Abstract
Background
This study investigates the prediction of mild cognitive impairment-to-Alzheimer's disease (MCI-to-AD) conversion based on extensive multimodal data with varying degrees of missing values.
Methods
Based on Alzheimer's Disease Neuroimaging Initiative data from MCI-patients including all available modalities, we predicted the conversion to AD within 3 years. Different ways of replacing missing data in combination with different classification algorithms are compared. The performance was evaluated on features prioritized by experts and automatically selected features.
Results
The conversion to AD could be predicted with a maximal accuracy of 73% using support vector machines and features chosen by experts. Among data modalities, neuropsychological, magnetic resonance imaging, and positron emission tomography data were most informative. The best single feature was the functional activities questionnaire.
Conclusion
Extensive multimodal and incomplete data can be adequately handled by a combination of missing data substitution, feature selection, and classification.
Keywords: Mild cognitive impairment, Alzheimer's dementia, Prognosis, Multimodal biomarker, Missing data, Feature selection
1. Background
Alzheimer's disease (AD) is the most common cause for dementia in the elderly and primarily diagnosed based on clinical symptoms such as memory loss and disorientation [1]. As an intermediate stage between normal age-related cognitive decline and dementia, mild cognitive impairment (MCI) has been identified [2]. Because not all MCI patients convert to AD and the MCI group is very heterogeneous, it is a highly relevant task to differentiate MCI subjects who will develop AD within the next years from those who will be stable or even improve.
Recent studies tried to solve this task by using a combination of biomarkers, e.g. obtained via positron emission tomography (PET) or magnetic resonance imaging (MRI), and algorithms adopted from machine learning [3], [4], [5]. Computer-based decision support systems are assumed to be not only more sensitive for the detection of early disease states, but also more objective and reliable than medical decisions made by single clinicians [6]. Those automatic diagnostic tools become especially important when data of different modalities are integrated into one diagnostic decision as recommended by the National Institute on Aging (NIA), because this requires expertise in more than one clinical field.
In this study we consider several generalizations with the aim (1) to make full use of databases such as the ADNI (Alzheimer's Disease Neuroimaging Initiative [7]) and (2) to optimize automatic multimodal classification for use in everyday clinical routine.
First, what is a good way to deal with missing data? Missing data are a severe problem in many medical databases and is usually solved by discarding all patients with missing data. However, for multimodal data it is very likely that most of the patients will lack data from one or the other domain and a requirement of complete cases results in very small data sets. Here, we compared three different approaches to replace (“impute”) missing data entries: mean imputation, imputation by the Expectation-Maximization (EM) algorithm, and a combined approach.
Second, most studies focus on a certain subset of domains for automatic multimodal classification (e.g., neuropsychology and MRI), not least because of missing data [8], [9], [10], [4]. By replacing missing values, we were able to take the multimodal approach further and include all modalities available in ADNI. In total we assessed 288 features from 10 different domains including clinical data, neuropsychology, genetics, biospecimen, MRI, and PET.
Third, if expert knowledge is not yet available or not yet complete, it is desirable to have a framework that can deal with features of different importance and even irrelevant features, namely by automatic feature selection. Here, we compared two methods for fully automatic feature selection (F-score and feedforward/backward selection) with manual feature selection by a group of experts.
Fourth, we compared three state-of-the-art classification algorithms: Support Vector Machines (SVMs), a single classification tree, and Random Forests. By not making any concrete assumptions about the scale or the distribution of the data, they are well suited for the analysis of data sets comprising many different features.
Fifth, what is a good way to deal with unbalanced data? Class frequency is often unbalanced and can lead to large discrepancies between sensitivity and specificity [8]. Here, we propose a way to balance sensitivity and specificity via the receiver operating characteristic (ROC).
2. Methods
2.1. Data
2.1.1. Subjects
Data used in this project were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was initiated in 2003 by the NIA, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration, pharmaceutical companies, and nonprofit organizations for the development of diverse biomarkers for the early detection of AD [7] (For more information on study procedures see http://adni.loni.usc.edu/methods/documents/).
For this study, patients with a baseline diagnosis of MCI and a follow-up time of at least 36 months were extracted from the ADNI database. Patients who were diagnosed with MCI, NL or MCI to NL at all visits during the 3-year follow-up were included in the MCI-stable group, whereas patients whose diagnosis changed to AD during the 3-year follow-up were regarded as MCI-converters. After this procedure, 237 patients were selected, 151 of which belonged to the MCI-stable group, and 86 to the MCI-converter group (see Table 1).
Table 1.
Baseline subject characteristics
| Characteristic | MCI-stable (n = 151) | MCI-converters (n = 86) | P-value |
|---|---|---|---|
| Age, mean (SD) | 74.12 (7.66) | 74.62 (6.90) | .61 |
| Gender | .76 | ||
| Females, n (%) | 48 (31.79) | 29 (33.72) | |
| Males, n (%) | 103 (68.21) | 57 (66.28) | |
| Education, y; mean (SD) | 15.82 (2.96) | 15.72 (3.02) | .80 |
| MMSE, score; mean (SD) | 27.59 (1.69) | 26.69 (1.72) | 1.1 × 10−4 |
Abbreviations: MCI, mild cognitive impairment; SD, standard deviation; y, years; MMSE, Mini-Mental State Examinations.
NOTE. P-values were calculated via a two-sided t-test. For baseline characteristics of other features, see Table B.4.
2.1.2. Features
Based on the ADNI database, features from 10 modalities were extracted including neuropsychological testing (NP, 15 features), medical history (MEDHIST, 21 features), medical symptoms at baseline (BLSYMP, 25 features), neurological and physical examinations (EXAMS, 28 features), MRI lesion load (LESION, 1 feature), MRI volume-based morphometry (VOLUME, 24 features), voxel-based morphometry (VOXEL, 117 features), laboratory data including cerebrospinal fluid (CSF) examinations (BIO, 47 features), PET scans (PET, 7 features), and demographic information about age, gender, and education (DEMO, 3 features). This resulted in a total of 288 features (see Table B.4). All features were obtained from the baseline visits of the patients.
Please note that we here only used the sum scores of the different neuropsychological tests because we assumed that they cover all important aspects of the test. However, because it might be also interesting to look at specific domains of cognition, we performed additional analyses on the subscores of the Alzheimer's Disease Assessment Score (ADAS) and the functional activities questionnaire (FAQ).
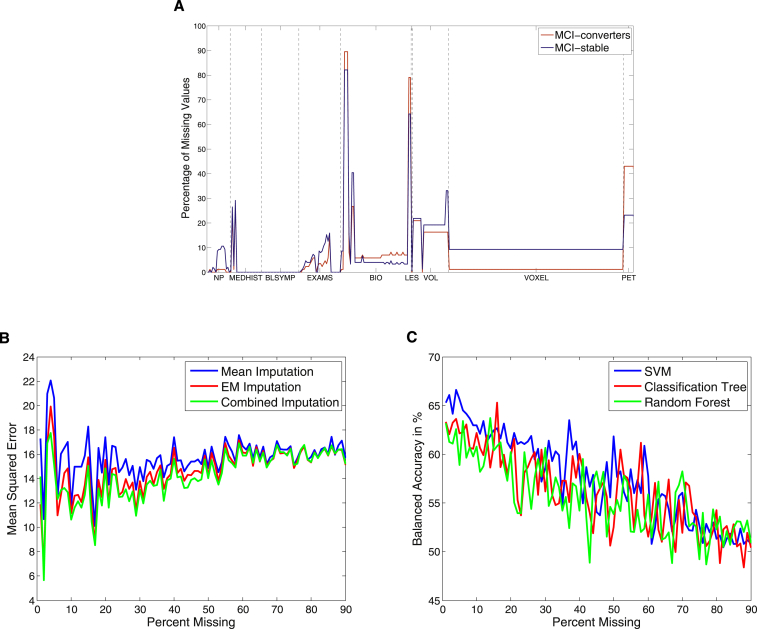
In our final feature set, 7.94% of data were missing (9.1% in the MCI-stable group and 5.9% in the MCI-converter group). The number of missing values per feature varied between 0% and 82.12% for MCI-stable patients and 0% and 89.53% for MCI-converters (see Fig. 1A for details). In about 20.14% of the features, no data were missing. In five features including CSF data, more than 50% were missing.
Fig. 1.
(A) Proportion of missing data for each feature, separately for mild cognitive impairment (MCI)-converters and MCI-stable patients. (B) Mean squared error between true and imputed values for different percentages of missing data. (C) Balanced accuracy for the different classification algorithms and different percentages of missing data.
2.2. Missing data handling
We compared three ways of missing data replacement: mean imputation, imputation by the EM algorithm, and a combined imputation.
In mean imputation, the mean value over all nonmissing data is calculated for each of the features individually. This value is then substituted for all missing values of the respective feature. To account for categorical and discrete variables we first determined the scale for each of the features and then substituted with the mode, median, or mean value accordingly. The main advantage of mean imputation is its low computational cost. However, the estimates are often biased [11].
A less biased treatment of missing data can be achieved by using the Expectation-Maximization (EM) algorithm [12]. This is an iterative procedure which switches between an expectation (E) and a maximization (M) step until the most likely values for the missing data are found. We used here a regularized version of the EM algorithm described in [13] (The code can be found at http://www.clidyn.ethz.ch/imputation/).
Because a considerable fluctuation of EM estimates has been found for categorical features [14], a third imputation technique was implemented which is a combination of mean and EM imputation: for all categorical features mean imputation was performed and the EM imputation was then only carried out on the remaining noncategorical features.
The performance of these three imputation techniques was evaluated on a complete version of the given data set, where only features without missing values (58 instead of 288 features) were contained. Subsequently, we randomly deleted 0% to 90% of the data and calculated the mean squared error between the imputed and true values. Because the combined method produced lower errors for almost all cases than mean imputation and EM algorithm alone (see Fig. 1B), we decided to use the combined method for all further analyses. Importantly, imputation values were calculated based only on training data and were then used to replace missing values in both training and test data.
Additionally, we investigated which classification algorithm comes closest to recovering the prediction accuracy of the complete data (as above 58 out of 288 features). We therefore performed the different classification analyses (see section 2.4) for the complete data set and a series of incomplete data sets, in which we randomly deleted 0% to 90% of the data (see Fig. 1C). Missing data were replaced via combined imputation.
2.3. Feature selection
To assess the importance of certain features or feature combinations, we looked at the following sets: all features together, single features and each feature domain (NP, MEDHIST, etc.). Additionally, we compared automatic with manual feature selection.
2.3.1. Automatic feature selection
For automatic feature selection, we implemented two different approaches: F-score and Forward/Backward Feature Selection. In both approaches, feature selection was performed on independent feature selection sets determined by a threefold nested cross-validation.
The F-score measures the ability of a feature to discriminate between two classes and is calculated as the between-class variance divided by the within-class variance [15]. It is fast and easy to calculate but ignores dependencies between features. Here, features were ranked by the F-score in the training set and the 10 best ranked features were used for classification.
In forward and backward feature selection, the accuracy of a classifier is used as a criterion for feature selection [16]. Here, we used an SVM with default parameter (C = 1) and 10-fold cross-validation.
In a forward feature selection the idea is to start with the single best feature, then add other features incrementally and keep only those which increases the classification performance. Backward feature selection starts with all features and then features are incrementally removed from the feature set. To achieve a more robust selection procedure [16], feature selection was repeated 20 times based on the feature selection set and only those features were included which were selected in a certain fraction of the cases (1 for backward selection and 0.2 for forward selection). The average number of features for forward selection was 9.49 and for backward selection 281.84.
The 10 most commonly chosen features for the F-score and forward selection are shown in Table 2. Please note that the feature ranking is averaged over the different analyses and therefore does not reflect a certain combination of features in a particular analysis. However, all three feature selection methods can lead to a feature set containing highly correlated variables (e.g. the ADAS-11 and ADAS-13). Whereas the F-score assesses each feature independently and thus do not take interactions between the features into account, forward and backward selection add or remove features incrementally and thus combinations of features are assessed. Counterintuitively, by reducing the noise the combination of highly correlated and even redundant variables can lead to a better class separation than when using the features alone (e.g., see [16]).
Table 2.
Feature ranking for F-score and forward feature selection
| Rank | F-score | Forward feature selection |
|---|---|---|
| 1 | FAQ (NP) | FAQ (NP) |
| 2 | ADAS 13 (NP) | ADAS 13 (NP) |
| 3 | ADAS 11 (NP) | RIGHTHIPPO (VOLUME) |
| 4 | AVEASSOC (PET) | X2SDSIGPXL (PET) |
| 5 | BCVOMIT (BLSYMP) | ADAS 11 (NP) |
| 6 | TAU (BIO) | SUMZ3 (PET) |
| 7 | X2SDSIGPXL (PET) | SUMZ2 (PET) |
| 8 | MIDTEMP (VOLUME) | LEFTHIPPO (VOLUME) |
| 9 | AVEREF (PET) | DIGITSCOR (NP) |
| 10 | NXHEEL (EXAMS) | AVEASSOC (PET) |
Abbreviations: FAQ, Functional Activities Questionnaire; ADAS, Alzheimer's Disease Assessment Score; AVEASSOC, average regional association cortex value; X2SDSIGPXL/X3SDSIGPXL, number of pixels with Z-scores ≥ 2/3 standard deviations; SUMZ2/SUMZ3, sum of pixel Z-scores ≥ 2/3 standard deviations; BCVOMIT, vomiting; NXHEEL, cerebellar—heel to shin; MIDTEMP, volume of middle temporal lobe; LEFTHIPPO/RIGHTHIPPO, volume of left and right hippocampus; AVEREF, average regional value of the reference region used for normalization; DIGITSCOR, Digit Symbol Substitution Test.
2.3.2. Manual feature selection
In manual feature selection, experts of the different fields (namely, R. Buchert A. Maeurer, A. Roberts, L. Spies, and P. Suppa) have chosen altogether 36 features. The feature selection procedure was as follows: For each domain an expert was appointed who was asked to select, from a list of possible features taken from the ADNI database, those features that according to their knowledge are most important in characterizing Alzheimer's disease. The only specification we made was that the number of features per domain should not exceed 10 features. In particular, the expert features included seven features from NP (Alzheimer's Disease Assessment Score 11 and 13 [ADAS 11 and 13], Clinical Dementia Rating Scale [CDR], FAQ, Geriatric Depression Scale [GDS], Neuropsychiatric Inventory [NPI], and Mini-Mental State Examination [MMSE]), seven features from MEDHIST (Family History Questionnaire, Medical History of neurological, psychiatric or cardiovascular disease), five features from BIO (ApoE4-alleles 1 and 2; Abeta, Tau, and Ptau), one feature from LESION (white matter lesion load), six features from VOLUME (volumes of left and right middle/inferior temporal lobe, left, and right hippocampus), seven features from PET (averaged uptake values in 18F-FDG images in frontal and association cortex and sum of pixel-wise Z-scores), and three features of DEMO (age, gender, and education).
These expert features were further divided into standard and advanced features. Standard features comprised all features from DEMO and MEDHIST, the neurospychological screening procedures MMSE, CDR, GDS, and NPI and gene data from BIO (in total 16 features). All other expert features belonged to advanced features (in total 20 features). Importantly, experts assessed the importance of the single features based on their general knowledge and did not use the ADNI data set at hand.
2.4. Classification
For the classification of features, we used three different supervised learning algorithms: SVMs [17], [18], a single classification tree [19], and Random Forests [20]. All three algorithms are used here to learn a model between baseline features and group membership (either MCI-converter or MCI-stable) based on training data, which is then evaluated on independent test data.
SVMs are a very popular classification method [17], [18] and have been used for disease classification, including Alzheimer's disease, before [21], [22], [23]. SVMs find a decision boundary which maximizes the margin between two groups. We used here the library for support vector machines (LIBSVM) LIBSVM toolbox for MATLAB [24] with a Radial Basis Function (RBF) kernel and default parameters (C = 1) based on standardized features (Software can be found at http://www.csie.ntu.edu.tw/∼cjlin/libsvm/). Nonlinear kernels such as the RBF kernel are often associated with an improvement in accuracy and have the advantage to account for complex interactions in the data. To assess the importance of nonlinear interactions, we additionally show results on a subset for a linear SVM.
Classification trees are a tree-based technique for partitioning complex decisions into a number of simpler decision rules [19]. The motivation using decision trees is the interpretability of decisions. However, a single classification tree strongly depends on the training data and even small variations in the input data can lead to a completely different tree structure [25]. Here, we used MATLAB's fit function from the ClassificationTree class.
Random Forest is an ensemble method, where a number of classification trees is grown and the output is determined by a majority vote among all trees [20], [26]. Because the classification is not based on only one tree, Random Forests are thought to produce more robust classification results. However, this is achieved at the cost of the interpretability of the decision process. Here, we used the software of L. Breiman and A. Cutler with 100 trees and m set to (http://www.stat.berkeley.edu/∼breiman/RandomForests/).
To estimate the generalization error for new data sets, we performed a nested cross-validation for all three classification methods and the different feature selection methods. The data are first split into three parts. Each of the three parts is once the feature selection set, the remaining two parts are the validation set. Based on the validation set, a 10-fold cross-validation was performed. To get more stable results this procedure was repeated 30 times and mean values were calculated. As measures of classification performance, we show sensitivity, specificity, and balanced accuracy (mean of sensitivity and specificity).
Because of the imbalanced class sizes, we observed that the classifiers performed with high specificity but low sensitivity (see Table A.3). Discrepancies in sensitivity and specificity have also been reported previously [27], [8], [28]. Therefore, we adjusted the classification procedure during the training process via the ROC such that the output of the classifier is optimized for balanced accuracy [29].
P-values were calculated via nonparametric permutation tests [30] as it has recently been shown that P-values based on parametric tests such as the binomial test are biased in combination with cross-validation [31]. The labels were randomly permuted 1000 times.
3. Results
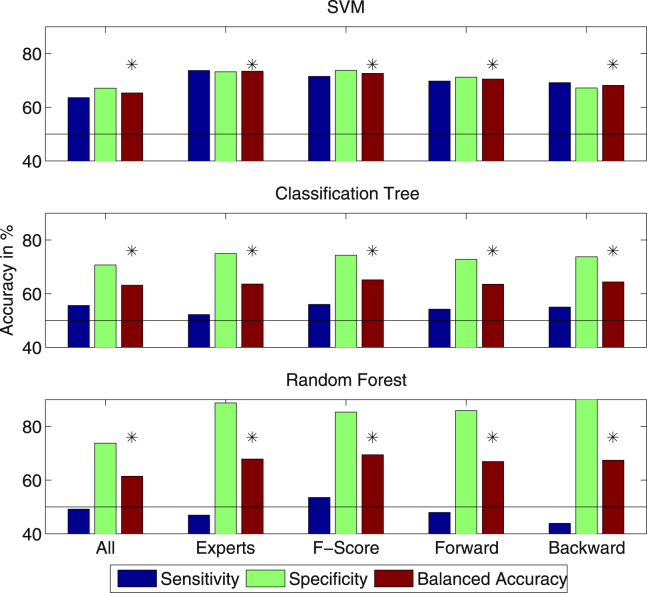
The conversion to AD was predicted with classification accuracies varying between 61.48% and 73.44% (for all: Pperm < .001) based on all features and feature subsets determined by either experts, F-score or forward/backward feature selection (see Fig. 2).
Fig. 2.
Accuracies for all features and different feature selection sets using Support Vector Machines (SVMs), a single classification tree and Random Forests (∗P < .001).
3.1. ROC-optimization
For Random Forests, we reported results without ROC optimization, because the optimization here lead to a more severe imbalance between sensitivity and specificity (see Table A.3). For single classification trees, it does not make a large difference whether the threshold is adjusted via ROC or not (see Table A.3). Only for SVMs, the adjustment of the threshold leads to a balance of sensitivity and specificity (see Table A.3 for results without ROC adjustment).
3.2. Effect of feature selection
Classification performance was improved by all feature selection methods. For SVMs, expert features with a balanced accuracy of 73.44% were significantly better than all other features sets (P < .05, evaluated via a two-sample t-test on the balanced accuracies from the 30 repetitions of cross-validation), with the exception of the F-score where the difference was not significant (P = .07). For a single classification tree and Random Forests, the balanced accuracy was highest for the feature set selected via the F-score (65.15% and 69.45%) and significantly better than all other feature sets (P < .05) with exception of backward selection for a single classification tree (P = .31). Within automatic feature selection, features selected based on the F-score gave better results than forward and backward selection for all three algorithms. Whereas the difference to forward selection was significant for all three methods, the difference to backward selection was only significant for SVMs and Random Forests (P < .05).
3.3. Classification based on single features and modalities
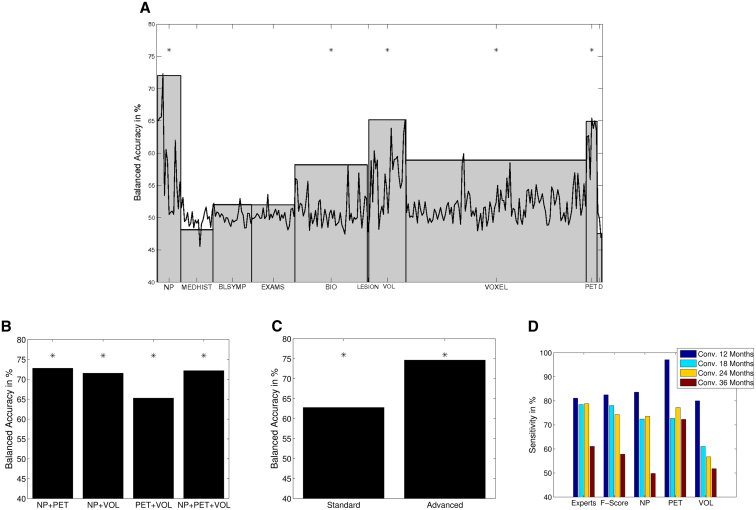
SVM classification results for single features and all features contained within one modality are shown in Fig. 3A and Table B.4. The best performing single feature was the FAQ (72.27%, Pperm < .001). Within MRI volume features, right hippocampus allowed for the best prediction of the conversion to AD (balanced accuracy 65.13%, Pperm < .001). Within PET features, X2SDSIGPXL was most discriminative (balanced accuracy 65.42%, Pperm < .001).
Fig. 3.
(A) Support Vector Machine (SVM) classification results for single features and each data modality (∗P < .001, only for modalities). (B) SVM classification results for different combinations of the data modalities neuropsychological testing (NP), positron emission tomography (PET), and VOLUME (∗P < .001). (C) SVM classification results for standard and advanced features (∗P < .001, standard: medical data, genes, and neuropsychological screening tests; advanced: extended neuropsychological testing, magnetic resonance imaging [MRI], cerebrospinal fluid [CSF], and PET). (D) Sensitivity for patients converting after different time frames.
Best performing modalities were neuropsychological testing (NP, balanced accuracy 72.01%, Pperm < .001), MRI volumes (VOLUMES, balanced accuracy 65.17%, Pperm < .001), and PET (balanced accuracy 64.92%, Pperm < .001). Biospecimen features (BIO, balanced accuracy 58.19%, Pperm < .005) and voxel-based measures (VOXEL, balanced accuracy 58.90%, Pperm < .005) also gave significantly above chance accuracy. In Fig. 3B, we show the balanced accuracy for all combinations of the best performing modalities NP, VOLUMES, and PET. The balanced accuracy for advanced features was significantly higher than for standard features (74.68%–62.80%, P < .01, see Fig. 3C).
3.4. Sensitivity with respect to conversion times
The sensitivity for patients converting after different time frames (i.e., 12–36 months) is shown in Fig. 3D. As expected, the onset of AD could be best predicted for patients converting after 12 months and worst for patients converting after 36 months.
3.5. Linear vs. nonlinear SVM
For expert features, classification accuracy was significantly lower for a linear SVM than for a nonlinear SVM (71.78%–73.44%, P < .05). For the F-score, the difference was not significant (P = .54).
3.6. Comparison of mean and combined imputation
Results of SVM analyses for expert features and features obtained via the F-score were not significantly different for mean imputation and the combined method (P = .36 and P = .70). In Fig. 1C, we show the balanced accuracy for SVM, classification tree, and Random Forest separately for different amounts of missing data. As expected the classification accuracy decreases for all three algorithms with higher percentages of missing data, and the variance is highest for missing values between 30% and 60%.
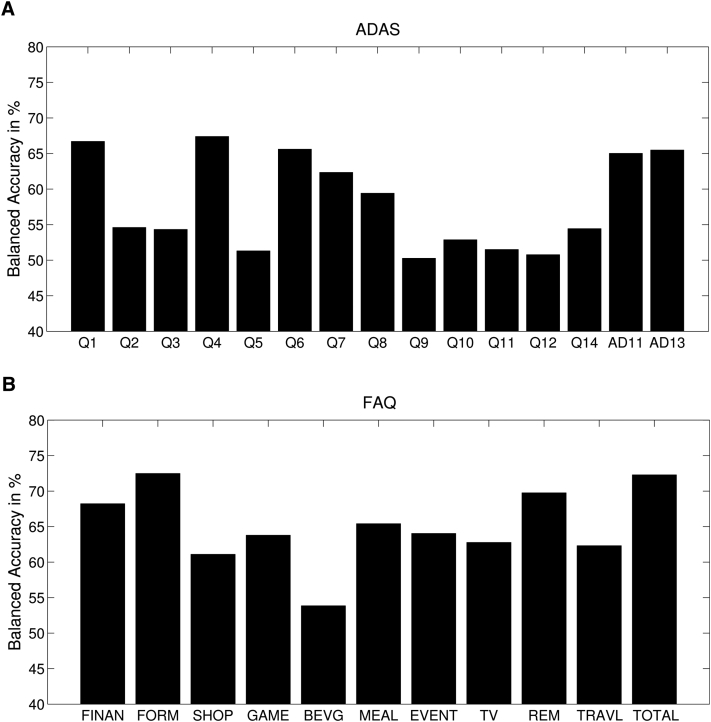
3.7. Prediction based on neuropsychological subscores
To determine the impact of cognitive subdomains, we performed additional SVM analyses on the subscores of ADAS and FAQ (see Fig. 4). For the ADAS, most successful subdomains were word recall (66.69%), naming (67.39%), and orientation (65.60%). For the FAQ, we found the subdomains of financing (68.22%), assembling of documents (72.48%), and remembering of appointments (69.76%) as most predictive.
Fig. 4.
(A) Support Vector Machine (SVM) accuracies for the cognitive subdomains of Alzheimer's Disease Assessment Score (ADAS, Q1 to Q12: Word recall, constructional praxis, delayed word recall, naming, ideational praxis, orientation, word recognition, remembering test instructions, comprehension, word finding difficulty, spoken language ability, number cancellation; AD11, total score on the 11-item ADAS; AD13, total score on the modified 13-item ADAS). (B) SVM accuracies for the individual functional activities of Functional Activities Questionnaire (FAQ) (FINAN, writing checks etc.; FORM, assembling tax records etc.; SHOP, shopping alone; GAME, playing a game of skill etc.; BEVG, making a cup of coffee etc.; MEAL, preparing a meal; EVENT, keeping track of current events; TV, understanding TV etc.; REM, remembering appointments etc.; TRAVL, traveling out of neighborhood).
4. Discussion
In this study we have shown that the conversion to AD within 3 years can be predicted with a comparably high accuracy based on a heterogeneous set of features, even when certain parts of data are missing.
4.1. Comparison of classification algorithms
Among the algorithms evaluated in the present study, nonlinear SVMs produced best classification results. The superiority of SVMs against other machine learning algorithms in terms of accuracy has been reported in many studies [32], [33]. SVMs generally seem to be quite tolerant toward irrelevant features, most likely because they successfully exploit dependencies among features [33]. Most classification algorithms have problems when dealing with large and noisy data sets comprising also collinearities in the data [34]. In a recent review, however, it has been shown that our proposed classification algorithms are quite robust with respect to collinearity [35]. Whereas SVMs alleviate the multicollinearity problem via regularization, in Random Forests it is alleviated via choosing a random subset of features for each tree. Dormann et al. [35] come to the conclusion that in terms of accuracy it does not make a big difference whether one “ignores” the collinearity in the data or apply diagnostic tools such as the variance inflation factor. Nevertheless, those tools might be helpful in interpreting the data.
4.2. Feature selection
Manual and automatic feature selection significantly improved the performance of all three classification algorithms. If expert features are available, the choice of these might be the preferable way. But if not, we demonstrated that automatic feature selection methods, in particular feature selection based on the F-score or forward selection, can achieve similar classification performance. Be eliminating on average only six to seven features, the backward feature selection provided worst results. Generally, it is said that forward selection selects much less features than backward feature selection and this way leads often to higher classification accuracies [16]. Because the effect of deleting single features especially from a large and noisy data set is expected to be relatively low, the classification accuracy in backward selection saturates at a quite early level. However, there are cases in which backward selection gives superior results, especially in cases where the interaction between several features should be taken into account (see [16]). Over forward and backward selection, the F-score has the additional advantage that it is easy to compute and does not depend itself on a classification procedure.
4.3. Missing data
In most studies, the problem of missing data is solved by restricting automatic classification to patients with complete data [26], [3]. However, this becomes a problem especially in the case of multimodal data, because the probability that test results are missing increases with the number of tests.
In this study, we substituted missing values to produce a complete data set. This has the advantage that every machine learning algorithm can be used for classification. In accordance with other studies the EM algorithm and the combined method produced a lower error between imputed and actual values [36]. However, in this study we found that in terms of accuracy, it does not make a large difference whether the combined method or mean imputation is used.
For all three classification algorithms, a similar pattern for different percentages of missing data has been observed. However, SVMs started with the highest accuracy and also provided the highest mean value over all analyses.
4.4. ROC optimization
Our proposed ROC optimization yielded the 50:50 balance of sensitivity and specificity only for SVMs. It is evident that the desirable balance of sensitivity and specificity depends on the task and might change when new treatment options become available. However, ROC adjustment can also be used to optimize for specific values of sensitivity or specificity depending on predefined costs.
4.5. Best single features
The best performing single feature was the FAQ, which is interesting since this measure, in contrast to the MMSE, is not used in everyday clinical routine. Its relevance for the prediction of conversion to AD has been previously shown [37]. Within the FAQ, the ability of assembling documents such as tax records had the strongest influence on the later diagnosis. Future studies might evaluate whether the FAQ or similar scores assessing cognitive and social functioning should play a stronger role in diagnostic guidelines.
4.6. Best modalities
In accordance with other studies mostly focusing on the domains neuropsychology, MRI, and PET [9], [38], [4] our analyses also identified these modalities as containing most useful predictive information. This shows that the proposed approach is capable of identifying disease-relevant modalities even in the presence of missing and potentially uninformative data.
4.7. Multimodal classification
There are a number of studies that also explored the use of multimodal data for either diagnosing AD/MCI (vs. healthy controls) or predicting the conversion from MCI to AD [28], [8], [9], [39], [4], [38]. All these studies come to the conclusion that multimodal prediction is superior to unimodal prediction (accuracy was typically increased by 2%–7%). In our case, the best multimodal accuracy only slightly exceeded the best unimodal accuracy. This may be explained by the simplicity of our approach: Features were just concatenated into one vector. By this, we did not make any use of the specific covariance structure between the modalities. However, all of the mentioned studies were based only on a certain combination of complete neuropsychological, MRI, CSF, gene, and PET data and not as in our study on an incomplete set comprising features from all modalities available in ADNI.
4.8. Limitations
First, the algorithms we used produce only dichotomous class labels and can therefore not directly be used for expressing uncertainty. Future studies might exploit probabilistic models for generating probabilistic outputs [40].
Second, we did training and testing both on baseline MCI-patients after the guideline that training and testing data should be of the same kind. However, other studies have shown that a classifier can benefit from training in AD and healthy controls [4], [8].
Third, one may criticize that the use of neuropsychological tests introduce some kind of circularity in the analysis. However, three of the most successfully discriminating scores were not used to make the diagnosis in ADNI, namely the FAQ, ADAS 11, and ADAS 13 (http://adni.loni.usc.edu/wp-content/uploads/2010/09/ADNI_GeneralProceduresManual.pdf, p. 20-21).
Fourth, another point for criticism might be the fact that the expert features are based on individual expert opinions only and by this do not reflect expert knowledge that is universally valid. Future studies might involve more sophisticated strategies such as a voting scheme over several experts or a Delphi review.
Fifth, our proposed models do not account for censoring in the data. When such models are intended to be brought into clinical practice, it is necessary to find ways to deal with missing data due to censoring. One possibility might be to combine SVM and Cox regression as suggested by [41].
4.9. Conclusion
Based on a large and heterogeneous set of incomplete ADNI data, the conversion from MCI to AD could be predicted with comparably high accuracy and balanced sensitivity and specificity. We recommend the substitution of missing values via a combination of mean imputation and EM algorithm and for classification SVMs because they are very flexible toward different data characteristics. The dimensionality of the data should be reduced to the most relevant features either by hand based on expert knowledge or by an automatic feature selection method. Future studies should explore the use of probabilistic models for disease prediction based on incomplete data and more sophisticated ways of combining the data of different modalities.
Research in context.
-
1.
Systematic review: A number of studies have used multimodal data and machine learning approaches for the prediction of the conversion to Alzheimer. However, they mostly focused on complete data sets and a small subset of data modalities.
-
2.
Interpretation: We optimized automatic multimodal classification for clinical routine regarding missing data, extensive multimodal data, different kinds of feature selection and classification algorithms and demonstrated comparable classification accuracies.
-
3.
Future directions: Future studies might explore the use of probabilistic models for disease prediction and might also include differential diagnoses of AD.
Acknowledgments
The authors would like to thank Catharina Lange, Anja Maeurer, Anna Roberts, Lothar Spies, and Per Suppa for their advice regarding the expert features, Fabian Wenzel and Daniel Gieschen for their advice regarding missing data and Patrik Bey, Idai Gertel, and Igor Merkulow for their general support. Finally, we would like to thank the reviewers for their valuable comments and their helpful suggestions.
Footnotes
This work was supported by the Bernstein Computational Program of the German Federal Ministry of Education and Research (01GQ1001C, 01GQ0851, GRK 1589/1), the European Regional Development Fund of the European Union (10153458 and 10153460), and Philips Research.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.dadm.2015.01.006.
Supplementary data
References
- 1.Dal-Bianco P. Alzheimer-Differentialdiagnostik. Facharzt. 2010;3:4–9. [Google Scholar]
- 2.Petersen R. Mild cognitive impairment clinical trials. Nat Rev Drug Discov. 2003;2:646–653. doi: 10.1038/nrd1155. [DOI] [PubMed] [Google Scholar]
- 3.Hinrichs C., Singh V., Xu G., Johnson S.C., ADNI Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. Neuroimage. 2011;55:574–589. doi: 10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young J., Modat M., Cardoso M.J., Mendelson A., Cash D., Ourselin S. Accurate multimodal probabilistic prediction of conversion to Alzheimer’s disease in patients with mild cognitive impairment. Neuroimage Clin. 2013;2:735–745. doi: 10.1016/j.nicl.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y., Mattila J., Ruiz M.A.M., Paajanen T., Koikkalainen J., van Gils M. Predicting AD conversion: comparison between prodromal AD guidelines and computer assisted predictAD tool. PLoS One. 2013;8:e55246. doi: 10.1371/journal.pone.0055246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kloeppel S., Stonnington C.M., Barnes J., Chen F., Chu C., Good C.D. Accuracy of dementia diagnosis—a direct comparison between radiologists and a computerized method. Brain. 2008;131:2969–2974. doi: 10.1093/brain/awn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiner M.W., Veitch D.P., Aisen P.S., Beckett L.A., Cairns N.J., Green R.C. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2013;9:e111–e194. doi: 10.1016/j.jalz.2013.05.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui Y., Liu B., Luo S., Zhen X., Fan M., Liu1 T. Identification of conversion from mild cognitive impairment to Alzheimer’s disease using multivariate predictors. PLoS One. 2011;6:e21896. doi: 10.1371/journal.pone.0021896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng B., Zhang D., Shen D. Domain transfer learning for MCI conversion prediction. Med Image Comput Comp Assist Interv. 2012;15:82–90. doi: 10.1007/978-3-642-33415-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westman E., Muehlboeck J.S., Simmons A. Combining MRI and CSF measures for classification of Alzheimer’s disease and prediction of mild cognitive impairment conversion. Neuroimage. 2012;62:229–238. doi: 10.1016/j.neuroimage.2012.04.056. [DOI] [PubMed] [Google Scholar]
- 11.Donders A.R.T., van der Heijden G.J., Stijnen T., Moons K.G. Review: a gentle introduction to imputation of missing values. J Clin Epidemiol. 2006;59:1087–1091. doi: 10.1016/j.jclinepi.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Dempster A.P., Laird N.M., Rubin D.B. Maximum likelihood from incomplete data via the em algorithm. J R Stat Soc Series B. 1977:1–38. [Google Scholar]
- 13.Schneider T. Analysis of incomplete climate data: estimation of mean values and covariance matrices and imputation of missing values. J Climate. 2001;14:853–871. [Google Scholar]
- 14.Kim S.H., Kim S.H. Fluctuation of estimates in an EM procedure for categorical data. J Stat Comput Simul. 2005;75:941–957. [Google Scholar]
- 15.Chen Y.W., Lin C.J. Combining SVMs with various feature selection strategies. In: Guyon I., Nikravesh M., Gunn S., Zadeh L., editors. Feature extraction; vol. 207 of studies in fuzziness and soft computing. Springer; Berlin Heidelberg: 2006. pp. 315–324. [Google Scholar]
- 16.Guyon I., Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157–1182. [Google Scholar]
- 17.Cortes C., Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–297. [Google Scholar]
- 18.Shawe-Taylor J., Christianini N. Cambridge University Press; 2000. Support vector machine and other Kernel-based learning methods. [Google Scholar]
- 19.Breiman L., Friedman J., Olshen R., Stone C. Wadsworth and Brooks; 1984. Classification and regression trees. [Google Scholar]
- 20.Breiman L. Random Forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 21.Kloeppel S., Stonnington C.M., Chu C., Draganski B., Scahill R.I., Rohrer J.D. Automatic classification of MR scans in Alzheimer’s disease. Brain. 2008;131:681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weygandt M., Hackmack K., Pfueller C., Bellmann-Strobl J., Paul F., Zipp F. MRI pattern recognition in multiple sclerosis normal—appearing brain areas. PLoS One. 2011;6:e21138. doi: 10.1371/journal.pone.0021138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hackmack K., Paul F., Weygandt M., Allefeld C., Haynes J.D. Multi-scale classification of disease using structural MRI and wavelet transform. Neuroimage. 2012;62:48–58. doi: 10.1016/j.neuroimage.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 24.Chang C.C., Chih-Jen L. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol. 2011;2:1–27. [Google Scholar]
- 25.Rokach L., Maimon O. World Scientific Publishing Company; Singapore: 2008. Data mining with decision trees: theory and applications. [Google Scholar]
- 26.Gray K.R., Aljabar P., Heckemann R.A., Hammers A., Rueckert D., ADNI Random forest-based similarity measures for multi-modal classification of Alzheimer’s disease. Neuroimage. 2013;65:167–175. doi: 10.1016/j.neuroimage.2012.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuingnet R., Gerardin E., Tessieras J., Auzias G., Lehericy S., Habert M.O. Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using the ADNI database. Neuroimage. 2011;56:766–781. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Zhang D., Wang Y., Zhou L., Yuan H., Shen D., ADNI Multimodal classification of Alzheimer’s disease and mild cognitive impairment. Neuroimage. 2011;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27:861–874. [Google Scholar]
- 30.Nichols T.E., Holmes A.P. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noirhomme Q., Lesenfants D., Gomez F., Soddu A., Schrouff J., Garraux G. Biased binomial assessment of cross-validated estimation of classification accuracies illustrated in diagnosis predictions. Neuroimage Clin. 2014;4:687–694. doi: 10.1016/j.nicl.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Statnikov A., Wang L., Aliferis C. A comprehensive comparison of random forests and support vector machines for microarray-based cancer classification. BMC Bioinformatics. 2008;9:319. doi: 10.1186/1471-2105-9-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotsiantis S. Supervised machine learning: a review of classification techniques. Informatica. 2007;31:249–268. [Google Scholar]
- 34.Morlini I. On multicollinearity and concurvity in some nonlinear multivariate models. Stat Methods Appt. 2006;15:3–26. [Google Scholar]
- 35.Dormann C.F., Elith J., Bacher S., Buchmann C., Carl G., Carre G. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography. 2013;36:27–46. [Google Scholar]
- 36.Musil C.M., Warner C.B., Yobas P.K., Jones S.L. A comparison of imputation techniques for handling missing data. West J Nurs Res. 2002;24:815–829. doi: 10.1177/019394502762477004. [DOI] [PubMed] [Google Scholar]
- 37.Devanand D.P., Liu X., Tabert M.H., Pradhaban G., Cuasay K., Bell K. Combining early markers strongly predicts conversion from mild cognitive impairment to Alzheimer’s disease. Biol Psychiatry. 2008;64:871–879. doi: 10.1016/j.biopsych.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., Liu Y., Gong P., Zhang C., Ye J., ADNI Hierarchical interactions model for predicting mild cognitive impairment (MCI) to Alzheimer’s disease (AD) conversion. PLoS One. 2014;9:e82450. doi: 10.1371/journal.pone.0082450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaffer J.L., Petrella J.R., Sheldon F.C., Choudhury K.R., Calhoun V.D., Coleman R.E. Predicting cognitive decline in subjects at risk for Alzheimer disease by using combined cerebrospinal fluid, MR imaging, and PET biomarkers. Radiology. 2013;266:583–591. doi: 10.1148/radiol.12120010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Filippone M., Marquandt A., Blain C., Williams S., Mourao-Miranda J., Girolami M. Probabilistic prediction of neurological disorders with a statistical assessment of neuroimaing data modalities. Ann Appl Stat. 2013;6:1883–1905. doi: 10.1214/12-aoas562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Da X., Toledo J.B., Zee J., Wolk D.A., Xie S.X., Ou Y. Integration and relative value of biomarkers for prediction of MCI to AD progression: spatial patterns of brain atrophy, cognitive scores, APOE genotype and CSF biomarkers. Neuroimage Clin. 2014;4:164–173. doi: 10.1016/j.nicl.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.