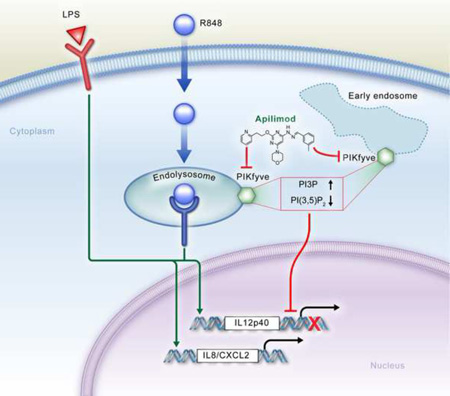
Summary
Toll-Like Receptor (TLR) signaling is a key component of innate immunity. Aberrant TLR activation leads to immune disorders via dysregulation of cytokine production, such as IL-12/23. Herein we identify and characterize PIKfyve, a lipid kinase, as a critical player in TLR signaling using apilimod as an affinity tool. Apilimod is a potent small molecular inhibitor of IL-12/23 with an unknown target and has been evaluated in clinical trials for patients with Crohn’s disease or rheumatoid arthritis. Using a chemical genetics approach, we show that it binds to PIKfyve and blocks its phosphotransferase activity, leading to selective inhibition of IL-12/23p40. Pharmacological or genetic inactivation of PIKfyve is necessary and sufficient for suppression of IL-12/23p40 expression. Thus, we have uncovered a novel phosphoinositide-mediated regulatory mechanism that controls TLR signaling.
Graphical Abstract
Introduction
Toll-Like Receptors (TLRs) recognize molecules that are broadly shared by pathogens yet are distinguishable from host molecules. Activation of TLR signaling induces expression of genes that orchestrate the inflammatory and anti-pathogen responses (Takeuchi and Akira, 2010). It is known that dysregulated TLR signaling, plays a role in a number of autoimmune diseases primarily due to dysregulation of cytokine production (Krieg and Vollmer, 2007). IL-12 and IL-23 are cytokines of particular importance; they share the common IL12p40 subunit and are key drivers for the development of T helper cell type 1 (Th1) and type 17 (Th17) cells, respectively (Langrish et al., 2004). Both cytokines are clinical targets for treatment of autoimmune disease (Abraham and Cho, 2009; Gately et al., 1998).
The activation of these cytokines is under tight control by the TLR signaling network, including NFκB, IRF, MAPK and PI3K pathways. In addition to these signaling pathways, many positive or negative regulators have also been recently discovered to play important roles in TLR-cytokine expression. Classical genetics has played a central role in the discovery of many key regulators in TLR biology. A number of critical nodes were successfully identified by forward genetics. For example, UNC93B, a key chaperone for endosomal TLRs, was identified from an N-ethyl-N-nitrosourea (ENU) mutagenesis-based screen (Tabeta et al., 2006). Hypothesis-driven reverse genetics has become a dominant approach over the past 10–15 years in elucidating TLR signaling network using specific gene deletion or mutation approaches (Medzhitov et al., 1997; Yamamoto et al., 2003).
In addition to these classical gene discovery approaches, forward chemical genetics has emerged as another powerful approach to illuminate the biological function of genes, particularly in the case where a gene is a multifunctional enzyme or its deletion/mutation leads to embryonic lethality. This is achieved by identifying the target of a small molecular compound that induces a phenotype of interest (Schreiber, 2000; Spring, 2005; Kung and Shokat, 2005). A number of proteins that govern fundamental cellular processes have been characterized using small molecular drugs. For example, the molecular target and mechanism of action for rapamycin, a widely used immunosuppressant during organ and bone marrow transplantation, was elucidated by forward chemical genetics (Sabatini et al., 1994; Kunz et al., 1993; Brown et al., 1994; Chiu et al., 1994). Rapamycin interacts with the FKBP-rapamycin-binding (FRB) domain of the mammalian target of rapamycin (mTOR) and inhibits its kinase activity within mTORC1 complex (containing mTOR, Raptor and mLST8) during acute administration. This discovery led to an explosion of studies revealing important roles of mTOR in multiple biological processes using rapamycin as an inhibitor, including its function in TLR9-induced IFNα production (Cao et al., 2008). However, the limitation of available potent small molecules which perturb interesting biological pathways has posed challenges in fully utilizing the potential of chemical genetics. Although there are very few disease-modifying compounds targeting the TLR-cytokine axis (Hennessy et al., 2010), apilimod emerges as an ideal tool for novel gene discovery in TLR signaling.
Apilimod is the first small molecule developed to specifically block TLR-mediated IL-12/23 production that has entered clinical trials (Wada et al., 2007; Wada et al., 2012). It has been tested in patients with Crohn’s disease (CD), rheumatoid arthritis (RA) (Billich, 2007) and psoriasis (Wada et al., 2012). While apilimod showed clinical improvement in patients with active CD in a phase I/II trial, no significant improvement over placebo was seen in a phase II trial (Sands et al., 2009), though it was generally well-tolerated. At the onset of these trials the therapeutic target(s) for apilimod were unknown, making the assessment of efficacy and toxicity difficult due to a lack of appropriate pharmacodynamic (PD) markers. Without knowledge of the target, further progress in the development or improvement of this drug is challenging.
In this study, using apilimod as an affinity probe, we found phosphatidylinositol-3-phosphate 5-kinase (PIKfyve) to be the molecular target of this drug. PIKfyve is a 240kD lipid kinase that phosphorylates the D-5 position in endosomal phosphatidylinositol-3-phosphate (PI3P) to yield the 3,5-bisphosphate (PI(3,5)P2) (Shisheva, 2008). This kinase binds to PI(3)P via its FYVE domain. PIKfyve is critical for maintaining the proper morphology of the endosome/lysosome. The enlarged endosome/lysosome structure was observed in cells expressing PIKfyve dominant negative or siRNA (Ikonomov et al., 2001; Rutherford et al., 2006). Vac14 and Sac3 were reported to form a regulatory complex with PIKfyve to control the endosomal phosphoinositide metabolism (Sbrissa et al., 2004; Sbrissa et al., 2007). The vacuolization and low PI(3,5)P2 levels in fibroblasts isolated from Vac14 and Sac3 null mice suggest that both are required for maximal PIKfyve activity (Jin et al., 2008; Zhang et al., 2007; Chow et al., 2007). PIKfyve-mediated PI(3,5)P2 signaling was reported to regulate endosomal trafficking and play a key role in multiple biological processes, such as GLUT4 translocation and retroviral budding (Ikonomov et al., 2002; Jefferies et al., 2008). Neurodegeneration was observed in both human and mice with Vac14 and Sac3 mutations (Zhang et al., 2007; Chow et al., 2007; Jin et al., 2008), possibly due to the deficiency of autophagy-mediated intracellular trafficking in cells lacking PI(3,5)P2 (Ferguson et al., 2009). Here we report that apilimod specifically binds to PIKfyve and inhibits its lipid kinase activity, and demonstrate PIKfyve’s novel function in controlling TLR-mediated cytokine expression. These novel findings unravel the critical role of PI(3,5)P2 in modulating TLR signaling and control of discrete immune cell functions.
Results
Apilimod selectively inhibits TLR-induced cytokine expression
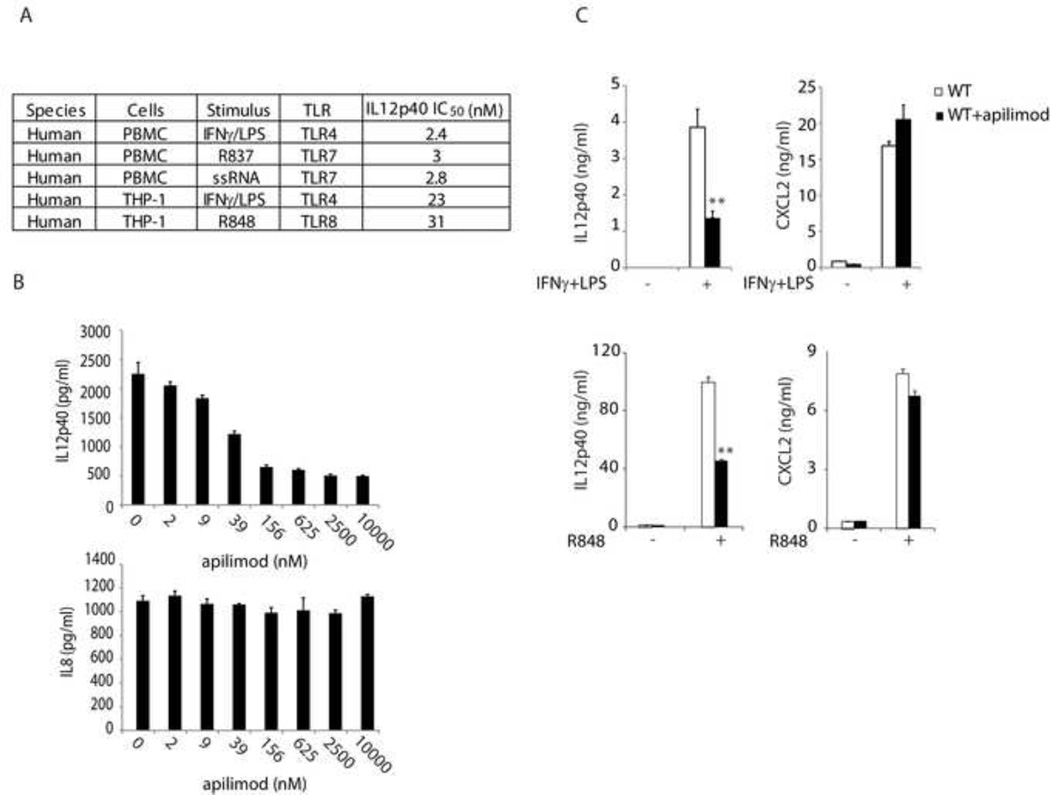
Apilimod is a 1,3,5-triazine derivative discovered in a cell-based screen aimed at identifying inhibitors of IL-12 production, using IFNγ/LPS-stimulated human peripheral blood mononuclear cells (PBMCs) (Wada et al., 2007). We found that in addition to this TLR4-dependent pathway, apilimod inhibited the expression of IL12p40 induced by other TLRs (Figure 1A and S1), TLR ligand/agonist pairs used in manuscript: LPS (TLR4), ssRNA (TLR7), R837 (TLR7), R848 (TLR7/8). This inhibitor thus regulates the expression of IL-12 induced by multiple TLRs. Consistent with the results in human PBMC (Wada et al., 2007), apilimod selectively inhibited the production of IL12p40, whereas it had little effect on IL8 production in THP-1 cells (Figure 1B). Apilimod thus exhibited potent yet selective cellular activity in TLR pathways. In addition, we also evaluated the activity of apilimod in mouse cells. As shown in Figure 1C, apilimod selectively inhibited IFNγ/LPS or R848-induced production of IL12p40 but not CXCL2 in mouse bone marrow-derived dentritic cells (BMDC). Apilimod thus exhibited selective cellular activity across multiple species. It would therefore be important to identify the molecular target(s) for apilimod and reveal a potential novel regulatory mechanism for TLR cytokine modulation.
Figure 1. Apilimod selectively inhibits TLR-induced cytokine expression.
The production of cytokines was measured by ELISA following overnight stimulation. Representative results were shown from three independent experiments. (A) Inhibition of IL12p40 by apilimod following stimulation of cells with IFNγ (50 ng/ml)/LPS (1 µg/ml), R837 (10 µg/ml) and R848 (10 µg/ml), or ssRNA (ORN 02, 5 µg/ml). (see also Figure S1) (B) THP-1 cells were treated with apilimod in the presence of IFNγ (50 ng/ml)/LPS (1 µg/ml). The data were analyzed using one-way Anova method (P<0.0001), indicating a significant effect of apilimod on TLR4-induced expression of IL12p40. (C) Mouse BMDCs were treated with apilimod (1 µM) and challenged with IFNγ (50 ng/ml)/LPS (1 µg/ml) or R848 (0.1 µM). **, P<0.01 using Student’s t-Test. Data represent mean values ± SD.
Apilimod binds to and inhibits PIKfyve kinase activity
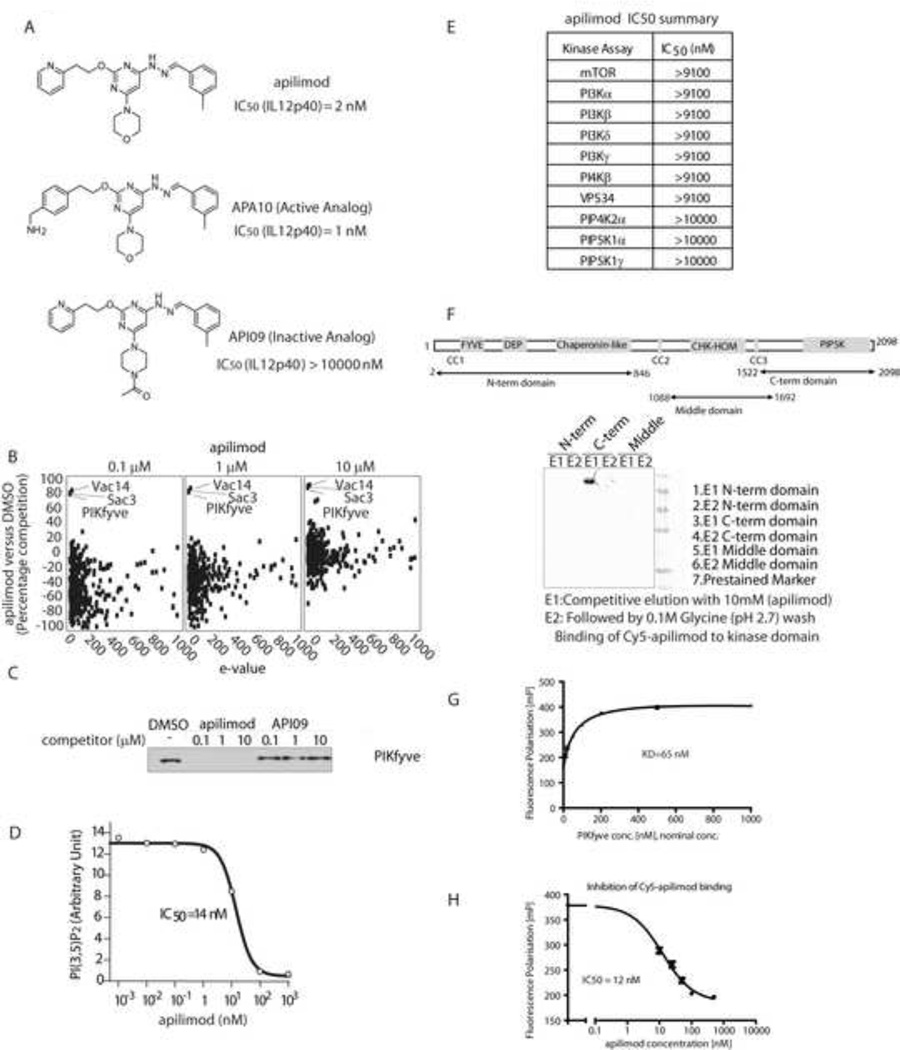
First, we aimed to identify the binding partners of apilimod to yield potential molecular target(s) by employing a quantitative chemical proteomics approach (Huang et al., 2009). A bioactive analogue of apilimod, APA10 (Figure 2A), was immobilized on a gel matrix and incubated with a THP-1 cell extract premixed with either DMSO, or with an excess of apilimod. Unbound proteins were removed by washing and specifically bound proteins were eluted, digested and identified using LC-MS/MS. While a total of 974 retained proteins were identified (Table S1), only three proteins were significantly competed from the matrix by increasing dose of apilimod. The three proteins identified comprise all known members of the PIKfyve regulatory complex, PIKfyve, Vac14 and Sac3 (Figure 2B). In an independent pull down experiment, PIKfyve protein was competed from the matrix with apilimod but not with an inactive analog API09 (Figure 2A and 2C). Vac14 and Sac3 form a complex with PIKfyve and both are required for maintaining PIKfyve activity in converting PI3P to PI(3,5)P2. We conclude that apilimod specifically interacts with the PIKfyve regulatory complex in cells.
Figure 2. Apilimod binds to and inhibits PIKfyve kinase activity.
(A) Structure of apilimod and its analogs and their IC50 for IFNγ (50 ng/ml)/LPS (1 µg/ml) induced-IL12p40 secretion in THP-1 cells. (B) Scatter plot depicting proteins identified in a quantitative chemical proteomics experiment. Proteins are plotted as a function of the percentage competition with apilimod relative to DMSO, on the y-axis, versus the interaction specificity (e-value) on the x-axis. (see also Table S1 for protein list for apilimod quantitative chemical proteomics) (C) U2OS cells lysates were pre-incubated with DMSO or indicated competitor compound for 30 mins before adding beads with immobilized APA10. The PIKfyve captured on the beads was detected by Western Blot. (D) The effect of apilimod on PIKfyve kinase activity was measured in vitro, by quantifying the ratio of synthesized PI(3,5)P2 to an internal standard. (E) Lipid kinase inhibition profiling for apilimod. (see also Table S2 for protein kinase inhibition profiling for apilimod) (F) The binding of apilimod with indicated PIKfyve truncants was assessed using a APA10-based Sepharose HP affinity resin. (G) The KD of Cy5-apilimod to PIKfyve kinase domain was determined. (H) The IC50 of apilimod for the interaction between Cy5-apilimod and PIKfyve kinase domain was determined.
To address the mode of action of apilimod on the PIKfyve complex, an in vitro kinase assay measuring the conversion of PI(3)P to PI(3,5)P2 was developed. Apilimod inhibited PIKfyve kinase activity with IC50=14 nM (Figure 2D). In contrast, apilimod had no activity toward other lipid kinases and protein kinases, including PIP4K, PIP5K, mTOR, PI3K and PI4K isoforms (Figure 2E and Table S2). In addition, we mapped the apilimod binding region to the PIKfyve kinase domain (amino acids 1522 to 2098, Figure 2F). Furthermore, in a competition assay using fluorescence polarization of a labeled apilimod analogue (Cy5-apilimod), unlabeled apilimod showed an IC50 of 12 nM, indicating that this interaction is responsible for the inhibition of the catalytic activity described above (Figure 2G and 2H). These results demonstrated that apilimod is a potent yet highly selective PIKfyve lipid kinase inhibitor.
Apilimod inhibits PIKfyve kinase activity in cells
To determine if apilimod affected PIKfyve function in cells, we quantified cellular phosphoinositides upon apilimod treatment. As shown in Figure 3A, HPLC analysis of deacylated lipids revealed a specific dose-dependent decrease of PI(3,5)P2 in HeLa cells treated with apilimod but not with the inactive analog API09 for 2 hrs. Additionally, a marked increase (up to 2.5-fold) of PI(3)P was observed, which clearly demonstrated aplimod inhibited PIKfyve activity in cells and blocked the conversion of PI(3)P to PI(3,5)P2. Strikingly, even a low dose treatment of apilimod (10 nM) resulted in a marked decrease of cellular PI(3,5)P2 levels. In contrast, all other phosphoinositides tested were largely unchanged even when cells were treated with 1 µM apilimod. These observations are consistent with apilimod being a highly selective and potent inhibitor of PIKfyve (Figure 2D and 2E).
Figure 3. Apilimod inhibits PIKfyve kinase activity in cells.
(A) HeLa cells metabolically labeled with [3H] inositol for 72 hrs and treated with two doses of apilimod or the inactive analog API09 for 120 mins. Lipids were extracted, deacetylated and analysised by HPLC. Data were normalized with cells treated with DMSO vehicle control, and analyzed using Student’s t-Test (*, P<0.05, **, P<0.01), showing a significant difference of the indicated phosphoinositide levels between API09 and apilimod-treated samples. (B) Images of RAW264.7 cells treated with DMSO, apilimod (10 nM) or API09 (10 nM) for 3 hrs. The results are representive from three independent experiments. (C) Electron microscope images of RAW264.7 cells treated with apilimod (200 nM) for 6 hrs. (D) RAW264.7 cells stably expressing GFP-FYVE (early endosome marker) or mCherry-CD63 (endolysosome marker) were treated with DMSO or apilimod (100 nM) for 60 mins. Images were acquired using a Zeiss LSM510 confocal microscope with 63X lens. Scale bar, 5 µm. (E) A549 cells transfected with GFP-PIKfyve or GFP-PIKfyve K1831E mutant were treated with 10 nM apilimod for 4 hrs and imaged using a Zeiss Axiovert microscope. Arrow indicates GFP positive cells. The results are representive from three independent experiments.
As PIKfyve is reported to be critical for maintaining the integrity of endosomes and lysosomes (Dove et al., 2009), we then examined cell morphology. Upon treatment of apilimod, we observed enlarged vacuoles in RAW264.7 cells (Figure 3B and 3C), which were also seen upon inactivation of PIKfyve using a dominant-negative PIKfyve mutant or RNAi (Ikonomov et al., 2001; Rutherford et al., 2006). In contrast, treatment with API09 did not induce vacuoles (Figure 3B). Using an early endosome (GFP-FYVE) or an endolysosome (mCherry-CD63) marker, we showed that the enlarged vacuoles originate from both early endosome and endolysosome, which is consitant with the findings in PIKfyve deficient cells (Rutherford et al., 2006) (Figure 3D). Hence, the vacuoles induced by apilimod are due to the disruption of PIKfyve activity. Indeed, overexpression of wild type GFP-PIKfyve caused the disappearance of vacuoles induced by 10 nM apilimod in A549 cells, whereas overexpression of the dominant negative GFP-PIKfyve-K1831E “kinase dead” mutant caused more extensive vacuole formation (Figure 3E). These data are fully consistent with PIKfyve being the cellular target of apilimod.
PIKfyve modulates TLR-induced IL12p40 expression
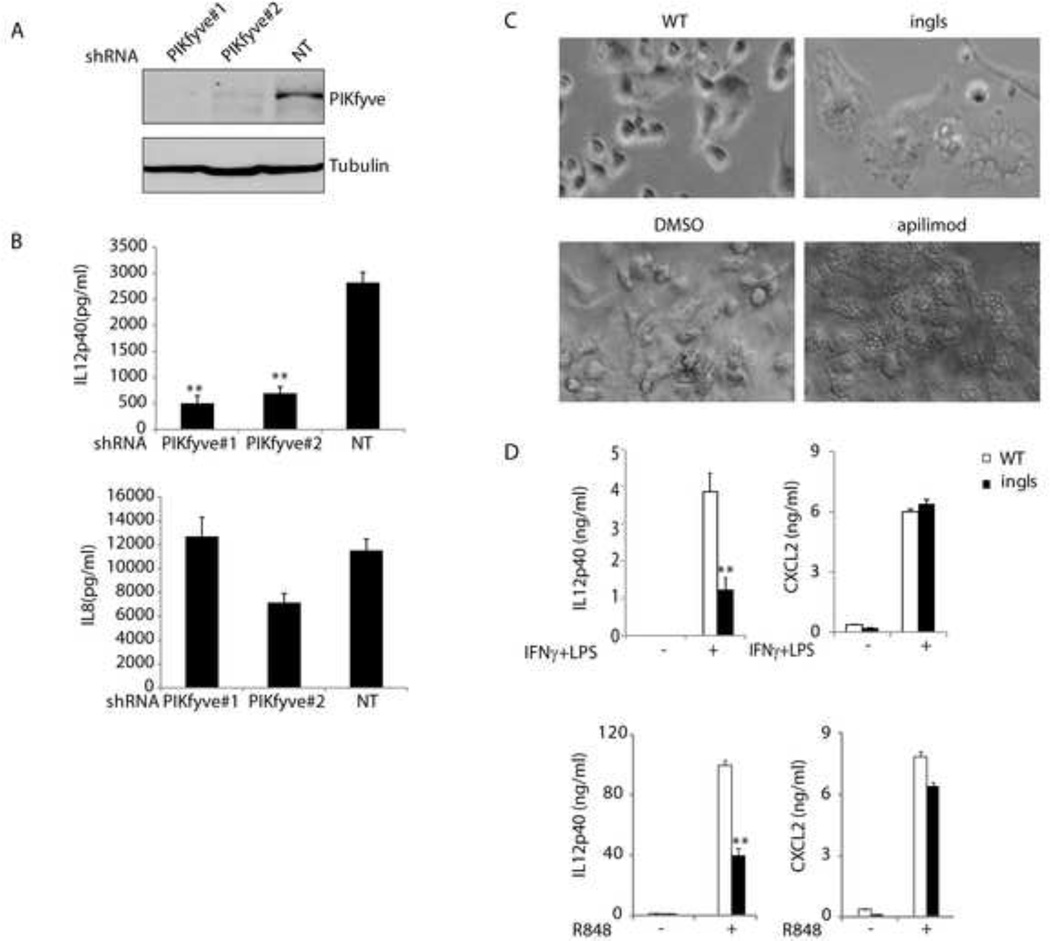
To establish a functional correlation between PIKfyve and TLR-cytokine production, we first analyzed THP-1 cells infected with PIKfyve shRNAs. As shown in Figures 4A and 4B, knockdown of PIKfyve in THP-1 cells led to decreased production of IL12p40, whereas it had little effect on IL8 production. This is consistent with the cytokine profile observed in apilimod-treated cells (Figure 1B).
Figure 4. PIKfyve modulates TLR-induced IL12p40 expression.
(A) THP-1 cells expressing indicated shRNAs were lysed and blotted with indicated antibodies. (B) THP-1 cells expressing indicated shRNAs were stimulated with IFNγ (50 ng/ml)/LPS (1 µg/ml) overnight. The cytokine production was measured by ELISA. **, P<0.01 using Student’s t-Test, indicating a significant difference on IL12p40 production between cells expressing control (NT) and PIKfyve shRNA. (C) Images of BMDCs from wild-type (WT) and ingls mice and those from WT BMDCs treated with DMSO or apilimod (1 µM). (D) BMDCs from WT or ingls mice were challenged with IFNγ (50 ng/ml)/LPS (1 µg/ml) or R848 (0.1 µM). The cytokine production was measured by ELISA following overnight stimulation. Representative results from three independent experiments. **, P<0.01 using Student’s t-Test, indicating a significant difference between the samples from WT and ingls mice with the same treatment.
Furthermore, dendritic cells from mutant mice with reduced PIKfyve activity were also defective in TLR signaling. As PIKfyve knock-out mice are embryonic lethal (Ikonomov et al., 2011), we utilized the spontaneous mutant mouse, ingls, which carries a missense mutation in Vac14 (L156R) (Jin et al., 2008). Ingls mutation interrupts the interaction of PIKfyve with Vac14, which abolishes the kinase activity of PIKfyve and reduces the level of PI(3,5)P2, a phenotype closely mimicking that seen during PIKfyve inhibitor application. Indeed, bone-marrow-derived DCs (BMDCs) from Vac14ingls/ingls mice exhibited profound vacuole formation as seen in apilimod treated BMDCs from wild-type mice (Figure 4C). As shown in Figure 4D, production of IL12p40 but not CXCL2 was impaired in CD11c+ DCs derived from Vac14ingls/ingls bone marrow upon treatment with IFNγ/LPS or R848. These data are consistent with the cytokine profiling patterns observed in cells treated with apilimod (Figure 1C). Taken together, our data suggest that PIKfyve is the molecular target of apilimod, revealing a new link between PIKfyve and cytokine expression induced by TLR signaling.
Discussion
In this study, we characterized a novel regulator of TLR signaling through identification of the target of a small-molecule compound. The results uncovered a novel mechanism for selective regulation of TLR-driven cytokine expression mediated by localized control of minor species of phosphoinositides in cells. By characterizing a selective TLR-cytokine inhibitor, our study has illustrated the power of chemical genetic approaches in provinding new insights into TLR signaling. Although a PIKfyve inhibitor YM201636 has been described previously (Jefferies et al., 2008), it is a weaker PIKfyve inhibitor that possesses activity toward PI3K family members (Jefferies et al., 2008; Ikonomov et al., 2009;). Therefore, apilimod is a more desirable tool to be used in elucidating the role of PIKfyve in important biological processes.
Using high content screen and cell-based high throughput function screen technology, a number of small molecular agonists or antagonists for specific phenotypic changes have been identified. While drug discovery pipeline is greatly expanded with the discovery of these compounds, the absence of efficacy target(s) and of a mechanism of action of these leads hindered the progress of pipeline development, such as compound optimization and safety evaluation. A practicable drug target “fishing” strategy is desperately needed to hunt for the “prey” of these compounds. Recently, chemoproteomics has been shown to be a powerful tool to address this issue (Huang et al., 2009). Our studies provided a successful case of chemoproteomics-driven target identification. The PIKfyve regulatory complex was the only proteins competed off from the affinity matrix in the presence of active analog among the 974 proteins quantified from total cell lysate using LC-MS/MS. This is the key step for identifying the binding partner of apilimod, and enabled a rapid target validation. Moreover, this approach could provide a rapid and exhaustive method to predict the specificity of a compound. As in this case, Apilimod was later to show has no activity towards other protein and lipid kinases (Figure 2E and Table S2).
The target elucidation of apilimod provides a number of benefits for drug development. First, a PIKfyve activity-based biochemical screen could be performed to find other inhibitors. Second, the safety issues, particularly “on-target” vs. “off-target” effects of compound, could be monitored and addressed during the development. More importantly, the target identification provides an opportunity to shed light on the molecular mechanism of PIKfyve inhibition mediated TLR signaling defect. This not only expands the therapeutic indications for Apilimod, also enable the expansion of PIKfyve network downstream of TLR signaling to reveal more druggable nodes which could potentially bypass the “on-target’ toxicity induced by PIKfyve inhibition.
It is well established that inositol phospholipids play a critical role in intracellular signaling, and several of these specifically regulate TLR pathways. For example, Kagan et al. proposed that PI(4,5)P2 mediated TIRAP recruitment to the plasma membrane is required for delivery of MyD88 to TLR4 (Kagan and Medzhitov, 2006). Also, class I PI3K inhibitors abolish endosomal TLR-induced type I IFN production in pDC by blocking nuclear translocation of IRF7 but not the uptake and endosomal trafficking of ligands (Guiducci et al., 2008). In contrast, PI3K could function as a negative regulator for TLR-induced IL-12 synthesis in DC and THP-1 cells (Fukao et al., 2002). Here we show that a mammalian PI kinase, PIKfyve, regulates TLR signaling via modulation of PI(3)P and PI(3,5)P2 levels. Interestingly, PI(3)P and PI(3,5)P2 are two of the least abundant phosphoinositide species in cells (~0.25% of cellular inositol lipids), yet are critical for maintenance of a normal endosomal compartment. Their critical role becomes clear in the presence of PIKfyve inhibitors: where treated cells form large vacuoles of endosomal/lysosomal origin (Figure 3D). It remains to be seen how PIKfyve-depednent modulation of endosomal PI(3)P and PI(3,5)P2 levels regulate specific TLR-cytokine expression. As the PIKfyve kinase knock-in mice are embryonic lethal (data not shown), apilimod will become an important tool to dissect the function of PIKfyve lipid kinase activity in TLR pathways as well as other biological processes.
In summary, we uncovered a new role of PIKfyve in selectively regulating TLR-signaling. Our results thus propose a new druggable node for selective regulation of TLR-induced IL-12/23 expression, and in turn, provide opportunities for pharmacological intervention in IL-12/23-mediated diseases.
Significance
Selective low-molecular-weight inhibitors of aberrant cytokine expression are a highly desirable therapy to treat autoimmune disorders (Dinarello, 2010). While several small-molecule signaling modulators are under investigation, such as p38 and IKKβ inhibitors, there are very few disease-modifying compounds that target the TLR-cytokine axis and none of which that have a known mechanism of action (Hennessy et al., 2010). Our results uncover the molecular and therapeutic target of a clinically evaluated anti-inflammation drug which inhibits TLR-driven IL-12/23 production. This is the first report that demonstrates a role for PIKfyve in TLR signaling, and illustrates the power of chemical genetics approaches. The identification of PIKfyve as the molecular target of apilimod yields a novel signaling node within the TLR signaling cascade that could be targeted in inflammatory conditions, and allows the optimization of the compound for further development. More importantly, we demonstrate apilimod is a potent and highly selective PIKfyve inhibitor and provide the scientific community with a powerful tool to elucidate the role of PIKfyve kinase activity in multiple cellular processes including endolysomal intergrity, receptor trafficking, autophagy, and neurodegeneration among others.
Experimental Procedures
Constructs and reagents
The lentivirus mCherry-CD63 were made as previously described (Kim et al., 2008). pENTR-PIKfyve (mouse) was purchased from Open Biosystems. pENTR-PIKfyve K1831E was created using QuikChange® XL Site-Directed Mutagenesis Kit (Stratagene). The PIKfyve wild type and kinase-dead mutant were subcloned into pcDNA6.2/N-EmGFP-DEST vector (Invitrogen) using LR clonase.
All the control and gene specific shRNAs used in the manuscript were ordered from Sigma MISSION® shRNA collection. The control NT shRNA is the pLKO.1-puro Non-Mammalian shRNA Control. This control contains a shRNA insert that does not target human and mouse genes.
PIKfyve antibody was purchased from Abnova. Tubulin antibody was purchased from Abcam. All TLR ligands were purchased from Invivogen. All ELISA kits were obtained from R&D systems.
Cell culture
THP-1, RAW264.7, and A549 cells were purchased from ATCC and maintained under standard conditions described in the ATCC instructions. Human PBMCs were isolated using Ficoll (GE Health). The isolation and in vitro differentiation of mouse bone marrow cells were performed as previously described (Gilliet et al., 2002). Cell differentiation was confirmed by CD11c staining.
Compound affinity pull down and mass spectrometry analysis
Compound affinity purification, mass spectrometry and data analysis were performed essentially as previously described (Huang et al., 2009), except that the cell lysate was made from THP-1 cells and apilimod bioactive analogue APA10 was coupled to NHS (N-hydroxysuccinimide)-activated Sepharose 4 beads. The e-value on x-axis indicates the frequency of each protein which was detected in prior proteomics experiments. Small e-values are considered advantageous because this suggests a specific protein interaction.
Expression of human PIKfyve
HEK293T cells were seeded in 6-well plates at a density of 5×105 cells per well in DMEM medium containing 10% FCS. After 24 hrs, when a confluency of about 50% had been reached, the cells were transfected with an expression plasmid encoding human PIKfyve as a fusion protein with GST. Transfection was done using Fugene (Roche; 3 µl per well) and 1 µg/well of the DNA. Forty-eight hours after the transfections, cells were washed with ice-cold PBS and then scraped into 0.5 ml ice-cold lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 10 mM β-glycerophosphate, protease inhibitor [Roche]). Following centrifugation, the supernatant was harvested, followed by addition of glutathione-Sepharose beads (GE Healthcare; equilibrated with lysis buffer; approx. 100 µl of packed beads per ml of lysate). After 1hr incubation with end-over-end agitation, the beads were washed 3-times with lysis buffer with 1% Nonidet P-40, then 3-times with lysis buffer without NP40. Beads were collected by centrifugation at 500xg and used directly in the enzyme assay.
In vitro kinase assay
5µL of the compound or its dilution series in the assay buffer (25 mM HEPES buffer, 1 mM DTT, 5mM glycerophosphate, 2.5 mM MgCl2, 2.5 mM MnCl2, 120 mM NaCl, 1 mM EDTA), 10 µL of human PIKfyve enzyme purified from HEK293 cell lysates and 5 µL 50 µM ATP solution were pre-incubated for 5 mins at the room temperature. Then 5 µL of the di-C8 PI(3)P substrate (Echelon Biosciences) was added into the wells to have the final concentration of the substrate at 10 mM to initiate the reaction. The reactions were allowed to proceed at room temperature for 2 hrs and then quenched by adding 25 µL Acetonitrile/Water (50:50v/v) solution, which also contained 25 mM EDTA and 1 µM di-C8-sPI(3,4)P2 (internal standard, Cayman Chemical). After centrifugation, the plate was sealed for LC/MS/MS analysis. The MS/MS Data was acquired on a Thermo TSQ triple quadruple MS system (Thermo Fisher Scientific) coupled to a Thermo LX2 HPLC system. The LC system was run in the reversed-phase chromatography mode using a Waters XBridge C18 column (2.1×30 mm, 3.5 mm). The LC mobile phase contained 0.1% dimethylisopropylamine (DMIPA) in either water (mobile A), or acetonitrile (mobile B). The flow rate was 0.8 ml/min with a rapid gradient from 5 to 95% B in 1.2 min. The injection volume was 5 ml. The MS/MS transition for PI(3,5)P2 was 745.06 -> 158.98 with 23 eV as collision energy while the transition for internal standards was 761.06 ->158.98.
All other kinase assays were performed internally by Novartis kinase profiling service except for assays for PIP4K and PIP5K. The effect of apilimod on PIP4K and PIP5K isoforms was determined by Millipore IC50 Profiler Service.
Measurement of cellular inositol lipids levels
HeLa cells were incubated for 72 hrs in inositol-free DMEM (MP Biomedicals) supplemented with 10% dialyzed FBS (Invitrogen) and 25 µCi/ml 3H-myo inositol (MP Biomedicals). Compounds were then added to the cells and incubated for 120 mins. Prior to lipid extraction, cells were washed twice with PBS and incubated 15 minutes in inositol-free DMEM supplemented with 10% dialyzed FBS and compounds. Cells were then treated with 0.7 ml 4.5% perchloric acid in ice for 15 mins, scraped and centrifuged. Pellets were washed twice with ice-cold 1 ml 0.1 M EDTA, and deacylated as described (C.J Kirk et al., 1990). Deacylated phosphoinositides were separated using high performance liquid chromatography (Shimadzu) using a flow rate of 0.5 ml/min and a gradient of degassed H2O (pump A) and 1 M (NH4)2HPHO4 pH 3.8 (pump B) as follows. 0% B for 5 mins; 0–4% B for 15 mins; 4% B for 80 mins; 4–12% B for 20 mins; 12% B for 60 mins, 12–80% B for 40 mins; 80% B for 35 mins, 80-0% B for 5 mins. Peaks were identified using deacylated 32P-standards of PI(3)P, PI(3,4)P2, PI(3,5)P2, and PI(3,4,5)P3 and internal standards. Radioactivity was detected by an online flow scintillation analyzer (B-RAM, IN/US).
Apilimod/PIKfyve truncants binding assay
The Sf9 insect cells expressing PIKfyve truncants with an N-terminal His tag were lysed in 50 mM Tris pH 8.0, 0.3M NaCl, 10% glycerol, 2 mM TCEP, 0.05% Tween, including a cocktail of protease inhibitors (0'Complete EDTA free). The supernatant was filtered and loaded on a SpinTrap device (GE Healthcare) containing NHS-activated Sepharose HP coupled to the apilimod derivative APA10. After 30 min incubation, the resin was washed with TBS. A first elution step was performed with 10 µM apilimod in TBS. A second elution step was performed with 100 mM glycine buffer (pH 2.7). The eluate was quantitatively precipitated using sodium deoxycholate and trichloroacetic acid. The pellet was dissolved in 30 µl sample buffer for SDS-PAGE and Western-Blot.
PIKfyve kinase domain/apilimod affinity fluorescence polarization assay
To measure the Cy5-apilimod/PIKfyve kinase domain binding affinity, an equal volume of 10 nM Cy5-apilimod and of different concentrations of PIKfyve kinase domain proteins were incubated in 384 black microtiter plates for 90 min. After a short spin, fluorescence polarization was measured on a Perkin Elmer Envision or a Molecular Devices Analyst GT plate reader. The KD was determined by fitting of a hyperbola using the program Prism (GraphPad Software, Inc.). Each data point was derived from n=4.
Competition by unlabeled apilimod was measured under the same conditions. Their IC50 was determined by fitting of a sigmoidal dose-response curve using the Prism.
Virus production and infection
Lentivirus shRNA was packaged in 293T cells via co-transfection of pLP1, pLP2 and pLP/VSVG. Retrovirus was packaged in 293T cells via co-transfection of Gag-Pol and VSVG. RAW cells were infected with supernatant containing virus plus polybrene (Sigma, final concentration 8 µg/ml) overnight. The stable cell lines were maintained in medium containing puromycin (6 µg/ml) or G418 (400 µg/ml). THP-1 cells were infected with supernatant containing lentivirus plus polybrene and HEPES (final concentration 10 mM) via spin-infection (90 min, 2100 RPM). The stable cell lines were maintained in medium containing puromycin (2 µg/ml).
Live cell imaging
For live cell imaging, cells were seeded into a P35 glass bottom dish (MatTek) (4×105 cells/dish) and maintained in phenol-red-free DMEM supplemented with 5% FBS and 25mM HEPES (pH 7.4). Images were acquired using a Zeiss LSM510 Meta confocal microscope with a Plan-Apochromat 63x/1.4 Oil DIC lens. Zeiss LSM Image Browser was used for imaging analysis.
Mouse breeding
Mouse strains Vac14 <ingls> and C57 BL/6 were purchased from Jackson Laboratory. All mice were maintained according to NIBR animal guidelines.
Supplementary Material
Highlights.
PIKfyve is the molecular and cellular target of IL12/IL23 antagonist apilimod.
Apilimod is a potent and highly selective PIKfyve inhibitor.
PIKfyve, a mammalian PI kinase, is required for TLR-induced production of IL12/23.
Acknowledgments
G. Michaud, N. Tao, J. Zhu, R. Kutil, P. Bergman, F. Harbinski, E. Triantafellow, J. Leighton-Davies, C. Ostermeier, D. Scherer-Becker, M. Duckely, F. Freuler, C. Xin, E. McWhinnie, S. Lehmann, G. Tavares, S. Rieffel, A. Saenger, K. Twesten, P. Graff, H. Wang, J. Trappe provided technical assistance for PIKfyve target identification and validation; K. Hoegenauer and B. Thai synthesized Cy5-labeled apilimod; J. Vyas (Mass General Hospital) provided mCherry-CD63 construct; N. Watson (Whitehead Institute for Biomedical Research) provided EM analysis; A. Ho and A. Szilvasi provided assistance in imaging and flow cytometry; A. Titelbaum provided assistance in mouse breeding. We also like to thank J. Solomon, A. Visintin, F. Martin and S. Szabo for comments and advice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Financial Interests Statement Except R.T, A.A, B.Z, G.J., H.P. and P. DC, all other authors are employees of Novartis Institutes for Biomedical Research, Inc. Novartis is not involved in the discovery and development of apilimod.
Reference List
- Abraham C, Cho JH. IL-23 and autoimmunity: new insights into the pathogenesis of inflammatory bowel disease. Annu. Rev Med. 2009;60:97–110. doi: 10.1146/annurev.med.60.051407.123757. [DOI] [PubMed] [Google Scholar]
- Billich A. Drug evaluation: apilimod, an oral IL-12/IL-23 inhibitor for the treatment of autoimmune diseases and common variable immunodeficiency. IDrugs. 2007;10:53–59. [PubMed] [Google Scholar]
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- Kirk CJ, Morris AJ, Shears SB. Peptide Hormone Action: a Practical Approach. Oxford: IRL Press; 1990. pp. 151–158. [Google Scholar]
- Cao W, Manicassamy S, Tang H, Kasturi SP, Pirani A, Murthy N, Pulendran B. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat. Immunol. 2008;9:1157–1164. doi: 10.1038/ni.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu MI, Katz H, Berlin V. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl. Acad. Sci. U. S. A. 1994;91:12574–12578. doi: 10.1073/pnas.91.26.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, Szigeti K, Shy ME, Li J, Zhang X, Lupski JR, Weisman LS, Meisler MH. Mutation of FIG 4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello CA. Anti-inflammatory Agents: Present and Future. Cell. 2010;140:935–950. doi: 10.1016/j.cell.2010.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SK, Dong K, Kobayashi T, Williams FK, Michell RH. Phosphatidylinositol 3,5-bisphosphate and Fab1p/PIKfyve underPPIn endo-lysosome function. Biochem. J. 2009;419:1–13. doi: 10.1042/BJ20081950. [DOI] [PubMed] [Google Scholar]
- Ferguson CJ, Lenk GM, Meisler MH. Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum. Mol. Genet. 2009;18:4868–4878. doi: 10.1093/hmg/ddp460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T, Tanabe M, Terauchi Y, Ota T, Matsuda S, Asano T, Kadowaki T, Takeuchi T, Koyasu S. PI3K–mediated negative feedback regulation of IL-12 production in DCs. Nat. Immunol. 2002;3:875–881. doi: 10.1038/ni825. [DOI] [PubMed] [Google Scholar]
- Gately MK, Renzetti LM, Magram J, Stern AS, Adorini L, Gubler U, Presky DH. The interleukin-12/interleukin-12-receptor system: role in normal and pathologic immune responses. Annu. Rev Immunol. 1998;16:495–521. doi: 10.1146/annurev.immunol.16.1.495. [DOI] [PubMed] [Google Scholar]
- Gilliet M, Boonstra A, Paturel C, Antonenko S, Xu XL, Trinchieri G, O'Garra A, Liu YJ. The development of murine plasmacytoid dendritic cell precursors is differentially regulated by FLT3-ligand and granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 2002;195:953–958. doi: 10.1084/jem.20020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiducci C, Ghirelli C, Marloie-Provost MA, Matray T, Coffman RL, Liu YJ, Barrat FJ, Soumelis V. PI3K is critical for the nuclear translocation of IRF-7 and type I IFN production by human plasmacytoid predendritic cells in response to TLR activation. J. Exp. Med. 2008;205:315–322. doi: 10.1084/jem.20070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy EJ, Parker AE, O'Neill LAJ. Targeting Toll-like receptors: emerging therapeutics? Nat Rev Drug Discov. 2010;9:293–307. doi: 10.1038/nrd3203. [DOI] [PubMed] [Google Scholar]
- Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, Hild M, Shi X, Wilson CJ, Mickanin C, Myer V, Fazal A, Tomlinson R, Serluca F, Shao W, Cheng H, Shultz M, Rau C, Schirle M, Schlegl J, Ghidelli S, Fawell S, Lu C, Curtis D, Kirschner MW, Lengauer C, Finan PM, Tallarico JA, Bouwmeester T, Porter JA, Bauer A, Cong F. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Delvecchio K, Xie Y, Jin JP, Rappolee D, Shisheva A. The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve−/− embryos but normality of PIKfyve+/− mice. J. Biol. Chem. 2011;286:13404–13413. doi: 10.1074/jbc.M111.222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Mlak K, Shisheva A. Requirement for PIKfyve enzymatic activity in acute and long-term insulin cellular effects. Endocrinology. 2002;143:4742–4754. doi: 10.1210/en.2002-220615. [DOI] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Shisheva A. Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 5-kinase PIKfyve. J. Biol. Chem. 2001;276:26141–26147. doi: 10.1074/jbc.M101722200. [DOI] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Shisheva A. YM201636, an inhibitor of retroviral budding and PIKfyve-catalyzed PtdIns(3,5)P2 synthesis, halts glucose entry by insulin in adipocytes. Biochem. Biophys. Res. Commun. 2009;382:566–570. doi: 10.1016/j.bbrc.2009.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies HB, Cooke FT, Jat P, Boucheron C, Koizumi T, Hayakawa M, Kaizawa H, Ohishi T, Workman P, Waterfield MD, Parker PJ. A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding. EMBO Rep. 2008;9:164–170. doi: 10.1038/sj.embor.7401155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N, Chow CY, Liu L, Zolov SN, Bronson R, Davisson M, Petersen JL, Zhang Y, Park S, Duex JE, Goldowitz D, Meisler MH, Weisman LS. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell. 2006;125:943–955. doi: 10.1016/j.cell.2006.03.047. [DOI] [PubMed] [Google Scholar]
- Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- Krieg AM, Vollmer J. Toll-like receptors 7, 8, and 9: linking innate immunity to autoimmunity. Immunol. Rev. 2007;220:251–269. doi: 10.1111/j.1600-065X.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- Kung C, Shokat KM. Small-molecule kinase-inhibitor target assessment. Chembiochem. 2005;6:523–526. doi: 10.1002/cbic.200400393. [DOI] [PubMed] [Google Scholar]
- Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- Langrish CL, McKenzie BS, Wilson NJ, de Waal MR, Kastelein RA, Cua DJ. IL-12 and IL-23: master regulators of innate and adaptive immunity. Immunol Rev. 2004;202:96–105. doi: 10.1111/j.0105-2896.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- Rutherford AC, Traer C, Wassmer T, Pattni K, Bujny MV, Carlton JG, Stenmark H, Cullen PJ. The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J. Cell Sci. 2006;119:3944–3957. doi: 10.1242/jcs.03153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- Sands BE, Jacobson EW, Sylwestrowicz T, Younes Z, Dryden G, Fedorak R, Greenbloom S. Randomized, double-blind, placebo-controlled trial of the oral interleukin-12/23 inhibitor apilimod mesylate for treatment of active Crohn's disease. Inflamm. Bowel. Dis. 2009 doi: 10.1002/ibd.21159. [DOI] [PubMed] [Google Scholar]
- Sbrissa D, Ikonomov OC, Fu Z, Ijuin T, Gruenberg J, Takenawa T, Shisheva A. Core Protein Machinery for Mammalian Phosphatidylinositol 3,5-Bisphosphate Synthesis and Turnover That Regulates the Progression of Endosomal Transport. Journal of Biological Chemistry. 2007;282:23878–23891. doi: 10.1074/jbc.M611678200. [DOI] [PubMed] [Google Scholar]
- Sbrissa D, Ikonomov OC, Strakova J, Dondapati R, Mlak K, Deeb R, Silver R, Shisheva A. A Mammalian Ortholog of Saccharomyces cerevisiae Vac14 That Associates with and Up-Regulates PIKfyve Phosphoinositide 5-Kinase Activity. Mol. Cell. Biol. 2004;24:10437–10447. doi: 10.1128/MCB.24.23.10437-10447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SL. Target-oriented and diversity-oriented organic synthesis in drug discovery. Science. 2000;287:1964–1969. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- Shisheva A. PIKfyve: Partners, significance, debates and paradoxes. Cell Biol. Int. 2008;32:591–604. doi: 10.1016/j.cellbi.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring DR. Chemical genetics to chemical genomics: small molecules offer big insights. Chem. Soc. Rev. 2005;34:472–482. doi: 10.1039/b312875j. [DOI] [PubMed] [Google Scholar]
- Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, Shamel L, Herskovits AA, Portnoy DA, Cooke M, Tarantino LM, Wiltshire T, Steinberg BE, Grinstein S, Beutler B. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat. Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Wada Y, Cardinale I, Khatcherian A, Chu J, Kantor AB, Gottlieb AB, Tatsuta N, Jacobson E, Barsoum J, Krueger JG. Apilimod inhibits the production of IL-12 and IL-23 and reduces dendritic cell infiltration in psoriasis. PLoS. One. 2012;7:e35069. doi: 10.1371/journal.pone.0035069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Lu R, Zhou D, Chu J, Przewloka T, Zhang S, Li L, Wu Y, Qin J, Balasubramanyam V, Barsoum J, Ono M. Selective abrogation of Th1 response by STA-5326, a potent IL-12/IL-23 inhibitor. Blood. 2007;109:1156–1164. doi: 10.1182/blood-2006-04-019398. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zolov SN, Chow CY, Slutsky SG, Richardson SC, Piper RC, Yang B, Nau JJ, Westrick RJ, Morrison SJ, Meisler MH, Weisman LS. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17518–17523. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.