Abstract
Background
Pseudomonas aeruginosa strains that are classed as extensively drug resistant (XDR-PA) are resistant to all antibiotics except for one or two classes and are frequently the cause of hard-to-treat infections worldwide. Our study aimed to characterize clinical XDR-PA isolates recovered during 2011–2012 at nine hospitals in the Hunan province of China.
Methods
Thirty-seven non-repetitive XDR-PA strains from 37 patients were investigated for genes encoding antimicrobial resistance determinants, efflux pumps, outer membrane proteins, and movable genetic elements using polymerase chain reaction (PCR). The expression of genes encoding the efflux pump component MexA and the outer membrane protein OprD was measured using real-time PCR. In addition, clonal relatedness of these XDR-PA isolates was analyzed by pulsed-field gel electrophoresis (PFGE).
Results
Various genes encoding antimicrobial resistance determinants were found in all isolates. In particular, the blaTEM-1, blaCARB, armA, blaIMP-4, blaVIM-2, and rmtB, were found in 100, 37.8, 22, 22, 19 and 5 % of the isolates, respectively. Remarkably, two isolates coharbored blaIMP-4, blaVIM-2, and armA. In all 37 antibiotic-resistant strains, the relative expression of oprD was decreased while mexA was increased compared to the expression of these genes in antibiotic-susceptible P. aeruginosa strains. All of the XDR-PA isolates harbored class I integrons as well as multiple other mobile genetic elements, such as tnpU, tnp513, tnpA (Tn21), and merA. A high genotypic diversity among the strains was detected by PFGE.
Conclusions
Multiple antibiotic-resistance mechanisms contributed to the drug resistance of the XDR-PA isolates investigated in this study. Thus, the XDR-PA isolates in this area were not clonally related. Instead, multiple types of movable genetic elements were coharbored within each XDR-PA isolate, which may have aided the rapid development of these XDR-PA strains. This is the first report of XDR-PA strains that coharbor blaIMP-4, blaVIM-2, and armA.
Keywords: Extensively drug-resistant Pseudomonas aeruginosa, Resistance mechanism, Movable genetic elements, Molecular epidemiology, PFGE
Background
Pseudomonas aeruginosa, a rod-shaped, non-fermenting gram-negative bacterium, causes nosocomial infections that can lead to sepsis, pneumonia, endocarditis, and urinary tract infections. The emergence of extensively drug-resistant P. aeruginosa (XDR-PA) strains showing resistant to all antimicrobial agents except for one or two classes is becoming a major public health concern [1–5].
That was found by previous studies the mechanisms of antibiotic resistance associated with clinical XDR-PA isolates are complex [6, 7]. The prevailing hypothesis is that XDR-PA isolates acquire numerous drug-resistance determinants through horizontal gene transfer that is mediated by mobile genetic elements [8–10]. In addition, genes encoding the antibiotic-inactivating enzymes β-lactamases, aminoglycoside-modifying enzymes (AMEs), and 16S rRNA methylases (16S-RMTases) are frequently associated with antibiotic resistance in XDR-PA strains. In recent studies, an over-expression of drug-efflux pumps and diminished expression of outer membrane proteins are suggested to play a role in drug resistance [11–14].
Although the prevalence of drug-resistance determinants in XDR-PA strains isolated in other countries has been determined [15, 16], few studies have investigated the resistance mechanisms and the epidemiological profiles of clinical XDR-PA isolates found in China. Before 2011, XDR-PA strains were rarely found in China; thereafter, a gradual emergence has occurred in some hospitals. Therefore, the purpose of this study was to analyze the resistance mechanisms and molecular epidemiology of clinical XDR-PA strains isolated earlier in our region recovered from 2011 to 2012.
Methods
Bacterial isolates
Thirty-seven out of 482 (7.7 %) P. aeruginosa isolates that were screened were XDR-PA strains displaying resistance to all antimicrobial agents except colistin. The P. aeruginosa strains were isolated from September 1, 2011 to June 30, 2012 from nine of fifteen different hospitals in the Hunan province of China. Drug susceptibility of XDR-PA isolates was determined by the Kirby-Bauer (K-B) disk diffusion method and two quality control strains (Escherichia coli ATCC25922 and P. aeruginosa ATCC27853) were included in the analyses. The results were analyzed and interpreted according to the guidelines of Clinical and Laboratory Standards Institute (CLSI) [17]. To prevent analysis of redundant strains, only the first strain was collected when duplicate strains were from the same patient.
Phenotypic tests for carbapenemase production
The modified Hodge test (MHT) was used to detect the production of carbapenemase using an imipenem disc (10 μg) as described by CLSI [17]. A combined-disc test was carried out to detect the production of metallo-β-lactamase (MBL). Two discs [One disc contained imipenem (10 μg) and 5 μL of 0.5 M EDTA (Sigma Chemicals), and the other disc contained only imipenem (10 μg)] were placed 20 mm apart on a Mueller–Hinton agar plate inoculated with each test strain. A strain was considered positive for metallo-β-lactamase production when the zone diameter around the imipenem-EDTA disc was more than 4 mm of the imipenem-only disc [18].
Detection of genes encoding antimicrobial resistance determinants and genes associated with movable genetic elements
The DNA templates used in polymerase chain reactions (PCR) to amplify genes encoding antimicrobial resistance determinants were obtained as follows: bacterial suspensions were incubated for 10 min at 95 °C followed by centrifugation at 10,000×g for 10 min to remove cellular debris. Genes coding for carbapenem β-lactamases (blaKPC, blaSME, blaGES, blaIMI/blaNMC, blaNDM-1, blaVIM-2, blaIMP-4, blaSIM-1, blaGIM, blaSPM, blaOXA-23, and blaOXA-51), extended-spectrum β-lactamases (ESBLs) (blaTEM-1, blaCTX-M, blaSHV, blaOXA-1, blaOXA-2, blaOXA-10, blaVEB, and blaPER), and AmpC β-lactamases (blaMOX, blaFOX, blaDHA, blaCIT, and blaEBC) were performed by PCR with previously described primers [18–20]. In addition, the isolates were screened by PCR for AME genes (aac(3)-IIa and ant(2′’)-Ia) [21], 16S-RMTases (armA, npmA, rmtA, rmtB, rmtC, rmtD, and rmtE) [22], a drug-efflux pump component (mexA), and an outer membrane protein (oprD). The PCR primers used to screen for these genes are listed in Table 1. To detect genes associated with the movable genetic elements, intI, traA, traF, trbC, tnp513, ISpa7, ISEcp1, tnpU, tnpA (Tn21), tnsA, and merA, PCR was performed with primers shown in Table 1. All amplified DNA fragments were sequenced and then analyzed using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST).
Table 1.
Primers used in this study for PCR and RT-PCR analyses
| Gene | Sequence (5′–3′) | Fragment length (bp) |
|---|---|---|
| intI | F:CCGAGGATGCGAACCACTTC R:CCGCCACTGCGCCGTTACCA |
789 |
| traA | F:AAGTGTTCAGGGTGCTTCTGCGC R:GTCATGTACATGATGACCATTT |
272 |
| traF | F:CGGTGATGATTTGCGAACGA R:AGCATTCCGGTCGGCCTGTA |
400 |
| trbC | F:CGGYATWCCGSCSACRCTGCG R:GCCACCTGYSBGCAGTCMCC |
255 |
| tnp513 | F:ATGTCGCTGGCAAGGAACGC R:GGGTTCGCTGCGAGGATTGT |
240 |
| ISpa7 | F:TCAGGCCTTCATCGCTGCCATCAGG R:TAGGCGTACAGTGCTCTTTCAACGCA |
300 |
| ISEcp1 | F:CTTCATTGGCATTGATAAGTTAG R:TGTAGCATCGGTTTCCCAGTTTC |
299 |
| tnpU | F:CCAACTGATGGCGGTGCCTT R:CGGTATGGTGGCTTTCGC |
403 |
| tnpA (Tn21) | F:ATGCCACGTCGTTCCATCCTGTCC R:CCGGGTCTGCTCCCGCTGGCC |
300 |
| tnsA | F:GCAGCAGCCTTACAAGACGAG R:GCCACATAGCGCAACTCCTCC |
416 |
| merA | F:GACCAGCCGCAGTTCGTCTA R:GCAGCASGAAAGCT GCTTCA |
462 |
| mexA | F:CGACCAGGCCGTGAGCAAGCAGC R:GGAGACCTTCGCCGCGTTGTCGC |
275 |
| oprD | F:ATGAAAGTGATGAAGTGGAGCG R:TTACAGGATCGACAGCGGATAG |
949 |
| mexA in RT-PCR | F:GGCGACAACGCGGCGAAGG R:CCTTCTGCTTGACGCCTTCCTGC |
202 |
| oprD in RT-PCR | F:CGGCGACATCAGCAACACC R:GGGCCGTTGAAGTCGGAGTA |
195 |
| 16S rRNA in RT-PCR | F:CCTACGGGAGGCAGCAG R:ATTACCGCGGCTGCTGG |
194 |
Quantifying expression of mexA and oprD
The expression of mexA and oprD in the XDR-PA isolates was determined by real-time PCR (RT-PCR). The experimental group consisted of the 37 XDR-PA isolates and the control group was made up of 31 P. aeruginosa isolates that were collected at the same time as the XDR-PA strains but were shown to be sensitive to all antimicrobial agents tested. The primers used for quantifying mexA and oprD expression are listed in Table 1. The RT-PCR reactions were carried out by a QuantiFast SYBR Green RT-PCR Kit from Qiagen and a Real-Time PCR System of LightCycler 2.0 from Roche, Burgess Hill, UK, according to the manufacturer protocols. All reactions were repeated three times using 10 ng of RNA template that was prepared using the RNeasy Mini Kit from Qiagen, Crawley, UK and treated with Dnase. Gene expression was normalized relative to that of the 16S rRNA gene using the 2−△△CT method [23]. The expression of 16S rRNA gene was determined by the primers shown in Table 1.
Pulsed-field gel electrophoresis (PFGE)
Clonal relatedness of the XDR-PA isolates was analyzed by PFGE. Preparation of genomic DNA was done in agarose blocks. The DNA was then digested by the restriction enzyme XbaI from Promega, USA, followed by embedding into 1 % PFGE agarose gel. PFGE was performed for 24 h using the GenePath System from Bio-Rad with the follow conditions: 5.5 V/cm, 12 °C, 120°, and a switch time from 4 to 40 s. The molecular size marker, Salmonella enterica strain H9812, was obtained from the respiratory laboratory of infectious diseases, CDC, China. The gel was stained for 30 min with ethidium bromide and the gel image was documented using the gel documentation system Gel Doc 2000 from Bio-Rad. Finally, analyses of the results were performed by the BioNumerics software platform (Applied Math, Sint-Maten-Latem, Belgium) and visual inspection with the criteria of relatedness proposed by Tenover et al. [24].
Statistical analysis
WHONET software (version 5.4, WHO) was used to analyze the patient demographic information and the antibiotic resistance data using the 2014 CLSI criteria for breakpoints for P. aeruginosa. Comparisons of different groups were analyzed by a two-sided Chi square (χ2) test with the SPSS13.0 software (SPSS Inc., USA). A P < 0.05 was regarded as statistically significant.
Results
Phenotypic screening and resistance determinants
Of the 37 XDR-PA isolates, 13 (35.1 %) isolates were positive for MTH and MBL, while all other isolates were negative (Fig. 1).
Fig. 1.

Clinical features, molecular characterization, and dendrogram based on PFGE of 37 XDR-PA isolates. The dendrogram was developed using the BioNumerics software platform. P patients, Ss specimens, RDs resistance determinants, xy Xiangya Hospital, f 3 the Third Xiangya Hospital, yy People’s Hospital of Liuyang, cs the Third Hospital of Changsha, se the Second People’s Hospital of Hunan province, xt Xiangtan Central Hospital, lz Chinese Medicine Hospital of Liuyang, cz People’s Hospital of Chenzhou, cy the First Hospital of Changsha, and A–U PFGE types
Diverse types of genes encoding antibiotic-inactivating enzymes were detected. Specifically, the genes blaTEM-1, blaCARB, armA, blaIMP-4, blaVIM-2, and rmtB were found in 100, 37.8, 22, 22, 19 and 5 % of the XDR-PA isolates, respectively. The ESBL gene blaTEM-1 was found in all isolates, while the other ESBL genes that were tested were not found in any of the isolates. Among the 37 XDR-PA isolates, 13 (35.1 %) isolates were positive for MBLs genes, 8 contained blaIMP-4, and 7 contained blaVIM-2. Two types of 16S-RMTases genes were detected. Eight isolates contained armA, while two isolates contained rmtB. Eight isolates coharbored blaIMP-4 and armA and two isolates coharbored blaIMP-4, blaVIM-2, and armA. No isolates harbored genes encoding AmpC β-lactamases or AMEs.
The efflux-pump gene mexA and the outer membrane protein gene oprD were detected in all 37 XDR-PA isolates. However, no mutations were detected in any of the oprD genes. Figure 1 lists the antibiotic resistance genes detected in each isolate.
Expression of mexA and oprD
The expression of mexA was significantly higher in the 37 XDR-PA isolates than in the control group consisting of 31 antibiotic-sensitive isolates (1.95 ± 0.48 and 0.70 ± 0.13, P = 0.018, respectively), while expression of oprD in the XDR-PA strains was significantly lower than the control group (3.18 ± 0.60 and 0.94 ± 0.08; P = 0.002, respectively; Fig. 2).
Fig. 2.
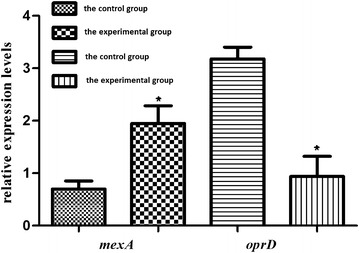
The relative expression of the mexA and oprD genes in the XDR-PA isolates. The relative expression levels are expressed as the mean ± standard error of the mean (SEM). *P values less than 0.05 are considered significant and are indicated by an asterisk
Distribution of genes associated with movable genetic elements
Of the movable genetic element genes tested, intI, merA, tnp513, tnpA (Tn21), and tnpU were detected in 100, 100, 81, 49, and 16 % of the isolates, respectively. Moreover, most of the 37 XDR-PA isolates coharbored three or more genes associated with movable genetic elements. For instance, isolates xy36 coharbored five types of movable genetic element genes (Fig. 1).
Clonal relatedness of the XDR-PA isolates
Using PFGE, the 37 XDR-PA isolates were divided into 21 PFGE types (Fig. 1). The main PFGE type found was A (five strains), including three strains from Xiangya Hospital and two strains from the Third Xiangya Hospital. In addition, an A1 subtype was found in both hospitals. The B- and C-type isolates were found in the Xiangya Hospital, while the D-type isolates were found in the Third Hospital of Changsha as well as Chinese Medicine Hospital of Liuyang. The E-type strains were isolated from the Second People’s Hospital of Hunan province and the Third Xiangya Hospital, and an E1 subtype was found in both hospitals. The F-type strains were isolated from the People’s Hospital of Liuyang; the G-type strains were isolated from the Third Xiangya Hospital; and the H-type isolates were found at Xiangya Hospital and Xiangtan Central Hospital. The remaining 13 types, from I to U, were collected from the Xiangya Hospital (eight strains), the People’s Hospital of Chenzhou (one strain), the First Hospital of Changsha (one strain), the Chinese Medicine Hospital of Liuyang (one strain), the Third Xiangya Hospital (one strain), and the Third Hospital of Changsha (one strain).
Discussion
Pseudomonas aeruginosa is considered one of the primary causes of hospital-acquired infections. An increase in the prevalence of clinical XDR-PA isolates correlates with a rise in mortality and morbidity rates. Consequently, XDR-PA strains pose a considerable threat to public health worldwide. In our study, 37 out of 482 (7.7 %) P. aeruginosa strains were found to be XDR-PA strains. The XDR-PA strains were isolated from nine of fifteen teaching hospitals detected in our region, suggesting that the occurrence of XDR-PA isolates in our region is low. To aid the prevention of the spread of XDR-PA strains, we have analyzed the resistance mechanisms and the molecular epidemiology of the XDR-PA strains.
Carbapenems are one of the most effective drugs against severe infections caused by gram-negative bacilli. Unfortunately, pathogens displaying resistance to carbapenems are increasing due to the following three main causes: production of carbapenemases, the over-expression of efflux pumps, and the diminished expression of the outer membrane porin OprD [15]. In P. aeruginosa, production of carbapenemases, especially MBLs, is an important antibiotic resistance mechanisms [25]. In this study, 13 (35.1 %) strains produced the metallo-enzymes IMP-4 and VIM-2, indicating that production of carbapenemases played role in carbapenem-resistant in the XDR-PA isolates. Notably, the prevalence of carbapenemases in the 37 XDR-PA isolates in our study were higher than that of other countries in the world [26, 27] and from other regions in China [28]. One isolate that coharbored two MBL genes was reported in previous studies [29]. This observation led to the emergence of a new drug-resistant model for P. aeruginosa. In our study, two isolates (xy35 and xy36) coharbored blaIMP-4 and blaVIM-2. To our knowledge, this is the first report of the co-existence of blaIMP-4 and blaVIM-2 in a P. aeruginosa strain.
Drug-efflux pumps have been correlated with bacterial resistance since the 1980s [30]. The first efflux pump found in P. aeruginosa is the MexAB-OprM pump, which has a broad range of substrates, including carbapenems, quinolones, aminoglycosides, tetracyclines, and macrolides. Therefore, the presence of this efflux pump in bacteria may lead to multi-drug resistance. In our study, the mexA gene, encoding a component of MexAB-OprM efflux pump, was found in all 37 XDR-PA isolates. Furthermore, a significant over-expression of mexA in the XDR-PA isolates was seen compared with the mexA levels in antibiotic-susceptible strains. Thus, over-expression of an efflux pump, especially MexAB-OprM, may play a key role in the antibiotic resistance of these XDR-PA isolates.
Decreased expression of OprD can significantly reduce the susceptibility of P. aeruginosa to carbapenems. Previous studies showed that decreased expression of oprD is primarily due to mutations in the OprD-encoding genes [31]. In our study, the expression of oprD in the 37 XDR-PA isolates was significantly lower than that in susceptible strains confirming that a decreased level of this porin plays a key role in carbapenem-resistance. However, no mutations or deletions were detected for the oprD gene in the 37 XDR-PA isolates. This finding was not consistent with previous studies [32, 33], suggesting that decreased expression of oprD in the 37 XDR-PA isolates might be due to regulation by small bioactive molecules, amino acids, or efflux pump expression [34]. Therefore, in-depth studies on the role of outer membrane proteins in bacterial drug resistance are needed in the future.
Production of AMEs and 16S-RMTases is the main cause of bacterial resistance to aminoglycoside agents [35, 36]. In contrast to these reports, none of the 37 XDR-PA isolates, which were resistant to aminoglycoside antibiotics, harbored genes encoding AMEs and only 27.0 % (n = 10) harbored either armA or rmtB 16S-RMTases genes. These data suggest that 16S-RMTases play an important role in aminoglycoside resistance in the XDR-PA isolates but also that some of these isolates may utilize an alternative, unknown resistance mechanism, such as new drug efflux pumps, new 16S-RMTases, or mutational activation of the AmgRS two-component system [37].
Movable genetic elements, including integrons, plasmids, transposons, and insertion sequences, play a key role in the horizontal transfer of resistance genes [38]. In this study, most of the XDR-PA isolates coharbored three or more genes associated with movable genetic elements. The most frequently detected genes were intI, merA tnp513, tnpA (Tn21), and tnpU, and their distribution percentage among the XDR-PA isolates was 100, 100, 81, 49, and 16 %, respectively. Furthermore, two isolates, cs15 and xy36, coharbored 5 types of genes associated with movable genetic elements.
The XDR-PA strains previously isolated in other countries were shown to be clonally related [1]. In contrast, the XDR-PA strains isolated from hospitals within the Hunan province of China were shown to be genetically diverse. The 37 XDR-PA isolates were divided into 21 different PFGE types and no one type predominated amongst the isolates suggesting that none of the XDR-PA isolates are considered to be an epidemic clone. Interestingly, the A-type, D-type, E-type, and H-type were detected in two different hospitals and the B-type, C-type, and F-type were found in two different wards of the same hospital during the same time period. Although prevalence of the XDR-PA isolates in this area was sporadic, dissemination of the same isolate was detected inter- and intra-hospital. The high heterogeneity of the PFGE types suggests that the mobile genetic elements may play a role in the emergence of clinical XDR-PA strains.
Conclusions
In conclusion, this study shows that the extreme antibiotic resistance of the clinical XDR-PA isolates is due to strains coharboring multiple antibiotic resistance genes, over-expressing drug-efflux pumps, and decreasing expression of oprD, which encodes an outer membrane porin. Moreover, this study has identified for the first time a P. aeruginosa isolate that coharbors the blaIMP-4, blaVIM-2, and armA antibiotic resistance genes. Although the XDR-PA isolates were not clonally related, the strains were shown to carry multiple genes encoding different types of movable genetic elements, which may aid the rapid development of XDR-PA isolates.
Authors’ contributions
MZ contributed to the conception of the study. JL contributed significantly to analysis and manuscript preparation; QD and YH performed the data analyses and wrote the manuscript; HW, QY and WL helped perform the analysis with constructive discussions. All authors read and approved the final manuscript.
Acknowledgements
We are thankful to the staff in the Laboratory of Xiangya Hospital of Central South University who helped us in the collection and identification of these XDR-PA isolates. In particular, we thank Xin Xia, a teacher at the Center for Disease Control (CDC) in the Hunan Province of China, for helping us with the PFGE experiments.
Competing interests
The authors declare that they have no competing interests.
Ethical approval
All experiments involving animals were performed according to the ethical standards of the institute. The experimental protocols were signed and approved by the ethics committee for each of the nine teaching hospitals in China.
Funding
This work was supported by the Hunan Development and Reform Investment [2012] No. 1493 from the Development and Reform Commission of the Hunan Province, a Grant (14JJ7003) from the Natural Science Foundation of the Hunan Province, and the Hunan Development and Reform Investment [2014] No. 658 from Development and Reform Commission of the Hunan Province.
Informed consent
A signed, informed consent was given by all participants included in the study.
Contributor Information
Jun Li, Email: lijun198412@126.com.
Mingxiang Zou, Phone: +86 73184327440, Email: zoumingxiang@126.com.
Qingya Dou, Email: 280764284@qq.com.
Yongmei Hu, Email: 626973094@qq.com.
Haichen Wang, Email: 337228840@qq.com.
Qun Yan, Email: yanqunhn@qq.com.
Wen’ en Liu, Email: wenenliu@163.com.
References
- 1.Ciofi Degli Atti M, Bernaschi P, Carletti M, Luzzi I, Garcia-Fernandez A, Bertaina A, Sisto A, Locatelli F, Raponi M. An outbreak of extremely drug-resistant Pseudomonas aeruginosa in a tertiary care pediatric hospital in Italy. BMC Infect Dis. 2014;14:494. doi: 10.1186/1471-2334-14-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fonseca EL, Freitas Fdos S, Vicente AC. The colistin-only-sensitive Brazilian Pseudomonas aeruginosa clone SP (sequence type 277) is spread worldwide. Antimicrob Agents Chemother. 2010;54(6):2743. doi: 10.1128/AAC.00012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin JS, Kwon KT, Moon DC, Lee JC. Emergence of 16S rRNA methylase rmtA in colistin-only-sensitive Pseudomonas aeruginosa in South Korea. Int J Antimicrob Agents. 2009;33(5):490–491. doi: 10.1016/j.ijantimicag.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 4.Lee YC, Ahn BJ, Jin JS, Kim JU, Lee SH, do Song Y, Lee WK, Lee JC. Molecular characterization of Pseudomonas aeruginosa isolates resistant to all antimicrobial agents, but susceptible to colistin, in Daegu, Korea. J Microbiology (Seoul, Korea) 2007;45(4):358–363. [PubMed] [Google Scholar]
- 5.Viedma E, Juan C, Acosta J, Zamorano L, Otero JR, Sanz F, Chaves F, Oliver A. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum beta-lactamases GES-1 and GES-5 in Spain. Antimicrob Agents Chemother. 2009;53(11):4930–4933. doi: 10.1128/AAC.00900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Livermore DM. Multiple mechanisms of antimicrobial resistance in, Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis. 2002;34(5):634–640. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 7.Poole K. Pseudomonas aeruginosa: resistance to the max. Front Microbiol. 2011;2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beige F, Baseri Salehi M, Bahador N, Mobasherzadeh S. Plasmid mediated antibiotic resistance in isolated bacteria from burned patients. Jundishapur J Microbiol. 2015;8(1):e13567. doi: 10.5812/jjm.13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiddee A, Henghiranyawong K, Yimsabai J, Tiloklurs M, Niumsup PR. Nosocomial spread of class 1 integron-carrying extensively drug-resistant Pseudomonas aeruginosa isolates in a Thai hospital. Int J Antimicrob Agents. 2013;42(4):301–306. doi: 10.1016/j.ijantimicag.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Stalder T, Barraud O, Casellas M, Dagot C, Ploy MC. Integron involvement in environmental spread of antibiotic resistance. Front Microbiol. 2012;3:119. doi: 10.3389/fmicb.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aghazadeh M, Hojabri Z, Mahdian R, Nahaei MR, Rahmati M, Hojabri T, Pirzadeh T, Pajand O. Role of efflux pumps: MexAB-OprM and MexXY(-OprA), AmpC cephalosporinase and OprD porin in non-metallo-beta-lactamase producing Pseudomonas aeruginosa isolated from cystic fibrosis and burn patients. Infect Genet Evol. 2014;24:187–192. doi: 10.1016/j.meegid.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Lee JY, KO KS. OprD mutations and inactivation, expression of efflux pumps and AmpC, and metallo-beta-lactamases in carbapenem-resistant Pseudomonas aeruginosa isolates from South Korea. Int J Antimicrob Agents. 2012;40(2):168–172. doi: 10.1016/j.ijantimicag.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Quale J, Bratu S, Gupta J, Landman D. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother. 2006;50(5):1633–1641. doi: 10.1128/AAC.50.5.1633-1641.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xavier DE, Picao RC, Girardello R, Fehlberg LC, Gales AC. Efflux pumps expression and its association with porin down-regulation and beta-lactamase production among Pseudomonas aeruginosa causing bloodstream infections in Brazil. BMC Microbiol. 2010;10:217. doi: 10.1186/1471-2180-10-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castanheira M, Deshpande LM, Costello A, Davies TA, Jones RN. Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseudomonas aeruginosa collected during 2009–11 in 14 European and Mediterranean countries. J Antimicrob Chemother. 2014;69(7):1804–1814. doi: 10.1093/jac/dku048. [DOI] [PubMed] [Google Scholar]
- 16.Polotto M, Casella T, de Lucca Oliveira MG, Rubio FG, Nogueira ML, de Almeida MT, Nogueira MC. Detection of P. aeruginosa harboring bla CTX-M-2, bla GES-1 and bla GES-5, bla IMP-1 and bla SPM-1 causing infections in Brazilian tertiary-care hospital. BMC Infect Dis. 2012;12:176. doi: 10.1186/1471-2334-12-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 24th informational supplement. M100-S24. Wayne: Clinical and Laboratory Standards Institute; 2014.
- 18.Fang H, Ataker F, Hedin G, Dornbusch K. Molecular epidemiology of extended-spectrum beta-lactamases among Escherichia coli isolates collected in a Swedish hospital and its associated health care facilities from 2001 to 2006. J Clin Microbiol. 2008;46(2):707–712. doi: 10.1128/JCM.01943-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soleimani N, Aganj M, Ali L, Shokoohizadeh L, Sakinc T. Frequency distribution of genes encoding aminoglycoside modifying enzymes in uropathogenic E. coli isolated from Iranian hospital. BMC Res Notes. 2014;7:842. doi: 10.1186/1756-0500-7-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doi Y, Arakawa Y. Ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis. 2007;45(1):88–94. doi: 10.1086/518605. [DOI] [PubMed] [Google Scholar]
- 23.Arabestani MR, Rajabpour M, Yousefi Mashouf R, Alikhani MY, Mousavi SM. Expression of efflux pump MexAB-OprM and OprD of Pseudomonas aeruginosa strains isolated from clinical samples using qRT-PCR. Arch Iranian Med. 2015;18(2):102–8. doi:015182/AIM.008. [PubMed]
- 24.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong DJ, Bae IK, Jang IH, Jeong SH, Kang HK, Lee K. Epidemiology and Characteristics of Metallo-β-Lactamase-Producing Pseudomonas aeruginosa. Infect Chemother. 2015;47(2):81–97. doi: 10.3947/ic.2015.47.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalantar E, Torabi V, Salimizand H, Soheili F, Beiranvand S. Soltan Dallal MM. First survey of metallo-β-lactamase producers in clinical isolates of Pseudomonas aeruginosa from a referral burn center in Kurdistan Province. Jundishapur J Nat Pharm. Prod. 2012;7(1):23–6. doi: 10.17795/jjnpp-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong JS, Kim JO, Lee H, Bae IK, Jeong SH, Lee K. Characteristics of metallo-β-lactamase-producing Pseudomonas aeruginosa in Korea. Infect Chemother. 2015;47(1):33–40. doi: 10.3947/ic.2015.47.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Gu B, Mei Y, Wen Y, Xia W. Increasing resistance rate to carbapenem among blood culture isolates of Klebsiella pneumoniae, Acinetobacter baumannii and Pseudomonas aeruginosa in a university-affiliated hospital in China, 2004–2011. J Antibiot (Tokyo). 2015;68(2):115–20. doi: 10.1038/ja.2014.119. [DOI] [PubMed] [Google Scholar]
- 29.Toval F, Guzmán-Marte A, Madriz V, Somogyi T, Rodríguez C, García F. Predominance of carbapenem-resistant Pseudomonas aeruginosa isolates carrying blaIMP and blaVIM metallo-β-lactamases in a major hospital in Costa Rica. J Med Microbiol. 2015;64(Pt 1):37–43. doi: 10.1099/jmm.0.081802-0. [DOI] [PubMed] [Google Scholar]
- 30.George AM, Levy SB. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol. 1983;155(2):531–540. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shu JC, Chia JH, Siu LK, Kuo AJ, Huang SH, Su LH, Wu TL. Interplay between mutational and horizontally acquired resistance mechanisms and its association with carbapenem resistance amongst extensively drug-resistant Pseudomonas aeruginosa (XDR-PA) Int J Antimicrob Agents. 2012;39(3):217–222. doi: 10.1016/j.ijantimicag.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen J, Pan Y, Fang Y. Role of the outer membrane protein OprD2 in carbapenem-resistance mechanisms of Pseudomonas aeruginosa. PLoS One. 2015;10(10):e0139995. doi: 10.1371/journal.pone.0139995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rojo-Bezares B, Estepa V, Cebollada R, de Toro M, Somalo S, Seral C, Castillo FJ, Torres C, Sáenz Y. Carbapenem-resistant Pseudomonas aeruginosa strains from a Spanish hospital: characterization of metallo-beta-lactamases, porin OprD and integrons. Int J Med Microbiol. 2014;304(3–4):405–414. doi: 10.1016/j.ijmm.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Li H, Luo YF, Williams BJ, Blackwell TS, Xie CM. Structure and function of OprD protein in Pseudomonas aeruginosa: from antibiotic resistance to novel therapies. Int J Med Microbiol. 2012;302(2):63–68. doi: 10.1016/j.ijmm.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashenafi M, Ammosova T, Nekhai S, Byrnes WM. Purification and characterization of aminoglycoside phosphotransferase APH(6)-Id, a streptomycin-inactivating enzyme. Mol Cell Biochem. 2014;387(1–2):207–216. doi: 10.1007/s11010-013-1886-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galani I, Souli M, Panagea T, Poulakou G, Kanellakopoulou K, Giamarellou H. Prevalence of 16S rRNA methylase genes in Enterobacteriaceae isolates from a Greek university hospital. Clin Microbiol Infect. 2012;18(3):E52–E54. doi: 10.1111/j.1469-0691.2011.03738.x. [DOI] [PubMed] [Google Scholar]
- 37.Lau CH, Fraud S, Jones M, Peterson SN, Poole K. Mutational activation of the AmgRS two-component system in aminoglycoside-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57(5):2243–2251. doi: 10.1128/AAC.00170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ibrahim ME, Magzoub MA, Bilal NE, Hamid ME. Distribution of Class I integrons and their effect on the prevalence of multi-drug resistant Escherichia coli clinical isolates from Sudan. Saudi Med J. 2013;34(3):240–247. [PubMed] [Google Scholar]


