Abstract
Background
Drought stress is one of the major causes of crop loss. WRKY transcription factors, as one of the largest transcription factor families, play important roles in regulation of many plant processes, including drought stress response. However, far less information is available on drought-responsive WRKY genes in wheat (Triticum aestivum L.), one of the three staple food crops.
Results
Forty eight putative drought-induced WRKY genes were identified from a comparison between de novo transcriptome sequencing data of wheat without or with drought treatment. TaWRKY1 and TaWRKY33 from WRKY Groups III and II, respectively, were selected for further investigation. Subcellular localization assays revealed that TaWRKY1 and TaWRKY33 were localized in the nuclei in wheat mesophyll protoplasts. Various abiotic stress-related cis-acting elements were observed in the promoters of TaWRKY1 and TaWRKY33. Quantitative real-time PCR (qRT-PCR) analysis showed that TaWRKY1 was slightly up-regulated by high-temperature and abscisic acid (ABA), and down-regulated by low-temperature. TaWRKY33 was involved in high responses to high-temperature, low-temperature, ABA and jasmonic acid methylester (MeJA). Overexpression of TaWRKY1 and TaWRKY33 activated several stress-related downstream genes, increased germination rates, and promoted root growth in Arabidopsis under various stresses. TaWRKY33 transgenic Arabidopsis lines showed lower rates of water loss than TaWRKY1 transgenic Arabidopsis lines and wild type plants during dehydration. Most importantly, TaWRKY33 transgenic lines exhibited enhanced tolerance to heat stress.
Conclusions
The functional roles highlight the importance of WRKYs in stress response.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-016-0806-4) contains supplementary material, which is available to authorized users.
Keywords: Drought tolerance, WRKY transcription factor, Stress response mechanisms, Thermotolerance, Triticum aestivum
Background
Being unable to move, plants have developed a series of complex mechanisms to cope with abiotic and biotic stresses. Recognition of stress cues and transduction of signals to activate adaptive responses and regulation of stress-related genes are key steps leading to plant stress tolerance [1–4].
Due to the potential impact on agricultural production much attention has been focused on abiotic stress factors. Abiotic stresses initiate the synthesis of different types of proteins, including transcription factors, enzymes, molecular chaperones, ion channels, and transporters [5]. Transcriptional regulation mechanisms play a critical role in plant development and responses to environmental stimuli [4, 6, 7]. Transcription factors, with specific DNA-binding domains (DBD) and trans-acting functional domains, can combine with specific DNA sequences to activate or inhibit transcription of downstream genes. Using transcription factors to improve the tolerance of plants to abiotic stresses is a promising strategy due to the ability of transcription factors to modulate a set of genes through binding to either promoter or enhancer region of a gene [8]. Overexpression of constitutive active DREB2A which had a transcriptional activation domain between residues 254 and 335 resulted in significant drought stress tolerance through regulates expression of many water stress-inducible genes [9]. In our previous study, GmHsf-34 gene improved drought and heat stresses tolerance in Arabidopsis plants [10]. These studies indicate the potential for improvement of abiotic stress tolerance in plants through transcriptional regulation.
WRKY transcription factors, one of the ten largest transcription factor families, are characterized by a highly conserved WRKYGQK heptapeptide at the N-terminus and a zinc finger-like motif at the C-terminus [11]. Conservation of the WRKY domain is mirrored by a remarkable conservation of its cognate binding site, the W box (TTGACC⁄T) [11–13]. A few WRKY proteins which show slight variations in the heptapeptide WRKYGQK motif can not bind the W box and may bind the WK box (TTTTCCAC) [14–17]. WRKYs are divided into three groups based on the number of WRKY domains and type of zinc finger motif. The first group has two WRKY domains. Groups II and III have a single WRKY domain and are distinguished according to the type of zinc finger motif [17]. Groups I and II share the same C2H2 zinc finger motif whereas group III contains a C2-HC-type motif [18]. Later, according to a more accurate phylogenetic analysis, Zhang and Wang divided WRKY factors into Groups I, IIa + IIb, IIc, IId + IIe, and III with Group II genes not being monophyletic [12].
Increasing data indicates that WRKY genes are rapidly induced by pathogen infection and exogenous phytohormones [19–25]. Forty nine of 72 Arabidopsis WRKY genes were differentially regulated after infection by Pseudomonas syringae or SA treatment [26]. Transcript abundance of 13 canola WRKY genes changed after pathogen infection [15]. Similarly, 28 grape WRKY genes showed various transcription expression in response to biotic stress caused by grape white rot and/or salicylic acid (SA). Among them 16 WRKY genes were upregulated by both pathogenic white rot bacteria and SA, indicating that these WRKY genes participated in the SA-dependent defense signal pathway [27]. Heterologous expression of OsWRKY6 activated defense-related genes and enhanced resistance to pathogens in Arabidopsis [28]. Recently, it was reported that the OsMKK4-OsMPK3/OsMPK6 cascade regulates transactivation activity of OsWRKY53, and a phospho-mimic mutant of OsWRKY53 resulted in further-enhanced disease resistance against the blast fungus in rice compared to native OsWRKY53 [24].
In comparison with research progress on biotic stresses, the functions of WRKYs in abiotic stresses are far less known [29–36]. Increasing numbers of reports are showing that WRKYs respond to abiotic stress and abscisic acid (ABA) signaling in plants [37–41]. Several Arabidopsis WRKY genes can be induced by drought and/or cold stress [42, 43]. AtWRKY46 regulated osmotic stress responses and stomatal movement independently in Arabidopsis [44]. OsWRKY08 improved the osmotic stress tolerance of transgenic Arabidopsis through positive regulation of the expression of ABA-independent abiotic stress responsive genes [45]. Overexpression and RNAi analysis demonstrated that GmWRKY27 improved salt and drought tolerance in transgenic soybean hairy roots by inhibits expression of a downstream gene GmNAC29 which was a negative factor of stress tolerance [46]. Therefore, WRKYs play a broad-spectrum regulatory role as positive and negative regulators in response to biotic and abiotic stresses, senescence, seed development and seed germination [17, 25, 47].
Drought stress is one of the most severe environmental factors restricting crop distribution and production. The molecular mechanisms underlying plant tolerance to drought stress are still not fully understood because of the complex nature [48]. Bread wheat (Triticum aestivum L.) is one of the most widely cultivated and important food crops in the world. Drought affects growth and productivity of wheat, and reduces yields worldwide. It was recently reported that wheat TaWRKY2 and TaWRKY19 conferred tolerance to drought stress in transgenic plants [49]. To investigate putative drought-mediated WRKY genes, we performed de novo transcriptome sequencing of drought-treated wheat, and identified 48 wheat drought-responsive WRKY genes. We further investigated stress tolerance conferred by TaWRKY1 and TaWRKY33 in transgenic Arabidopsis. The present study investigated the possibility of improving stress tolerance in plants by screening stress responsive candidate genes.
Results
Identification of drought-responsive WRKY genes in wheat
In order to identify WRKY genes regulated by drought, we compared wheat de novo transcriptome sequencing data with or without drought treatment. A pairwise comparison of drought vs. without drought treatments revealed 48 WRKYs showing significant up- or down-regulation in transcription level (more than a twofold change) (Table 1). Nucleic acid sequences of 48 WRKYs in wheat were listed in Additional file 1: Table S1.
Table 1.
Drought-induced responsive WRKY genes in wheat
| Gene name | Gene ID | CK | Drought | Log fold change | Up/Down | FDR |
|---|---|---|---|---|---|---|
| TaWRKY1 | Unigene50292_All | 1 | 16 | 4.17 | Up | 1.51E-04 |
| TaWRKY2 | CL2151.Contig2_All | 1 | 18 | 4.00 | Up | 4.90E-04 |
| TaWRKY3 | CL7466.Contig1_All | 65 | 818 | 3.65 | Up | 1.51E-165 |
| TaWRKY4 | Unigene9495_All | 7 | 80 | 3.51 | Up | 4.36E-16 |
| TaWRKY5 | Unigene24182_All | 43 | 485 | 3.50 | Up | 1.16E-94 |
| TaWRKY6 | CL15640.Contig3_All | 66 | 609 | 3.21 | Up | 7.22E-110 |
| TaWRKY7 | Unigene23958_All | 35 | 311 | 3.15 | Up | 2.17E-55 |
| TaWRKY8 | CL7466.Contig2_All | 108 | 928 | 3.10 | Up | 3.97E-162 |
| TaWRKY9 | CL2311.Contig1_All | 46 | 350 | 2.93 | Up | 6.18E-58 |
| TaWRKY10 | CL2960.Contig4_All | 11 | 78 | 2.83 | Up | 6.39E-13 |
| TaWRKY11 | CL9014.Contig1_All | 375 | 2331 | 2.64 | Up | 0 |
| TaWRKY12 | CL2311.Contig2_All | 17 | 98 | 2.53 | Up | 2.58E-14 |
| TaWRKY13 | CL15640.Contig5_All | 125 | 643 | 2.36 | Up | 1.07E-83 |
| TaWRKY14 | CL2151.Contig1_All | 35 | 179 | 2.35 | Up | 1.25E-23 |
| TaWRKY15 | CL15640.Contig2_All | 136 | 675 | 2.31 | Up | 1.51E-85 |
| TaWRKY16 | CL321.Contig3_All | 58 | 276 | 2.25 | Up | 3.32E-34 |
| TaWRKY17 | Unigene47896_All | 14 | 64 | 2.19 | Up | 3.05E-08 |
| TaWRKY18 | CL2151.Contig3_All | 11 | 49 | 2.16 | Up | 2.22E-06 |
| TaWRKY19 | CL4329.Contig1_All | 695 | 2919 | 2.07 | Up | 0 |
| TaWRKY20 | CL3634.Contig1_All | 26 | 106 | 2.03 | Up | 5.50E-12 |
| TaWRKY21 | CL14217.Contig1_All | 731 | 2775 | 1.92 | Up | 1.73E-277 |
| TaWRKY22 | Unigene45898_All | 13 | 48 | 1.88 | Up | 2.04E-05 |
| TaWRKY23 | CL9014.Contig2_All | 294 | 1079 | 1.88 | Up | 1.71E-104 |
| TaWRKY24 | Unigene23130_All | 874 | 2872 | 1.72 | Up | 1.76E-245 |
| TaWRKY25 | CL9014.Contig3_All | 70 | 230 | 1.72 | Up | 3.05E-20 |
| TaWRKY26 | CL213.Contig2_All | 43 | 126 | 1.55 | Up | 5.17E-10 |
| TaWRKY27 | CL9014.Contig6_All | 41 | 118 | 1.53 | Up | 3.20E-09 |
| TaWRKY28 | Unigene27690_All | 86 | 242 | 1.49 | Up | 1.56E-17 |
| TaWRKY29 | CL14321.Contig2_All | 210 | 549 | 1.39 | Up | 7.32E-35 |
| TaWRKY30 | CL15640.Contig4_All | 332 | 844 | 1.35 | Up | 6.48E-51 |
| TaWRKY31 | CL15640.Contig8_All | 50 | 127 | 1.34 | Up | 2.53E-08 |
| TaWRKY32 | CL213.Contig3_All | 61 | 153 | 1.33 | Up | 1.14E-09 |
| TaWRKY33 | Unigene22134_All | 2632 | 6548 | 1.31 | Up | 0 |
| TaWRKY34 | CL1516.Contig3_All | 336 | 833 | 1.31 | Up | 3.13E-48 |
| TaWRKY35 | CL15191.Contig2_All | 211 | 519 | 1.30 | Up | 6.40E-30 |
| TaWRKY36 | CL9113.Contig1_All | 87 | 196 | 1.17 | Up | 3.61E-10 |
| TaWRKY37 | CL9910.Contig2_All | 98 | 214 | 1.13 | Up | 2.07E-10 |
| TaWRKY38 | Unigene13575_All | 126 | 275 | 1.13 | Up | 3.97E-13 |
| TaWRKY39 | CL16569.Contig1_All | 125 | 268 | 1.10 | Up | 2.23E-12 |
| TaWRKY40 | CL9014.Contig5_All | 498 | 1013 | 1.02 | Up | 8.10E-40 |
| TaWRKY41 | CL1681.Contig3_All | 108 | 51 | -1.07 | Down | 2.59E-05 |
| TaWRKY42 | CL14934.Contig1_All | 1552 | 412 | -1.90 | Down | 1.85E-152 |
| TaWRKY43 | CL14934.Contig2_All | 700 | 184 | -1.92 | Down | 1.02E-69 |
| TaWRKY44 | CL8633.Contig1_All | 172 | 39 | -2.13 | Down | 4.13E-20 |
| TaWRKY45 | Unigene25087_All | 66 | 8 | -3.03 | Down | 9.61E-12 |
| TaWRKY46 | Unigene39119_All | 26 | 2 | -3.69 | Down | 8.13E-06 |
| TaWRKY47 | Unigene32932_All | 16 | 1 | -3.99 | Down | 5.15E-04 |
| TaWRKY48 | Unigene33182_All | 20 | 1 | -4.31 | Down | 4.81E-05 |
CK, mean of sample without drought treatment
Log fold change, log2 (Drought/CK)
FDR false discovery rate
To investigate the evolutionary relationships of the drought-induced wheat WRKYs with previously reported WRKYs, a phylogenic tree was constructed using MEGA5.1. Twenty four drought-induced wheat WRKYs belonged to Group II, 15 to Group III, and nine to Group I (Fig. 1).
Fig. 1.
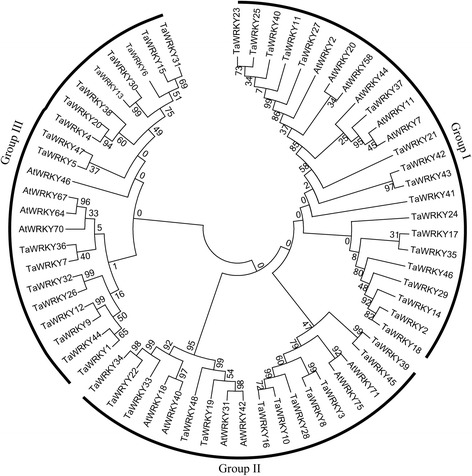
Maximum likelihood phylogenetic tree of drought-responsive WRKY genes in wheat and 16 AtWRKY proteins. The phylogenetic tree was based on comparisons of amino acid sequences and produced by MEGA 5.1 software
Sequence analysis of TaWRKY1 and TaWRKY33
Among the 48 drought-induced wheat WRKY genes, TaWRKY1 to TaWRKY8 showed the largest transcript differences, being up-regulated more than three-log fold (log2 (Drought/CK)) and TaWRKY21/24/33/42 showed the largest background transcript levels among all WRKY genes regulated by drought (Table 1). The drought stress expression patterns of these 12 wheat WRKY genes were further investigated. As shown in Fig. 2, TaWRKY1 and TaWRKY33 gave high responses to drought stress, peaking at more than 30-fold at one and two h, respectively. These genes were selected for further investigation.
Fig. 2.
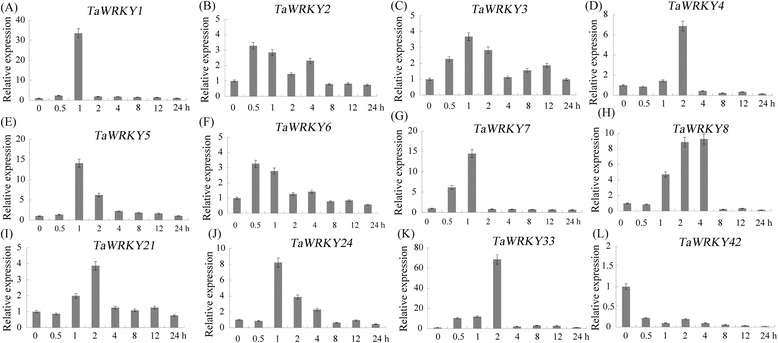
Expression patterns of 12 wheat WRKY genes under drought stress. These 12 wheat WRKY genes include TaWRKY1 (a), TaWRKY2 (b), TaWRKY3 (c), TaWRKY4 (d), TaWRKY5 (e), TaWRKY6 (f), TaWRKY7 (g), TaWRKY8 (h), TaWRKY21 (i), TaWRKY24 (j), TaWRKY33 (k) and TaWRKY42 (l). The ordinates are fold changes, and the horizontal ordinate is treatment time. The actin gene was used as an internal reference. The data are representative of three independent experiments
TaWRKY1 contained a 912 bp open reading frame (ORF) encoding a 303 amino acid protein of 32.41 kDa with pI 4.68. The ORF of TaWRKY33 was 1071 bp encoding a 38.8 kDa protein with pI 8.17. The predicted amino acid sequences of TaWRKY1 and TaWRKY33 possessed one WRKY domain with the highly conserved WRKYGQK motif, but two different deduced zinc finger motifs (C–X7–C–X23–H–X1–C and C–X5–C–X23–H–X1–H), respectively. TaWRKY1 contained an N-terminal CUT domain (amino acids 36 to 112) and a C-terminal NL domain (amino acids 271 to 292) according to SMART (Fig. 3a). TaWRKY33 contained an N-terminal basic region leucin zipper (BRLZ) domain (amino acids 40 to 94) and a C-terminal E-Z type HEAT Repeat (EZ_HEAT) domain (amino acids 314 to 345) (Fig. 3a). A four-stranded β-sheet with a zinc-binding pocket formed by conserved Cys/His residues was present in WRKY domains in the tertiary structures of TaWRKY1 and TaWRKY33 (Fig. 3b). We searched for WRKY homologies in NCBI using TaWRKY1 as a query. Amino acid sequence alignment showed that TaWRKY1 shared the highest identity (100 %) with AetWRKY70 (Aet07853) from the wild diploid Aegilops tauschii (2n = 14; DD), a progenitor of hexaploid wheat (T. aestivum; 2n = 6 × = 42; AABBDD) [50], suggesting that TaWRKY1 was located in a D-genome chromosome. No candidate with complete identity to TaWRKY33 was found in the genomic databases of A. tauschii and Triticum urartu (2n = 14; AA), the A-genome Progenitor. Therefore, TaWRKY33 might be located in a B chromosome.
Fig. 3.
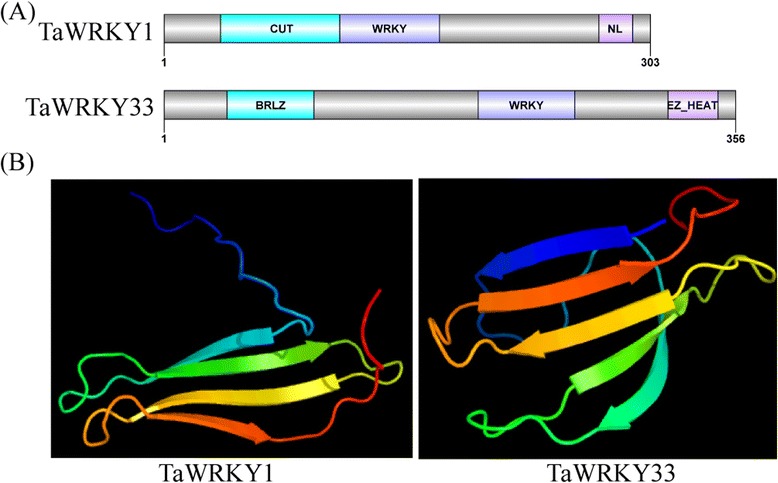
Domain organization (a) and tertiary structures (b) of TaWRKY1 and TaWRKY33
TaWRKY1 and TaWRKY33 were localized in the nucleus
To further investigate their biological activities TaWRKY1 and TaWRKY33 were fused to the N-terminus of the green fluorescent protein (GFP) reporter gene under control of the CaMV 35S promoter and transferred into wheat mesophyll protoplasts. The 35S::GFP vector was transformed as the control. Fluorescence of TaWRKY1-GFP and TaWRKY33-GFP were specifically detected in the nucleus, whereas fluorescence of the control GFP was distributed throughout the cells (Fig. 4). Therefore, TaWRKY1 and TaWRKY33 likely function in the nucleus.
Fig. 4.
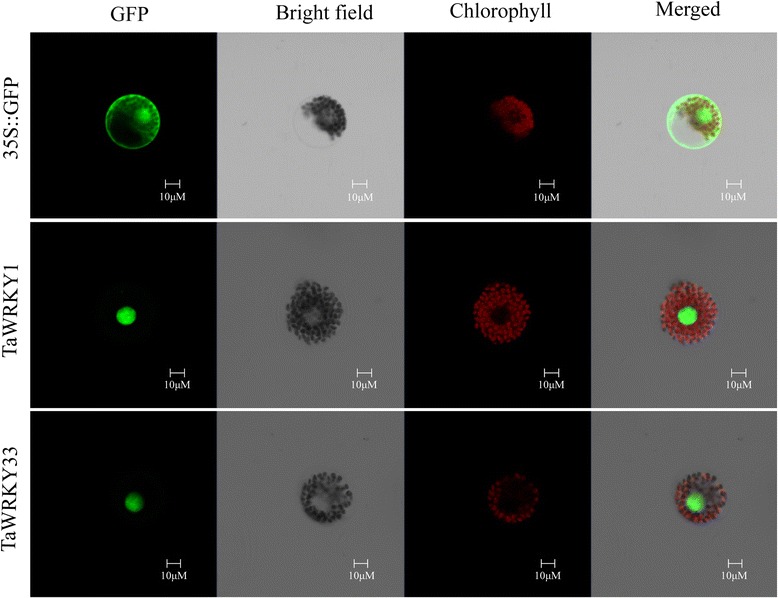
Subcellular localization of the TaWRKY1 and TaWRKY33 proteins. 35S::TaWRKY1-GFP, 35S::TaWRKY33-GFP and 35S::GFP control vectors were transiently expressed in wheat protoplasts. Scale bars = 10 μm
Stress-related regulatory elements in the TaWRKY1 and TaWRKY33 promoters
To gain further insight into the mechanism of transcriptional regulation we isolated 2.0 kb promoter regions upstream of the TaWRKY1 and TaWRKY33 ATG start codons. We searched for putative cis-acting elements in the promoter regions using the databases Plant Cis-acting Elements, and PLACE (http://www.dna.affrc.go.jp/PLACE/) (Tables 2 and 3). A number of regulatory elements responding to drought, salt, low-temperature and ABA were recognized in both promoters, including ABA-responsive elements (ABREs), dehydration-responsive elements (DREs), W-box elements, and MYB and MYC binding sequences. In addition, gibberellin responsive elements (GAREs) and several elicitor responsive elements (ELREs) were identified (Tables 2 and 3).
Table 2.
Putative cis-acting elements in the TaWRKY1 and TaWRKY33 promoters
| Gene | ABRE | CBFHV | CCAAT-Box | DRE | DRE/CRT | DPBF | ELRE | GARE | LTRE | MYB | MYC | PYR | W-box | WRKY |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Element | ||||||||||||||
| TaWRKY1 | 17 | 2 | 2 | 0 | 1 | 3 | 2 | 4 | 2 | 26 | 26 | 0 | 9 | 9 |
| TaWRKY33 | 9 | 5 | 3 | 2 | 2 | 3 | 4 | 2 | 4 | 24 | 8 | 3 | 23 | 16 |
Table 3.
Functions of elements in the TaWRKY1 and TaWRKY33 promoters
| Elements | Core sequence | Function |
|---|---|---|
| ABRE | ACGTG/ACGTSSSC/MACGYGB | ABA- and drought-responsive elements |
| CBFHV | RYCGAC | Drought- and cold-responsive elements |
| CCAAT-Box | CCAAT | Heat-responsive element |
| DRE | ACCGAGA/ACCGAC | ABA- and drought-responsive elements |
| DRE/CRT | RCCGAC | Drought-, high salt- and cold-responsive elements |
| DPBF | ACACNNG | Dehydration-responsive element |
| ELRE | TTGACC | Elicitor-responsive element |
| GARE | TAACAAR | GA-responsive element |
| LTRE | CCGAAA/CCGAC | Low-temperature responsive element |
| MYB | WAACCA/YAACKG/CTAACCA/CNGTTR/AACGG/TAACAAA/TAACAAA/MACCWAMC/CCWACC/GGATA | ABA- and drought-responsive elements |
| MYC | CATGTG/CANNTG | ABA- and drought-responsive elements |
| PYR | TTTTTTCC/CCTTTT | GA- and ABA-responsive elements |
| W-Box | TTTGACY/TTGAC/CTGACY/TGACY | SA-responsive element |
| WRKY | TGAC | Wound-responsive element |
Response mechanisms of TaWRKY1 and TaWRKT33 under abiotic stress
In order to clarify potential functions, the responses of TaWRKY1 and TaWRKY33 under various abiotic stress conditions were analyzed by qRT-PCR (Fig. 5). The TaWRKY1 gene was slightly induced by high-temperature and exogenous ABA at a maximum level of about three-fold. Transcription of TaWRKY1 was not affected by jasmonic acid methylester (MeJA), but was down-regulated by low-temperature.
Fig. 5.
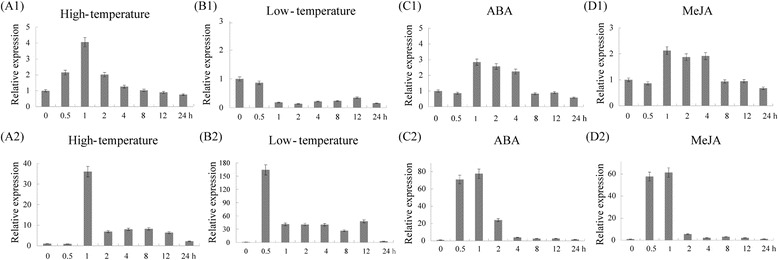
Expression patterns of TaWRKY1 (a1–d1) and TaWRKY33 (a2–d2) under abiotic stresses. The vertical ordinate is fold change; the horizontal ordinate is treatment time. The actin gene was used as an internal reference. The data are representative of three independent experiments
By comparison, TaWRKY33 rapidly responded to high-temperature, ABA and MeJA, with peak levels (more than 35-fold) occurring after one h of treatment. Low-temperature also activated transcription of TaWRKY33, with peak transcription levels earlier than those for drought, high-temperature, ABA and MeJA.
Improved drought and ABA tolerance and decreased rates of water loss in transgenic Arabidopsis
WRKY transcription factors might be involved in plant stress signaling [51–53]. TaWRKY1 and TaWRKY33 under the control of CaMV35S were transformed into Arabidopsis plants to further investigate their functions. Semi-quantitative RT-PCR was used to confirm transgenic Arabidopsis plants carrying TaWRKY1 and TaWRKY33 genes (Additional file 2: Figure S1A). Progenies from transgenic lines were used for analysis of seed germination under osmotic stress. There was no difference in seed germination between transgenic lines and WT plants grown on Murashige and Skoog (MS) media (Fig. 6a and d). In comparison more than 88.7 % of TaWRKY1 and TaWRKY33 transgenic seeds germinated in 4 % polyethylene glycol 6000 (PEG6000)-supplemented MS media after four days compared to 72.4 % for WT seeds (Fig. 6b and e). In 6 % PEG6000-supplemented MS media (Fig. 6c) TaWRKY1 transgenic seeds showed clear differences in germination rates compared to WT; nevertheless, TaWRKY33 transgenic lines had higher germination rates than TaWRKY1 transgenic lines and WT (Fig. 6f).
Fig. 6.
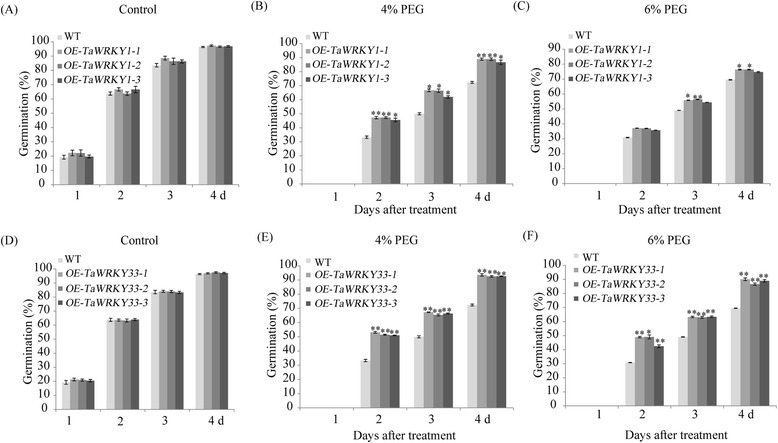
Germination of transgenic Arabidopsis lines under mock drought stress. Seed germinations of WT and TaWRKY1 transgenic Arabidopsis lines on MS medium with or without 4 and 6 % PEG6000 (a-c). Seed germinations of WT and TaWRKY33 transgenic Arabidopsis lines on MS medium with or without 4 and 6 % PEG6000 (d-f). Seeds were incubated at 4 °C for three days followed by 22 °C for germination. Seeds from three independent transgenic lines with TaWRKY1 and TaWRKY33 were grown on MS medium with or without 4 and 6 % PEG6000. WT seeds were grown in the same conditions as a control. Data are means ± SD of three independent experiments and * above the error bars or different letters above the columns indicate significant differences at p <0.05
ABA tolerance of TaWRKY1 and TaWRKY33 transgenic lines was identified by seed germination rates of Arabidopsis on MS media containing ABA. Average germination rates of TaWRKY1 transgenic lines were about 82 % compared to 75 % for WT in 0.5 μM ABA-supplemented MS media, meanwhile the germination rates of TaWRKY33 transgenic lines were higher than those of the TaWRKY1 transgenic lines and WT (Additional file 2: Figure S1C and S1F). Treated with 1 μM ABA, TaWRKY33 transgenic lines exhibited obviously higher seed germination rates than those of WT, and TaWRKY1 transgenic lines shared almost the same germination rates with WT (Additional file 2: Figure S1D and S1G).
Transgenic lines and WT Arabidopsis seeds were grown on MS medium for 5 days at 22 °C, and then transferred to MS medium containing 4 and 6 % PEG6000, respectively (Fig. 7 and Additional file 3: Figure S2). The TaWRKY1 and TaWRKY33 transgenic lines had similar phenotypes to WT seedlings under normal conditions. Total root lengths of the transgenic lines were longer than those of WT plants under both PEG6000 treatments after seven days, although PEG6000 stress reduced the growth of both transgenic and WT plants. TaWRKY33 significantly promoted root growth in transgenic lines compared with TaWRKY1 transgenic lines under PEG6000 treatment.
Fig. 7.
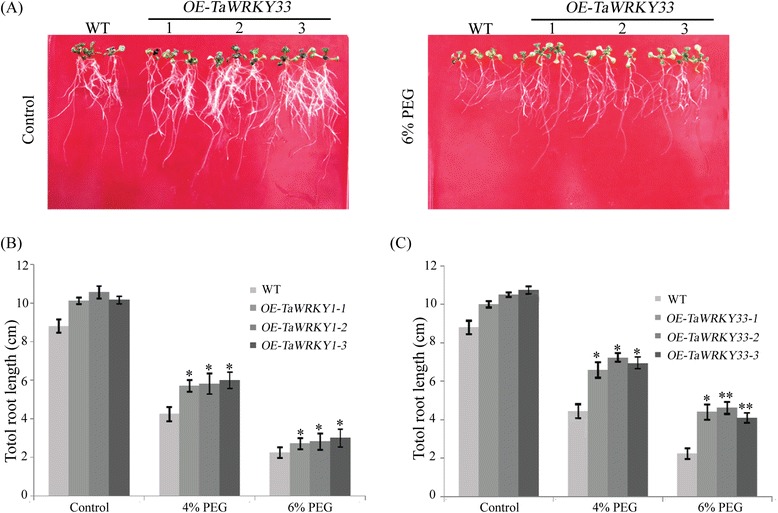
Total root lengths of transgenic Arabidopsis lines under mock drought stress. Phenotypes of WT and TaWRKY33 transgenic Arabidopsis seedlings under MS medium with or without 6 % PEG6000 (a). Root lengths of WT and TaWRKY1 transgenic Arabidopsis seedlings grown on MS medium with or without 4 and 6 % PEG6000 (b). Root lengths of WT and TaWRKY33 transgenic Arabidopsis seedlings grown on MS medium with or without 4 and 6 % PEG6000 (c). Five-day-old Arabidopsis seedlings were planted on MS medium with or without 4 and 6 % PEG6000 for seven days. Data are means ± SD of three independent experiments and * above the error bars or different letters above the columns indicate significant differences at p <0.05
The transgenic lines showed lower rates of water loss compared with WT plants during dehydration treatment (Fig. 8). For example, rates of water loss of the TaWRKY33 transgenics were less than 20.3 %, but TaWRKY1 transgenic lines and WT plants lost 22.1 and 27.8 % after two h of dehydration, respectively (Fig. 8). These results showed that TaWRKY33 transgenic lines had stronger water retaining capacity than WT plants.
Fig. 8.
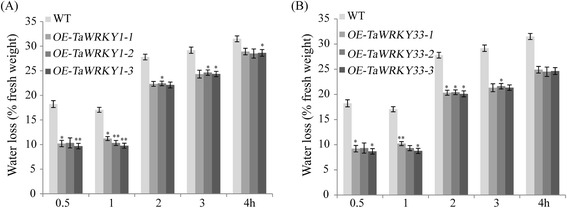
Determination of water loss of excised leaves from four-week-old Arabidopsis. The water loss of excised leaves of WT and TaWRKY1 transgenic Arabidopsis lines (a). The water loss of excised leaves of WT and TaWRKY33 transgenic Arabidopsis lines (b). Leaves at a similar stage from each line were used for the experiments. Data are means ± SD of three independent experiments and * above the error bars or different letters above the columns indicate significant differences at p <0.05
Enhanced thermotolerance of TaWRKY33 transgenic lines
Following earlier results on response to high-temperature (Fig. 5) the functions of transgenic lines under high-temperature stress were investigated (Fig. 9). TaWRKY33 transgenic lines exhibited high survival rates after exposure to 45 °C for five h, whereas TaWRKY1 transgenic lines showed no clear differences from WT (Fig. 9). The survival rates of the TaWRKY33 transgenic lines were more than 50 % after heat treatment compared to less than 30 % for TaWRKY1 transgenics and WT. This suggested that TaWRKY33 had a positive role in thermotolerance.
Fig. 9.
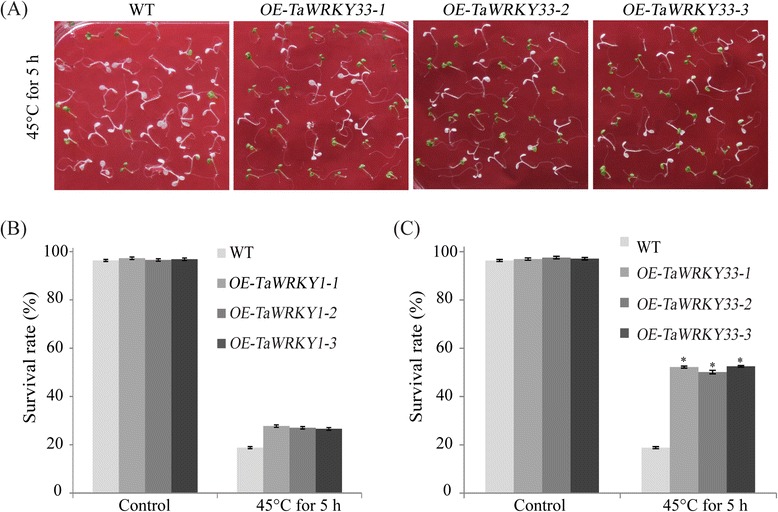
Analysis of transgenic Arabidopsis lines under heat stress. Phenotypes of WT and TaWRKY33 transgenic Arabidopsis lines under heat stress (a). Sruvival rates of WT and TaWRKY1 transgenic Arabidopsis lines under heat stress (b). Sruvival rates of WT and TaWRKY33 transgenic Arabidopsis lines under heat stress (c). Seeds from three independent transgenic lines of TaWRKY1 and TaWRKY33 were grown on MS medium. Five-day-old seedlings were heat-treated at 45 °C for five h before returning them to 22 °C to continue to grow vertically for 2 days. Data are means ± SD of three independent experiments and * above the error bars or different letters above the columns indicate significant differences at p <0.05
Changed transcripts of stress-responsive genes
TaWRKY1 and TaWRKY33 conferred stress tolerance in Arabidopsis. To investigate the tolerance mechanism we analyzed several stress-related genes possibly activated by TaWRKY1 and TaWRKY33. Compared to WT, transcripts of ABA1, ABA2, ABI1, ABI5 and RD29A were increased in TaWRKY1 transgenics whereas DREB2B expression was not significantly changed under normal conditions (Fig. 10a). Similarly, overexpression of TaWRKY33 regulated transcripts of ABA1, ABA2, ABI1, ABI5, DREB2B and RD29A, especially ABA2 and ABI5 to extremely high levels (Fig. 10b). As shown in Fig. 11, the LUC/REN ratio was increased significantly when the ABA2 and ABI5 pro-LUC reporter constructs were co-transfected with TaWRKY33, compared with the control that was co-transfected with the empty construct. These results indicated that overexpression of the TaWRKY1 and TaWRKY33 genes activated stress-responsive downstream genes.
Fig. 10.
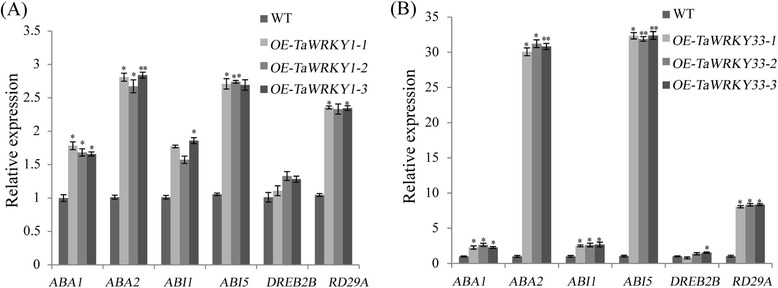
Expression levels of stress-responsive genes under regulation of TaWRKY1 (a) and TaWRKY33 (b). The vertical ordinates are fold changes, and the horizontal coordinates are gene names. Data are means ± SD of three independent experiments and * above the error bars or different letters above the columns indicate significant differences at p <0.05
Fig. 11.
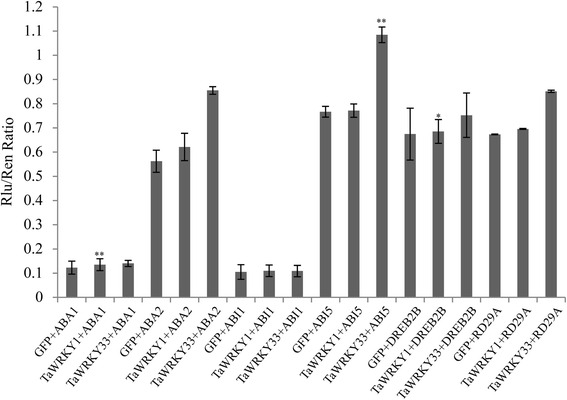
The activation of Arabidopsis promoters in transient Luciferase Assay. Co-operative activation of ABA1, ABA2, ABI1, ABI5, RD29A and DREB2B promoters from affected by TaWRKY1 and TaWRKY33 in a dual luciferase transient Arabidopsis transformation assays. Data are means ± SD of three independent experiments and * above the error bars or different letters above the columns indicate significant differences at p <0.05
Discussion
The functions of WRKYs have been extensively explored in various plant species over the past ten years, especially in Arabidopsis and rice. Little information existed about the role of wheat WRKYs in mediating abiotic responses. Recently, Sezer et al. characterized 160 TaWRKYs according to sequence similarity, motif type and phylogenetic relationships, improving knowledge of WRKYs in wheat [54]. In the present study, 48 putative drought-responsive WRKY genes were identified from de novo transcriptome sequencing data of drought-treated wheat. The phylogenic tree revealed that most drought-responsive WRKYs belonged to Groups II and III (Fig. 1). Recent investigations showed that most WRKYs in these groups function in drought tolerance in many plant species. For example, WRKY63/ABO3, belonging to Group III, mediated responses to ABA and drought tolerance in Arabidopsis [55]. Similarly, AtWRKY57 and GmWRKY54, which were identified as group II, were induced by drought and their expression conferred drought tolerance in Arabidopsis [48, 56]. In the present study TaWRKY1 and TaWRKY33, members of Groups II and III, conferred drought tolerance in Arabidopsis (Figs. 6, 7, 8 and 9). Therefore, it was supposed that WRKYs in these groups might be involved in drought stress response.
WRKYs are important in many aspects of plant defense, including MAMP- (MTI) or PAMP-triggered (PTI) immunity, effector-triggered immunity (ETI) and systemin acquired resistance [56–64]. Increasing evidence shows that WRKYs are activated not only by disease-related stimuli and pathogen infection, but also by multiple abiotic stresses [17, 18, 52]. For example, 10 of 13 rice and 8 of 15 wheat WRKY genes responded to PEG6000, salt, cold or heat stresses [65, 66]. TaWRKY44 may act as a positive regulator in drought, salt and osmotic stress responses [67]. Overexpression of GhWRKY25 conferred tolerance to salt stress in tobacco [68]. In the present study, except for drought response, TaWRKY33 was involved in strong responses to high- and low-temperature and ABA, possibly related to cis-elements in the promoter (Tables 2 and 3). For instance, the TaWRKY33 promoter contained multiple ABRE and LTRE elements that might be responsible for low-temperature and exogenous ABA. The ELRE might induce large responses of TaWRKY33 to abiotic stresses. In addition, TaWRKY33 was highly induced by MeJA although there is no MeJA-related element. This could be the reason why MeJA-related elements had not been identified previously.
ABA is regarded to play a crucial role in plant abiotic stress response and development and is considered to be a negative regulator of biotrophic pathogen resistance [27, 69]. It has been reported that ABA-dependent and ABA-independent pathways exist in stress response [67]. DREB2s play important roles in ABA-independent pathway and often as marker genes in stress responses [70]. A number of transcription factors and their target genes are involved in mediating ABA signal transduction and have been shown to regulate many molecular and cellular responses [71]. Previous studies show that ABI1/2 and AtWRKY40 are key regulatory components of ABA receptors RCARs and ABAR, respectively. ABI5, a positive regulator of ABA signaling, exists in the downstream of ABI1/2 and AtWRKY40. They are key players in ABA signal transduction and act by negatively regulating ABA response. ABA synthesis genes ABA1 and ABA2 were both detected in these studies, implying an acceleration of ABA production. Consistent with that, transcript abundance of ABI5 also increased (Fig. 10), demonstrating that TaWRKY33 likely increased the level of drought tolerance by increasing traffic through the ABA synthesis and transduction pathways.
It was reported that RD29A was induced by dehydration, low-temperature, high salinity or exogenous ABA. The promoter region of RD29A contains the cis-acting DRE that is involved in expression of RD29A rapidly responding to dehydration and high salinity stresses in Arabidopsis. Here, RD29A was up-regulated in TaWRKY33 transgenic lines (Fig. 10), suggesting that TaWRKY33 acts as a positive regulator in hyperosmotic stress response in Arabidopsis. These studies collectively demonstrated that TaWRKY33 might play a key role in ABA- and drought-responsive signaling networks. Overexpression of wheat TaWRKY2 enhanced STZ expression, whereas TaWRKY19 promoted DREB2A-mediated activation of RD29A, RD29B and Cor6.6, resulting in tolerance to salt and drought in transgenic plants [49]. Therefore, wheat WRKYs affected stress tolerance through regulation of different downstream genes. Taken together, a model was proposed in which TaWRKY1 and TaWRKY33 transcription was activated under abiotic stress (Additional file 4: Figure S3). The MeJA-mediated signaling pathway is relevant to resistance to necrotrophic pathogens, wounding and insect herbivores [72–74]. The ABA and the JA could jointly modulate stress-related gene expression despite antagonistic interactions between the ABA and the JA/ET signaling pathways, they also [75]. In the present study, TaWRKY33 was moderately and highly responsive to ABA and MeJA, respectively (Fig. 5). These results indicated that TaWRKY33 could coordinately integrate the ABA and the MeJA pathways, but not antagonize them. Therefore, we speculate that TaWRKY33 might have roles in interaction of the ABA and MeJA signaling pathways and might be related to both abiotic stress tolerance and disease responses in plants.
Conclusions
Forty-eight putative drought-responsive WRKY genes were identified from de novo transcriptome sequencing data of drought-treated wheat. They were classified into three groups, according to sequence similarity and motif identity. TaWRKY1 and TaWRKY33, belonging to Groups II and III, were selected for further investigation. Both TaWRKY1 and TaWRKY33 responded to multiple stresses. Overexpression of TaWRKY1 and TaWRKY33 activated several stress-related downstream genes, increased germination rates and promoted root growth in Arabidopsis under stresses. These studies provide candidate genes for future functional analysis of TaWRKYs involved in the drought- and heat-related signal pathways in wheat.
Methods
De novo sequencing of drought-treated wheat
Total RNA was isolated using TRIzol reagent (Invitrogen) and treated with RNase free DNase I (Qiagen). Poly (A) mRNA was purified from total RNA using oligo (dT) magnetic beads and fragmented into small pieces using divalent cations. First-strand cDNA was generated using reverse transcriptase and random primers. This was followed by synthesis of the second-strand cDNA. Then, single-end and paired-end RNA-seq libraries were prepared following Illumina’s protocols and sequenced on the Illumina GA II platform [76, 77].
De novo assembly of the short reads was performed using SOAPdenovo software (http://soap.genomics.org.cn), which adopts the de Bruijn graph data structure that is sensitive to the sequencing error to construct contigs [78]. According to the overlap information in the short reads, the reads were then realigned to the contig sequence with high coverage, and the paired-end relationship between reads was transferred to linkage between contigs. Unreliable linkages between two contigs were filtered and the remaining contigs with compatible connections to each other, and having at least three read-pairs, were constructed into scaffolds. We constructed scaffolds starting with short paired-ends and then iterated the scaffolding process, step by step, using longer insert size paired-ends. To fill the intra-scaffold gaps, we used paired-end information to retrieve read pairs that had one read with one end mapped to the contigs and another read located in the gap region, and then did a local assembly with the unmapped end to extend the contig sequence in the small gaps in the scaffolds.
Gene expression profiling was measured by mapping reads to assembled sequences using SOAP [79]. The most widely used approach is to count uniquely mapped reads. Then the FPKM value for each transcript was measured in Fragments Per kb per Million fragments [80]. We then used the False Discovery Rate (FDR) method to determine the threshold of the p-value in multiple tests. FDR ≤ 0.001 and a relative change threshold of two-fold were used to judge the significance of differentiated gene expression. The analysis firstly maps all differentially expressed genes (DEGs) to GO terms in the database by virtue of calculating gene numbers for every term, followed by an ultra-geometric test to find significantly enriched GO terms in DEGs compared to the transcriptome background. The calculated p- value was subjected to a Bonferroni Correction, taking a corrected p-value of 0.05 as a threshold. GO terms fulfilling this condition were defined as significantly enriched GO terms in DEGs [81]. For pathway enrichment analysis, we mapped all DEGs to terms in KEGG database.
Plant materials and stress treatments
Wheat (T. aestivum cv. Xiaobaimai) seedlings which were provided by Dr Rui-Lian Jing (Institute of Crop Science, Chinese Academy of Agricultural Sciences) were grown in Hoagland’s liquid medium at 22 °C under a 16 h light/8 h darkness photoperiod. Ten-day-old seedlings were used for dehydration, high-temperature, low-temperature, MeJA and ABA treatments. Seedlings on filter paper were exposed to air for induction of rapid drought conditions, or placed in 4 and 42 °C chambers for low and high-temperature treatments, respectively. For dehydration treatment, seedlings were transferred to filter paper and dried at 25 °C under normal conditions. For MeJA and ABA treatments, seedling roots were immersed in solutions containing 100 μM MeJA and 100 μM ABA, respectively. The samples were harvested at 0, 0.5, 1, 2, 4, 8, 12 and 24 h.
RNA extraction and qRT-PCR analyses
Total RNA was extracted using Trizol reagent according to the manufacturer’s protocol (TIANGEN, China) and treated with DNase I (TaKaRa, Japan) to remove genomic DNA contamination. First strand cDNA was synthesized using a PrimeScript First-Strand cDNA Synthesis Kit (TaKaRa) following the manufacturer’s instructions. qRT-PCR was conducted using an ABI Prism 7500 system (Applied Biosystems, Foster City, CA). The actin gene was used as an internal control for normalization of template cDNA. Each PCR was repeated three times in total volumes of 20 μl containing 2 × Taq PCR Master Mix (TIANGEN). Validation experiments were performed to demonstrate that amplification efficiencies of the TaWRKY1- and TaWRKY33-specific primers were approximately equal to the amplification efficiency of the endogenous reference primers. Quantitative and data analyes were performed as previously described [82].
Gene isolation and sequence analysis
Open reading frames of TaWRKY1 (Genbank No. KT285206) and TaWRKY33 (Genbank No. KT285207) were amplified by PCR using specific primers. PCR products were cloned into pEASY-T1 vectors (TransGen, China) and sequenced with an ABI 3730XL 96-capillary DNA analyzer (Lifetech, America).
Maximum likelihood was used to construct phylogenetic trees by the MEGA5.1 program, and the confidence levels of monophyletic groups were estimated using bootstrap analyses of 1000 replicates [83].
Predicted protein domains of TaWRKY1 and TaWRKY33 were identified by the SMART tool (http://smart.embl-heidelberg.de/), and their tertiary structures were obtained using the Phyre2 tool (http://www.sbg.bio.ic.ac.uk/phyre2).
Plasmid construction for subcellular localization analysis
The open reading frames of TaWRKY1 and TaWRKY33 were inserted into N-terminal GFP protein driven by the CaMV 35S promoter of subcellular localization vector p16318 [83]. For transient expression assays, mesophyll protoplasts were isolated, transfected with p16318::TaWRKY1 and p16318::TaWRKY33, and GFP fluorescence signals were observed with a confocal laser scanning microscope (Nikon, Japan). FM4-64 dye (Molecular Probes, Carlsbad, CA) was excitated at 543 nm and fluorescence was recorded using a 650 nm long pass filter. All transient expression experiments were repeated three times [84].
Transient luciferase assay in Arabidopsis
For the analysis of transcription activities of TaWRKY1 and TaWRKY33 in response to ABA1, ABA2, ABI1, ABI5, RD29A and DREB2B promoters, the 2.5 kb promoter regions were cloned into the transient expression reporter vector pGreenII 0800-LUC which contains the CaMV 35S promoter-REN cassette and the promoterless-LUC cassette, respectively [85, 86]. The TaWRKY1 and TaWRKY33 genes were cloned into N-terminal GFP protein driven by the CaMV 35S promoter. The constructed effectors and reporter plasmids were transfected into mesophyll protoplasts of Arabidopsis Columbia-0 which were collected by our own laboratory. Transfected protoplasts were incubated in darkness at 22 °C. Firefly luciferase and renilla luciferase were assayed using the dual luciferase assay reagents (Promega, USA). Data was collected as the ratio of LUC/REN. All transient expression experiments were repeated three times.
Generation, and stress treatments of transgenic Arabidopsis
The coding sequences of TaWRKY1 and TaWRKY33 were cloned into pBI121 under control of the CaMV 35S promoter, resulting in 35S::TaWRKY1 and 35S::TaWRKY33 constructs. These constructs were confirmed by sequencing and then separately used in transformation mediated by Agrobacterium (Agrobacterium tumefaciens) to obtain three transgenic Arabidopsis lines. Kanamycin-resistant Arabidopsis transformants carrying TaWRKY1 and TaWRKY33 were generated using the vacuum infiltration method [86]. Transformed plants were cultured on MS medium containing 0.8 % agar and 50 mM Kanamycin in a day/night regime of 16/8 h under white light (with 50 photons m-1 s-1) at 22 °C for 2 weeks and then transferred to soil.
Homozygous T3 seeds of transgenic lines were used for phenotypic analysis. Arabidopsis seeds were grown on 10 × 10 cm MS agar plates that were routinely kept for three days in darkness at 4 °C to break dormancy and transferred to a tissue culture room under a day/night regime of 16/8 h under white light (with 50 photons m-1 s-1) at 22 °C for five days. For the germination assay, seeds were subjected to 4 or 6 % (w/v) PEG6000, and 0.5 or 1 μM ABA treatments. For drought treatment, 5-day-old seedlings were transferred to MS agar plates containing 4 and 6 % PEG6000 for seven days. Total root lengths of the Arabidopsis plants were measured [87]. Five-day-old seedlings were heat-treated at 45 °C for five h before returning them to 22 °C to continue to grow vertically for two days. Seeds were considered germinated when radicles had completely emerged from the seed coat. All measurements were repeated three times.
Abbreviations
ABA, abscisic acid; ABRE, ABA-responsive element; DEGs, differentially expressed genes; DRE, dehydration-responsive elements; ELRE, elicitor responsive element; FDR, false discovery rate; GFP, green fluorescent protein; MeJA, jasmonic acid methylester; PEG6000, polyethylene glycol 6000; qRT-PCR, quantitative real-time PCR; WT, wild type.
Acknowledgments
We thank Dr. Dongying Gao (Department of Plant Sciences, University of Georgia, USA) for suggestions on the manuscript. We acknowledge Beijing Genomics Institute (BGI) at Shenzhen for the transcriptome sequencing.
Funding
This research was financially supported by the National Natural Science Foundation of China (31371620) and the National Transgenic Key Project of the Ministry of Agriculture (2014ZX08009-016B).
Availability of data and material
The transcriptome data was available in the Sequence Read Achive (SRA) under accession number SRP071191. Nucleic acid sequences of WRKYs in wheat have been deposited in NCBI GenBank under accessions number KU892127 to KU892155 and KU892157 to KU892167, respectively. The accessions number of TaWRKY1 and TaWRKY33 were Genbank No. KT285206 and Genbank No. KT285207, respectively.
Authors’ contributions
ZSX coordinated the project, conceived and designed experiments, and edited the manuscript; GYH conducted the bioinformatic work, performed experiments and wrote the first draft; JYX performed experiments; YXW provided analytical tools; JML and PSL generated and analyzed data; MC managed reagents; YZM contributed with valuable discussions. All authors have read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent to publish
This research is not applicable to the consent for publication.
Ethics
This research is not applicable to the ethics approval and consent.
Additional files
Nucleic acid sequences of drought-induced responsive WRKY genes in wheat. (XLSX 24 kb)
Detection of the expression levels of TaWRKY1 and TaWRKY33 transgenic Arabidopsis lines (A). Germination of transgenic Arabidopsis lines under ABA stress (B-G). (TIF 527 kb)
Phenotypes of TaWRKY1 transgenics. (TIF 928 kb)
Probable modes of action of TaWRKY1 and TaWRKY33. (TIF 228 kb)
References
- 1.Xu ZS, Chen M, Li LC, Ma YZ. Functions of the ERF transcription factor family in plants. Botany. 2008;86:969–77. doi: 10.1139/B08-041. [DOI] [Google Scholar]
- 2.Chinnusamy V, Schumaker K, Zhu JK. Molecular genetics perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot. 2004;55:225–36. doi: 10.1093/jxb/erh005. [DOI] [PubMed] [Google Scholar]
- 3.Rajendra B, Jones JD. Role of plant hormones in plant defence responses. Plant Mol Biol. 2009;69:473–88. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 4.Xu ZS, Chen M, Li LC, Ma YZ. Functions and application of the AP2/ERF transcription factor family in crop improvement. J Integr Plant Biol. 2011;53:570–85. doi: 10.1111/j.1744-7909.2011.01062.x. [DOI] [PubMed] [Google Scholar]
- 5.Mukhopadhyay A, Vij S, Tyagi AK. Overexpression of zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgentic tobacco. Proc Natl Acad Sci U S A. 2004;101:6309–14. doi: 10.1073/pnas.0401572101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu ZS, Ni ZY, Liu L, Nie LN, Li LC, Chen M, Ma YZ. Characterization of the TaAIDFa gene encoding a CRT/DRE-binding factor responsive to drought, high-salt, and cold stress in wheat. Mol Genet Genomics. 2008;280:497–508. doi: 10.1007/s00438-008-0382-x. [DOI] [PubMed] [Google Scholar]
- 7.Guo RY, Yu FF, Gao Z, An HL, Cao XC, Guo XQ. GhWRKY3, a novel cotton (Gossypium hirsutum L.) WRKY gene, is involved in diverse stress responses. Mol Biol Rep. 2011;38:49–58. doi: 10.1007/s11033-010-0076-4. [DOI] [PubMed] [Google Scholar]
- 8.Yang SJ, Vanderbeld B, Wan JX, Huang YF. Narrowing down the targets: toward successful genetic engineering of drought-tolerant crops. Mol Plant. 2010;3:469–90. doi: 10.1093/mp/ssq016. [DOI] [PubMed] [Google Scholar]
- 9.Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006;18:1292–309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li PS, Yu TF, He GH, Chen M, Zhou YB, Chai SC, Xu ZS, Ma YZ. Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genomics. 2014;15:1009. doi: 10.1186/1471-2164-15-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 12.Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996;15:5690–700. [PMC free article] [PubMed] [Google Scholar]
- 13.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15:247–58. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Wang L. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol. 2005;5:1–12. doi: 10.1186/1471-2148-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, Shen QJ. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005;137:176–89. doi: 10.1104/pp.104.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mangelsen E, Kilian J, Berendzen KW, Kolukisaoglu UH, Harter K, Jansson C, Wanke D. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics. 2008;9:194. doi: 10.1186/1471-2164-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang B, Jiang Y, Rahman MH, Deyholos MK, Kav NN. Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol. 2009;9:68. doi: 10.1186/1471-2229-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Song Y, Li S, Zhang L, Zou C, Yu D. The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta. 1819;2012:120–8. doi: 10.1016/j.bbagrm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Schiff M, Dinesh-Kumar SP. Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J. 2004;38:800–9. doi: 10.1111/j.1365-313X.2004.02085.x. [DOI] [PubMed] [Google Scholar]
- 20.Fan H, Wang F, Gao H, Wang L, Xu J, Zhao Z. Pathogen induced MdWRKY1 in ‘Qinguan’ apple enhances disease resistance. J Plant Biol. 2011;54:150–8. doi: 10.1007/s12374-011-9151-1. [DOI] [Google Scholar]
- 21.Lim JH, Park CJ, Huh SU, Choi LM, Lee GJ, Kim YJ. Capsicum annuum WRKYb transcription factor that binds to the CaPR-10 promoter functions as a positive regulator in innate immunity upon TMV infection. Biochem Biophys Res Commun. 2011;411:613–9. doi: 10.1016/j.bbrc.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Peng XX, Hu YJ, Tang XK, Zhou PL, Deng XB, Wang HH, Guo ZJ. Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta. 2012;236:1485–98. doi: 10.1007/s00425-012-1698-7. [DOI] [PubMed] [Google Scholar]
- 23.Hsu FC, Chou MY, Chou SJ, Li YR, Peng HP, Shih MC. Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. Plant Cell. 2013;25:2699–713. doi: 10.1105/tpc.113.114447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chujo T, Miyamoto K, Ogawa S, Masuda Y, Shimizu T, Kishi-Kaboshi M, Takahashi A, Nishizawa Y, Minami E, Nojiri H, Yamane H, Okada K. Overexpression of phosphomimic mutated OsWRKY53 leads to enhanced blast resistance in rice. PLoS One. 2014;9:e98737. doi: 10.1371/journal.pone.0098737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Fu Q, Huang W, Yu D. Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep. 2009;28:683–693. doi: 10.1007/s00299-008-0666-y. [DOI] [PubMed] [Google Scholar]
- 26.Dong J, Chen C, Chen Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol. 2003;51:21–37. doi: 10.1023/A:1020780022549. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Feng JC. Identification and characterization of the grape WRKY family. Biomed Res Int. 2014;2014:787680. doi: 10.1155/2014/787680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang SH, Yie SW, Hwanga DJ. Heterologous expression of OsWRKY6 gene in Arabidopsis activates the expression of defense related genes and enhances resistance to pathogens. Plant Sci. 2011;181:316–23. doi: 10.1016/j.plantsci.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 29.Huang T, Duman JG. Cloning and characterization of athermal hysteresis (antifreeze) protein with DNA-binding activity from winter bittersweet nightshade, Solanum dulcamara. Plant Mol Biol. 2002;48:339–50. doi: 10.1023/A:1014062714786. [DOI] [PubMed] [Google Scholar]
- 30.Pnueli L, Hallak-Herr E, Rozenberg M, Cohen M, Goloubinoff P, Kaplan A, Mittler R. Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. Plant J. 2002;31:319–30. doi: 10.1046/j.1365-313X.2002.01364.x. [DOI] [PubMed] [Google Scholar]
- 31.Rizhsky L, Liang H, Mittler R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 2002;130:1143–51. doi: 10.1104/pp.006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mare C, Mazzucotelli E, Crosatti C, Francia E, Stanca AM, Cattivelli L. Hv-WRKY38: a new transcription factor involved in cold- and drought-response in barley. Plant Mol Biol. 2004;55:399–416. doi: 10.1007/s11103-004-0906-7. [DOI] [PubMed] [Google Scholar]
- 33.Zou X, Seemann JR, Neuman D, Shen QJ. A WRKY gene from creosote bush encodes an activator of the abscisic acid signaling pathway. J Biol Chem. 2004;279:55770–9. doi: 10.1074/jbc.M408536200. [DOI] [PubMed] [Google Scholar]
- 34.Yan Y, Jia H, Wang F, Wang C, Liu S, Guo X. Overexpression of GhWRKY27a reduces tolerance to drought stress and resistance to Rhizoctonia solani infection in transgenic Nicotiana benthamiana. Front Physiol. 2015;6:265. doi: 10.3389/fphys.2015.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X, Shiroto Y, Kishitani S, Ito Y, Toriyama K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009;28:21–30. doi: 10.1007/s00299-008-0614-x. [DOI] [PubMed] [Google Scholar]
- 36.Li S, Fu Q, Chen L, Huang W, Yu D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta. 2011;233:1237–52. doi: 10.1007/s00425-011-1375-2. [DOI] [PubMed] [Google Scholar]
- 37.Qin Y, Tian Y, Han L, Yang X. Constitutive expression of a salinity-induced wheat WRKY transcription factor enhances salinity and ionic stress tolerance in transgenic Arabidopsis thaliana. Biochem Biophys Res Commun. 2013;441:476–81. doi: 10.1016/j.bbrc.2013.10.088. [DOI] [PubMed] [Google Scholar]
- 38.Yan H, Jia H, Chen X, Hao L, An H, Guo X. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signalling and the modulation of reactive oxygen species production. Plant Cell Physiol. 2014;55:2060–76. doi: 10.1093/pcp/pcu133. [DOI] [PubMed] [Google Scholar]
- 39.Jiang Y, Duan Y, Yin J, Ye S, Zhu J, Zhang F, Lu W, Fan D, Luo K. Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J Exp Bot. 2014;65:6629–44. doi: 10.1093/jxb/eru381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kayum MA, Jung HJ, Park JI, Ahmed NU, Saha G, Yang TJ, Nou IS. Identification and expression analysis of WRKY family genes under biotic and abiotic stresses in Brassica rapa. Mol Genet Genomics. 2014;290:79–95. doi: 10.1007/s00438-014-0898-1. [DOI] [PubMed] [Google Scholar]
- 41.Wang S, Wang J, Yao W, Zhou B, Li R, Jiang T. Expression patterns of WRKY genes in di-haploid Populus simonii × P. nigra in response to salinity stress revealed by quantitative real-time PCR and RNA sequencing. Plant Cell Rep. 2014;33:1687–96. doi: 10.1007/s00299-014-1647-y. [DOI] [PubMed] [Google Scholar]
- 42.Seki M, Umezawa T, Urano K, Shinozaki K. Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol. 2007;10:296–302. doi: 10.1016/j.pbi.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 43.Ma T, Li M, Zhao A, Xu X, Liu G, Cheng L. LcWRKY5: an unknown function gene from sheepgrass improves drought tolerance in transgenic Arabidopsis. Plant Cell Rep. 2014;33:1507–18. doi: 10.1007/s00299-014-1634-3. [DOI] [PubMed] [Google Scholar]
- 44.Ding ZJ, Yan JY, Xu XY, Yu DQ, Li GX, Zhang SQ, Zheng SJ. Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 2014;79:13–27. doi: 10.1111/tpj.12538. [DOI] [PubMed] [Google Scholar]
- 45.Song Y, Jing S, Yu D. Overexpression of the stress-induced OsWRKY08 improves osmotic stress tolerance in Arabidopsis. Chin Sci Bull. 2009;54:4671–8. [Google Scholar]
- 46.Zhang JS, Chen SY. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 2015;83:224–36. doi: 10.1111/tpj.12911. [DOI] [PubMed] [Google Scholar]
- 47.Shekhawat UK, Ganapathi TR. MusaWRKY71 overexpression in banana plants leads to altered abiotic and biotic stress responses. PLoS One. 2013;8:e75506. doi: 10.1371/journal.pone.0075506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang Y, Liang G, Yu D. Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol Plant. 2012;5:1375–88. doi: 10.1093/mp/sss080. [DOI] [PubMed] [Google Scholar]
- 49.Niu CF, Wei W, Zhou QY, Tian AG, Hao YJ, Zhang WK, Ma B, Lin Q, Zhang ZB, Zhang JS, Chen SY. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012;35:1156–70. doi: 10.1111/j.1365-3040.2012.02480.x. [DOI] [PubMed] [Google Scholar]
- 50.Salamini F, Ozkan H, Brandolini A, Schäfer-Pregl R, Martin W. Genetics and geography of wild cereal domestication in the near east. Nature Rev Genet. 2002;3:429–41. doi: 10.1038/nrg817. [DOI] [PubMed] [Google Scholar]
- 51.Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315:1098–103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- 52.Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, Zhang XF, Zhao R, Sun HL, Liu R, Yu YT, Zhang DP. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA responsive genes of inhibition. Plant Cell. 2010;22:1909–35. doi: 10.1105/tpc.110.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, Langum TJ, Smidt L, Boomsma DD, Emme NJ, Chen X, Finer JJ, Shen QJ, Rushton PJ. WRKY transcription factors: key components in abscisic acid signaling. Plant Biotechnol J. 2012;10:2–11. doi: 10.1111/j.1467-7652.2011.00634.x. [DOI] [PubMed] [Google Scholar]
- 54.Okay S, Derelli E, Unver T. Transcriptome-wide identification of bread wheat WRKY transcription factors in response to drought stress. Mol Genet Genomics. 2014;289:765–81. doi: 10.1007/s00438-014-0849-x. [DOI] [PubMed] [Google Scholar]
- 55.Ren XZ, Chen ZZ, Liu Y, Zhang HR, Zhang M, Liu Q, Hong XH, Zhu JK, Gong ZZ. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010;63:417–29. doi: 10.1111/j.1365-313X.2010.04248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, Wang CM, Wang HW, Zhang JS, Chen SY. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J. 2008;6:486–503. doi: 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- 57.Xu X, Chen C, Fan B, Chen Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell. 2006;18:1310–26. doi: 10.1105/tpc.105.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol. 2007;10:366–71. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 59.Encinas-Villarejo S, Maldonado AM, Amil-Ruiz F, Santos B, Romero F, Pliego-Alfaro F, Muñoz-Blanco J, Caballero JL. Evidence for a positive regulatory role of strawberry (Fragaria × ananassa) FaWRKY1 and Arabidopsis AtWRKY75 proteins in resistance. J Exp Bot. 2009;60:3043–65. doi: 10.1093/jxb/erp152. [DOI] [PubMed] [Google Scholar]
- 60.Jing S, Zhou X, Song Y, Yu D. Heterologous expression of OsWRKY23 gene enhances pathogen defense and dark-induced leaf senescence in Arabidopsis. Plant Growth Regul. 2009;58:181–90. doi: 10.1007/s10725-009-9366-z. [DOI] [Google Scholar]
- 61.Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150:1648–55. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tao Z, Liu H, Qiu D, Zhou Y, Li X, Xu C, Wang S. A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol. 2009;151:936–48. doi: 10.1104/pp.109.145623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abbruscato P, Nepusz T, Mizzi L, Del Corvo M, Morandini P, Fumasoni I, Michel C, Paccanaro A, Guiderdoni E, Schaffrath U, Morel JB, Piffanelli P, Faivre-Rampant O. OsWRKY22, a monocot WRKY gene, plays a role in the resistance response to blast. Mol Plant Pathol. 2012;13:828–41. doi: 10.1111/j.1364-3703.2012.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ishihama N, Yoshioka H. Post-translational regulation of WRKY transcription factors in plant immunity. Curr Opin Plant Biol. 2012;15:431–7. doi: 10.1016/j.pbi.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 65.Qiu Y, Jing S, Fu J, Li L, Yu D. Cloning and analysis of expression profiles of 13 WRKY genes in rice. Chin Sci Bull. 2004;49:2159–68. [Google Scholar]
- 66.Wu HL, Ni ZF, Yao YY, Guo GG, Sun QX. Cloning and expression profiles of 15 genes encoding WRKY transcription factor in wheat (Triticum aestivem L.) Prog Nat Sci. 2008;18:697–705. doi: 10.1016/j.pnsc.2007.12.006. [DOI] [Google Scholar]
- 67.Wang X, Zeng J, Li Y, Rong X, Sun J, Sun T, Li M, Wang L, Feng Y, Chai R, Chen M, Chang J, Li K, Yang G, He G. Expression of TaWRKY44, a wheat WRKY gene, in transgenic tobacco confers multiple abiotic stress tolerances. Front Plant Sci. 2015;6:615. doi: 10.3389/fpls.2015.00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X, Song Y, Xing F, Wang N, Wen F, Zhu C. GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana. Protoplasma. 2015. Epub ahead of print, doi:10.1007/s00709-015-0885-3. [DOI] [PubMed]
- 69.Chinnusamy V, Gong ZZ, Zhu JK. Abscisic acid-mediated epigenetic processes in plant development and stress responses. J Integr Plant Biol. 2008;50:1187–95. doi: 10.1111/j.1744-7909.2008.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–7. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 71.Luo X, Bai X, Sun XL, Zhu D, Liu BH, Ji W, Cai H, Cao L, Wu J, Hu MR, Liu X, Tang LL, Zhu YM. Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signaling. J Exp Bot. 2013;8:2155–69. doi: 10.1093/jxb/ert073. [DOI] [PubMed] [Google Scholar]
- 72.Robert-Seilaniantz A, Navarro L, Bari R, Jones DG. Pathological hormone imbalances. Curr Opin Plant Biol. 2007;10:372–9. doi: 10.1016/j.pbi.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 73.Kessler A, Baldwin IT. Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol. 2002;53:299–328. doi: 10.1146/annurev.arplant.53.100301.135207. [DOI] [PubMed] [Google Scholar]
- 74.Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD. Systemic acquired resistance. Plant Cell. 1996;8:1809–19. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006;9:436–42. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 76.Chen S, Yang P, Jiang F, Wei Y, Ma Z, Kang L. De novo analysis of transcriptome dynamics in the migratory locust during the development of phase traits. PLoS One. 2010;5:e15633. doi: 10.1371/journal.pone.0015633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shen GM, Dou W, Niu JZ, Jiang HB, Yang WJ, Jia FX, Hu F, Cong L, Wang JJ. Transcriptome analysis of the oriental fruit fly (Bactrocera dorsalis) PLoS One. 2011;6:e29127. doi: 10.1371/journal.pone.0029127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pevzner PA, Tang H, Waterman MS. An Eulerian path approach to DNA fragment assembly. Proc Natl Acad Sci U S A. 2001;98:9748–53. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–7. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 80.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 81.Liu B, Jiang GF, Zhang YF, Li JL, Li XJ, Yue JS, Chen F, Liu HQ, Li HJ, Zhu SP, Wang JJ, Ran C. Analysis of transcriptome differences between resistant and susceptible strains of the citrus Red mite Panonychus citri (Acari: Tetranychidae) PLoS One. 2011;6:e28516. doi: 10.1371/journal.pone.0028516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu P, Xu ZS, Pan-Pan L, Hu D, Chen M, Li LC, Ma YZ. A wheat plasma membrane-localized PI4K gene possessing threonine autophophorylation activity confers tolerance to drought and salt in Arabidopsis. J Exp Bot. 2013;64:2915–27. doi: 10.1093/jxb/ert133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu ZS, Xia LQ, Chen M, Cheng XG, Zhang RY, Li LC, Zhao YX, Lu Y, Ni ZY, Liu L, Qiu ZG, Ma YZ. Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol Biol. 2007;65:719–32. doi: 10.1007/s11103-007-9237-9. [DOI] [PubMed] [Google Scholar]
- 85.Shan W, Chen JY, Kuang JF, Lu WJ. Banana fruit NAC transcription factor MaNAC5 cooperates with MaWRKYs to enhance the expression of pathogenesis-related genes against Colletotrichum musae. Mol Plant Pathol. 2015; doi: 10.1111/mpp.12281. [DOI] [PMC free article] [PubMed]
- 86.Huang W, Sun W, Lv H, Luo M, Zeng S, Pattanaik S, Yuan L, Wang Y. A R2R3-MYB transcription factor from Epimedium sagittatum regulates the flavonoid biosynthetic pathway. PLoS One. 2013;8:e70778. doi: 10.1371/journal.pone.0070778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang AL, Xu ZS, Zhao ZY, Cui XY, Chen M, Li LC, Ma YZ. Genome-wide analysis of C3H zinc finger transcription factor family and their drought responses in Aegilops tauschii. PMB Reporter. 2014;32:1241–56. [Google Scholar]


