Abstract
Duchenne muscular dystrophy (DMD) is a genetic disorder caused by mutations in the dystrophin-encoding DMD gene. The DMD gene, spanning over 2.4 megabases along the short arm of the X chromosome (Xp21.2), is the largest genetic locus known in the human genome. The size of DMD, combined with the complexity of the DMD phenotype and the extent of the affected tissues, begs for the development of novel, ideally complementary, therapeutic approaches. Genome editing based on the delivery of sequence-specific programmable nucleases into dystrophin-defective cells has recently enriched the portfolio of potential therapies under investigation. Experiments involving different programmable nuclease platforms and target cell types have established that the application of genome-editing principles to the targeted manipulation of defective DMD loci can result in the rescue of dystrophin protein synthesis in gene-edited cells. Looking towards translation into the clinic, these proof-of-principle experiments have been swiftly followed by the conversion of well-established viral vector systems into delivery agents for DMD editing. These gene-editing tools consist of zinc-finger nucleases (ZFNs), engineered homing endoculeases (HEs), transcription activator-like effector nucleases (TALENs), and RNA-guided nucleases (RGNs) based on clustered, regularly interspaced, short palindromic repeats (CRISPR)–Cas9 systems. Here, we succinctly review these fast-paced developments and technologies, highlighting their relative merits and potential bottlenecks, when used as part of in vivo and ex vivo gene-editing strategies.
Background
Duchenne muscular dystrophy (DMD) is a lethal X-linked genetic disorder (affecting approximately 1 in 5000 boys) [1] caused by mutations in the ~2.4-megabase DMD gene [2] which lead to irrevocable muscle wasting owing to the absence of dystrophin in the striated muscle cell lineage [3]. Although dystrophin-disrupting mutations can be of different types, 68 % of them consist of intragenic large deletions [4]. These deletions can be found along the entire length of the enormous DMD locus, with 66 % nested within a major, recombination-prone, hotspot region spanning exons 45 through 55 [4]. The resulting joining of exons flanking DMD-causing mutations by pre-mRNA splicing yields transcripts harboring out-of-frame sequences and premature stop codons, which are presumably degraded by nonsense-mediated mRNA decay mechanisms.
In muscle cells, the long rod-shaped dystrophin protein anchors the intracellular cytoskeleton to the extracellular matrix via a large glycoprotein complex embedded in the plasma membrane called the dystrophin-associated glycoprotein complex (DGC). This structural link is fundamental for proper cellular signaling and structural integrity. Indeed, in the absence of dystrophin, a relentless degenerative process is initiated that consists of the substitution of muscle mass by dysfunctional fibrotic and fat tissues [3]. As time elapses, patients with DMD become dependent on a wheelchair for ambulation and, later on, require breathing assistance. Crucially, with the aid of palliative treatments, which include supportive respiratory and cardiac care, the life expectancy of patients with DMD is improving and a greater proportion of these patients now reach their late 30s [5].
Targeting the root cause of DMD
The complexity of DMD, combined with the extent of affected tissue, demands the development of different, ideally complementary, therapeutic approaches. The goal of pursuing parallel approaches is to target different aspects and stages of the disease and hence maximize the length and quality of patients’ lives. Towards this end, various candidate therapies are currently under intense investigation [3, 5, 6]. These research lines include: (1) mutation-specific exon skipping via modulation of pre-mRNA splicing by antisense oligonucleotides; (2) compensatory upregulation of dystrophin’s autosomal paralog utrophin by small-molecule drugs or artificial transcription factors; (3) cell therapies involving allogenic myogenic stem/progenitor cell transplantation; and (4) gene therapies based on the delivery of shortened versions of dystrophin (for example, microdystrophins) to affected tissues. Of note, these recombinant microdystrophins are devoid of centrally located motifs that are, to some extent, dispensable. The miniaturization bypasses the fact that the full-length 11-kilobase (kb) dystrophin coding sequence is well over the packaging limit of most viral vector systems.
More recently, genome-editing strategies based on sequence-specific programmable nucleases have been proposed as another group of therapies for DMD [7–10]. Programmable nucleases are tailored to induce double-stranded DNA breaks (DSBs) at predefined positions within complex genomes [11–13]. In chronological order of appearance, these enzymes are: zinc-finger nucleases (ZFNs) [14], engineered homing endonucleases (HEs) [15], transcription activator-like effector nucleases (TALENs) [16–18], and RNA-guided nucleases (RGNs) based on dual RNA-programmable clustered, regularly interspaced, short palindromic repeat (CRISPR)–Cas9 systems [19–22] (Fig. 1). HEs, also known as meganucleases, from the LAGLIDADG family can be engineered to cleave DNA sequences other than those of their natural target sites. The designing of new substrate specificities depends, however, on complex protein engineering efforts involving the screening of large combinatorial assemblies of HE parts [15]. Regardless, redesigned HE were shown to create indel footprints at intronic DMD sequences, albeit at very low frequencies (<1 % of target alleles in human myoblasts) [23]. In contrast to the construction of redesigned HEs, the modular nature of the DNA-binding motifs of ZFNs and TALENs makes them more amenable to protein engineering [14, 16–18]. Of note, the assembly of highly specific TALENs is particularly straightforward owing to a simple one-to-one relationship between the binding of each of their DNA-binding modules, that is, transcription activator-like effectors (TALE) repeats, and a specific nucleotide [16, 17]. Among other features, ZFNs and TALENs differ from RGNs in that they are chimeric enzymes that assemble at their target nucleotide sequences as catalytically active dimers through protein–DNA binding, whereas RGNs are ribonucleoprotein complexes whose DNA cutting specificities are ultimately governed by DNA–RNA hybridization. Indeed, RGNs consist of a Cas9 endonuclease and a sequence-customizable single-guide RNA (sgRNA) moiety that addresses the protein component to a specific target site. Typically, the target site consists of 18–20 nucleotides complementary to the 5′ end of the sgRNA and a protospacer adjacent motif (PAM; NGG and NNGRRT in the case of the prototypic Streptococcus pyogenes Cas9 and its smaller orthologue Staphylococcus aureus Cas9, respectively) [19, 24]. Hence, in comparison with the strictly protein-based systems, RGNs are more versatile owing to their mode of construction, which does not involve protein engineering [11–13].
Fig. 1.
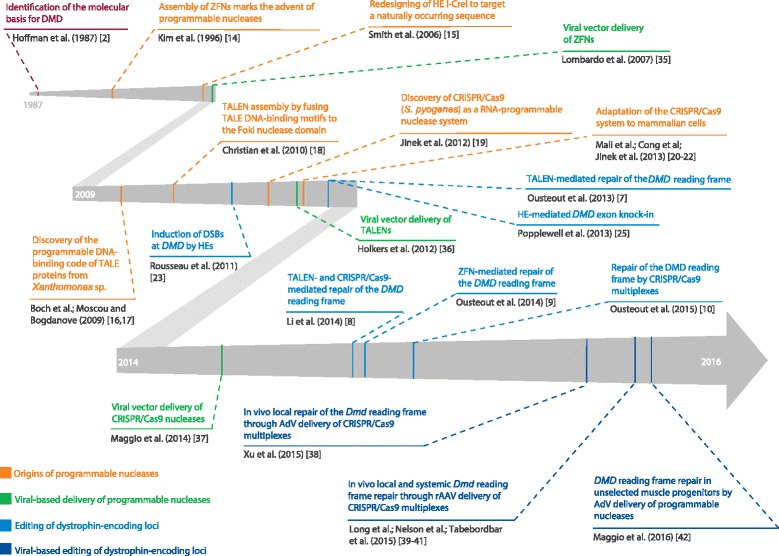
Milestones on the path towards somatic genetic therapies for Duchenne muscular dystrophy that rely on viral-based DMD editing. The time marks correspond to the first release date of the referenced articles (for example, advanced online publication). AdV adenoviral vector, CRISPR–Cas9 clustered regularly interspaced short palindromic repeat-associated Cas9 nuclease, DMD Duchenne muscular dystrophy, DSB double-stranded DNA break, HE homing endonuclease, rAAV recombinant adeno-associated virus, TALE transcription activator-like effector
Regardless of the DNA cutting system that is selected, the repair of the ensuing DSBs by different endogenous cellular DNA repair processes can yield specific genome editing outcomes. For example, the engagement of homologous recombination (HR) and non-homologous end joining (NHEJ) mechanisms can result in targeted exogenous DNA additions and endogenous DNA deletions, respectively [11–13]. The incorporation of small insertions and deletions (indels) following the repair of DSBs by NHEJ can also be exploited for knocking out trans-acting and cis-acting genomic elements [11–13]. By operating at the DNA level, such interventions can potentially lead to the correction of disease-causing mutations on a permanent basis.
DMD gene editing
DMD editing based on targeted addition of “exon patches” corresponding to missing or disrupted coding sequences might become ideal therapeutic options as they result in the synthesis of full-length dystrophin [8, 25]. Proof-of-principle experiments demonstrated that combining DMD-repairing exon patches with engineered meganucleases [25], RGNs, or TALENs [8] can indeed restore full-length message coding for dystrophin. At present, however, most DMD editing approaches under investigation are based on inducing NHEJ to disrupt or delete specific sequences [7–10]. These strategies exploit the fact that, in contrast to HR, NHEJ is active in both dividing and post-mitotic cells [26, 27], which makes these approaches more amenable to both ex vivo and in vivo applications (Table 1). The NHEJ-based strategies also capitalize on the fact that internally truncated in-frame DMD transcripts, despite being shorter than the full-length DMD transcript, often yield functional dystrophins [28–30]. Indeed, such dystrophins are characteristic of patients with Becker muscular dystrophy, whose disease phenotypes are milder than those of their counterparts with DMD [28–30]. Therefore, programmable nucleases have been tailored for correcting defective DMD alleles by targeting: (1) splicing sites for inducing DNA-borne exon skipping; (2) exonic sequences for resetting reading frames and “overwriting” downstream premature stop codons; and (3) flanking intronic sequences for directly excising mutations through the use of pairs of programmable nucleases (multiplexing) [7–10]. DNA-borne exon skipping by NHEJ-mediated splicing motif knockout and reading-frame resetting by frame shifting are mutation-specific and rely on the fraction of indel footprints that yield in-frame sequences. Importantly, the resulting indels might introduce immunogenic epitopes into de novo-synthesized dystrophin molecules. Depending on certain variables (for example, revertant mutation backgrounds), these epitopes might be recognized as foreign by the immune system. Related to this potential issue, T-cell immunity to dystrophin epitopes was detected in two patients undergoing a clinical trial based on recombinant adeno-associated viral vector (rAAV) delivery of a microdystrophin construct [31].
Table 1.
Comparison of ex vivo and in vivo viral-based DMD editing strategies under investigation
| ✓ Pros × Cons |
Viral-based DMD editing | |
|---|---|---|
| Ex vivo | In vivo | |
| Background | × Knowledge about the grafting of different types of myogenic cells into recipient human muscles is scarce | ✓ Builds upon an increasing amount of knowledge about the in vivo administration of viral vectors into recipient human muscles (for example, microdystrophin-encoding rAAVs) |
| Production | ✓ Potentially less dependent on large-scale production of viral vectors × Reliant on the upscaling of cell culture systems × The required numbers of certain myogenic cell types might not be achievable owing to senescence (for example, myoblasts) × The current protocols do not permit culturing bona fide skeletal muscle stem cells (that is, satellite cells) in vitro |
✓ Independent from the upscaling of cell culture systems × More reliant on large-scale production of viral vectors |
| Delivery | ✓ Well-defined genetic modification environment that enables careful monitoring of procedures, events, and outcomes ✓ Lower stringency for monitoring the biodistribution (for example, gonads and shedding of vector elements) × Protocols for effective myogenic cell engraftment, migration, and differentiation need to be improved (for example, via signaling gradients and cell-autonomous reprogramming of iPSCs) × Local and locoregional administration of myogenic cells might be difficult to apply to a broad range of muscle groups × Protocols for the systemic delivery and tissue homing of myogenic cells need to be developed |
✓ Direct exposure to gene-editing tools facilitates in situ correction of differentiated striated muscle tissues ✓ Possible in situ transduction of resident tissue-specific stem cells might generate a long-term source of gene-edited muscle progenitor cells ✓ Expanding range of viral vector pseudotypes enables the investigation of different transduction patterns—for example, tropism for affected tissues—while avoiding APCs. Such transductional targeting can be combined with transcriptional targeting (that is, use of tissue-specific promoters) × Local and locoregional administration of viral vector particles might be difficult to apply to a broad range of muscle groups × Protocols for the systemic delivery of viral vectors to affected tissues need to be improved Higher stringency for monitoring the biodistribution (for example, gonads and shedding of vector elements) |
| Strategy | ✓ Relies mostly on targeting replicating cells that are amenable to gene-editing approaches based on NHEJ as well as HR | × Relies mostly on targeting post-mitotic cells, which are less amenable to HR-based gene-editing principles |
| Immunology | ✓ Minimizes the exposure of the patient to immunogenic components of viral vectors and gene-editing tools ✓ Possibly compatible with the re-administration of gene-edited autologous cells ✓ Avoids the blocking of viral vector particles by neutralizing antibodies present in the majority of the human population |
× Patient exposure to immunogenic components of vector particles and gene-editing tools. Possible mounting of cellular responses to transduced cells displaying foreign epitopes × Anti-vector neutralizing antibodies in the majority of the human population. Serotype cross-neutralizing activity might render vector pseudotyping and vector re-administration ineffective |
In vivo approaches entail the direct administration of gene-editing viral vectors to the patient. Ex vivo approaches encompass the in vitro transduction of patient-derived cells (for example, myogenic stem or progenitor cells) with gene-editing viral vectors, which is followed by cell culture and autologous transplantation back into the patient. Both treatment modalities can, in principle, be applied either locally or systemically. APCs antigen-presenting cells, HR homologous recombination, iPSCs induced pluripotent stem cells, NHEJ non-homologous end joining, rAAVs recombinant adeno-associated viruses
In contrast to those triggering single-exon deletions, the DMD correction approaches based on targeted multi-exon deletions do not give rise to indel-derived epitopes and are applicable to a wider range of DMD-causing genotypes, with de novo-generated intronic junctions leading to predictable in-frame mRNA templates [10, 32]. However, multiplexing approaches carry increased risks for unwarranted, possibly deleterious, genome-modifying events (for example, off-target DSBs, inversions, and translocations), owing to their dependency on two programmable nucleases rather than one [12]. These increased risks will be present despite the fact that targeted DSBs in boys with DMD will be restricted to a single allele.
Viral-based DMD editing
The clinical application of DMD-editing concepts will require improved methods for delivering large and complex molecular tools into target cells, as well as increasing the efficiency, specificity, and fidelity of the ensuing DNA modifications [12]. Similarly to their effective contribution to “classic” gene replacement therapies [33], viral vectors are expected to become instrumental tools for investigating and developing therapeutic gene-editing approaches ex vivo and in vivo (for a recent review on the adaptation and testing of viral vector systems for genome editing purposes, see [34]). Indeed, ZFNs, TALENs, and RGNs have all been shown to be amenable to viral vector delivery [35–37] (Fig. 1). More recently, adenoviral vectors (AdVs) and rAAVs have been successfully converted into DMD-editing agents in both patient-derived cells and mouse models of DMD [38–42] (Fig. 1).
In vivo
The Dmdmdx mouse model has a (mild) dystrophic phenotype that is due to a nonsense mutation located in exon 23 of the Dmd gene; historically, this has been the principal animal model for investigating DMD-targeted therapies and certain pathophysiological aspects of the disease [43]. In one study, conventional, commonly used, serotype-5 AdVs constructed to encode either S. pyogenes Cas9 or sgRNAs that targeted sequences flanking Dmd exons 21 through 23 were co-injected into the gastrocnemius muscles of newborn Dmdmdx mice [38]. At 3 weeks post-injection, dystrophin synthesis was readily detected in transduced muscle fibers. A semi-quantitative assay based on western blot analysis estimated that these fibers contained ~50 % of the wild-type levels of dystrophin. The gene-edited muscle regions displayed reduced Evans blue dye uptake under rest and force-generating conditions, indicating improved muscle fiber integrity.
A notorious characteristic of prototypic serotype-5 AdVs is their immunogenicity and, although they can be made without viral genes [34, 44], capsid-cell interactions can still trigger strong innate immune responses [45, 46]. In addition, the high prevalence of neutralizing antibodies directed against the capsids of serotype-5 AdVs in the human population has contributed to spurring the development of AdVs based on alternative serotypes [45]. Historically, these immunological determinants have in fact precluded the efficacious deployment of AdV technologies in “classic” gene therapy settings in which long-term maintenance of transduced cells is a prerequisite. AdVs are currently mostly used in human individuals either as oncolytic or vaccination agents [47]. The use of AdVs in translational in vivo gene editing will require dampening their immunogenicity and improving their targeting to specific cell types or organs. These efforts will be heavily guided by insights into the biology of host–vector interactions [45, 46]. For example, while serotype-5 AdVs bind through their fibers to the coxsackievirus and adenovirus receptor (CAR) to enter cells in vitro [48], their uptake by liver cells after intravenous administration in vivo is CAR-independent and governed by the interaction of their hexons with blood coagulation factors [49].
Three other studies investigated the in vivo delivery of RGN components (that is, sgRNA and Cas9 nucleases) by capsid-pseudotyped rAAVs for inducing the in-frame deletion of Dmd exon 23. These rAAV particles consist of rAAV DNA from serotype 2 packaged in capsids from AAV serotype 8 (rAAV-8) [40] or serotype 9 (rAAV-9) [39, 41], whose tropism for striated mouse muscle had previously been established [50, 51]. Pairs of these vectors encoding sgRNAs and either S. pyogenes Cas9 [39] or the smaller S. aureus Cas9 [40, 41] were co-administered into newborn and adult Dmdmdx mice. Nelson and colleagues detected abundant dystrophin protein synthesis 8 weeks after co-injecting a mixture of rAAV-8 particles encoding S. aureus Cas9 and cognate sgRNAs into tibialis anterior muscles [40]. Importantly, treated muscles had improved contractibility and force-generating functions. Finally, by capitalizing on the well-established performance of rAAV-8 after systemic administration in mice [50], Nelson and colleagues were able to detect dystrophin in cardiac muscle tissue after a single intravenous injection [40].
Instead of rAAV-8, Long and colleagues used rAAV-9 to introduce S. pyogenes RGN complexes into striated muscle tissues of newborn Dmdmdx mice [39]. Dystrophin was detected in striated muscle tissues after local and systemic administration of the engineered viral vectors [39]. Consistent with the slow kinetics of gene expression from rAAVs, which might in part be related to the processes underlying the conversion of vector DNA from a single-stranded to a transcriptionally active double-stranded form [52], a time-dependent increase in dystrophin buildup was observed. For instance, tibialis anterior muscles of postnatal day 12 Dmdmdx mice subjected to direct intramuscular injections with the engineered viral vector contained approximately 8 and 26 % of dystrophin-positive fibers at 3 and 6 weeks post-administration, respectively [39].
In the third study, Tabebordbar and coworkers used rAAV-9 pairs for delivering S. aureus Cas9 and sgRNAs to the tibialis anterior muscle of dystrophin-defective Dmdmdx mice [41]. Similarly to the results of the two other studies on rAAV-mediated Dmd exon 23 deletion experiments [39, 40], administration of the rAAV-9 pairs led to robust rescue of dystrophin protein synthesis in transduced muscles and to a concomitant measurable improvement in functional parameters (that is, specific force and force drop) compared with those in unedited controls [41]. In addition, intraperitoneal co-injection of rAAV-9 particles into dystrophic mice led to frequencies of Dmd exon 23 excision in cardiac and skeletal muscle tissues ranging from 3 to 18 %, as determined by RT-PCR, depending on the muscle groups analyzed [41]. Importantly, Dmd-editing rAAV-9 particles were also administered intramuscularly or systemically to Pax7-ZsGreen Dmdmdx mice whose satellite cells are marked by green fluorescence. Subsequently, after isolating, expanding, and inducing myogenic differentiation of the Pax7-ZsGreen-positive cells, the authors reported in-frame Dmd exon 23 deletions in myotubes derived from these cells [41]. The population of Pax7-positive satellite cells harbors the resident mononuclear stem cell population of skeletal muscle and is typically lodged between the sarcolemma of muscle fibers and the basal lamina [53]. The “stemness” qualities of self-renewal and lifelong differentiation capacity make these tissue-specific stem cells ideal substrates for regenerative medicine approaches for treating muscular dystrophies as, in contrast to their committed progenitor offspring, these cells support robust long-term tissue homeostasis and repair [54, 55]. Recent experiments in transgenic Dmdmdx mice showed that, in addition to its other functions, dystrophin has a transient but critical regulatory role in activated Pax7-positive satellite cells, which further supports the therapeutic relevance of this cell population. In particular, the 427-kilodalton dystrophin isoform is expressed at very high levels in these cells, where it governs asymmetric cell division, a process that is indispensable for maintaining the stem cell pool and for generating committed Myf5-positive myoblast progenitors for muscle repair [56]. Among other processes, this mechanism presumably involves interactions between the spectrin-like repeats R8 and R9 of dystrophin and Mark2, a protein that regulates cell polarity [56, 57]. If conserved in humans, this cell-autonomous mechanism would be evidence that DMD is also a stem cell disease, which would strengthen the view that satellite cells should be preferential targets for DMD therapies, in addition to differentiated cells. Interestingly, the very high amounts of dystrophin seen in activated Pax7-positive satellite cells are followed by very low and intermediate levels of the protein in myoblasts and differentiated muscle cells, respectively [56]. Such differentiation-stage-specific oscillations in dystrophin amounts strengthen the rationale for repairing the genetic defects by direct endogenous DMD editing, as this strategy is expected to restore proper regulation of dystrophin synthesis.
Taken together, these findings demonstrate that rAAV delivery of RGN complexes can result in the structural improvement of treated striated tissues and also lead to the partial rescue of specific muscle functions in dystrophic mice. Although dystrophin synthesis was detected at 6 months after a single injection in one experiment [40], no long-term detailed assessments of these approaches were done. Regardless, the available data do support the potential of these vectors as in vivo DMD-repairing agents, thus warranting further research. Future developments should include assuring the transient presence of programmable nucleases in post-mitotic tissues, preclinical testing in large outbred animal models [43], and identifying or engineering rAAV capsids that have preferential tropism for human striated muscle cells, including satellite cells, while bypassing the host’s humoral immunity against prevalent AAV serotypes [58].
The administration of rAAVs to some human individuals resulted in clinical endpoints that had not been predicted on the basis of the available preclinical data. These findings are simultaneously sobering and illuminating. An example is provided by the elimination of transduced hepatocytes in patients with hemophilia B, which was due to the development of a dose-dependent T-cell response to capsid epitopes from an rAAV-2-encoding human factor IX [59]. This type of dose-dependent cellular immune response has also been documented in human skeletal muscle cells transduced with rAAVs [60], although it is of note that the emergence of T-cell responses directed against rAAV capsid epitopes does not always equate with the elimination of transduced muscle cells [61]. In addition, short-term immune suppression might help to dampen cellular immune responses in muscular dystrophy patients subjected to high-doses of rAAV particles [62]. It is worth mentioning, however, that the altered immune cell composition and inflammatory environment that characterize dystrophic muscle tissue might introduce potential confounding factors associated with in vivo rAAV delivery. Knowledge about these issues and preclinical data obtained from canine models of DMD [63–65] are guiding the design of new clinical trials based on the administration of rAAVs to patients with DMD [66]. Further insights are being gathered from the application of rAAVs to patients suffering from other muscular disorders such as Limb-girdle muscular dystrophy caused by α-sarcoglycan deficiency [67]. In particular, there is mounting evidence for the importance of restricting transgene expression to muscle cells by using tissue-specific promoters [67]. In the future, muscle-restricted transgene expression might be further improved by combining transcriptional with transductional targeting through rAAVs with capsids with a strict tropism for human muscle tissue. The recently discovered pan-AAV receptor AAVR [68] is likely to have an important role in this research; for instance, by shedding light on rAAV transduction profiles in different cell types, including immune-related cells. Therefore, although rAAVs have a substantially milder immunogenic profile than that of AdVs, they also need to be adapted for translational in vivo gene-editing purposes, which, as for AdVs, will be rooted in an increasing knowledge about vector-host interactions and biodistribution at the organismal level. Finally, in the context of future clinical protocols for in vivo DMD editing, the synthesis of programmable nucleases should be restricted not only spatially but also temporally.
Ex vivo
Ex vivo DMD editing strategies to generate genetically corrected human cells with myoregenerative capacity for autologous transplantation can also be envisaged (Table 1). These approaches offer a controlled genome-modification environment, bypass vector-neutralizing antibodies, and minimize direct contact between the patient and immunogenic components, such as those from vector particles, gene-editing tools, and allogenic donor cells (Table 1). Importantly, provided that clinically applicable delivery vehicles of gene-editing tools become available, ex vivo DMD editing can naturally build upon the numerous investigations that are being conducted on the isolation, characterization, and testing of human myogenic cells isolated from different tissues for treating muscular dystrophies [69–73]. These cellular substrates include satellite cells [53, 54] and their committed myoblast progeny [74], induced pluripotent stem cells [75], mesenchymal stromal cells [76, 77], vasculature-associated mesoangioblasts/pericytes [78], and blood-derived CD133+ cells [79]. Of note, the latter two cell types have been shown to be amenable to systemic administration in animal models and, to some extent, can colonize their satellite cell niche [80–82]. In addition, mesoangioblasts/pericytes and CD133+ cells have entered early stage clinical testing in the context of allogenic cell therapies for DMD [83, 84]. These clinical investigations complement earlier and ongoing testing of allogenic myoblast transplantation that are based on intramuscular injections [71–73, 85, 86].
Despite these encouraging developments, the hurdles towards the clinical application of ex vivo DMD cell therapies remain numerous and complex. Preeminent examples of such hurdles include achieving sufficient numbers of undifferentiated cells in vitro, as well as robust cell engraftment, migration, and differentiation of the transplanted graphs in vivo. Ideally, the transplanted cells should also be capable of homing to damaged tissue after systemic administration and should dedifferentiate or transdifferentiate (when belonging to muscle and non-muscle lineages, respectively) into satellite cells (Table 1). Therefore, although certain therapeutic-cell candidates are well positioned to fulfil some of these criteria, none of them fulfils all of the criteria yet [69, 72]. For example, CD133+ blood-derived cells and mesoangioblasts/pericytes have been shown to be compatible with systemic administration procedures in preclinical models of muscular dystrophies [78, 79] but their contribution to effective myoregeneration requires further investigation. In contrast, the features of human satellite cells make them natural, highly potent, muscle-repairing entities. Besides being available in diverse human muscle groups, satellite cells have the capacity to readily engraft as functional stem cells and robustly contribute to de novo muscle repair in xenotransplantation experiments [72]. However, harvested satellite cells are not amenable to systemic administration or current ex vivo culture conditions, as they readily differentiate into myoblasts with reduced regenerative capacity [87]. Importantly, the latter hurdle might not be insurmountable, as ongoing research indicates that extrinsic factors such as the composition and elasticity of culture vessels can be modulated to mimic the rigidity of the native satellite cell niche (that is, 12 instead of ~106 kilopascals) and, in doing so, enable the in vitro survival and self-renewal of bona fide satellite cells [88]. The development of such biomimetic tissue-engineering technologies directed to the in vitro expansion of human satellite cells is in demand.
In addition to that of skeletal muscle, cardiac muscle impairment is a key component of DMD that also needs to be tackled in future therapies. Despite intense research on the isolation and characterization of stem and progenitor cells for the repair of damaged heart tissue (for example, after ischemia), so far there is no evidence for a significant functional improvement of the myocardium through the cell-autonomous differentiation of the transplanted cells into mature, electrically coupled cardiomyocytes [89, 90].
Other equally important areas for further research in the field of DMD-targeted regenerative medicine are: (1) deepening our knowledge about the origins and biology of the various cell therapy candidates and their interaction(s) with their respective niches; (2) gathering all possible information on the fate and behavior of transplanted cells from ongoing and future cell therapy trials; (3) moving forward with gene replacement approaches involving stable transduction of recombinant constructs; and (4) testing different gene-editing reagents and strategies for developing autologous cell transplantation approaches. Regarding the latter research avenue, it will be crucial to efficiently introduce different gene-editing tools into human muscle progenitor cells and non-muscle cells with myogenic capacity. AdVs outperform rAAVs in ex vivo settings owing to their higher functional vector particle titers, larger packaging capacity (up to 37 kb), and faster kinetics of transgene expression [34, 52]. Our laboratory has recently reported that tropism-modified AdVs are particularly efficient and versatile vehicles for introducing RGNs and TALENs into CAR-negative myoblasts from patients with DMD [42]. The strict episomal nature of the transduced AdV genomes enabled transient high-level expression of programmable nucleases that corrected native DMD alleles and yielded permanent and regulated dystrophin synthesis. In this work, we exploited targeted NHEJ-mediated correction of DMD-causing intragenic deletions by reading-frame resetting, DNA-borne exon skipping, and in-frame excision of single or multiple exons [42]. The rescue of dystrophin synthesis could be readily detected in unselected populations of target cells [42]. Bypassing the need for cell selection expedients is expected to simplify and help translate ex vivo DMD editing protocols to the clinic. Moreover, AdV-based delivery systems will aid with assessing and comparing different DMD editing reagents and strategies in panels of human myogenic cells harboring the various DMD mutations, which are not represented in the currently available animal models. In addition, the well-defined in vitro conditions permit the straightforward monitoring of intended as well as unwarranted or potentially deleterious interactions between the gene-editing reagents and the human genome (Table 1). Prominent examples of such quality controls will include the genome-wide tracking of adverse DNA-modifying events directly in patient cells (for example, off-target activities of programmable nucleases).
Conclusions and future directions
The application of genome-editing principles for DMD repair purposes is expanding the range of genetic therapy options for tackling DMD. In this context, the coopting of viral vector systems as carriers of programmable nucleases is set to have an important role in the path to DNA-targeted DMD therapies and, along the way, in defining the best strategies and optimizing the corresponding reagents. In view of the complexity of the DMD phenotype and the extent of the affected tissues, it is sensible to consider that future DMD therapies will profit from integrating complementary approaches. For example, the simultaneous treatment of skeletal and cardiac tissues from patients with DMD might be approached by combining ex vivo and in vivo gene-editing strategies, respectively. Such schemes can potentially address the skeletal and heart components of DMD while circumventing the current lack of cell entities capable of differentiating into functional cardiomyocytes. Regardless of the particular therapy or combination of therapies ultimately selected, there is widespread agreement that they should preferably be applied as early as possible so that most striated musculature is still in place and the degeneration process can be halted or, ideally, reversed in the treated muscle groups. Finally, the insights gained from these DMD-targeted research efforts will probably also be useful for devising advanced genetic therapies for addressing other neuromuscular disorders for which, at present, there are no therapeutic options available.
Acknowledgments
The authors are grateful to Josephine M. Janssen and Jin Liu for their excellent support and input to the research activities carried out in our research group. The authors also thank Rob Hoeben for his critical reading of the manuscript (Department of Molecular Cell Biology, Leiden University Medical Center, Leiden, the Netherlands). XC is the recipient of a PhD research fellowship from the China Scholarship Council-Leiden University Joint Scholarship Programme. This work was in part funded by grants from the Dutch Prinses Beatrix Spierfonds (W.OR11-18) and the French AFMTéléthon (16621).
Authors’ contributions
MAFVG conceived the idea for the review. IM, XC, and MAFVG wrote the article and approved the final manuscript. IM and XC contributed equally to this work.
Abbreviations
- AdV
adenoviral vector
- APC
antigen-presenting cell
- CAR
coxsackievirus and adenovirus receptor
- CRISPR
clustered, regularly interspaced, short palindromic repeats
- DGC
dystrophin-associated glycoprotein complex
- DMD
Duchenne muscular dystrophy
- DSB
double-stranded DNA break
- HR
homologous recombination
- indel
insertion and deletion
- iPSC
induced pluripotent stem cell
- kb
kilobase
- NHEJ
non-homologous end joining
- PAM
protospacer adjacent motif
- rAAV
recombinant adeno-associated viral vector
- RGN
RNA-guided nuclease
- sgRNA
single-guide RNA
- TALE
transcription activator-like effector
- TALEN
transcription activator effector-like nuclease
- ZFN
zinc-finger nuclease
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mendell JR, Shilling C, Leslie ND, Flanigan KM, Al-Dahhak R, Gastier-Foster J, et al. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann Neurol. 2012;71:304–13. doi: 10.1002/ana.23528. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–28. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 3.Guiraud S, Aartsma-Rus A, Vieira NM, Davies KE, van Ommen GJ, Kunkel LM. The pathogenesis and therapy of muscular dystrophies. Annu Rev Genomics Hum Genet. 2015;16:281–308. doi: 10.1146/annurev-genom-090314-025003. [DOI] [PubMed] [Google Scholar]
- 4.Bladen CL, Salgado D, Monges S, Foncuberta ME, Kekou K, Kosma K, et al. The TREAT-NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum Mutat. 2015;36:395–402. doi: 10.1002/humu.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guiraud S, Chen H, Burns DT, Davies KE. Advances in genetic therapeutic strategies for Duchenne muscular dystrophy. Exp Physiol. 2015;100:1458–67. doi: 10.1113/EP085308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konieczny P, Swiderski K, Chamberlain JS. Gene and cell-mediated therapies for muscular dystrophy. Muscle Nerve. 2013;47:649–63. doi: 10.1002/mus.23738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ousterout DG, Perez-Pinera P, Thakore PI, Kabadi AM, Brown MT, Qin X, et al. Reading frame correction by targeted genome editing restores dystrophin expression in cells from Duchenne muscular dystrophy patients. Mol Ther. 2013;21:1718–26. doi: 10.1038/mt.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T, et al. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Rep. 2015;4:143–54. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ousterout DG, Kabadi AM, Thakore PI, Perez-Pinera P, Brown MT, Majoros WH, et al. Correction of dystrophin expression in cells from Duchenne muscular dystrophy patients through genomic excision of exon 51 by zinc finger nucleases. Mol Ther. 2015;23:523–32. doi: 10.1038/mt.2014.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H, Kim J-S. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–34. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 12.Maggio I, Gonçalves MA. Genome editing at the crossroads of delivery, specificity, and fidelity. Trends Biotechnol. 2015;33:280–91. doi: 10.1016/j.tibtech.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Chandrasegaran S, Carroll D. Origins of programmable nucleases for genome engineering. J Mol Biol. 2016;428:963–89. doi: 10.1016/j.jmb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996;93:1156–60. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith J, Grizot S, Arnould S, Duclert A, Epinat J-C, Chames P, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–12. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 17.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 18.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–61. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rousseau J, Chapdelaine P, Boisvert S, Almeida LP, Corbeil J, Montpetit A, Tremblay JP. Endonucleases: tools to correct the dystrophin gene. J Gene Med. 2011;13:522–37. doi: 10.1002/jgm.1611. [DOI] [PubMed] [Google Scholar]
- 24.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–91. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popplewell L, Koo T, Leclerc X, Duclert A, Mamchaoui K, Gouble A, et al. Gene correction of a duchenne muscular dystrophy mutation by meganuclease-enhanced exon knock-in. Hum Gene Ther. 2013;24:692–701. doi: 10.1089/hum.2013.081. [DOI] [PubMed] [Google Scholar]
- 26.Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–8. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–15. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.England SB, Nicholson LV, Johnson MA, Forrest SM, Love DR, Zubrzycka-Gaarn EE, et al. Very mild muscular dystrophy associated with the deletion of 46 % of dystrophin. Nature. 1990;343:180–2. doi: 10.1038/343180a0. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura A, Yoshida K, Fukushima K, Ueda H, Urasawa N, Koyama J, et al. Follow-up of three patients with a large in-frame deletion of exons 45–55 in the Duchenne muscular dystrophy (DMD) gene. J Clin Neurosci. 2008;15:757–63. doi: 10.1016/j.jocn.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 30.Taglia A, Petillo R, D’Ambrosio P, Picillo E, Torella A, Orsini C, et al. Clinical features of patients with dystrophinopathy sharing the 45–55 exon deletion of DMD gene. Acta Myol. 2015;34:9–13. [PMC free article] [PubMed] [Google Scholar]
- 31.Mendell JR, Campbell K, Rodino-Klapac L, Sahenk Z, Shilling C, Lewis S, et al. Dystrophin immunity in Duchenne’s muscular dystrophy. N Engl J Med. 2010;363:1429–37. doi: 10.1056/NEJMoa1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Echigoya Y, Yokota T. Skipping multiple exons of dystrophin transcripts using cocktail antisense oligonucleotides. Nucleic Acid Ther. 2014;24:57–68. doi: 10.1089/nat.2013.0451. [DOI] [PubMed] [Google Scholar]
- 33.Naldini L. Gene therapy returns to centre stage. Nature. 2015;526:351–60. doi: 10.1038/nature15818. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Gonçalves MA. Engineered viruses as genome editing devices. Mol Ther. 2016;24:447–57. doi: 10.1038/mt.2015.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 36.Holkers M, Maggio I, Liu J, Janssen JM, Miselli F, Mussolino C, et al. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013;41:e63. doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maggio I, Holkers M, Liu J, Janssen JM, Chen X, Gonçalves MA. Adenoviral vector delivery of RNA-guided CRISPR/Cas9 nuclease complexes induces targeted mutagenesis in a diverse array of human cells. Sci Rep. 2014;4:5105. doi: 10.1038/srep05105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu L, Park KH, Zhao L, Xu J, El Refaey M, Gao Y, et al. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol Ther. 2016;24:564–9. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–3. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–7. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabebordbar M, Zhu K, Cheng JK, Chew WL, Widrick JJ, Yan WX, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–11. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maggio I, Stefanucci L, Janssen JM, Liu J, Chen X, Mouly V, et al. Selection-free gene repair after adenoviral vector transduction of designer nucleases: rescue of dystrophin synthesis in DMD muscle cell populations. Nucleic Acids Res. 2016;44:1449–70. doi: 10.1093/nar/gkv1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGreevy JW, Hakim CH, McIntosh MA, Duan D. Animal models of Duchenne muscular dystrophy: from basic mechanisms to gene therapy. Dis Model Mech. 2015;8:195–213. doi: 10.1242/dmm.018424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gonçalves MA, de Vries AA. Adenovirus: from foe to friend. Rev Med Virol. 2006;16:167–86. doi: 10.1002/rmv.494. [DOI] [PubMed] [Google Scholar]
- 45.Alonso-Padilla J, Papp T, Kajan GL, Benko M, Havenga M, Lemckert A, et al. Development of novel adenoviral vectors to overcome challenges observed with HAdV-5-based constructs. Mol Ther. 2016;24:6–16. doi: 10.1038/mt.2015.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hendrickx R, Stichling N, Koelen J, Kuryk L, Lipiec A, Greber UF. Innate immunity to adenovirus. Hum Gene Ther. 2014;25:265–84. doi: 10.1089/hum.2014.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Majhen D, Calderon H, Chandra N, Fajardo CA, Rajan A, Alemany R, et al. Adenovirus-based vaccines for fighting infectious diseases and cancer: progress in the field. Hum Gene Ther. 2014;25:301–17. doi: 10.1089/hum.2013.235. [DOI] [PubMed] [Google Scholar]
- 48.Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, et al. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–3. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 49.Waddington SN, McVey JH, Bhella D, Parker AL, Barker K, Atoda H, et al. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132:397–409. doi: 10.1016/j.cell.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z, Zhu T, Qiao C, Zhou L, Wang B, Zhang J, et al. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat Biotechnol. 2005;23:321–8. doi: 10.1038/nbt1073. [DOI] [PubMed] [Google Scholar]
- 51.Inagaki K, Fuess S, Storm TA, Gibson GA, McTiernan CF, Kay MA, et al. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol Ther. 2006;14:45–53. doi: 10.1016/j.ymthe.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferrari FK, Samulski T, Shenk T, Samulski RJ. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol. 1996;70:3227–34. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–5. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu X, Wilschut KJ, Kouklis G, Tian H, Hesse R, Garland C, et al. Human satellite cell transplantation and regeneration from diverse skeletal muscles. Stem Cell Rep. 2015;5:419–34. doi: 10.1016/j.stemcr.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin H, Price F, Rudnicki MA. Satellite cells and the muscle stem cell niche. Physiol Rev. 2013;93:23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dumont NA, Wang YX, von Maltzahn J, Pasut A, Bentzinger CF, Brun CE, et al. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat Med. 2015;21:1455–63. doi: 10.1038/nm.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamashita K, Suzuki A, Satoh Y, Ide M, Amano Y, Masuda-Hirata M, et al. The 8th and 9th tandem spectrin-like repeats of utrophin cooperatively form a functional unit to interact with polarity-regulating kinase PAR-1b. Biochem Biophys Res Commun. 2010;391:812–7. doi: 10.1016/j.bbrc.2009.11.144. [DOI] [PubMed] [Google Scholar]
- 58.Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nat Rev Genet. 2014;15:445–51. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manno CS, Arruda VR, Pierce GF, Glader B, Ragni M, Rasko J, et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med. 2006;12:342–7. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 60.Mingozzi F, Meulenberg JJ, Hui DJ, Basner-Tschakarjan E, Hasbrouck NC, Edmonson SA, et al. AAV-1-mediated gene transfer to skeletal muscle in humans results in dose-dependent activation of capsid-specific T cells. Blood. 2009;114:2077–86. doi: 10.1182/blood-2008-07-167510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brantly ML, Chulay JD, Wang L, Mueller C, Humphries M, Spencer LT, et al. Sustained transgene expression despite T lymphocyte responses in a clinical trial of rAAV1-AAT gene therapy. Proc Natl Acad Sci U S A. 2009;106:16363–8. doi: 10.1073/pnas.0904514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mendell JR, Rodino-Klapac LR, Rosales-Quintero X, Kota J, Coley BD, Galloway G, et al. Limb-girdle muscular dystrophy type 2D gene therapy restores α-sarcoglycan and associated proteins. Ann Neurol. 2009;66:290–7. doi: 10.1002/ana.21732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Z, Storb R, Halbert CL, Banks GB, Butts TM, Finn EE, et al. Successful regional delivery and long-term expression of a dystrophin gene in canine muscular dystrophy: a preclinical model for human therapies. Mol Ther. 2012;20:1501–7. doi: 10.1038/mt.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shin JH, Pan X, Hakim CH, Yang HT, Yue Y, Zhang K, Terjung RL, Duan D. Microdystrophin ameliorates muscular dystrophy in the canine model of duchenne muscular dystrophy. Mol Ther. 2013;21:750–7. doi: 10.1038/mt.2012.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kornegay JN, Li J, Bogan JR, Bogan DJ, Chen C, Zheng H, et al. Widespread muscle expression of an AAV9 human mini-dystrophin vector after intravenous injection in neonatal dystrophin-deficient dogs. Mol Ther. 2010;18:1501–8. doi: 10.1038/mt.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bengtsson NE, Seto JT, Hall JK, Chamberlain JS, Odom GL. Progress and prospects of gene therapy clinical trials for the muscular dystrophies. Hum Mol Genet. 2016;25(R1):R9–R17. doi: 10.1093/hmg/ddv420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mendell JR, Rodino-Klapac LR, Rosales XQ, Coley BD, Galloway G, Lewis S, et al. Sustained alpha-sarcoglycan gene expression after gene transfer in limb-girdle muscular dystrophy, type 2D. Ann Neurol. 2010;68:629–38. doi: 10.1002/ana.22251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Y, et al. An essential receptor for adeno-associated virus infection. Nature. 2016;530:108–12. doi: 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bentzinger CF, Wang YX, von Maltzahn J, Rudnicki MA. The emerging biology of muscle stem cells: implications for cell-based therapies. Bioessays. 2013;35:231–41. doi: 10.1002/bies.201200063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Briggs D, Morgan JE. Recent progress in satellite cell/myoblast engraftment—relevance for therapy. FEBS J. 2013;280:4281–93. doi: 10.1111/febs.12273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Negroni E, Gidaro T, Bigot A, Butler-Browne GS, Mouly V, Trollet C. Invited review: stem cells and muscle diseases: advances in cell therapy strategies. Neuropathol Appl Neurobiol. 2015;41:270–87. doi: 10.1111/nan.12198. [DOI] [PubMed] [Google Scholar]
- 72.Negroni E, Vallese D, Vilquin JT, Butler-Browne G, Mouly V, Trollet C. Current advances in cell therapy strategies for muscular dystrophies. Expert Opin Biol Ther. 2011;11:157–76. doi: 10.1517/14712598.2011.542748. [DOI] [PubMed] [Google Scholar]
- 73.Skuk D, Tremblay JP. Intramuscular cell transplantation as a potential treatment of myopathies: clinical and preclinical relevant data. Expert Opin Biol Ther. 2011;11:359–74. doi: 10.1517/14712598.2011.548800. [DOI] [PubMed] [Google Scholar]
- 74.Huard J, Verreault S, Roy R, Tremblay M, Tremblay JP. High efficiency of muscle regeneration after human myoblast clone transplantation in SCID mice. J Clin Invest. 1994;93:586–99. doi: 10.1172/JCI117011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–86. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.De Bari C, Dell’Accio F, Vandenabeele F, Vermeesch JR, Raymackers JM, Luyten FP. Skeletal muscle repair by adult human mesenchymal stem cells from synovial membrane. J Cell Biol. 2003;160:909–18. doi: 10.1083/jcb.200212064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de la Garza-Rodea AS, van der Velde I, Boersma H, Gonçalves MA, van Bekkum DW, de Vries AA, et al. Long-term contribution of human bone marrow mesenchymal stromal cells to skeletal muscle regeneration in mice. Cell Transplant. 2011;20:217–31. doi: 10.3727/096368910X522117. [DOI] [PubMed] [Google Scholar]
- 78.Dellavalle A, Sampaolesi M, Tonlorenzi R, Tagliafico E, Sacchetti B, Perani L, et al. Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nat Cell Biol. 2007;9:255–67. doi: 10.1038/ncb1542. [DOI] [PubMed] [Google Scholar]
- 79.Torrente Y, Belicchi M, Sampaolesi M, Pisati F, Meregalli M, D’Antona G, et al. Human circulating AC133+ stem cells restore dystrophin expression and ameliorate function in dystrophic skeletal muscle. J Clin Invest. 2004;114:182–95. doi: 10.1172/JCI20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sampaolesi M, Torrente Y, Innocenzi A, Tonlorenzi R, D’Antona G, Pellegrino MA, et al. Cell therapy of α-sarcoglycan null dystrophic mice through intra-arterial delivery of mesoangioblasts. Science. 2003;301:487–92. doi: 10.1126/science.1082254. [DOI] [PubMed] [Google Scholar]
- 81.Tedesco FS, Hoshiya H, D’Antona G, Gerli MF, Messina G, Antonini S, et al. Stem cell-mediated transfer of a human artificial chromosome ameliorates muscular dystrophy. Sci Transl Med. 2011;3:96ra78. doi: 10.1126/scitranslmed.3002342. [DOI] [PubMed] [Google Scholar]
- 82.Benchaouir R, Meregalli M, Farini A, D’Antona G, Belicchi M, Goyenvalle A, et al. Restoration of human dystrophin following transplantation of exon-skipping-engineered DMD patient stem cells into dystrophic mice. Cell Stem Cell. 2007;1:646–57. doi: 10.1016/j.stem.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 83.Torrente Y, Belicchi M, Marchesi C, D’Antona G, Cogiamanian F, Pisati F, et al. Autologous transplantation of muscle-derived CD133+ stem cells in Duchenne muscle patients. Cell Transplant. 2007;16:563–77. doi: 10.3727/000000007783465064. [DOI] [PubMed] [Google Scholar]
- 84.Cossu G, Previtali SC, Napolitano S, Cicalese MP, Tedesco FS, Nicastro F, et al. Intra-arterial transplantation of HLA-matched donor mesoangioblasts in Duchenne muscular dystrophy. EMBO Mol Med. 2015;7:1513–28. doi: 10.15252/emmm.201505636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Skuk D, Tremblay JP. Confirmation of donor-derived dystrophin in a Duchenne muscular dystrophy patient allotransplanted with normal myoblasts. Muscle Nerve. 2016 doi: 10.1002/mus.25129. [DOI] [PubMed] [Google Scholar]
- 86.Law PK, Bertorini TE, Goodwin TG, Chen M, Fang QW, Li HJ, et al. Dystrophin production induced by myoblast transfer therapy in Duchenne muscular dystrophy. Lancet. 1990;336:114–5. doi: 10.1016/0140-6736(90)91628-N. [DOI] [PubMed] [Google Scholar]
- 87.Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–7. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 88.Gilbert PM, Havenstrite KL, Magnusson KE, Sacco A, Leonardi NA, Kraft P, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–81. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Balsam LB, Wagers AJ, Christensen JL, Kofidis T, Weissman IL, Robbins RC. Haematopoietic stem cells adopt mature haematopoietic fates in ischaemic myocardium. Nature. 2004;428:668–73. doi: 10.1038/nature02460. [DOI] [PubMed] [Google Scholar]
- 90.Murry CE, Soonpaa MH, Reinecke H, Nakajima H, Nakajima HO, Rubart M, et al. Haematopoietic stem cells do not transdifferentiate into cardiac myocytes in myocardial infarcts. Nature. 2004;428:664–8. doi: 10.1038/nature02446. [DOI] [PubMed] [Google Scholar]


