Abstract
Background
Biodegradation of rubber (polyisoprene) is initiated by oxidative cleavage of the polyisoprene backbone and is performed either by an extracellular rubber oxygenase (RoxA) from Gram-negative rubber degrading bacteria or by a latex clearing protein (Lcp) secreted by Gram-positive rubber degrading bacteria. Only little is known on the biochemistry of polyisoprene cleavage by Lcp and on the types and functions of the involved cofactors.
Results
A rubber-degrading bacterium was isolated from the effluent of a rubber-processing factory and was taxonomically identified as a Rhodococcus rhodochrous species. A gene of R. rhodochrous RPK1 that coded for a polyisoprene-cleaving latex clearing protein (lcpRr) was identified, cloned, expressed in Escherichia coli and purified. Purified LcpRr had a specific activity of 3.1 U/mg at 30 °C and degraded poly(1,4-cis-isoprene) to a mixture of oligoisoprene molecules with terminal keto and aldehyde groups. The pH optimum of LcpRr was higher (pH 8) than for other rubber-cleaving enzymes (≈ pH 7). UVvis spectroscopic analysis of LcpRr revealed a cytochrome-specific absorption spectrum with an additional feature at long wavelengths that has not been observed for any other rubber-cleaving enzyme. The presence of one b-type haem in LcpRr as a co-factor was confirmed by (i) metal analysis, (ii) solvent extraction, (iii) bipyridyl assay and (iv) detection of haem-b specific m/z values via mass-spectrometry.
Conclusions
Our data point to substantial differences in the active sites of Lcp proteins obtained from different rubber degrading bacteria.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-016-0703-x) contains supplementary material, which is available to authorized users.
Keywords: Latex clearing protein (Lcp), Rubber oxygenase, Dioxygenase, Rhodococcus, Biodegradation
Background
Natural rubber is an important biopolymer that has been produced for more than a century by cultivating the rubber tree (Hevea brasiliensis). Natural rubber obtained by tapping of the rubber trees is used for countless applications, for example for the production of tires, sealings, latex gloves and many, many other items. The main component of rubber latex is the hydrocarbon poly(cis-1,4-isoprene). Chemosynthetic rubber is also produced at a scale that is almost comparable to that of the natural compound.
Despite the economic importance of rubber and the enormous amounts of rubber waste materials that are permanently released into the environment, complete degradation in nature is rarely detected and wastes continue to accumulate. Knowledge of the reasons for this is limited. In fact, application is made of this extremely slow natural degradation for example in the use of rubber tyres to provide attachment sites for creating artificial coral reefs. However, microorganisms that can attack rubber have been detected in many ecosystems in which the physical parameters (temperature, pH, salinity) are moderate [1–7]. It is also well known that the initial microbial attack on rubber depends on the ability to produce and secrete rubber-cleaving enzymes into the environment. Only two types of rubber-cleaving enzymes are known. One is the rubber oxygenase RoxA that was first isolated from Xanthomonas sp. 35Y [8, 9] and so far has been found only in Gram-negative bacteria [10]. RoxA of Xanthomonas sp. 35Y is a c-type dihaem dioxygenase and cleaves poly(cis-1,4-isoprene) into a C15 compound with a terminal keto and aldehyde group (12-oxo-4,8-dimethyl-trideca-4,8-diene-1-al, ODTD) as the main product [11–13]. The other rubber cleaving enzyme is a protein designated as latex clearing protein (Lcp) [1]. It shares no significant sequence homology with RoxA, with cytochrome c peroxidases or with dihaeme 7,10-diol synthases [14] and is present in Gram-positive rubber degrading bacteria such as Streptomyces sp. K30 [1] and other Actinobacteria. G. polyisoprenivorans VH2 and Streptomyces sp. K30, two well-studied Gram-positive rubber degraders, oxidatively cleave poly(cis-1,4-isoprene) to products of different sizes but with the same keto and aldehyde end groups as in RoxA-generated ODTD [15–17]. There have been different reports published for the co-factor and metal-contents of the Lcps from Streptomyces sp. K30 and of G. polyisoprenivorans VH2 [15, 17, 18], and at present there are currently only two biochemically characterized Lcp proteins.
In this study, we used a waste pond at a rubber-processing factory in Thailand as a natural enrichment environment for rubber-degrading microorganisms and a source for the isolation of new rubber degrading strains. Taxonomic analysis revealed that one isolated strain was a member of the genus Rhodococcus, a taxon that had not been previously identified as having the ability to utilise rubber as a sole source of carbon and energy but that is well known for its members to have a high potential for the biodegradation of recalcitrant compounds [19]. Biochemical and biophysical characterization of the purified recombinant Lcp protein of Rhodococcus rhodochrous strain RPK1 revealed some unexpected properties not previously described for any other rubber-degrading enzyme in addition to properties shared with the two other characterized Lcp proteins.
Results and discussion
Taxonomic identification of isolate RPK1
Isolate RPK1 had a high rubber-degrading activity compared to other rubber degraders in liquid culture, as revealed by pronounced disintegration of rubber pieces (Fig. 1a). However, isolate RPK1 did not form clearing zones on an opaque polyisoprene latex mineral salts agar while known clear zone formers such as Xanthomonas sp. 35Y [8] or Streptomyces coelicolor strain 1A [3] formed large clearing zones. Isolate RPK1 developed colonies with an intense red colour upon growth and prolonged incubation on NB agar (Fig. 1b). Microscopic examination revealed non-motile cells. Depending on the growth phase the cells were coccoid (cells from late stationary phase), rod-shaped (cells from early and late log phase) or long rods (up to 1 ×5 μm), partially branched and star-like in exponentially growing cultures (Fig. 1c-e). Isolate RPK1 was catalase positive and Gram-positive. It grew well at 43 °C but no colonies developed at 45 °C. Strain RPK1 tolerated the presence of 3 % NaCl (in NB). It accumulated storage compounds that were stainable by Nile red (polyhydroxyalkanoates or triacylglycerols) and strain RPK1 synthesised polyphosphate granules as shown by staining with DAPI (4’,6-diamidine-2-phenylindole) and the use of DAPI-polyphosphate-specific emission filters in fluorescence microscopy (Fig. 1e, f). Isolate RPK1 utilised complex media (NB, LB medium) and grew with mineral salts media containing D-mannitol, fructose, acetate, benzoate or octane as a single carbon source. Glucose, sucrose, gluconate, pentane, petroleum or pyridine (excluding Rhodococcus pyridinivorans) were not used for growth. Polymers such as polyhydroxybutyrate (PHB), casein or starch were also not utilised by RPK1. These characteristics, in combination with the red colour of the colonies and the variable morphology of the cells indicated that the isolate RPK1 could be a member of the genus Rhodococcus. To verify this assumption we determined the DNA-sequence of the PCR-amplified 16S rRNA gene (accession No KU140418) and compared the sequences to the database by BLAST search. The 16S rRNA gene was 99.7 and 99.2 % identical to Rhodococcus MK3027 and to R. rhodochrous MTCC11081, respectively. Together with the biochemical and morphological data we concluded that isolate RPK1 is a member of the species R. rhodochrous. It differed from the rubber degrading Xanthomonas sp. 35Y [8, 9], Streptomyces sp. K30 [1], and other rubber degrading streptomycetes [3] by its inability to produce clearing zones on opaque polyisoprene latex agar plates. Previously, bacteria with a strong rubber-degrading activity but with no ability to form clearing zones had been isolated and identified as Gordonia polyisoprenivorans or Gordonia westfalica [20].
Fig. 1.
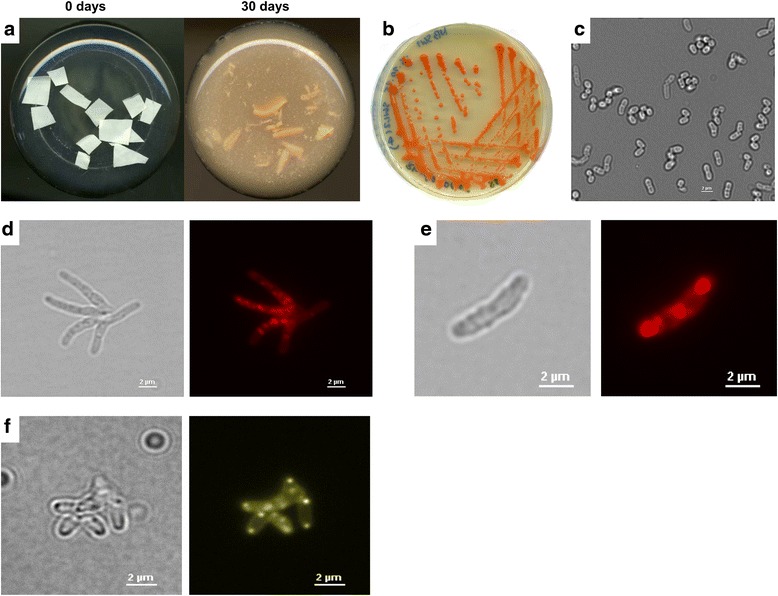
Features of R. rhodochrous RPK1. (a) Degradation of rubber pieces by R. rhodochrous RPK1 after 0 and 30 days of incubation in shaking flasks with mineral salts medium at 30 °C; (b) formation of red-coloured colonies of R. rhodochrous RPK1 during growth on NB agar; (c) morphology of stationary R. rhodochrous RPK1 cells in bright field microscopy, note almost coccoid cells; (d) R. rhodochrous RPK1 cells during growth on NB medium supplemented with acetate (bright field and fluorescent image stained with Nile red). Note, star-like branched cells typical for R. Rhodochrous; (e) R. rhodochrous RPK1 cells during growth on NB medium supplemented with acetate (bright field and fluorescent image stained with Nile red, note, presence of Nile-red-stainable granules, possibly representing PHB granules or triacylglycerol bodies; (f) R. rhodochrous RPK1 cells during growth on NB medium supplemented with acetate (bright field and fluorescent image stained with DAPI and examined for presence of polyphosphate granules using DAPI-polyphosphate-specific emission filters). Note, presence of cell-pole localized polyphosphate granules in most cells
Identification of the gene coding for the latex clearing protein in R. rhodochrous strain RPK1
BLAST analysis revealed that many Actinobacteria and all known rubber-degrading Actinobacteria for which the genome sequences have been determined have at least one gene that codes for a so-called latex clearing protein (lcp) that is suspected to be responsible for the initial oxidative attack on the polyisoprene carbon backbone [1, 21–23]. Remarkably, non clearing zone formers such as G. polyisoprenivorans also have functional lcp genes [20]. This indicates that the Lcp protein apparently is not directly responsible for the formation of clearing zones during growth on opaque latex-agar. Alignment of the amino acid sequences of the Lcp proteins from different species (most of them annotated as Lcp protein but without verified function or biochemical characterization) revealed conserved regions within the Lcp amino acid sequences [15]. We identified a hypothetical lcp gene in the genomes of R. rhodochrous strain MTCC11081, Rhodococcus sp. MK3027 and Rhodococcus sp. ARG-BN062 by screening of the published genome sequences for the presence of lcp-like sequences. The deduced amino acid sequence of these hypothetical Lcp proteins included the DUF2236 domain that constitutes the central part of most if not all Lcp proteins [15]. Two oligonucleotides based on the upstream and downstream regions of the lcp genes of these Rhodococcus strains were generated (LcpRr-PstI_for and LcpRr-HindIII_rev) and a PCR reaction was performed with the chromosomal DNA of R. rhodochrous strain RPK1. A 1.5 kbp DNA fragment was obtained and its DNA sequence was determined (accession number KU140417). Analysis of the DNA sequence revealed one large open reading frame of 1227 bp that coded for a peptide of 408 amino acids (45.2 kDa, Table 1). The deduced amino acid sequence revealed strong similarities to the postulated Lcp proteins of R. rhodochrous strains and of several other Rhodococcus sp. strains (81 to 99 % identical amino acids). High degrees of similarities were also detected to many other genome-deduced sequences of putative Lcp proteins from bacteria including those from many streptomycetes and other Actinobacteria. When we compared the overall amino acid sequence of LcpRr with that of the only two other biochemically characterized Lcp proteins, a 70 % (76 %) identity (similarity) and a 57 % (66 %) identity (similarity) was determined to the LcpVH2 from G. polyisoprenivorans [20] and LcpK30 protein from Streptomyces sp. K30 [1], respectively (Additional file 1). A 30 amino acid long sequence at the N-terminus of LcpRr was predicted to code for a signal peptide that enabled the secretion of the protein. The molecular mass of the predicted mature protein amounted to 42.2 kDa (Table 1).
Table 1.
Properties of biochemically characterized rubber oxygenases
| LcpRr | LcpK30 | LcpVH2 a | RoxAXsp | |
|---|---|---|---|---|
| Gene length [bp] | 1227 | 1224 | 1224 | 2037 |
| Residues (pre-/mature enzyme) | 408/378 | 407/377 | 407/371a | 678/648 |
| Molecular mass (apo-/mature protein) [kDa] | 45.2/42.2 | 44.0/41.0 | 45.5/41.7a | 74.7/71.5 |
| strep tagged [kDa] | 44.5 | 43.3 | ||
| pH optimum | 8 | 7 | 7 | 7 |
| Molar extinction coefficient [104 M−1 cm−1] | 9.5 (407 nm) | 8.0 (412 nm) | n.d. | 20.6 (406 nm) |
| Metal atoms per protein molecule | ||||
| Fe | 0.98 | 1.16/1.05 | n.d. | 2.3 |
| Cu | 0.36 | 0.056/- | sub-stoichiometric | - |
| Mn | - | - | n.d. | - |
| Ni | - | 0.12/0.05 | n.d. | - |
| Zn | - | - | n.d. | - |
| Mg, Ca | - | n.d. | n.d. | n.d. |
| Haeme type | b-type | b-type | not known/no haeme | c-type |
| Oxidation state of haeme iron | Fe3+ | Fe3+ | Fe3+---O2- | |
| UVvis effect upon addition of CO | no | no | yes | |
| Specific activity [U/mg] (23/30/37 °C) | 0.9/3.1/b | 1.5/n.d./4.6 | 1.3/n.d./n.d. | n.d./0.48/n.d. |
| Melting point | n.d. | 61.5 °C | n.d. | 54.3 °C |
| Conserved DUF2236 residuesc | ||||
| R | 163 | 164 | 161 | |
| T | 167 | 168 | 165 | |
| H | 197 | 198 | 195 |
Expression and purification of LcpRr
The DNA sequence coding for the LcpRr signal peptide was replaced by a Strep-tag coding sequence and the modified gene was cloned under the control of an L-rhamnose-dependent promoter into p4782.1 and subsequently transformed to E. coli JM109. The LcpRr protein was purified from the combined cells of a 4.8 L E. coli (p4782.1::lcpRr) culture as described in the method section. A yield of approximately 7.7 mg of purified LcpRr protein (5.3 mg/mL in BCA assay) was obtained after the final purification step. The Lcp proteins of Streptomyces sp. K30 (LcpK30) and of the rubber oxygenase RoxA of Xanthomonas sp. strain 35Y (RoxAXsp) were also purified and were used for comparison purposes. All purified proteins were separated by SDS-PAGE and checked for purity. As shown in Fig. 2, the LcpRr, LcpK30 and RoxAXsp proteins were almost homogenous. Both LcpRr, and LcpK30 proteins migrated at a slightly higher apparent molecular mass (50 and 47 kDa) as deduced from the gene sequences (44.5 and 43.3 kDa, respectively). Most remarkably, concentrated LcpRr had a brownish colour. This was in sharp contrast to the red colour of the concentrated solutions of LcpK30 or of RoxAXsp and indicated that there were substantial differences of the LcpRr protein in comparison to the other rubber-cleaving enzymes.
Fig. 2.
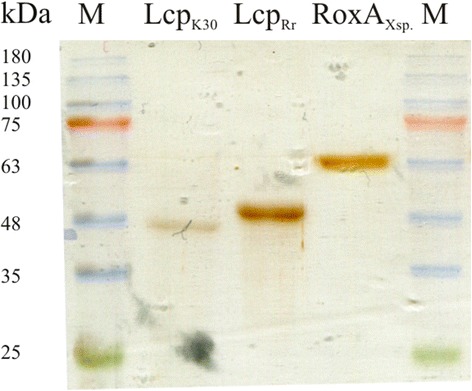
SDS-PAGE of purified LcpK30, LcpRr and RoxAXsp. Purified proteins were separated by reducing SDS-PAGE and subsequently stained with silver. The kDa values of marker proteins (M) are indicated
Biochemical properties of LcpRr
The purified LcpRr protein was tested for its rubber cleaving activity using both the oxygen consumption and the HPLC-based rubber cleavage product assay. The oxygen consumption assay (Fig. 3a) confirmed that LcpRr cleaved poly(cis-1,4-isoprene) latex in an oxygen-dependent manner; specific activities of 0.9 U/mg and of 3.1 U/mg were determined for LcpRr at pH 8 and at 23 °C and 30 °C, respectively (Table 1). Variable data were determined for the specific activity of LcpRr at 37 °C possibly because of the decreasing stability of the LcpRr protein at higher temperatures (see below). The HPLC (Fig. 3b) and Fuchsin assay (Additional file 2) revealed that LcpRr produced the same mixture of polyisoprene cleavage products (C20 and higher oligo-isoprenoids with terminal keto and aldehyde groups) that had been determined for LcpK30. ODTD was only detectable in trace amounts for LcpRr or for LcpK30 but was the main product of the RoxAXsp-derived rubber cleavage products. Determination of the activities of purified LcpRr at different pH values using the HPLC-based product assay revealed a pH optimum of around pH 8 (Fig. 4) that was about one pH unit higher than the pH optimum that had been previously determined for RoxAXsp or for LcpK30 and Lcp1VH2 [9, 15, 17]. The stability of all the Lcp preparations decreased upon incubation in buffer at 37 °C (Fig. 5a). In accordance with this, the concentration of rubber degrading products in an in vitro latex cleavage assay with LcpRr or LcpK30 increased for only 4–8 h (Fig. 5b). RoxAXsp, on the other hand, was much more stable and continuously produced ODTD molecules for up to 70 h [24].
Fig. 3.
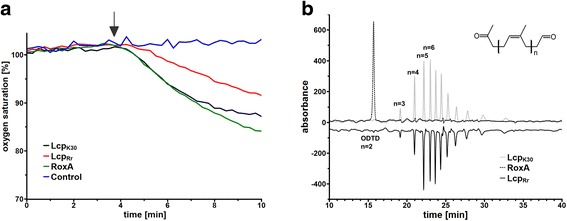
Assays for oxidative polyisoprene cleavage. (a) Oxygen consumption assay: 4 μg of the indicated enzyme (control without enzyme) was added after 3.75 min (arrow) and oxygen saturation was recorded. Activities were calculated from the slopes of the graphs during the first minutes. The mixtures were extracted with ethylacetate after 90 min of incubation at 23 °C and the formed cleavage products were analysed by HPLC (b). ODTD was the main cleavage product of RoxAXsp but was present only in trace amounts in LcpRr and LcpK30 preparations. The graph for LcpRr is given in its inverse orientation. Assays with each enzyme were performed at least three times; one typical dataset is shown
Fig. 4.
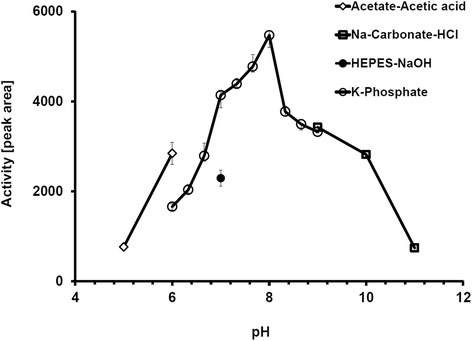
pH optimum of LcpRr. The pH optimum was determined using the HPLC-based product assay in a pH range of 5 to 11 using acetate buffer (pH 5 - pH 6, diamonds), phosphate buffer (pH 6 - pH 9, open circles), carbonate buffer (pH 9 – pH 11, squares), or HEPES (pH 7, closed circle). Assays were performed with two biological and two technical replicates. Error bars indicate standard deviation
Fig. 5.
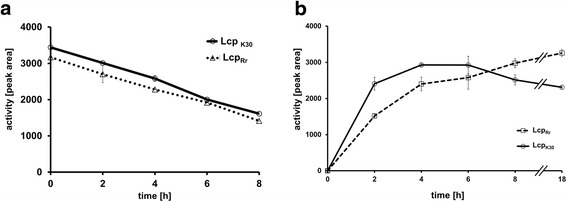
Stability of LcpRr and LcpRr and product formation. Lcp proteins were incubated in the presence of polyisoprene latex for 0 to 8 h at room temperature and the amount of formed products was determined by HPLC (a). Lcp proteins were incubated at 37 °C for up to 18 h before the standard activity assay was performed (b). Assays were performed with two biological and two technical replicates
LcpRr is a b–type cytochrome and revealed remarkable differences to LcpK30
Concentrated solutions of LcpRr had a brown colour while the LcpK30 solutions were red. Figure. 6 shows a comparison of the UVvis spectra of the purified LcpK30 and LcpRr proteins in the as isolated (oxidised) and in the dithionite-reduced state. LcpRr (and LcpK30) as isolated both showed similar absorption maxima at 407 (412) nm and at 535 (544) nm that are typical for haem-containing proteins in the oxidised form. However, purified LcpRr had an additional broad absorption maximum around 645 nm. The absorption band at 645 nm was absent in LcpK30 and in other biochemically characterized RoxA proteins such as RoxAXsp and RoxACco [10] and was responsible for the different (brown) colour of LcpRr. When the Lcp preparations were chemically reduced by the addition of sodium dithionite, the absorption bands at 407 (412) nm and 535 (544) nm shifted to 428 (430) nm and 560 (562) nm. A comparison of the reduced spectra of both Lcp proteins showed differences in the Q-bands (500 – 600 nm). Apparently, LcpRr is far less pronounced in this region than LcpK30. Nevertheless, these data corresponded to the Soret and Q-bands that are typical for haem-containing proteins and strongly indicated that LcpRr is a haem-containing protein. The band around 645 nm, however, was not changed by the addition of dithionite.
Fig. 6.
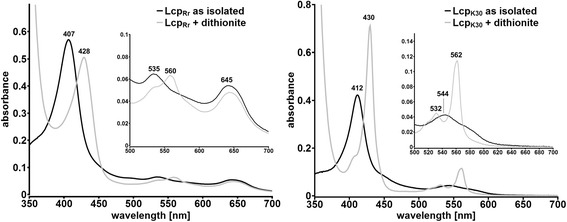
UVvis spectrum of LcpRr and LcpK30 as isolated (black lines) and after reduction with dithionite (grey lines). Both Lcp proteins show a prominent band at 407 (412 nm in case of LcpK30) that is characteristic for porphyrines. After reduction with dithionite a characteristic shift of the α-band to 428 (430) nm as well as an increase in the Q-band region (560/562 nm) was observable. Assays were repeated at least three times with two separate protein batches. A typical experiment is shown
To confirm that LcpRr is a haem-containing protein and to determine its haem type, a metal analysis and a spectral analysis by the haem-bipyridyl assay were performed. 6.5 μg Fe/mL LcpRr protein solution (5.3 mg protein/mL) were determined. This corresponded almost perfectly with a stoichiometry of one atom Fe per one LcpRr molecule. It was of interest that low amounts of copper (2.8 μg/mL) were also identified and corresponded to 0.36 atoms Cu per one LcpRr molecule. Zinc was detected at the detection limit (0.1 μg/mL) and Nickel was below the detection limit (<0.1 μg/mL); other metals (vanadium to zinc tested) were also not detected in significant amounts. Divalent cations such as magnesium or calcium were not present (below the detection limit of 0.1 μg/mL) (Table 1). The presence of approximately one third of an atom% Cu per LcpRr molecule was unexpected because only traces of copper had been previously detected in LcpK30 [18]. The determined amount of copper in LcpRr, however, was too high to be explained by an error in the determination of the metal or protein concentration. One possibility could be that the amount of copper was due to a contamination of the protein by traces of copper present in either the growth medium or in the buffer ingredients. For example, the used batch of NaCl that was present in some purification buffers was only of 98 % purity and could contain traces of heavy metals. However, sub-stoichiometric amounts of copper (precise concentration not known) had been also detected in Lcp1VH2 of G. polyisoprenivorans [15]. Addition of an equimolar concentration or of a 10-fold molar excess of copper ions [Cu(II)Cl2] to the assay mixture with purified LcpRr had no detectable effect on the UVvis spectrum or on the activity of LcpRr. The addition of 50 μM CuSO4 to the LcpRr-expression culture produced no increased activity or yield of LcpRr. At present, there is no convincing explanation for the finding of variable sub-stoichiometric amounts of copper in the purified Lcp proteins from R. rhodochrous RPK1.
An absorption maximum of 556 nm was determined using the bipyridyl assay for LcpRr and for haemoglobin that was used as a b-type cytochrome control protein (Additional file 3). This result indicated the presence of a b-type haem in LcpRr. In contrast to the covalently linked c-type cytochromes, the haem groups of the b-type cytochromes are not covalently linked to the peptide chain and can be therefore extracted by an acid solvent extraction [25]. Acid solvent extraction of the purified LcpRr yielded a coloured supernatant and a non-coloured precipitate. In contrast, solvent extraction of the c-type cytochromes such as RoxAXsp or of other commercially available cytochrome c enzymes yielded a non-coloured supernatant and a red precipitate which is in agreement with the covalent attachment of porphyrin to the polypeptide. MALDI-ToF analysis of the purified LcpRr resulted in the identification of ions with m/z values of 616 (data not shown) which is typical for haem b [26]. Taken together, all these results indicated that LcpRr is a b-type cytochrome similar to LcpK30 [18] Notably, MALDI-ToF analysis of LcpRr also revealed an ion species with m/z values of 619 besides that of 616 which could correspond to a verdo-haem [27]. As the activity of purified LcpRr rapidly and substantially decreased during storage, the haem species with m/z value of 619 could represent a haem degradation product of the inactivated LcpRr.
LcpRr is insensitive to most chelating inhibitors
Metal-dependent proteins are often inhibited by chelating compounds. Therefore, a variety of known chelator compounds was tested for their effects on the activity of LcpRr using the HPLC-based activity assay. EDTA, tiron, or phenanthroline had no significant effect on the activity (Fig. 7). Ethyl xanthogenate partially inhibited LcpRr by ≈ 40 % similar to that for LcpK30 but was different from the Lcp purified from G. polyisoprenivorans (Lcp1VH2) that completely inhibited LcpVH2 at 2 mM xanthogenate [15]. The only compound that had a strong effect on the activity of LcpRr was the metal chelator diethyl dithiocarbamate (82 % inhibition, Fig. 7). However, diethyl dithiocarbamate had no effect on the UVvis spectrum of LcpRr and this excluded a direct effect of the inhibitor at the haem site. Carbon monoxide, that completely inactivated RoxA and led to a prominent band at 415 nm by UVvis spectroscopy [18], had no effect on the absorption spectrum of LcpRr or LcpK30as isolated and this was in agreement with the presence of an oxidised (Fe3+) haem centre. Carbon monoxide had no inhibitory effect on the polyisoprene cleavage during the assay for the HPLC-based product when sufficient oxygen was also present in the assay mixture. However, when LcpRr was incubated in carbon monoxide-saturated and oxygen-free buffer before it was added to an oxygenated polyisoprene latex assay solution, a lag phase of LcpRr in its ability to consume oxygen was observed. The oxygen consumption and polyisoprene-cleaving activities were recovered within 30 to 50 min of incubation and exposure of the assay solution to air. The same result was obtained when LcpK30 was exposed to carbon monoxide. Addition of carbon monoxide to the dithionite-reduced LcpRr or LcpK30 had visible effects on the UVvis spectra as revealed by the increase of the α-band of LcpRr and LcpK30. The effect of carbon monoxide on the UVvis spectrum of Lcp was reversible by addition of a dioxygen atmosphere and indicated that the binding of carbon monoxide to the chemically reduced haem group in Lcp was reversible. This is different to RoxAXsp that binds carbon monoxide more strongly and completely inhibits the activity. An apparent consequence of these data is that the haems of the Lcp proteins undergo a reversible Fe3+ to Fe2+ reduction during oxidative polyisoprene-cleavage and that the reduced Lcp proteins were the carbon monoxide-sensitive molecular species. This is the first evidence for a switch in the oxidation state of the active haem site of an Lcp protein during catalysis.
Fig. 7.
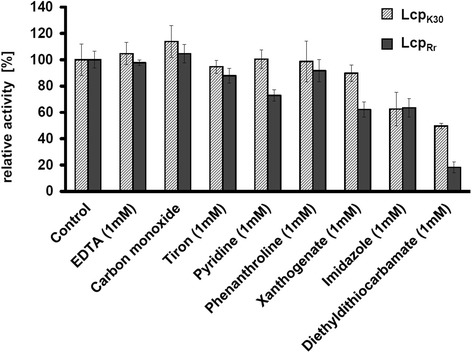
Inhibition of LcpRr by potential inhibitors. Activity was determined using the HPLC-based detection of polyisoprene cleavage products after 2 h of incubation of polyisoprene latex with Lcp at room temperature and subsequent solvent extraction of the products. The final concentration of inhibitors was 1 mM. The 100 % value corresponded to the area of the C35 (23 min) product peak (see Fig. 3b). Sensitivity to carbon monoxide was tested by replacing 0.5 volumes of (oxygenated) assay buffer by a deoxygenated and CO-saturated assay buffer. Cleavage products were solvent-extracted after 2 h of incubation time. At least two repetitions with two technical replicates were performed for each inhibitor. Error bars indicate the upper and lower values of one experiment
LcpRr but not LcpK30 is accessible for external ligands
Previous studies on rubber oxygenase RoxA had revealed that the active haem site in RoxA had only one axial amino acid ligand. The other axial ligand was a dioxygen molecule that was stably bound to haem in a Fe3+----O2− transition state. The oxygen molecule in RoxA could be partially removed by the addition of imidazole thereby moving the negative charge from the oxygen molecule to the iron atom (Fe2+). This charge transfer resulted in a small visible change of the UVvis spectrum as revealed by an increase of the absorption of the Q-bands at 549 nm [12, 15–17]. When imidazole was added to the dithionite-reduced RoxA, substantial increases in the Soret and Q-bands were determined compared to the reduced RoxA bands without imidazole [18, 24]. This increase in absorption was interpreted as the result of the binding of the imidazole molecule to the (now) free sixth (axial) coordination site of the haem iron. When we performed an analog experiment with purified LcpRr and with purified LcpK30 we found remarkable differences between both Lcp proteins: addition of imidazole to the dithionite-reduced LcpK30 had no effect on the UVvis spectrum and there was no detectable increase of the Q-bands. This indicated that the 6th coordination site of the haem apparently was not accessible for imidazole and the LcpK30 protein was present in a “closed state”. Binding of the substrate (polyisoprene) would therefore require a conformational change of the LcpK30 structure. In contrast, addition of imidazole to the dithionite-reduced LcpRr protein resulted in a substantial increase of the Soret- and Q-bands (Fig. 8) and this can be explained by the binding of imidazole to the reduced haem. Similar results were obtained when both the Lcp proteins were treated with mercaptoethanol: no change of the UVvis spectrum was determined for LcpK30 while prominent changes were detected for the LcpRr protein (Additional file 4). In conclusion, LcpK30 and LcpRr seem to rest in a different conformation in their as isolated states. While LcpK30as isolated was in a six-fold coordinated “closed” state, the haem group of LcpRr was readily accessible to external ligands and substrates, and this indicated a five-fold coordinated “open” state. Further evidence for this can be found in the UVvis spectra of five-fold coordinated myoglobin in the oxidised (met myoglobin) and reduced (desoxy-myoglobin) state. The UVvis spectra of the latter proteins showed similarities to the corresponding spectra of LcpRr, particularly in the region of the less pronounced Q-bands of reduced LcpRr compared to LcpK30 as well as in the 645 nm region in the oxidised (as isolated) state [28]. The presence of the 645 nm absorption band in LcpRr might be also explained by a charge transfer phenomenon of a charged residue/ion in close neighbourhood to the haem group in LcpRr and in its absence in LcpK30 [29]. Unfortunately, only the RoxA structure [30] but no Lcp structure was available to obtain direct support for our assumption.
Fig. 8.
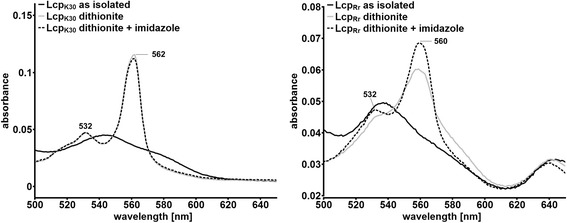
UVvis spectrum of LcpRr and LcpK30 before and after the addition of 1 mM imidazole (as isolated/after reduction) in the area of the Q-band. No significant change was observable for LcpK30 whereas for LcpRr a substantial increase at 560 nm was determined upon addition of imidazole. Assays were repeated two times with two separate protein batches. A typical experiment is shown
lcp genes are frequently present in the genomes of Actinobacteria [22] and many rubber degrading species have been described for members from this group [3–5, 23, 31, 32]. Most of the rubber degrading actinomycetes such as Streptomyces sp. K30 [1], Streptomyces coelicolor 1A, and many others [3, 6, 33] produce clearing zones on opaque polyisoprene latex agar. However several of the most potent rubber degraders do not produce clearing zones and apparently need a close contact to the rubber material they degrade. Two well known rubber degrading Gordonia species (G. polyisoprenivorans and G. westfalica) [20, 22] and also the strain from this study (R. rhodochrous RPK1) belong to this group of non-clearing zone formers. One might speculate that the Lcp proteins of non-clearing zone formers constitute a group that have an open conformation with free access to the active site (no conformational change is needed) and that the other Lcp proteins that have an ability to form a clearing zone have a closed form. The prototype of the first group would be LcpRr and the prototype of the latter would be LcpK30. It will be necessary to biochemically investigate more Lcp proteins and to solve the structure of Lcp proteins to find evidence for or against this hypothesis.
Conclusions
This study extends the list of biochemically characterized rubber-degrading non-clearing zone formers by latex clearing proteins (Lcp) to the genus Rhodococcus (besides Gordonia). The detection of rubber-cleaving activity with purified LcpRr and the absence of clearing zones during growth on polyisoprene latex agar raises the question of whether the designation “latex clearing protein” has been well-chosen. Rubber oxygenase B (RoxB) would be an appropriate alternative. However, the designation Lcp has been used is several previous publications and has also been used for many annotated genes in genome-sequenced Actinobacteria. Re-classification of Lcp as RoxB therefore could lead to the confusion of other research workers.
The isolation and characterization of the Lcp protein of R. rhodochrous RPK1 in this study showed that all the so far studied Lcp proteins can differ in some spectroscopic features and/or in spatial arrangements of their metal ions/cofactors and indicate the presence of two or even more subgroups of Lcp proteins. It will be necessary to study more Lcp proteins to reveal the complete variability of rubber degrading enzymes present in rubber-degrading organisms.
Methods
Bacterial strains, plasmids and culture conditions
Table 2 shows the bacterial strains, plasmids and oligonucleotides that were used in this study. R. rhodochrous strain RPK1 was grown with nutrient broth (NB) medium or in mineral salts medium (MSM, 9 g/L Na2HPO4 ×12 H2O, 1.5 g/L KH2PO4, 1 g/L NH4NO3, 0.2 g/L MgSO4 ×7 H2O, 0.02 g/L CaCl2 ×2 H2O, 1.2 mg/L Fe(III)ammonium citrate with solid rubber pieces or with water-soluble carbon sources as indicated at 30 °C. Pieces (1 cm ×1 cm) of heat-sterilised vulcanised rubber (commercial but not-powdered rubber protecting gloves) were added to the sterile mineral salts medium during the enrichment and growth of R. rhodochrous on rubber (0.6 % [wt/vol]). Plasmid-carrying recombinant E. coli strains were grown with LB medium at 22 °C or 37 °C in the presence of the appropriate antibiotic (ampicillin or kanamycin). Polyisoprene latex was kindly provided by Weber and Schaer, Hamburg (Germany) and was used after 3 washing steps in 0.1 % (wt/vol) Nonidet P40. For purification of LcpRr, recombinant E. coli cells were grown in LB medium supplemented with 0.1 % (wt/vol) L-rhamnose at 22 °C. Utilization of carbon sources was tested on mineral salts agar with separately filter-sterilised carbon sources at the following end concentrations (sugars, sugar alcohols and sugar acids at 0.5 % [wt/vol], sodium acetate [0.25 %, wt/vol], sodium benzoate [0.1 %, wt/vol]). Volatile compounds (alkanes) were applied by adding a quantity of 100 μL to a sterile filter paper placed in the lid of a petri disk. The plates were sealed with parafilm and incubated separately at 30 °C. Utilization of PHB was tested on PHB overlay plates as described previously [34]. Growth at different temperatures was tested on NB agar plates.
Table 2.
Bacterial strains, plasmids and oligonucleotides used in this study
| Strain or plasmid | Relevant characteristics | Reference |
| E. coli JM109 | Plasmid storage and expression of lcp | |
| E. coli XL1-blue | Transformation strain | Stratagene |
| Rhodococcus rhodochrous RPK1 | Wild type strain, degrades rubber | this study |
| pUC9::strep-lcp K30 (SN5339) | Cloning vector for lcp K30, Apr | [18] |
| pUC9::strep-lcp Rr (SN5759) | Cloning vector for lcp Rr, Apr | this study |
| p4782.1 (SN3513) | Mobilizable broad host range | [37] |
| Expression vector, Kmr | ||
| p4782.1::strep-lcp K30 (SN5496) | Coding sequence of strep-lcp K30 under | [18] |
| Rhamnose promoter control, Kmr | ||
| p4782.1::strep-lcp Rr (SN5760) | Coding sequence of strep-lcp Rr under | this study |
| Rhamnose promoter control, Kmr | ||
| Oligonucleotides | ||
| LcpRr-complete_for | GCAGAATCCACATGTCCT | |
| LcpRr-complete_rev | CGACAAACCCACAGATGA | |
| LcpRr-mature-PstI_for | GGGCCTGCAGCGGCCCTGGAGGTGGTCGCC | |
| LcpRr-mature-HindIII_rev | CCGGTAAGCTTTCAGGGATAGTTGGG | |
| 16S-universal-for | GAGTTTGATC(A/C)TGGCTCAG | |
| 16S-universal-rev | GG(C/T)TACCTTGTTACGACT | |
| 16S-Rr-complete_for | CTGGCGCGGTGCTTAAC | |
| 16S-Rr-complete_rev | CAGTAATTCCGGACAACG |
Kanamycin resistance (Kmr), Ampicillin resistance (Apr)
Enrichment and isolation of rubber-degrading microorganisms
Liquid from a waste pond at a rubber latex processing factory in Thailand (Namom rubber factory at Namom, Songkhla) was used as an inoculum to enrich for rubber-degrading microorganisms in a mineral salts medium (MSM) that had been supplemented with 1×1 cm pieces of rubber gloves as a sole source of carbon and energy. After two weeks of incubation at 30 °C, 0.1 volumes (without pieces of rubber) were transferred to fresh medium and incubated for an additional month. Substantial disintegration of the new rubber pieces became visible and indicated that active rubber-degrading microorganisms were present. Several bacterial strains were isolated from this enrichment culture by repeated purification from streaks onto NB and LB agar plates. Each isolate was subsequently tested for its ability to degrade rubber in liquid MSM with rubber pieces as carbon source. One isolate (designated as isolate RPK1) with strong rubber-degrading activity was selected for this study.
Cloning and heterologous expression of lcpRr, and determination of the 16S rRNA gene sequence of isolate RPK1
The lcpRr gene was amplified using the chromosomal DNA from R. rhodochrous strain RPK1 as template and the oligonucleotides LcpRr-complete_for and LcpRr-complete_rev as PCR primers and Takara Primestar DNA polymerase as the proof-reading polymerizing enzyme. The DNA sequence of the product was determined and is available under the accession no KU140417. Alternatively, the coding sequence of mature LcpRr was amplified from chromosomal DNA using LcpRr-mature-PstI_for and LcpRr-mature-HindIII_rev as primers. The DNA products were purified via agarose gel electrophoresis, cleaved with restriction enzymes PstI and HindIII and ligated into plasmid pUC9::lcpK30 that had been cleaved by the same restriction enzymes. The coding sequence for strep-tagged lcpRr was cut out using HindIII and NdeI and was subsequently ligated into the expression plasmid p4782.1 and transformed to competent E. coli JM109 cells.
A part of the 16S rRNA gene of the isolate RPK1 was PCR-amplified using the primers 16S-universal_for and 16S-universal_rev. The DNA sequence of the resulting PCR product was determined (1412 bp) and revealed a strong similarity to the 16S rRNA genes of several Rhodococcus sp. strains. The 16S rRNA gene sequence of isolate RPK1 was determined after PCR amplification using the primers (16S-Rr-complete_for and 16S-Rr-complete_rev), that were specific for the ends of the known 16S rRNA gene sequences of R. rhodochrous strains taken form the NCBI data base, and is now available under the accession no KU140418.
Purification of LcpRr, LcpK30 and of RoxAXsp
Purification of the rubber oxygenase of Xanthomonas sp. 35Y (RoxAXsp) and latex clearing protein LcpK30 was performed as described previously [12, 18] LcpRr was purified as follows: eight individual 600 mL LB cultures in 3 litre Erlenmeyer flasks were inoculated each with 0.02 volumes of a seed culture of E. coli JM109 harbouring the plasmid p4782.1::lcpRr that had been grown with the same medium. It was important that L-rhamnose (0.1 %, wt/vol) was present right from the beginning in the main cultures to maximise the yield of the expressed LcpRr protein. Cells of the main culture were harvested by centrifugation after ≈ 20 h of growth at 22 °C and were immediately used for protein purification. The cell pellet was resuspended in 100 mM potassium phosphate buffer, pH 7.7, containing 150 mM sodium chloride (KPN, 2 mL KPN/g cell wet weight). A soluble cell extract was prepared by two French press steps and subsequent centrifugation at 40,000 g for 40 min. The supernatant (≈ 60 mL) was directly applied to a 10 mL Strep-Tactin HC gravity flow column that had been equilibrated with KPN buffer. The column was washed with at least five volumes of KPN buffer before the LcpRr protein was eluted by ≈ 30 mL of 5 mM desthiobiotin dissolved in KPN. LcpRr-containing fractions were combined, desalted by running through a G25 Sephadex (26/160) Hiprep desalting column (53 mL bed volume) that had been equilibrated with 1 mM potassium phosphate (KP) buffer, pH 7.0 and subsequently concentrated to 1–2 mL via ultrafiltration (10 kDa cut-off). Remaining impurities were removed by chromatography on a Superdex 200 column (16/600, equilibrated with 1 mM KP, pH 7) at a flow rate of 1 mL/min. Combined LcpRr-containing fractions were ultrafiltrated (10 kDa cut-off) and concentrated to ≈ 1.5 mL. Aliquots of the purified LcpRr protein were stored on ice for about 3 days (LcpK30 up to 1 week) or shock-frozen with liquid nitrogen and stored at -70 °C.
Determination of the cytochrome type of LcpRr
The haem type of LcpRr was determined by the bi-pyridyl assay as described elsewhere [35]. Purified RoxAXsp, cytochrome c (horseheart, type III, Sigma, St. Louis, USA) (both c-type cytochromes) and haemoglobin (b-type) (bovine, Sigma, St. Louis, USA) were used as controls for known c-type and b-type cytochromes, respectively. 25 μL of the respective protein stock solution (4–8 mg/mL) was added to 975 μL solution A (100 mM sodium hydroxide, 20 % (v/v) pyridine, 0.3 mM potassium ferricyanide). Subsequently, 2–5 mg sodium dithionite were added and the spectrum of the reduced cytochrome was recorded. The absorption maxima of the resulting α-bands were characteristic for b-type (556 nm) and c-type (550 nm) cytochromes. Bi-pyridyl-haem complexes of α-type cytochromes absorb at 584–588 nm. Additional assays for determination of the haem type were performed via extraction of haem by acidic acetone and by a matrix assisted laser desorption ionization time of flight (MALDI-ToF) analysis as previously described in detail [18].
Assay of Lcp activity
An HPLC-based assay for LcpRr-derived polyisoprene degradation products was used for most routine assays: poly(cis-1,4-isoprene) latex was diluted with 100 mM KP buffer, pH 7, to 0.2 % (assay volume 0.7 mL) and incubated in the presence of purified Lcp protein for 2 h at a temperature as indicated (for routine assay at room temperature [23 °C]). In the case of inhibition studies, the corresponding compound was added and gently solubilised in the reaction mix before the enzyme was added (final inhibitor concentration 1 mM). The products were extracted with 1 mL ethyl acetate (in a 2 mL Eppendorf tube), dried, and dissolved in 100 μL methanol. Aliquots were applied to an RP8 HPLC column (12 ×4 mm, 5 μm particle size, 0.7 mL/min) with water (A) and methanol (B) as mobile phases. The concentration of B was increased from 50 % (v/v) to 100 % (v/v) within 15 min; products were detected at 210 nm. The C35 product peak (at ≈ 23 min) was used for quantification and compared to a control without inhibitor. Alternatively, activity of LcpRr was assayed by determination of the rate of oxygen consumption in an OXY-4 mini apparatus (PreSens, Regensburg, Germany) as described previously [18]. Triplicates and controls without LcpRr or with heat-inactivated LcpRr were recorded simultaneously. A stability assay was performed by incubation of the purified LcpRr protein in the assay buffer at 37 °C for variable time periods. The remaining activity of the protein was determined as described above.
Other techniques
The concentration of protein solutions was determined by the bicinchoninic acid (BCA) method. The concentrations of purified rubber-cleaving enzymes were also determined from the molar extinction coefficients of LcpRr, LcpK30 and RoxAXsp: LcpRr, ε407 = 9.5 104 M−1 cm−1, LcpK30, ε412 = 8.0 104 M−1 cm−1, and RoxAXsp, ε406 = 2.06 ×105 M−1 cm−1. Separation of proteins was performed by polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulphate (SDS-PAGE) under reducing (2-mercaptoethanol) conditions. The metal content of the purified Lcp protein was determined using inductively coupled plasma-MS (ICP-MS) by the Spuren-Analytisches Laboratorium Dr. Baumann (Germany). Fuchsin staining of polyisoprene degradation products was performed by addition of a 1 % Fuchsin solution (0.5 g Fuchsin, 12.5 mL acetic acid, 2.5 g Na2S2O3, 0.2 mL HCl (37 %) and 37.5 mL H2O) to the Lcp assay mixture. Staining of the cells for PHB and polyphosphate was performed as described previously [36].
Ethics approval
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
The DNA sequence of the R. rhodochrous RPK1 lcp and 16S rRNA gene have been submitted to NCBI and are available under the accession No KU140417 and KU140418. The R. rhodochrous RPK1 strain has been deposited at the Deutsche Sammlung für Mikroorganismen und Zellkulturen (DSMZ) and can be obtained once the Nagoya protocol regulations have been finished. Until then, strain RPK1 can be obtained from the lab of S.W. and K.U. upon request.
Abbreviations
BCA, bicinchoninic acid; BLAST, basic local alignment search tool; DAPI, 4’,6-diamidine-2-phenylindol; HPLC, high pressure liquid chromatography; ICP, inductively coupled plasma; KPN, potassium phosphate-sodium chloride; LB, lysogeny broth; Lcp, latex clearing protein; MALDI-ToF, matrix-assisted laser desorption ionisation time of flight; MS, mass spectrometry; MSM, mineral salts medium; NB, nutrient broth; ODTD, 12-oxo-4,8-dimethyl-trideca-4,8-diene-1-al; PHB, polyhydroxybutyrate; RoxA, rubber oxygenase A; SDS-PAGE, sodium dodecyl sulphate polyacrylamide gel electrophoresis; UVvis, ultra violet visible.
Acknowledgements
The help of T. Jurkowski (Institute of Biochemistry, University Stuttgart) in MALDI-MS analysis and the work of Simone Reinhardt and Anna Schweter during some experiments is greatly acknowledged.
Funding
This work was funded by grants of the Deutsche Forschungsgemeinschaft to D.J. and of the Strategic Scholarships Fellowships Frontier Research Networks, Thailand to S.W.
Authors’ contributions
SW, and WR. carried out most experiments. JB, KU. and DJ. designed the study; JB and DJ. supervised the experiments; all authors analysed the data, DJ. wrote the manuscript. BH. checked and improved language. All authors read and approved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Additional files
Identity and similarity of biochemically characterized Lcp proteins and amino acid sequences alignment of biochemically characterized Lcp proteins. (DOCX 61 kb)
Detection of aldehyde products of Lcp-degraded polyisoprene by Fuchsin assay. (DOCX 1.91 kb)
Bipyridyl assay of LcpRr in comparison to LcpK30. (DOCX 128 kb)
UVvis spectra of LcpK30 and LcpRr in the presence of mercaptoethanol. (DOCX 356 kb)
Footnotes
Sirimaporn Watcharakul and Wolf Röther, shared first authorship
References
- 1.Rose K, Tenberge KB, Steinbüchel A. Identification and characterization of genes from Streptomyces sp. strain K30 responsible for clear zone formation on natural rubber latex and poly(cis-1,4-isoprene) rubber degradation. Biomacromolecules. 2005;6:180–188. doi: 10.1021/bm0496110. [DOI] [PubMed] [Google Scholar]
- 2.Heisey RM, Papadatos S. Isolation of microorganisms able to metabolize purified natural rubber. Appl Environ Microbiol. 1995;61:3092–3097. doi: 10.1128/aem.61.8.3092-3097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jendrossek D, Tomasi G, Kroppenstedt RM. Bacterial degradation of natural rubber: a privilege of actinomycetes? FEMS Microbiol Lett. 1997;150:179–188. doi: 10.1016/S0378-1097(97)00072-4. [DOI] [PubMed] [Google Scholar]
- 4.Linos A, Berekaa MM, Reichelt R, Keller U, Schmitt J, Flemming HC, Kroppenstedt RM, Steinbüchel A. Biodegradation of cis-1,4-polyisoprene rubbers by distinct actinomycetes: microbial strategies and detailed surface analysis. Appl Environ Microbiol. 2000;66:1639–1645. doi: 10.1128/AEM.66.4.1639-1645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imai S, Ichikawa K, Muramatsu Y, Kasai D, Masai E, Fukuda M. Isolation and characterization of Streptomyces, Actinoplanes, and Methylibium strains that are involved in degradation of natural rubber and synthetic poly(cis-1,4-isoprene) Enzyme Microb Technol. 2011;49:526–531. doi: 10.1016/j.enzmictec.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 6.Chia KH, Nanthini J, Thottathil GP, Najimudin N, Haris MRHM, Sudesh K. Identification of new rubber-degrading bacterial strains from aged latex. Polym Deg Stab. 2014;109:354–361. doi: 10.1016/j.polymdegradstab.2014.07.027. [DOI] [Google Scholar]
- 7.Imai S, Yoshida R, Endo Y, Fukunaga Y, Yamazoe A, Kasai D, Masai E, Fukuda M. Rhizobacter gummiphilus sp. nov. a rubber-degrading bacterium isolated from the soil of a botanical garden in Japan. J Gen Appl Microbiol. 2013;59:199–205. [DOI] [PubMed]
- 8.Tsuchii A, Takeda K. Rubber-degrading enzyme from a bacterial culture. Appl Environ Microbiol. 1990;56:269–274. doi: 10.1128/aem.56.1.269-274.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braaz R, Fischer P, Jendrossek D. Novel type of heme-dependent oxygenase catalyzes oxidative cleavage of rubber (poly-cis-1,4-isoprene) Appl Environ Microbiol. 2004;70:7388–7395. doi: 10.1128/AEM.70.12.7388-7395.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birke J, Röther W, Schmitt G, Jendrossek D. Functional identification of rubber oxygenase (RoxA) in soil and marine myxobacteria. Appl Environ Microbiol. 2013;79:6391–6399. doi: 10.1128/AEM.02194-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braaz R, Armbruster W, Jendrossek D. Heme-dependent rubber oxygenase RoxA of Xanthomonas sp. cleaves the carbon backbone of poly(cis-1,4-Isoprene) by a dioxygenase mechanism. Appl Environ Microbiol. 2005;71:2473–2478. doi: 10.1128/AEM.71.5.2473-2478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitt G, Seiffert G, Kroneck PMH, Braaz R, Jendrossek D. Spectroscopic properties of rubber oxygenase RoxA from Xanthomonas sp., a new type of dihaem dioxygenase. Microbiology. 2010;156:2537–2548. doi: 10.1099/mic.0.038992-0. [DOI] [PubMed] [Google Scholar]
- 13.Birke J, Hambsch N, Schmitt G, Altenbuchner J, Jendrossek D. Phe317 is essential for rubber oxygenase RoxA activity. Appl Environ Microbiol. 2012;78:7876–7883. doi: 10.1128/AEM.02385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estupiñán M, Álvarez-García D, Barril X, Diaz P, Manresa A. In silico/in vivo insights into the functional and evolutionary pathway of Pseudomonas aeruginosa oleate-diol synthase. Discovery of a new bacterial di-heme cytochrome c peroxidase subfamily. PLoS ONE. 2015;10:e0131462. doi: 10.1371/journal.pone.0131462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiessl S, Böse D, Oetermann S, Eggers J, Pietruszka J, Steinbüchel A. Latex clearing protein-an oxygenase cleaving poly(cis-1,4-isoprene) rubber at the cis double bonds. Appl Environ Microbiol. 2014;80:5231–5240. doi: 10.1128/AEM.01502-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibrahim EMA, Arenskotter M, Luftmann H, Steinbüchel A. Identification of poly(cis-1,4-isoprene) degradation intermediates during growth of moderately thermophilic actinomycetes on rubber and cloning of a functional lcp homologue from Nocardia farcinica strain E1. Appl Environ Microbiol. 2006;72:3375–3382. doi: 10.1128/AEM.72.5.3375-3382.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birke J, Jendrossek D. Rubber oxygenase (RoxA) and latex clearing protein (Lcp) cleave Rubber to different products and use different cleavage mechanisms. Appl Environ Microbiol. 2014;80:5012–5020. doi: 10.1128/AEM.01271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birke J, Röther W, Jendrossek D. Latex clearing protein (Lcp) of Streptomyces sp. strain K30 is a b-type cytochrome and differs from rubber oxygenase A (RoxA) in its biophysical properties. Appl Environ Microbiol. 2015;81:3793–3799. doi: 10.1128/AEM.00275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínková L, Uhnáková B, Pátek M, Nesvera J, Kren V. Biodegradation potential of the genus Rhodococcus. Environ Int. 2009;35:162–177. doi: 10.1016/j.envint.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 20.Broker D, Dietz D, Arenskotter M, Steinbüchel A. The genomes of the non-clearing-zone-forming and natural-rubber-degrading species Gordonia polyisoprenivorans and Gordonia westfalica harbor genes expressing Lcp activity in Streptomyces strains. Appl Environ Microbiol. 2008;74:2288. doi: 10.1128/AEM.02145-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiessl S, Schuldes J, Thürmer A, Halbsguth T, Broker D, Angelov A, Liebl W, Daniel R, Steinbüchel A. Involvement of two latex-clearing proteins during rubber degradation and insights into the subsequent degradation pathway revealed by the genome sequence of Gordonia polyisoprenivorans strain VH2. Appl Environ Microbiol. 2012;78:2874–2887. doi: 10.1128/AEM.07969-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yikmis M, Steinbüchel A. Historical and recent achievements in the field of microbial degradation of natural and synthetic rubber. Appl Environ Microbiol. 2012;78:4543–4551. doi: 10.1128/AEM.00001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nanthini J, Chia K-H, Thottathil GP, Taylor TD, Kondo S, Najimudin N, Baybayan P, Singh S, Sudesh K. Complete genome sequence of Streptomyces sp. strain CFMR 7, a natural rubber degrading actinomycete isolated from Penang, Malaysia. J Biotechnol. 2015;214:47–48. doi: 10.1016/j.jbiotec.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Schmitt G: Spektroskopische Charakterisierung der Rubber Oxygenase RoxA aus Xanthomonas sp. 35Y. Doctoral thesis, University of Stuttgart, Germany, 2012.
- 25.Morrison M, Horie S. Determination of heme a concentration in cytochrome preparations by hemochromogen method. Anal Biochem. 1965;12:77–82. doi: 10.1016/0003-2697(65)90144-2. [DOI] [PubMed] [Google Scholar]
- 26.Yang HJ, Park KH, Sin S, Lee J, Park S, Kim HS, Kim J. Characterization of heme ions using MALDI-TOF MS and MALDI FT-ICR MS. Int J Mass Spectrom. 2013;343–344:37–44. doi: 10.1016/j.ijms.2013.03.014. [DOI] [Google Scholar]
- 27.Andreoletti P, Mouesca J-M, Gouet P, Jaquinod M, Capeillère-Blandin C, Jouve HM. Verdoheme formation in Proteus mirabilis catalase. Biochim Biophys Acta. 2009;1790:741–753. doi: 10.1016/j.bbagen.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 28.Schenkman KA, Marble DR, Burns DH, Feigl EO. Myoglobin oxygen dissociation by multiwavelength spectroscopy. J Appl Physiol. 1997;82:86–92. doi: 10.1152/jappl.1997.82.1.86. [DOI] [PubMed] [Google Scholar]
- 29.Pond AE, Roach MP, Sono M, Rux AH, Franzen S, Hu R, Thomas MR, Wilks A, Dou Y, Ikeda-Saito M, Ortiz de Montellano PR, Woodruff WH, Boxer SG, Dawson JH. Assignment of the heme axial ligand(s) for the ferric myoglobin (H93G) and heme oxygenase (H25A) cavity mutants as oxygen donors using magnetic circular dichroism. Biochemistry. 1999;38:7601–8. [DOI] [PubMed]
- 30.Seidel J, Schmitt G, Hoffmann M, Jendrossek D, Einsle O. Structure of the processive rubber oxygenase RoxA from Xanthomonas sp. Proc Natl Acad Sci U S A. 2013;110:13833–13838. doi: 10.1073/pnas.1305560110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linos A, Berekaa MM, Steinbüchel A, Kim KK, Sproer C, Kroppenstedt RM. Gordonia westfalica sp. nov. a novel rubber-degrading actinomycete. Int J Syst Evol Microbiol. 2002;52:1133–9. [DOI] [PubMed]
- 32.Arenskotter M, Baumeister D, Berekaa MM, Pötter G, Kroppenstedt RM, Linos A, Steinbüchel A. Taxonomic characterization of two rubber degrading bacteria belonging to the species Gordonia polyisoprenivorans and analysis of hyper variable regions of 16S rDNA sequences. FEMS Microbiol Lett. 2001;205:277–282. doi: 10.1016/S0378-1097(01)00497-9. [DOI] [PubMed] [Google Scholar]
- 33.Bode HB, Kerkhoff K, Jendrossek D. Bacterial degradation of natural and synthetic rubber. Biomacromolecules. 2001;2:295–303. doi: 10.1021/bm005638h. [DOI] [PubMed] [Google Scholar]
- 34.Jendrossek D, Müller B, Schlegel HG. Cloning and characterization of the poly(hydroxyalkanoic acid)-depolymerase gene locus, phaZ1, of Pseudomonas lemoignei and its gene product. Eur J Biochem. 1993;218:701–710. doi: 10.1111/j.1432-1033.1993.tb18424.x. [DOI] [PubMed] [Google Scholar]
- 35.Berry EA, Trumpower BL. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 36.Tumlirsch T, Sznajder A, Jendrossek D. Formation of polyphosphate by polyphosphate kinases and its relationship to PHB accumulation in Ralstonia eutropha H16. Appl Environ Microbiol. 2015;81:8277–8293. doi: 10.1128/AEM.02279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altenbuchner J, Viell P, Pelletier I. Positive selection vectors based on palindromic DNA sequences. Meth Enzymol. 1992;216:457–466. doi: 10.1016/0076-6879(92)16042-I. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The DNA sequence of the R. rhodochrous RPK1 lcp and 16S rRNA gene have been submitted to NCBI and are available under the accession No KU140417 and KU140418. The R. rhodochrous RPK1 strain has been deposited at the Deutsche Sammlung für Mikroorganismen und Zellkulturen (DSMZ) and can be obtained once the Nagoya protocol regulations have been finished. Until then, strain RPK1 can be obtained from the lab of S.W. and K.U. upon request.


