A, F8; type B, F9 |
Preimplantation genetic diagnosis is increasingly available in the United States and Europe. However, its practice is relatively unregulated, although professional societies (American Society for Reproductive Medicine and European Society of Human Reproduction and Embryology) have issued guidelines and recommended the accreditation of laboratories performing such genetic diagnosis.29,30
Prenatal Genetic and Genomic Testing
For all pregnancies, the baseline risk of some type of birth defect is 3 to 4%. The severity of such defects varies widely, reflecting the wide range of inherited mutations or genetic variants; spontaneous mutations arising in the gametes, embryo, or fetus; epigenetic alterations; and environmental influences. Maternal factors that increase the chance of having a child with a genetic condition or congenital anomaly include advancing age, health conditions such as diabetes and obesity, and exposures to teratogenic factors, such as alcohol and viral infections.
Prenatal genetic diagnostic testing currently requires the collection of a sample of fetal cells, either by aspirating chorionic villi by a transcervical or transabdominal approach under ultrasonographic guidance at 10 to 14 weeks of gestation or withdrawing amniotic fluid and collecting and culturing exfoliated fetal cells (amniocentesis) around 15 weeks of gestation. Prenatal diagnosis by chorionic villus sampling or amniocentesis is an option for high-risk pregnancies. These procedures generally carry rates of postprocedure miscarriage of approximately 1% or less. The information gained from the traditional cytogenetic or FISH analysis of chorionic villus samples or cultured fetal cells can be enhanced by DNA-array techniques, including array comparative genomic hybridization and SNP arrays. Such methods can detect genetic variation and abnormalities that usually escape lower-resolution cytogenetics, including copy-number variation.31
Although this information can be useful when specific copy-number variations that are known to be associated with a disorder are detected, the clinical significance of many structural variations is unknown. Many diseases are genetically heterogeneous, with some cases caused by copy-number variations and others caused by different factors. Although DNA array–based methods will probably be increasingly used in genetic diagnosis, the clinical guidelines for the appropriate use of this technology, especially in prenatal diagnosis, are debated. Guidance offered by the ACOG and other professional organizations will continue to evolve.32
The detection of a fetal anomaly on ultrasonography presents the opportunity to discuss with the family possible determination of a genetic basis for the malformation. However, the application of genetic and genomic testing in this situation should be carefully weighed because of the cost and complexities in evaluating the results, especially if there has not been a previous genetic analysis in a family member to guide interpretation of the findings.
Noninvasive Prenatal Diagnosis
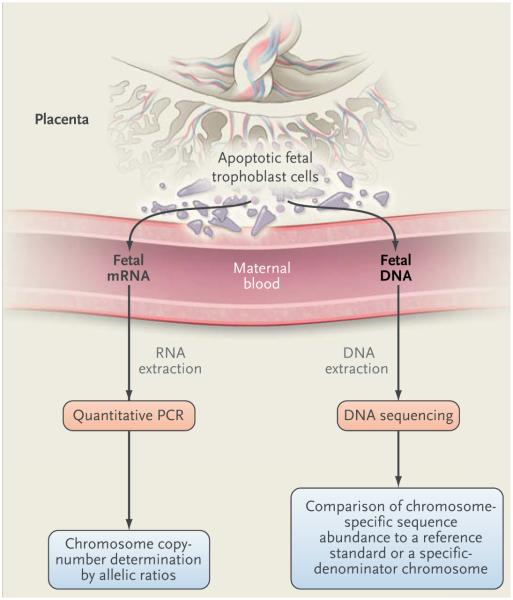
It has long been recognized that nucleated fetal cells reach the maternal circulation, but attempts to isolate these rare cells from maternal blood (which typically number 1 to 6 cells per milliliter of maternal blood) and use them for genetic testing have been disappointing because of low sensitivity. Cell-free fetal RNA and DNA, released from apoptotic placental trophoblast cells (and not from the fetus per se), hold greater promise for genetic testing as a result of advances in DNA sequencing methods and informatics (Table 2).33,34 In 2007, Down’s syndrome was detected by the quantitative assay of maternal blood cell-free RNA for PLAC4,35 a trophoblast-specific gene located in the Down’s syndrome region of chromosome 21 (Fig. 2).36 The PLAC4 coding sequence has a SNP that allows determination of allelic ratios when the fetus is heterozygous for the SNP. Euploid embryos have an allelic ratio of 1:1. A ratio of 2:1 indicates a strong likelihood of trisomy 21. The analysis of mRNAs encoded by different genes on chromosome 21 could improve the sensitivity of this method but has not been widely pursued.
Table 2.
Perinatal Genomic Tests.
| Type of Test* | Chorionic Villi |
Amniocytes | Nucleated Fetal Cells |
Cell-free Fetal DNA/RNA |
Polar Bodies | Biopsy of Blastomere or Blastocyst |
Potential for Genomewide Analysis† |
|---|---|---|---|---|---|---|---|
| Karyotype (cytogenetic) | Yes | Yes | No | No | No | No | Yes |
| FISH | Yes | Yes | Yes | No | Yes | Yes | No |
| Quantitative PCR | Yes | Yes | Yes | Yes | Yes | Yes | No |
| SNP or comparative genomic hybrid- ization array |
Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Shotgun sequencing or massively parallel sequencing |
Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Exome or whole-genome sequencing | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Mutation detection | Yes | Yes | Yes | Yes | Yes | Yes | No |
Listed are techniques that are in use or feasible for perinatal genomic testing. FISH denotes fluorescence in situ hybridization, PCR poly-merase chain reaction, and SNP single-nucleotide polymorphism.
Genomewide analyses include comparative genomic hybridization, 24-chromosome SNP arrays, whole-exome sequencing, and whole-genome sequencing.
Figure 2. Noninvasive Prenatal Diagnosis with the Use of Plasma Cell-free Fetal RNA or DNA in Maternal Blood Derived from Dying Trophoblast Cells of the Placenta.
For cell-free fetal RNA, target RNA molecules containing SNPs are quantified with the use of a PCR assay. The allelic ratio, which is determined on quantitative PCR, is used to determine the chromosome copy number when the fetus is heterozygous for the SNP. A 1:1 ratio of amplified allelic variants is expected in the euploid state, whereas a ratio of 2:1 indicates trisomy. Cell-free fetal DNA in plasma can be sequenced directly because it is smaller than maternal cell-free DNA; it can also be enriched by means of size fractionation before DNA sequence analysis. The abundance of specific chromosome sequences can be compared with normal reference samples or with another chromosome as a specified denominator in the sample to determine variation in chromosome copy number or structural changes in chromosomes. Additional methods of analysis of cell-free fetal DNA to determine chromosome copy number have been described, including methods to quantify differentially methylated regions of specific fetal chromosomes with the use of PCR. These methods await validation.
Cell-free fetal DNA is currently the material of choice for noninvasive prenatal genomic diagnosis. It represents 3 to 6% of circulating cell-free DNA in maternal plasma, and it can be detected in the first trimester of pregnancy, increasing in abundance as the placenta grows. Cell-free fetal DNA fragments are much smaller than cell-free maternal DNA, which facilitates DNA sequence analysis. Although fetal DNA is detectable at 5 weeks of gestation, current methods of analysis are unreliable before 7 weeks of gestation.
Since the presence of the Y chromosome defines male sex, its detection or lack thereof in maternal blood can be used to infer fetal sex. A recent review and meta-analysis37 of fetal sex determination with the use of maternal cell-free fetal DNA reported very good but imperfect results for testing after 7 weeks of gestation. The greatest sensitivity and specificity in the use of Y-chromosome sequences to determine sex are obtained after 20 weeks of gestation, at which time ultrasonography can do the job.
In addition to sex determination, detection of paternal genomic contributions to cell-free fetal DNA can be used to determine fetal RhD status with high accuracy in the pregnancy of an RhD-negative woman. This approach can also be used to detect paternally transmitted, dominant single-gene disorders, including Huntington’s disease, achondroplasia, and myotonic dystrophy.38 Carrier status for cystic fibrosis, hemoglobinopathies, and 21-hydroxylase deficiency has also been determined.38,39
In 2008, DNA sequencing to detect so-called chromosome dosage, which is reflected as either underrepresentation or overrepresentation of chromosome-specific sequences, was successfully used to identify trisomies of chromosomes 13, 18, and 21.40 Sequencing-based measurements of the proportion of small DNA fragments derived from chromosome 21 that exceed a threshold value relative to sequences from euploid reference samples have been reported to have a positive predictive value of 96.6% and a negative predictive value of 100%.41
Theoretically, this approach, which is based on shotgun sequencing of the small cell-free fetal DNA fragments, could identify less common, more complex aneuploid states resulting from unbalanced translocations or partial chromosome duplication. Detection of a fetal microdeletion syndrome from sequence analysis of cell-free fetal DNA in maternal plasma was recently reported.42 At present and in this context, it remains an experimental technology. So too does the prospect of broader genetic analyses, including whole-genome sequencing (Table 2).
If these approaches become technically feasible, it is not clear whether they would be used as a screening method or as a diagnostic test. They would need to be cost-effective with sufficiently rapid reporting of results in order to have a meaningful effect on decision making. Table 3 provides an overview of potential timing for the genetic diagnosis of cystic fibrosis, illustrating the challenges of informed consent across multiple medical conditions and genetic variations.
Table 3.
Potential Timing and Related Issues for Genetic Diagnosis of Cystic Fibrosis.*
| Timing of Genetic Test |
Screening for Mutations | Testing for Mutation in Family | Blood Testing for Immunoreactive Trypsinogen |
Ethical Issues | Social Issues | Technological and Biologic Issues |
|---|---|---|---|---|---|---|
| Preconception | General recommendation of professional societies with informed consent |
Genetic testing for high-risk condition |
NA | Meaningful reproductive choice |
Unplanned pregnancy | Changing technologies and variability of genotype- phenotype correlation |
| Preimplantation | Embryo selection | Genetic testing for high-risk condition |
NA | Moral status of embryo | Fiscal resources, access issues |
Potential risks of assisted reproductive technology |
| Prenatal | If not done before conception, general recommendation of professional societies with informed consent |
Available; may be influenced by prenatal findings (e.g., echogenic bowel) |
NA | Pregnancy management choices, autonomy |
Entry times into prenatal care |
Risk of invasive procedure |
| Newborn | Growing use in newborn screening |
Available; may be influenced by newborn findings (e.g., meconium ileus) |
Available | Informed consent | When to tell carrier status, access to clinical trials |
Range of severity with >1000 mutations |
NA denotes not applicable.
Newborn Genetic Screening
Every year, approximately 4 million infants in the United States undergo newborn blood-sample screening. Many observers consider this type of screening to be a classic example of a population health benefit derived from the application of genetic discoveries. Most states have an opt-out system for consent for newborn screening. There is much current discussion about individual choices, risks, benefits, and cost, especially with additional disorders under consideration for inclusion in newborn screening and the expansion of testing options to the preconception and prenatal periods.43 Biochemically based newborn screening for phenylketonuria (PKU) began in the 1960s after it became clear that the introduction of a phenylalanine-restricted diet could improve outcomes for children with PKU. By 2006, the collaborative efforts of advocacy groups, along with those of professional pediatric, public health, and genetic organizations, resulted in a uniform screening panel to identify 29 conditions through newborn screening. The conditions include hemoglobinopathies, endocrinopathies, cystic fibrosis, hearing loss, and disorders of metabolism. A mechanism for proposing and evaluating other disorders for inclusion in the uniform screening panel has been established.44,45 The Advisory Committee on Heritable Disorders in Newborns and Children recommended adding severe combined immunodeficiency to the uniform screening panel in 201046,47 and continues to consider evidence supporting the addition of other genetic conditions, such as spinal muscular atrophy. However, in the United States, uptake of the uniform screening panel varies according to state, resulting in a somewhat piecemeal approach to the challenge of diagnosing heritable disorders in newborns.
In practice, the availability of newborn screening and follow-up may figure in decision making about prenatal screening for conditions such as cystic fibrosis, in which a diagnosis shortly after birth may be considered a personally acceptable alternative to prenatal diagnosis. For many couples, particularly those with a known genetic risk, the decision as to whether to carry out genetic testing is determined after discussion with their physician and a genetic counselor. Reproductive benefit (i.e., access to genetic information at a time when it can be used to guide reproductive choices) has not historically been part of the rationale for including specific conditions in newborn screening, but it is now increasingly a part of public discussion.48 The availability of clinical trials involving potential health improvement for children with rare conditions has also become an argument for early identification of genetic conditions through newborn screening, although no known beneficial treatment may be available for some conditions.
Newborn Genetic Diagnosis
Guidelines from professional societies have begun to recommend that array comparative genomic hybridization be used for the rapid multiplex detection of genomic imbalances in the evaluation of patients with developmental delay or intellectual disability, congenital anomalies, or dysmorphic features, unless a clear phenotypic diagnosis can be more simply confirmed with routine karyotyping (e.g., Down’s syndrome).49 However, such testing is generally more expensive than other methods and may detect variation of uncertain clinical significance.
Advances in genetics have increased the potential to identify a growing number of conditions at a presymptomatic stage and have raised many ethical, legal, and social issues.50 One advance in particular, the presymptomatic diagnosis of an adult genetic condition early in life, raises the practical issue of how best to store and then retrieve this information later in life. Presymptomatic diagnosis of later-onset conditions such as familial cancer syndromes or Huntington’s disease is possible at preimplantation, prenatally, or after birth. Electronic health records and record linkages are being discussed as a tool to improve coordinated lifetime health and care, as is the case with the controversial National Collegiate Athletic Association recommendations on screening athletes for mutations that cause the sickle cell trait.51 Most U.S. athletes had newborn screening as infants, but the records have not accompanied the child into adulthood.
Direct-to-Consumer Analyses
Improvements in the speed and accuracy of DNA testing and the use of easily obtained material for analysis (e.g., desquamated cells in saliva) have enabled commercial scaling of genetic testing. Such developments have spawned a proliferation of Internet-based offerings of genetic tests, many of which are based on genomewide association studies for complex traits.52 Genetic testing for several hundred different traits is typically advertised.53,54 These commercial offerings have outpaced the development of public policy and regulatory oversight, in part because the criteria by which the clinical utility of personal genomic information is evaluated are subject to debate.55 Some companies indicate that they provide information on genetic disposition and minimize its direct implication for medical care. Other companies employ genetic counselors or indicate that they provide information about carrier or other genetic status. At least two relatively new companies target preconception testing for couples who want to know about potential recessive conditions.
An expert advisory panel of the Food and Drug Administration has recently recommended that direct-to-consumer genetic tests be subject to medical supervision, which might include both the interpretation of results and ordering of tests by doctors rather than by lay consumers.56 The ACMG and ACOG have similar and congruent perspectives that generally discourage direct-to-consumer testing until the resolution of several issues, including the limited knowledge of genetic testing by both patients and providers, difficulty in test interpretation, lack of federal oversight, and issues of privacy and confidentiality.57,58 Enhanced provider education and point-of-care tools, as well as increased involvement of genetic counselors and genetic medical specialists including those in maternal and fetal medicine, will be required.59,60
Genomics and Maternal and Child Health
Preconception health is increasingly recognized as a critical component in improving birth outcomes and reducing health disparities.61 Genetic variation and individual choices present an ongoing challenge to resource allocation in health care and the translation of genomic progress to improved health. Although the cost of genetic sequencing has generally continued to decrease, health disparities may be exacerbated by uneven access to preconception and prenatal care and the varied availability of access to assisted reproductive technology and to genetic specialists and testing among different populations. Issues regarding the patenting of uses of gene sequences, currently under court review, may also have an effect on cost and access. In addition, it is unclear who will pay for developing the evidence of benefit for screening, diagnosis, and treatment for the growing number of individually rare but collectively common genetic conditions.
Genetic tests can have a major effect on choices such as whom to marry, whether to have children, and whether to continue a pregnancy. The globalization of infertility practices with varied regulation makes possible choices of gametes and embryos, sperm, and uterus with varying degrees of genetic testing along the process. The technological challenges to having enough tissue to test from the pre-embryo for full sequencing are being addressed, as well as the integrated analysis of a complete adult human genome in a clinical context.62 Rapid advances in genetics and genomics will change genetic testing and screening. For example, personal genome testing on the basis of genomewide SNP scans will become outdated as whole-genome or whole-exome sequencing becomes affordable to consumers and their providers. These sequencing approaches offer simultaneous testing for many monogenic diseases, as well as for numerous mutations with an unknown effect. Moreover, there will be major challenges in interpreting the clinical significance of the large amount of data provided by whole-genome or whole-exome sequencing.
Conclusions
All new genomic technologies are potentially applicable to preconception, prenatal, and newborn care, but whether and how they will be used are subject to debate. Although we are now able to interrogate the human genome with exquisite precision, genotype may not predict phenotype. The development and implementation of guidelines involve questions of input from consumers and advocates, conflict of interest, and cost-effectiveness analysis. Yet the development of outcome measures to evaluate clinical genetic services is in a nascent state. Clinicians are encouraged to keep up with advances and national recommendations. Each clinician is an important educator of patients and a key member of the referral network for specialized services to bridge the gap between the worlds of personalized medicine and evidence-based medicine.
Supplementary Material
Acknowledgments
We thank Colleen Jackson-Cook, Ph.D., Virginia Commonwealth University; Michael T. Mennuti, M.D., University of Pennsylvania; Joe Leigh Simpson, M.D., Florida International University; Anna J.B. Smith, M.P.H., London School of Hygiene and Tropical Medicine; and Thomas J. Smith, M.D., Johns Hopkins University, for their helpful review and comments during the preparation of the manuscript.
Glossary
- Amplification
Production of multiple copies of a gene sequence, usually through the process of polymerase chain reaction.
- Aneuploidy
The occurrence of one or more extra or missing chromosomes, leading to an unbalanced chromosome complement, or any chromosome number that is not an exact multiple of the haploid number.
- Array comparative genomic hybridization
A technique that allows the detection of losses and gains in DNA copy number across the entire genome without previous knowledge of specific chromosomal abnormalities.
- Balanced translocation
The positional change of one or more chromosome segments in cells or gametes without alteration of the normal amount of genetic material.
- Copy-number variation
Variation from one person to the next in the number of copies of a particular gene or DNA sequence. The full extent to which copy-number variation contributes to human disease is not yet known.
- Epigenetic change
A change in the regulation of the expression of gene activity without alteration of genetic structure.
- Fluorescence in situ hybridization
A laboratory technique for detecting and locating a specific DNA sequence on a chromosome. The technique relies on exposing chromosomes to a small DNA sequence called a probe that has a tag (usually a fluorescent molecule) attached to it. The probe sequence binds to its corresponding sequence on the chromosome.
- Genomewide association study
An approach used in genetics research to look for associations between many specific genetic variations (typically hundreds of thousands of single-nucleotide polymorphisms) and particular diseases.
- Heterozygosity
The presence of different alleles at one or more loci on homologous (paired) chromosomes.
- Polymerase chain reaction
A laboratory technique used to amplify DNA sequences. Short, synthetic complementary DNA sequences called primers are used to select the portion of the genome to be amplified. The temperature of the sample is repeatedly raised and lowered to facilitate the copying of the target DNA sequence by a DNA-replication enzyme. The technique can produce a billion copies of the target sequence in just a few hours.
- Primer
A molecule (in the form of a short strand of RNA or DNA) whose presence is required for the formation of another molecule (in the form of a longer chain of DNA).
- Probe
A specific prefabricated sequence of DNA or RNA that is labeled by one of several methods and used to detect the presence of a complementary sequence by binding to that site.
- Single-nucleotide polymorphism
A single-nucleotide variation in a genetic sequence, a common form of variation in the human genome.
- Size fractionation
Separation according to the length of the nucleotide sequence.
- Whole-genome sequencing
Determination of the primary nucleotide sequence of the entire genome of an organism.
Footnotes
Contributor Information
Joann Bodurtha, McKusick–Nathans Institute of Genetic Medicine, Johns Hopkins Medical Institutions, Baltimore
Jerome F. Strauss, III, Department of Obstetrics and Gynecology, Virginia Commonwealth University, Richmond
References
- 1.American College of Obstetricians and Gynecologists Committee on Genetics Committee opinion no. 478: family history as a risk assessment tool. Obstet Gynecol. 2011;117:747–50. doi: 10.1097/AOG.0b013e318214780e. [DOI] [PubMed] [Google Scholar]
- 2.Genetic services policy project final report. Washington State Department of Health; Seattle: 2008. ( http://depts.washington.edu/genpol/docs/FinalReport.pdf) [Google Scholar]
- 3.Sharp RR, Goldlust ME, Eng C. Addressing gaps in physician education using personal genomic testing. Genet Med. 2011;13:750–1. doi: 10.1097/GIM.0b013e318228821f. [DOI] [PubMed] [Google Scholar]
- 4.de Jong A, Dondorp WJ, Frints SGM, de Die-Smulders CEM, de Wert GMWR. Advances in prenatal screening: the ethical dimension. Nat Rev Genet. 2011;12:657–63. doi: 10.1038/nrg3036. [DOI] [PubMed] [Google Scholar]
- 5.American College of Obstetricians and Gynecologists Committee on Genetics ACOG practice bulletin no. 77: screening for fetal chromosomal abnormalities. Obstet Gynecol. 2007;109:217–27. doi: 10.1097/00006250-200701000-00054. [DOI] [PubMed] [Google Scholar]
- 6.Idem ACOG practice bulletin no. 486: update on carrier screening for cystic fibrosis. Obstet Gynecol. 2011;117:1028–31. doi: 10.1097/AOG.0b013e31821922c2. [DOI] [PubMed] [Google Scholar]
- 7.Idem ACOG committee opinion no. 338: screening for fragile X syndrome. Obstet Gynecol. 2006;107:1483–5. doi: 10.1097/00006250-200606000-00059. [DOI] [PubMed] [Google Scholar]
- 8.Idem ACOG practice bulletin no. 78: hemoglobinopathies in pregnancy. Obstet Gynecol. 2007;109:229–37. doi: 10.1097/00006250-200701000-00055. [DOI] [PubMed] [Google Scholar]
- 9.Idem ACOG committee opinion no. 442: preconception and prenatal carrier screening for genetic diseases in individuals of Eastern European Jewish descent. Obstet Gynecol. 2009;114:950–3. doi: 10.1097/AOG.0b013e3181bd12f4. [DOI] [PubMed] [Google Scholar]
- 10.Idem ACOG committee opinion no. 432: spinal muscular atrophy. Obstet Gynecol. 2009;113:1194–6. doi: 10.1097/AOG.0b013e3181a6d03a. [DOI] [PubMed] [Google Scholar]
- 11.Driscoll DA, Gross SJ. Screening for fetal aneuploidy and neural tube defects. Genet Med. 2009;11:818–21. doi: 10.1097/GIM.0b013e3181bb267b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grody WW, Cutting GR, Klinger KW, Richards CS, Watson MS, Desnick RJ. Laboratory standards and guidelines for population-based cystic fibrosis carrier screening. Genet Med. 2001;3:149–54. doi: 10.1097/00125817-200103000-00010. [DOI] [PubMed] [Google Scholar]
- 13.Watson MS, Cutting GR, Desnick RJ, et al. Cystic fibrosis population carrier screening: 2004 revision of American College of Medical Genetics mutation panel. Genet Med. 2004;6:387–91. doi: 10.1097/01.GIM.0000139506.11694.7C. [DOI] [PMC free article] [PubMed] [Google Scholar]; Genet Med. 2004;6:548. Errata. 2005;7:286. [Google Scholar]
- 14.Sherman S, Pletcher BA, Driscoll DA. Fragile X syndrome: diagnostic and carrier testing. Genet Med. 2005;7:584–7. doi: 10.1097/01.GIM.0000182468.22666.dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross SJ, Pletcher BA, Monaghan KG. Carrier screening in individuals of Ashkenazi Jewish descent. Genet Med. 2008;10:54–6. doi: 10.1097/GIM.0b013e31815f247c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prior TW. Carrier screening for spinal muscular atrophy. Genet Med. 2008;10:840–2. doi: 10.1097/GIM.0b013e318188d069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell CJ, Dinwiddie DL, Miller NA, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med. 2011;3:65ra4. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaback MM, Desnick RJ. Hexosaminidase A deficiency. In: Pagon RA, Bird TD, Dolan CR, Stephens K, editors. GeneReviews. University of Washington; Seattle: 1993–1999. (updated Aug. 11, 2011) ( http://www.ncbi.nlm.nih.gov/books/NBK1218) [Google Scholar]
- 19.Anderson RA, Pickering S. The current status of preimplantation genetic screening: British Fertility Society policy and practice guidelines. Hum Fertil (Camb) 2008;11:71–5. doi: 10.1080/14647270802041607. [DOI] [PubMed] [Google Scholar]
- 20.Munné S, Howles CM, Wells D. The role of preimplantation genetic diagnosis in diagnosing embryo aneuploidy. Curr Opin Obstet Gynecol. 2009;21:442–9. doi: 10.1097/GCO.0b013e32832fad73. [DOI] [PubMed] [Google Scholar]
- 21.Cooper AR, Jungheim ES. Preimplantation genetic testing: indications and controversies. Clin Lab Med. 2010;30:519–31. doi: 10.1016/j.cll.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mastenbroek S, Twisk M, van der Veen F, Repping S. Preimplantation genetic screening: a systematic review and metaanalysis of RCTs. Hum Reprod Update. 2011;17:454–66. doi: 10.1093/humupd/dmr003. [DOI] [PubMed] [Google Scholar]
- 23.Treff NR, Levy B, Su J, Northrop LE, Tao X, Scott RT., Jr SNP microarray-based 24 chromosome aneuploidy screening is significantly more consistent than FISH. Mol Hum Reprod. 2010;16:583–9. doi: 10.1093/molehr/gaq039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spits C, Sermon K. PGD for monogenic disorders: aspects of molecular biology. Prenat Diagn. 2009;29:50–6. doi: 10.1002/pd.2161. [DOI] [PubMed] [Google Scholar]
- 25.Alfarawati S, Fragouli E, Colls P, Wells D. First births after preimplantation genetic diagnosis of structural chromosome abnormalities using comparative genomic hybridization and microarray analysis. Hum Reprod. 2011;26:1560–74. doi: 10.1093/humrep/der068. [DOI] [PubMed] [Google Scholar]
- 26.Bredenoord A, Dondorp W, Pennings G, de Die-Smulders C, Smeets B, de Wert G. Preimplantation genetic diagnosis for mitochondrial DNA disorders: ethical guidance for clinical practice. Eur J Hum Genet. 2009;17:1550–9. doi: 10.1038/ejhg.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuliev A, Rechitsky S. Polar body based preimplantation genetic diagnosis for Mendelian disorders. Mol Hum Reprod. 2011 Feb 14; doi: 10.1093/molehr/gar012. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 28.Geraedts JP, De Wert GM. Preimplantation genetic diagnosis. Clin Genet. 2009;76:315–25. doi: 10.1111/j.1399-0004.2009.01273.x. [DOI] [PubMed] [Google Scholar]
- 29.The Practice Committee of the American Society for Reproductive Medicine, Practice Committee of the Society for Assisted Reproductive Technology Revised guidelines for human embryology and andrology laboratories. Fertil Steril. 2008;90(Suppl):S45–S59. doi: 10.1016/j.fertnstert.2008.08.099. [DOI] [PubMed] [Google Scholar]
- 30.Harton G, Braude P, Lashwood A, et al. ESHRE PGD Consortium best practice guidelines for organization of a PGD centre for PGD/preimplantation genetic screening. Hum Reprod. 2011;26:14–24. doi: 10.1093/humrep/deq229. [DOI] [PubMed] [Google Scholar]
- 31.Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–46. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Committee on Genetics, American College of Obstetricians and Gynecologists ACOG committee opinion no. 446: array comparative genomic hybridization in prenatal diagnosis. Obstet Gynecol. 2009;114:1161–3. doi: 10.1097/AOG.0b013e3181c33cad. [DOI] [PubMed] [Google Scholar]
- 33.Go AT, van Vugt JM, Oudejans CB. Non-invasive aneuploidy detection using free fetal DNA and RNA in maternal plasma: recent progress and future possibilities. Hum Reprod Update. 2011;17:372–82. doi: 10.1093/humupd/dmq054. [DOI] [PubMed] [Google Scholar]
- 34.Ehrich M, Deciu C, Zwiefelhofer T, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol. 2011;204(3):205.e1–205.e11. doi: 10.1016/j.ajog.2010.12.060. [DOI] [PubMed] [Google Scholar]
- 35.Tsui NB, Akolekar R, Chiu RW, et al. Synergy of PLAC4 RNA concentration and measurement of the RNA single nucleotide polymorphism for the noninvasive prenatal detection of trisomy 21. Clin Chem. 2010;56:73–81. doi: 10.1373/clinchem.2009.132662. [DOI] [PubMed] [Google Scholar]
- 36.Kido S, Sakuragi N, Bronner MP, et al. D21S418E identifies a cAMP-regulated gene located on chromosome 21q22.3 that is expressed in placental syncytiotrophoblast and choriocarcinoma cells. Genomics. 1993;17:256–9. doi: 10.1006/geno.1993.1317. [DOI] [PubMed] [Google Scholar]
- 37.Devaney SA, Palomaki GE, Scott JA, Bianchi DW. Noninvasive fetal sex determination using cell-free fetal DNA: a systematic review and meta-analysis. JAMA. 2011;306:627–36. doi: 10.1001/jama.2011.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright CF, Burton H. The use of cell-free fetal nucleic acids in maternal blood for non-invasive prenatal diagnosis. Hum Reprod Update. 2009;15:139–51. doi: 10.1093/humupd/dmn047. [DOI] [PubMed] [Google Scholar]
- 39.Lazaros L, Hatzi E, Bouba I, Paraskevaidis E, Georgiou I. Non-invasive prenatal detection of paternal origin Hb Lepore in a male fetus at the 7th week of gestation. Fetal Diagn Ther. 2006;21:506–9. doi: 10.1159/000095662. [DOI] [PubMed] [Google Scholar]
- 40.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008;105:16266–71. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu RWK, Akolekar R, Zheng YWL, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ. 2011;342:c7401. doi: 10.1136/bmj.c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peters D, Chu T, Yatsenko SA, et al. Noninvasive prenatal diagnosis of a fetal microdeletion syndrome. N Engl J Med. 2011;365:1847–8. doi: 10.1056/NEJMc1106975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levy PA. An overview of newborn screening. J Dev Behav Pediatr. 2010;31:622–31. doi: 10.1097/DBP.0b013e3181eedf01. [DOI] [PubMed] [Google Scholar]
- 44.Watson MS, Mann MY, Lloyd-Puryear MA, Rinaldo P, Howell RR. Newborn screening: toward a uniform panel and system: executive summary. Genet Med. 2006;8(Suppl 1):1S–252S. doi: 10.1097/01.gim.0000223891.82390.ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Green NS, Rinaldo P, Brower A, et al. Committee report: advancing the current recommended panel of conditions for newborn screening. Genet Med. 2007;9:792–6. doi: 10.1097/gim.0b013e318159a38e. [DOI] [PubMed] [Google Scholar]
- 46.Lipstein EA, Vorona S, Browning MF, et al. Systematic evidence review of new-born screening and treatment of severe combined immunodeficiency. Pediatrics. 2010;125(5):e1226–e1135. doi: 10.1542/peds.2009-1567. [DOI] [PubMed] [Google Scholar]
- 47.Recommended uniform screening panel of the Secretary’s advisory committee on heritable disorders in newborns and children. Department of Health and Human Services, Health Resources and Services Administration, Maternal and Child Health Bureau; Washington, DC: 2011. ( http://www.hrsa.gov/advisorycommittees/mchbadvisory/heritabledisorders/recommendedpanel/index.html) [Google Scholar]
- 48.Bombard Y, Miller FA, Hayeems RZ, Avard D, Knoppers BM. Reconsidering reproductive benefit through newborn screening: a systematic review of guidelines on preconception, prenatal and new-born screening. Eur J Hum Genet. 2010;18:751–60. doi: 10.1038/ejhg.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manning M, Hudgins L. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010;12:742–5. doi: 10.1097/GIM.0b013e3181f8baad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hudson KL. Genomics, health care, and society. N Engl J Med. 2011;365:1033–41. doi: 10.1056/NEJMra1010517. [DOI] [PubMed] [Google Scholar]
- 51.Anderson SA, Doperak J, Chimes GP. Recommendations for routine sickle cell trait screening for NCAA Division I athletes. PMR. 2011;3:168–74. doi: 10.1016/j.pmrj.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 52.Jostins L, Barrett JC. Genetic risk prediction in complex disease. Hum Mol Genet. 2011;20:R182–R188. doi: 10.1093/hmg/ddr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borry P, Henneman L, Lakeman P, ten Kate LP, Cornel MC, Howard HC. Preconceptional genetic carrier testing and the commercial offer directly-to-consumers. Hum Reprod. 2011;26:972–7. doi: 10.1093/humrep/der042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vashlishan Murray AB, Carson MJ, Morris CA, Beckwith J. Illusions of scientific legitimacy: misrepresented science in the direct-to-consumer genetic-testing marketplace. Trends Genet. 2010;26:459–61. doi: 10.1016/j.tig.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 55.Foster MW, Mulvihill JJ, Sharp RR. Evaluating the utility of personal genomic information. Genet Med. 2009;11:570–4. doi: 10.1097/GIM.0b013e3181a2743e. [DOI] [PubMed] [Google Scholar]
- 56.Summary from the Molecular and Clinical Genetics Panel meeting — March 8–9, 2011. Department of Health & Human Services; Washington, DC: ( http://www.fda.gov/downloads/AdvisoryCommittees/CommitteesMeetingMaterials/MedicalDevices/MedicalDevicesAdvisoryCommittee/MolecularandClinicalGneticsPanel/UCM246907.pdf) [Google Scholar]
- 57.American College of Medical Genetics Board of Directors ACMG statement on direct-to-consumer genetic testing. Genet Med. 2004;6:60. doi: 10.109701.GIM.0000106164.59722.CE. [DOI] [PubMed] [Google Scholar]
- 58.Committee on Genetics, American College of Obstetricians and Gynecologists, Committee on Ethics, American College of Obstetricians and Gynecologists ACOG committee opinion no. 409: direct-to-consumer marketing of genetic testing. Obstet Gynecol. 2008;111:1493–4. doi: 10.1097/AOG.0b013e31817d250e. [DOI] [PubMed] [Google Scholar]
- 59.Family history, prenatal care, and women’s health: development of a family history and genetic screening tool and educational materials for health professionals and the public. National Coalition for Health Professional Education in Genetics; Lutherville, MD: 2010. ( http://www.nchpeg.org/index.php?option=com_docman&task=doc_download&grid=59&Itemid=135) [Google Scholar]
- 60.Feero WG, Green ED. Genomics education for health care professionals in the 21st century. JAMA. 2011;306:989–90. doi: 10.1001/jama.2011.1245. [DOI] [PubMed] [Google Scholar]
- 61.Broussard DL, Sappenfield WB, Fussman C, Kroelinger CD, Grigorescu V. Core state preconception health indicators: a voluntary, multi-state selection process. Matern Child Health J. 2011;15:158–68. doi: 10.1007/s10995-010-0575-x. [DOI] [PubMed] [Google Scholar]
- 62.Ashley EA, Butte AJ, Wheeler MT, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–35. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.