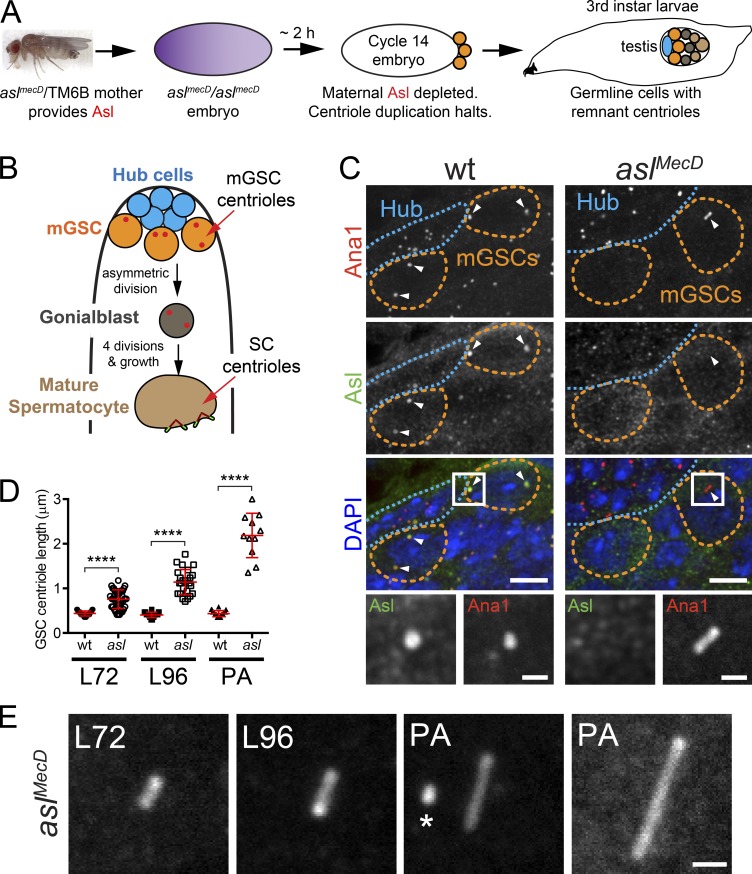
Loss of the centriole protein Asterless (Asl) prevents centriole duplication, which has limited the study of its function at centrioles. Here, Galletta et al. show that Asl controls centriole length and ensures proper basal body functions during spermatogenesis.
Abstract
Centrioles are the foundation of two organelles, centrosomes and cilia. Centriole numbers and functions are tightly controlled, and mutations in centriole proteins are linked to a variety of diseases, including microcephaly. Loss of the centriole protein Asterless (Asl), the Drosophila melanogaster orthologue of Cep152, prevents centriole duplication, which has limited the study of its nonduplication functions. Here, we identify populations of cells with Asl-free centrioles in developing Drosophila tissues, allowing us to assess its duplication-independent function. We show a role for Asl in controlling centriole length in germline and somatic tissue, functioning via the centriole protein Cep97. We also find that Asl is not essential for pericentriolar material recruitment or centrosome function in organizing mitotic spindles. Lastly, we show that Asl is required for proper basal body function and spermatid axoneme formation. Insights into the role of Asl/Cep152 beyond centriole duplication could help shed light on how Cep152 mutations lead to the development of microcephaly.
Introduction
The centrosome is the major microtubule organizing center (MTOC) of many cells, serving as a site of microtubule (MT) nucleation and minus end organization. These nonmembrane bound organelles are critical for a variety of cellular processes including cell migration, immune cell function, neuronal pathfinding, and axon selection, among others (Bettencourt-Dias et al., 2011; Bornens, 2012; Angus and Griffiths, 2013; Sakakibara et al., 2013; Elric and Etienne-Manneville, 2014). In many cells, centrosomes serve as spindle poles and help construct and organize a bipolar mitotic spindle (Vitre and Cleveland, 2012; Helmke et al., 2013). Centrosome number and activity are tightly regulated to guarantee proper MTOC function and avoid detrimental effects at the cellular, tissue, and organismal level, which have been linked to human diseases, including microcephaly and cancer (Noatynska et al., 2012; Korzeniewski et al., 2013; Nigg et al., 2014).
Centrosome number is controlled by limiting its duplication to once per cell cycle. Building a centrosome involves assembling its two major components, centrioles and pericentriolar material (PCM). Centrioles are barrel-like structures composed of nine triplet MTs and many highly conserved proteins that are recruited and arranged in a stepwise assembly process that ensures proper centriole size and function (Pelletier et al., 2006). For example, Sas-6 and Ana2/STIL are recruited early in the process to build the cartwheel structure that sets the diameter and radial symmetry of the centriole (Kitagawa et al., 2011; van Breugel et al., 2011; Dzhindzhev et al., 2014; Ohta et al., 2014). Cep97, CP110, and Sas-4/CPAP are then recruited to ensure proper centriole length (Spektor et al., 2007; Kohlmaier et al., 2009; Schmidt et al., 2009; Tang et al., 2009; Franz et al., 2013). When serving as MTOCs, the mother centriole recruits and organizes the PCM, from which MTs are nucleated. Some of the major PCM components include Pericentrin-like protein (PLP)/Pericentrin, Centrosomin (Cnn)/Cdk5Rap2/Cep215, and Spd2/Cep192, which then recruit gamma tubulin (γ-tub; Pelletier et al., 2004; Zimmerman et al., 2004; Fong et al., 2008; Giansanti et al., 2008; Zhu et al., 2008).
A major challenge to understanding the role of many centrosome proteins is that their loss leads to a loss of centrosome duplication and the subsequent dilution of centrosomes from the cell population (Goshima et al., 2007; Dobbelaere et al., 2008; Gönczy, 2012; Balestra et al., 2013). This precludes analysis of the potential roles of multifunctional centrosome proteins in PCM assembly, MTOC function, and ciliogenesis, among other processes. It is likely that proteins critical for centrosome duplication have unappreciated functions in other aspects of centrosome biology. One such protein is Asterless (Asl), the Drosophila melanogaster orthologue of vertebrate Cep152. In the absence of Asl or Cep152, centriole duplication halts because of its role in recruiting and stabilizing the master centriole duplication kinase Plk4 (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Hatch et al., 2010; Kim et al., 2013; Sonnen et al., 2013; Klebba et al., 2015). Asl may also “license” centrioles for their first duplication in Drosophila embryos (Novak et al., 2014). Although it is clear that Asl is critical for centriole duplication, little is known about other roles it may play in centrosome biology.
Several lines of evidence suggest Asl has other critical roles. Asl localizes along the length of the outer centriole surface (Varmark et al., 2007; Blachon et al., 2008). This differs from Plk4, which is found as a ring or spot on the centriole’s proximal end (Kim et al., 2013), suggesting Asl could do more than recruit Plk4, like structurally organizing the centriole. Also, Asl and Cep152 adopt a radially extended conformation with their C termini adjacent to the centriole wall and N termini extending into the PCM (Mennella et al., 2012; Sonnen et al., 2012). This suggests Asl/Cep152 could link the centriole wall with PCM, possibly directly scaffolding PCM proteins. There is evidence both supporting (Bonaccorsi et al., 1998; Varmark et al., 2007; Dzhindzhev et al., 2010; Conduit et al., 2014) and opposing (Blachon et al., 2008) the hypothesis that Asl functions in PCM organization. Finally, the removal of Asl from basal bodies during late spermatogenesis is important for zygotic development after fertilization (Khire et al., 2015).
Given this evidence and the critical nature of Asl/Cep152 for development and human health (Kalay et al., 2011; Poulton et al., 2014), we sought to identify functions for Asl beyond centriole duplication. We examined an Asl-free centriole population in the testes of asl mutant flies, focusing primarily on the male germline stem cells (mGSCs). We identified two novel roles for Asl. Most striking, we show that Asl plays a role in controlling centriole length, primarily via Cep97. We find that Asl-free centrioles recruit PCM and can organize mitotic spindles, indicating that Asl is not essential for PCM function. Finally, we show that asl mutant centrioles have defects in assembling into sperm, indicating a novel role for Asl in basal body function.
Results
Asl-free centrioles in asl mutant mGSCs
To elucidate roles for Asl beyond centriole duplication, we examined a population of cells containing centrioles, but lacking Asl, found in Drosophila testes. We refer to these centrioles as “remnants,” because they were originally built during early embryogenesis using maternally provided Asl (Fig. 1 A; Blachon et al., 2008). Using this strategy, we generated aslmecD zygotic mutants and performed a detailed examination of these Asl-free, remnant centrioles. We confirmed that aslmecD testes showed complete loss of full-length Asl protein (Fig. S1, A and B; Blachon et al., 2008).
Figure 1.
Asl is required for proper centriole length control. (A) asl embryos receive maternal Asl protein and mRNA (purple) used for the first 2 h of development. When this protein runs out, centriole duplication halts. Centrioles present at this point are reliably inherited by the developing testes (remnant centrioles). (B) Cell types in the Drosophila testis (centrioles, red). (C) Remnant centrioles (Ana1::tdTomato, red) in larval asl mGSCs do not have Asl (green) and are longer than in wt. Arrowheads, mGSC centrioles. Boxed region enlarged. DAPI, blue. (D) Centriole length (Ana1::tdTomato length) in mGSCs in wt (aslmecD/TM6B) and asl mutants in larvae 72 h after egg laying (L72), larvae 96 h after egg laying (L96), or PAs. Mean ± SD, red. Comparison: ordinary one-way analysis of variance followed by Tukey’s multiple comparisons test. ****, P < 0.0001. (E) Representative centrioles in mGSCs at indicated stages. The longest centriole observed is on the right. Asterisk, centriole in adjacent hub cell. Bars: (C) 5 µm; (insets) 1 µm; (E) 1 µm.
The testis contains several cell types including mGSCs, the hub (stem cell niche of somatic origin), somatic cyst stem cells, and the progeny of these stem cells (Fig. 1 B). In wild-type (wt) larval testes, all of these cell types contain Asl-positive centrioles (Figs. 1 C and S1 C). In testes from aslmecD homozygous mutant flies, remnant centrioles were located in four distinct cell populations (Fig. S1 D), two at the proximal tip (zones 1 and 2), one at the distal end (zone 4), and one in the central region (zone 3; Fig. S1, D vs. the even distribution in controls in C). The majority of remnant centrioles in the proximal tip are in the hub (Fig. 1 C, blue outline; and Fig. S1 D, zone 1). These cells have been quiescent since early development. The presence of these Asl-free centrioles reinforces that Asl is dispensable for centriole maintenance. The centrioles in zone 2 were in the mGSCs (Fig. 1 C, orange outline; and Fig. S1 D, zone 2), which are actively proceeding through the cell cycle. We found that 42% of mGSCs in aslmecD third-instar larvae (larvae 96 h after egg laying) contained Asl-free remnant centrioles (Figs. 1 C and S1 E). This was expected, as aslmecD mutants produce no full-length protein and a small amount of a ∼60 kD, C-terminally truncated protein lacking the centriole localization region (Fig. S1, A and B; Blachon et al., 2008; Klebba et al., 2015). The mGSCs of aslmecD are actively proceeding through the cell cycle and reproducibly contain Asl-free centrioles. They are therefore ideal for studying the nonduplication roles of Asl.
Asl is required for proper centriole length control
Examining aslmecD mGSC remnant centrioles revealed a striking result: by the late larval stage, all centrioles were longer than wt (Fig. 1 C). These centrioles get longer over time, averaging >2 µm long by the pharate adult (PA) stage. Some are almost 3 µm long (Fig. 1, D and E), 30-fold longer than the 100 nm length observed in wt mGSCs (Gottardo et al., 2015). This length increase is not simply a consequence of the time since Asl protein was exhausted during embryogenesis, because centrioles do not grow long in the hub. The major difference between these cells is that unlike hub cells, mGSCs have undergone multiple cell cycles since early embryogenesis. A model consistent with these observations is that centrioles retained in the asl mutant mGSCs get longer with each cell cycle. Interestingly, we found that the frequency with which centrioles are found in mGSCs decreases as larval and pupal development proceed (Fig. S1 F). This indicates that centrioles are being lost from the mGSCs over time, presumably as a consequence of cell division. We also observed remnant centrioles lost from the mGSCs in another centriole duplication mutant, sas-4S2214 (Fig. S1 G), indicating that this is a general phenomenon, not something specific to loss of Asl.
To ensure that the long centriole phenotype was a consequence of Asl loss, we examined other asl allelic combinations. Both aslmecD/Df(3R)ED5177 and asl2/Df(3R)ED5177, an independently derived, severe loss-of-function allele (Blachon et al., 2008) that produces no full-length protein (Fig. S1 B), also have abnormally long centrioles in mGSCs (Fig. S2, A–B′). These long centrioles are not a result of centriole duplication failure, because remnant centrioles in mGSCs of another centriole duplication mutant, sas-4S2214, were indistinguishable in length from wt (Fig. S2, C and D). These results strongly support that Asl controls centriole length. For all subsequent experiments, we used aslmecD homozygotes, which we refer to as asl.
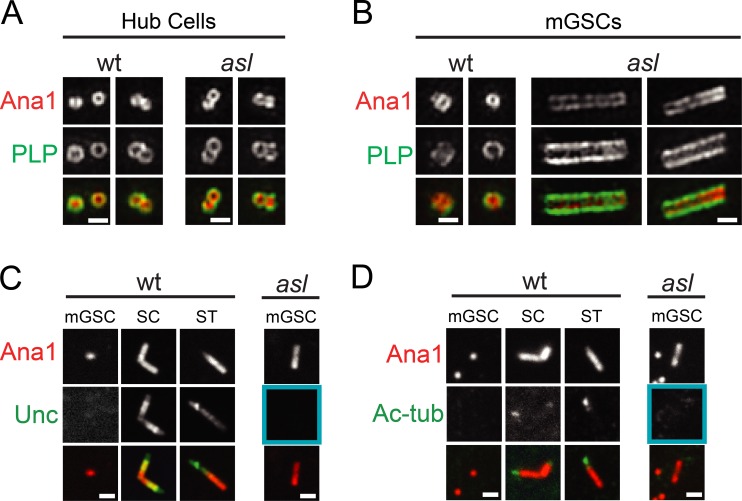
To understand the role of Asl in controlling mGSC centriole length, we examined the structure and composition of asl long centrioles using structured illumination microscopy (SIM). Centriole diameter and length in wt and asl hub cells were indistinguishable (Fig. 2 A), confirming that hub centrioles do not grow longer in asl. The radial distribution of Ana1 and PLP was normal on asl mGSC centrioles, confirming they are indeed centrioles and not unusual protein aggregates (Fig. 2 B). This is again consistent with the increased centriole length of Asl-free centrioles requiring passage through multiple cell cycles, a common circumstance in frequently dividing mGSC, but not in the quiescent hub.
Figure 2.
Loss of Asl does not trigger premature centriole to basal body transition in mGSCs. (A and B) SIM of hub (A) and mGSC (B) centrioles in wt (aslmecD/TM6B; left) and asl (right) larval testes. Ana1::tdTomato, red; PLP, green. Two examples of each. (C and D) Confocal images of wt and asl centrioles. (C) Unc (green) was not on asl (blue box) or wt mGSC centrioles (Ana1, red) but was on wt SC and ST centrioles. (D) Ac-tub was on centrioles in 75% of SC and 100% of ST. Ac-tub was not on centrioles in wt or asl (blue box) mGSCs. Bars, 0.5 µm.
One formal possibility is that the centrioles in asl mGSCs are long as a result of a premature execution of the normal developmental program of spermatogenesis in which centrioles elongate during basal body (BB) formation. To investigate this, we compared the “giant” centrioles in wt spermatocytes (SCs) to the long centrioles in asl mGSCs. Two defining characteristics of SC centrioles are the recruitment of the Uncoordinated (Unc) protein and the formation of small cilia at their distal tips marked by acetylated tubulin (Ac-tub; Riparbelli et al., 2012). The long centrioles in asl mGSCs did not contain Unc or Ac-tub (Fig. 2, C and D, blue boxes), indicating that asl long centrioles are a consequence of loss of Asl protein function and not an ectopic execution of the SC centriole elongation program. The absence of Ac-tub also suggests that centriole elongation in asl mutants arises by a mechanism distinct from the one in cp110 mutants, where centrioles form extensive Ac-tub projections at their distal ends (Franz et al., 2013). Thus, Asl normally functions to prevent precocious centriole elongation and its loss does not accelerate BB development.
Centrioles in asl mGSCs are proximalized
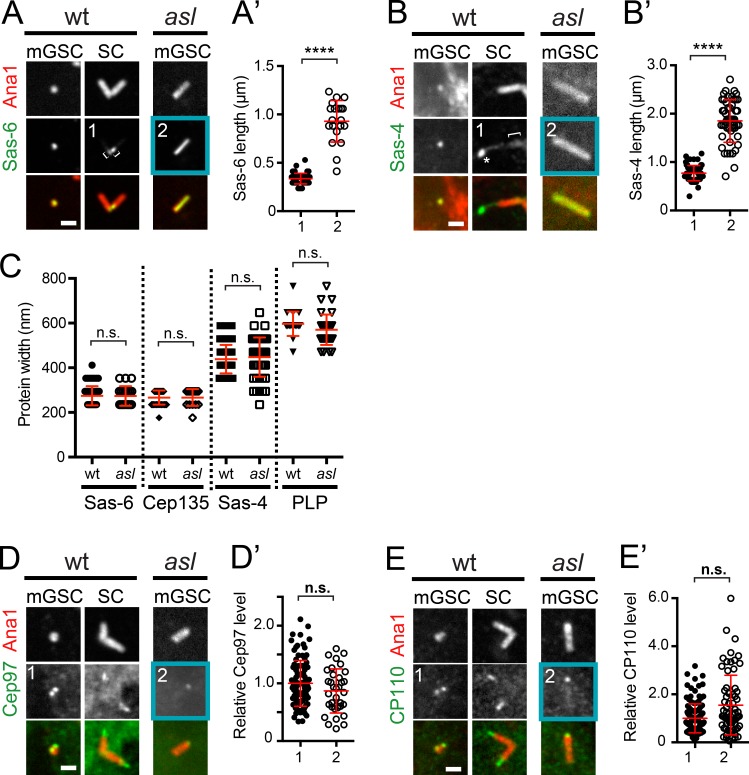
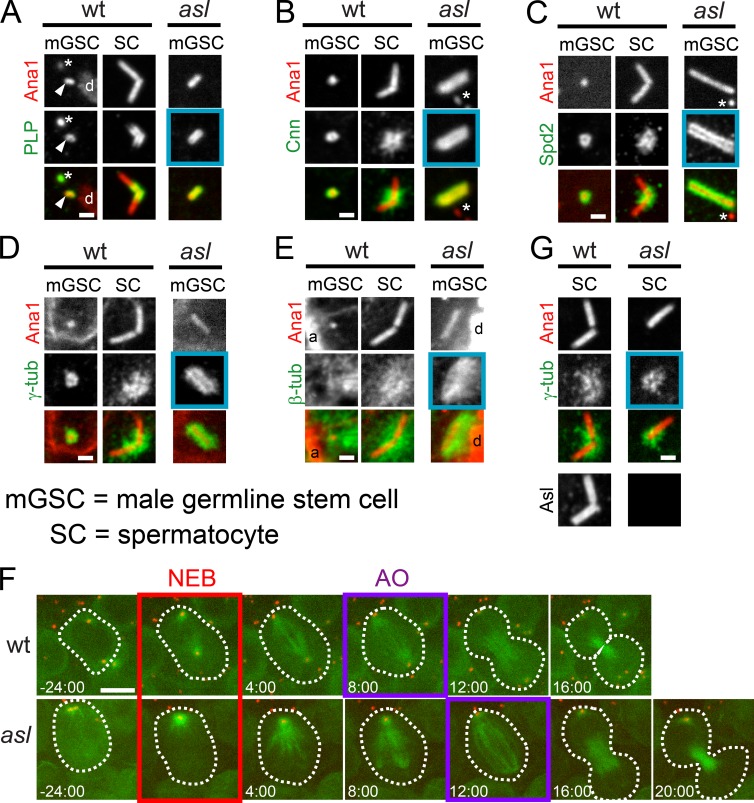
To understand the nature of the extension of centriole length and to identify possible causes, we examined localization of key centriole proteins. Sas-6, a component of the internal centriole cartwheel structure, and Sas-4, localize to diffraction-limited spots in wt mGSCs and to the proximal ends of wt SC giant centrioles (Fig. 3, A and B, left and middle). In asl mGSC long centrioles, Sas-6 and Sas-4 localize uniformly along the entire length (Fig. 3, A and B, right columns, blue boxes, and A′ and B′). The elongation of Sas-6 and Sas-4 indicates that Asl controls both overall centriole length and the distribution of proximal centriole characteristics.
Figure 3.
Asl regulates the position of centriole proteins. (A–E) Centrioles from interphase wt (aslmecD/TM6B) and asl mGSCs or wt mature SCs. (A) GFP::Sas-6 (green) and Ana1::tdTomato (red). Sas-6 extends the length of the centriole in asl mGSCs but is only proximal in wt SC centrioles (brackets). (A′) Length of Sas-6 signal in wt SCs (1) and asl mGSCs (2). (B) Sas-4::GFP (green) and Ana1::tdTomato (red). In SCs, Sas-4::GFP localizes proximally (bracket), where endogenous protein localizes (Gopalakrishnan et al., 2011; Fu and Glover, 2012). Additional localization beyond Ana1 (asterisk) is a consequence of overexpression. In asl mGSCs, Sas-4 is uniformly along the entire length of Ana1. (B′) Lengths of the Sas-4 signal in wt SCs (1, only the proximal signal) and asl mGSCs (2). (C) The radial distribution (diameter) of centriole proteins is unaltered in asl mGSCs. (D) GFP::Cep97 (green) and Ana1::tdTomato (red). (D′) Intensity of GFP::Cep97, normalized to the mean intensity of Cep97, at centrioles in wt (1) and asl (2) mGSCs. Cep97 levels are unaffected in asl. (E) CP110::GFP (green) and Ana1::tdTomato (red). (E′) Intensity of CP110::GFP, normalized as in D, in wt (1) and asl (2) mGSCs. CP110 levels are unaffected in asl. Bars, 1 µm. Mean ± SD, red. Comparison by unpaired t tests, with Welch’s corrections when appropriate. ****, P < 0.0001; n.s., not significant.
We next investigated the radial organization of Sas-6, Cep135, Sas-4, and PLP on asl long centrioles in mGSCs and wt SC giant centrioles (Fig. 3 C). Within the limits of this technique, the radial distribution of proteins on asl centrioles in mGSCs is normal. This provides additional evidence that these structures are centrioles, not protein aggregates or tubular extensions similar to the effects of Sas-6 and Ana2 overexpression (Gopalakrishnan et al., 2010; Stevens et al., 2010).
To determine if asl mGSC long centrioles retained distal characteristics, we examined Cep97 and CP110, proteins normally localized to the distal end of centrioles that function to control length (Spektor et al., 2007; Schmidt et al., 2009; Delgehyr et al., 2012; Franz et al., 2013). The antibodies we raised against these proteins were not suitable for immunofluorescence, so we generated animals expressing GFP-tagged versions of these proteins. As expected, Cep97 and CP110 localize as one or two dots in wt mGSCs and SCs (Fig. 3, D and E, left and middle columns). In asl mutant mGSCs, both Cep97 and CP110 are recruited to one end of long centrioles at almost normal levels (Fig. 3, D′ and E′), with the caveat that both are overexpressed. Thus, asl centrioles, although long and mostly of proximal character, form a distal end.
Asterless regulates centriole length in other proliferative cells
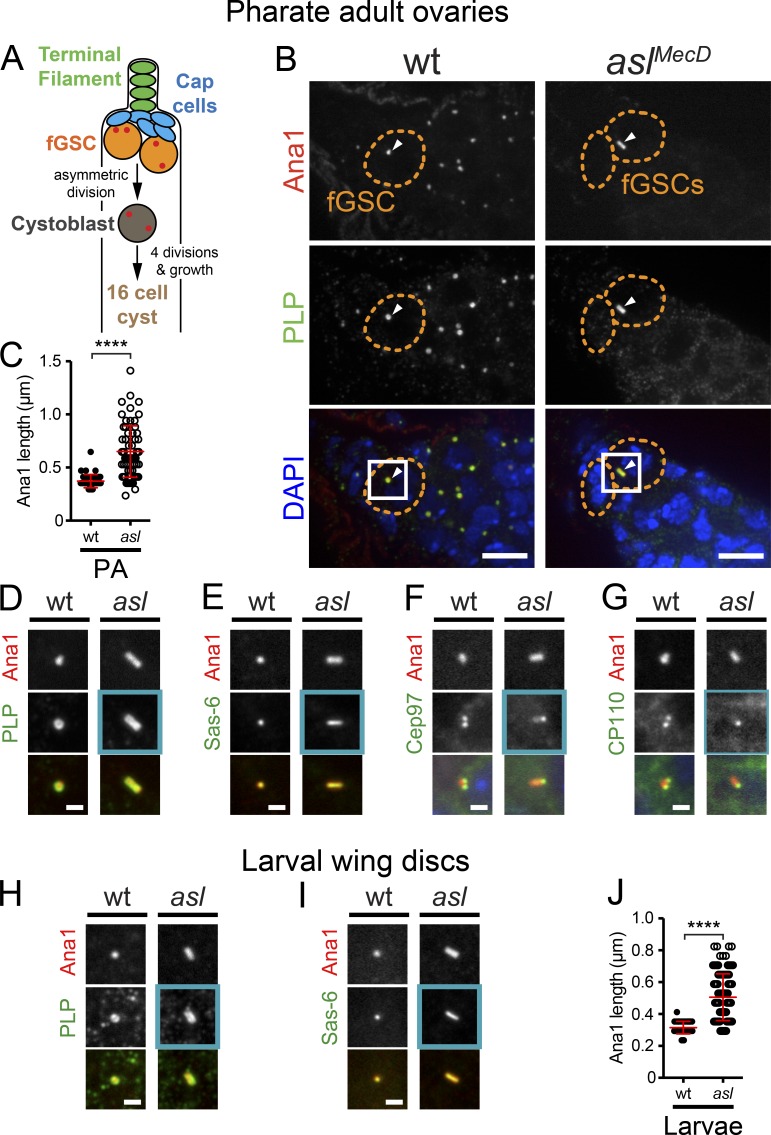
To determine if Asl controls centriole length in other cell types, we examined additional tissues in asl mutants. Like the testis, the ovary is complex with many cell types (Fig. 4 A). Similar to mGSCs, remnant centrioles in female germline stem cells (fGSCs) were frequently too long (Fig. 4, B and C; 47/65 centrioles greater than wt mean plus two SDs). We also found that PLP (Fig. 4 D) and Sas-6 (Fig. 4 E) were localized along the entire length of fGSCs centrioles, indicating that centrioles are “proximilized.” These remnant centrioles had a distal end as indicated by the presence of Cep97 and CP110 (Fig. 4, F and G).
Figure 4.
Asl controls centriole length in female germline and somatic cells. (A–G) Remnant centrioles in wt (aslmecD/TM6B) and asl fGSCs from PAs. (A) Cell types in Drosophila ovaries (centrioles, red). (B) Remnant centrioles (Ana1::tdTomato, red; PLP, green) in asl fGSCs are longer than in wt. Arrowheads, centrioles in fGSCs. DAPI, blue. Boxes are enlarged in D. (C) Measurements of centriole length (Ana1::tdTomato), in fGSCs. (D) Boxed region from B. PLP is along the length of asl centrioles. (E) GFP::Sas-6 (green) extends the length of asl centrioles. (F and G) GFP::Cep97 (F, green) and CP110::GFP (G, green) localize to the proximal tip of asl fGSC centrioles. (H–J) Centrioles in wt (aslmecD/TM6B) and asl larval wing discs. (H) Remnant asl centrioles (Ana1::tdTomato, red) are longer than wt. PLP (red) is along their entire length. (I) GFP::Sas-6 (green) extends the entire length of asl centrioles. (J) Centriole length (Ana1::tdTomato) in wing discs. Mean ± SD, red. Comparison: unpaired t tests, with Welch’s correction. ****, P < 0.0001. Bars: (B) 5 µm; (D–I) 1 µm.
To determine if Asl controls centriole length in somatic tissues, we examined remnant, centrioles in asl larval wing discs. Long, proximalized centrioles were also identified in these cells (Fig. 4, H–J; 62/118 centrioles greater than wt mean plus two SDs), indicating that centriole length control by Asl is a conserved mechanism among different cell types. There is significant proliferation in the wing disc during larval development (Milán et al., 1996), supporting a model where Asl is required for regulating the length of centrioles that transit through multiple cell cycles. We attempted to examine the affect of loss of Asl on centriole length outside the animal by knocking down Asl in S2 cells. We located a few remnant centrioles in these cells but did not see centrioles that were longer than normal by light microscopy (unpublished data). However, we note that previous studies of core centriole length did not see an effect from knockdown of cp110 in S2 cells, even by EM (Franz et al., 2013), although they did see an effect on centriole length in the animal in wing discs and we show an effect in mGSCs (see Fig. 6).
Figure 6.
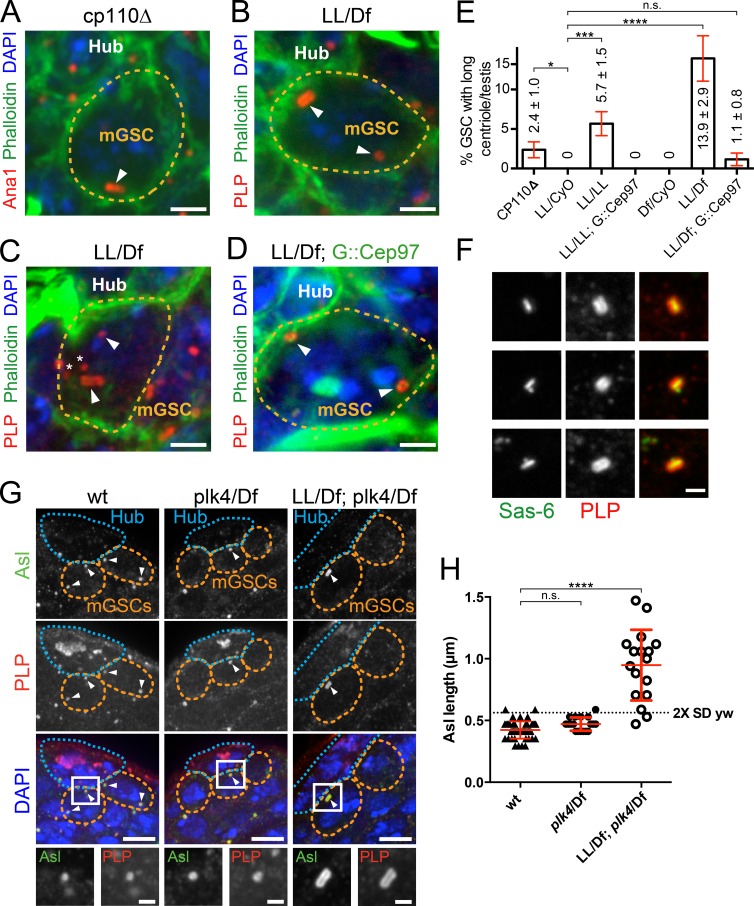
Cep97 controls mGSC centriole length. (A–D) mGSCs in adult testes of indicated genotypes. Ana1::tdTomato, red (in A); PLP, red (in B–D); phalloidin, green; DAPI, blue. Arrowheads represent centrioles in mGSCs. Asterisks represent centrioles from the hub. (A) An mGSC from a cp110Δ testis with a long centriole. The other centriole is not visible in this projection. (B) cep97LL/Df(2L)A1 (LL/Df) mGSC with a long apical centriole. (C) LL/Df mGSC with a long nonapical centriole. (D) An LL/Df mGSC expressing GFP::Cep97 has normal-length centrioles. (E) Percentage of mGSCs with long centrioles per testis ± SEM for indicated genotypes. Testes counted include cp110Δ, 36; LL/CyO, 17; LL/LL, 34; LL/CyO; GFP::Cep97, 23; LL/Df, 32; and LL/Df; GFP::Cep97, 27. Comparison: unpaired t tests, with Welch’s correction. ****, P < 0.0001; ***, P < 0.001; *, P ≤ 0.05; n.s., not significant. (F) GFP::Sas-6 (green) and PLP (red) extend along the length of cep97 centrioles. Procentriole can position at one end (middle row) or the middle (bottom row) of the elongated centriole. (G) Remnant centrioles (Asl, green; PLP, red) in cep97;plk4 double-mutant adult mGSCs are longer than in wt (yw) or plk4. mGSCs, orange line; DAPI, blue; arrowheads, centrioles in mGSCs. Boxed region enlarged (bottom). (H) Centriole (Ana1::tdtomato) length (red line, mean ± SD) in mGSCs in adults of the indicated genotypes. Dashed line is the mean of wt plus two SDs. Almost all centrioles in mGSCs are long in cep97;plk4. Comparison: ordinary one-way analysis of variance followed by Tukey’s multiple comparisons test. ****, P < 0.0001; n.s., not significant. Bars: (A–D) 2 µm; (F and G, bottom row) 1 µm; (G) 5 µm.
Asterless is not required for PCM recruitment and MTOC activity
Asl adopts an extended conformation with its C terminus adjacent to the centriole and its N terminus extended outward (Mennella et al., 2012; Sonnen et al., 2012), ideally positioned to organize PCM around the centriole. However, there are conflicting studies regarding the requirement for Asl in PCM recruitment (see Introduction). To directly test this, we examined several PCM proteins, beginning with PLP, which also extends from the centriole wall. Asl-free centrioles recruit PLP in both mitosis and interphase (Figs. 5 A and S2 E, right columns, blue box), consistent with RNAi studies in S2 cells (Mennella et al., 2012). Importantly, unlike SC centrioles, where PLP is exclusively proximal, PLP localizes along the length of asl long centrioles (Figs. 5 A and S2 E, right column, blue box). Because PLP, in Drosophila, often behaves more similarly to a centriole protein, we examined the distributions of the more canonical PCM proteins Cnn, Spd2, and γ-tub. Loss of Asl did not prevent the localization of any of these proteins to mGSC mitotic or interphase centrosomes (Fig. 5, B–D; and Fig. S2, F–H, right column, blue boxes), inconsistent with a role for Asl in recruiting PCM (Varmark et al., 2007; Dzhindzhev et al., 2010; Conduit et al., 2014). Furthermore, Cnn, Spd2, and γ-tub were localized along the length of the asl long centriole, in contrast to their strict proximal localization on giant SC centrioles (Fig. 5, B–D). These results expose an intimate, possibly physical, link between the core centriole structure and the position of PCM assembly (see Discussion).
Figure 5.
Asl is not required for PCM recruitment during mitosis. (A–E) Centrioles (Ana1::tdTomato, red) from mitotic wt (aslmecD/TM6B) and asl mGSCs or wt mature SCs. Asterisk represents centrioles in adjacent hub cells. (A) PLP (green) localizes to centrioles in wt mGSCs, wt SCs, and asl mGSCs (blue box, n = 7/7). mGSC centriole, arrowhead. DNA, d (phosphohistone, not depicted). (B) Cnn (green) localizes to centrioles in wt mGSCs, wt SCs, and asl mGSCs (blue box, n = 3/3). (C) Spd2 (green) localizes to centrioles in wt mGSCs, wt SCs, and asl mGSCs (blue box, n = 4/4). (D) γ-Tubulin (green) localizes to centrioles in wt mGSCs, wt SCs, and asl mGSCs (blue box, n = 10/10). (E) MTs (green) organized around centrioles in wt mGSCs, wt SCs, and asl mGSCs (blue box, n = 8/8). DNA, d (phosphohistone); actin, a (phalloidin). (F) Frames from Videos 1 and 2 of mGSCs in wt (top row) and asl (bottom row) expressing GFP::α-tubulin (MTs, green) and Ana1::tdTomato (red). Time (min) relative to nuclear envelope breakdown (NEB; red box). Anaphase onset (AO; purple box). (G) γ-Tubulin (green) localizes to centrioles in meiotic asl SCs (blue box, n = 15/15). Asl is absent from centrioles in asl (bottom right). Bars: (A–E and G) 1 µm; (F) 5 µm.
The presence of all of the PCM components on asl centrioles suggests that these centrosomes could function as MTOCs. We found that asl centrioles in mGSCs organize a modest MT array in interphase and a robust array during mitosis along the entire centriole (Figs. 5 E and S2 I, right column, blue box), mirroring the extended PCM localization. Live imaging of asl mGSCs during mitosis revealed that long centrioles can effectively organize a mitotic spindle pole (Fig. 5 F and Videos 1 and 2). However, because asl mGSCs only contain one centriole, they first build a half spindle focused at the long centriole, then an acentrosomal half-spindle on the other side of the chromosomes. These spindles properly proceed through mitosis with only slight delay (Fig. 5 F, AO), comparable mGSCs with no centrosomes (Fig. S2 J and Video 3). Most likely, this delay is a consequence of constructing the acentrosomal half-spindle, not the presence of an unusually long centriole.
To determine if Asl has a role in PCM recruitment in other cells types, we examined remnant centrioles in dividing SCs (Fig. S1 D, zone 4) of asl testis. We found that Asl-free centrioles in SCs recruit γ-tub during meiosis I (Fig. 5 G), consistent with previous findings (Blachon et al., 2008). Note that the example presented does not contain γ-tub along the entire centriole (Fig. 5 G), as it was selected from zone 4 (Fig. S1). These centrioles were constructed before Asl loss and thus do not have an extended luminal (cartwheel) structure (see section Asl is required for basal body function for further details). Together, our results indicate that Asl is not essential to recruit the major PCM components and its loss does not affect the ability of these centrosomes to participate in mitotic or meiotic spindle function.
Asl’s binding partner Cep97 is required for mGSC centriole length control
As mentioned, Cep97 and CP110 have been implicated in controlling centriole length, but both are present on asl long centrioles (Fig. 3, D and E, right column, blue box). However, previous studies have shown that Cep97 and CP110 remain at the distal ends of centrioles in other cases where centrioles grow too long (Kohlmaier et al., 2009; Schmidt et al., 2009; Tang et al., 2009; Korzeniewski et al., 2010; Lin et al., 2013). Thus the localization of these proteins to centrioles alone is not sufficient to ensure proper length control. Therefore, we hypothesized that Asl is required for proper function, not recruitment, of Cep97 and/or CP110 at the centriole. This hypothesis predicts that loss of either of these proteins would result in a similar long centriole phenotype to loss of Asl.
Long centrioles in mGSCs of cp110 null (cp110Δ) flies have not been reported (Franz et al., 2013). We reexamined cp110Δ flies and found that ∼2.4% of mGSCs/testes contain a long centriole similar to asl mutants and never observed in >1,000 control wt mGSCs (Fig. 6, A and E). To examine Cep97 loss, we obtained a transposable element insertion in the first Cep97 intron, which we refer to as cep97LL (Schuldiner et al., 2008). We confirmed that the level of Cep97-PB, the longest isoform, was reduced by >85% in cep97LL (Fig. S3 A; LL/LL). Examining homozygous cep97LL larval testes showed ∼6% of mGSCs/testis contain long centrioles, whereas controls had none (Fig. 6 E and Fig. S3, B–D, LL/Cyo vs. LL/LL). To confirm that this was a consequence of Cep97 loss, we examined cep97LL as hemizygotes over a small chromosomal deletion (Materials and methods; Df(2L)A1). cep97LL/Df(2L)A1 flies have long centrioles in ∼14% of mGSCs/testis (Fig. 6, B, C, and E, LL/Df), a phenotype abrogated by the introduction of a GFP::Cep97 transgene (Fig. 6, D and E; and Fig. S3 E).
All of the long centrioles in cep97LL had PLP and Sas6 decorating their entire length, indicating that they have an extended proximal organization similar to centrioles in asl mutants (Fig. 6, B, C, and F). Interestingly, we find that nascent procentrioles (indicated by Sas6) in the cep97LL were not always located at one end of the centriole (Fig. 6 F, bottom row). This supports a model where the entire long centriole is of proximal character, as procentriole formation is normally reserved for the proximal end. These data show that Cep97, and to a lesser extent CP110, work to regulate centriole length in mGSCs in a manner similar to Asl. Because the long centriole phenotype was significantly stronger in cep97, we focused subsequent studies on Cep97.
One inconsistency between the asl and cep97 mutant phenotypes is the frequency of long centrioles in mGSCs, with 100% of remnant centrioles being long in asl mutant and only 14% being long in the cep97. One main difference is that centriole duplication is blocked in asl, but not in cep97 mutants. To test if this accounted for the frequency difference, we blocked centriole duplication in the cep97 background by introducing a plk4 mutation. By blocking centriole duplication, the only centrioles that will remain will be those present in mGSC since early embryogenesis. Although remnant centrioles in the mGSCs of plk4 mutants were normal length (Fig. 6, G and H), in cep97 plk4 double mutants, 82% of remnant centrioles were long (longer than wt mean plus two SDs; Fig. 6, G and H). This is very similar to the 100% seen in asl mutants, and is consistent with Cep97 and Asl functioning in the same pathway to control centriole length. In both plk4 and cep97;plk4 mutants, centrioles are not found in every mGSC (Fig. 6 G). Combined with our findings that centrioles are not retained in all asl and sas-4 mutant mGSCs (Fig. S1, F and G), it suggests that long centrioles were seen at a low frequency in cep97 mutants because centrioles are normally lost from mGSCs at a certain frequency and the longest centrioles were the few retained by the mGSC over multiple cell cycles. This is contrary to the prominent model where mGSCs retain their mother centrioles indefinitely (Yamashita et al., 2007) but in agreement with analysis of remnant sas-4 mutant centrioles occasionally found in mGSCs (Riparbelli and Callaini, 2011).
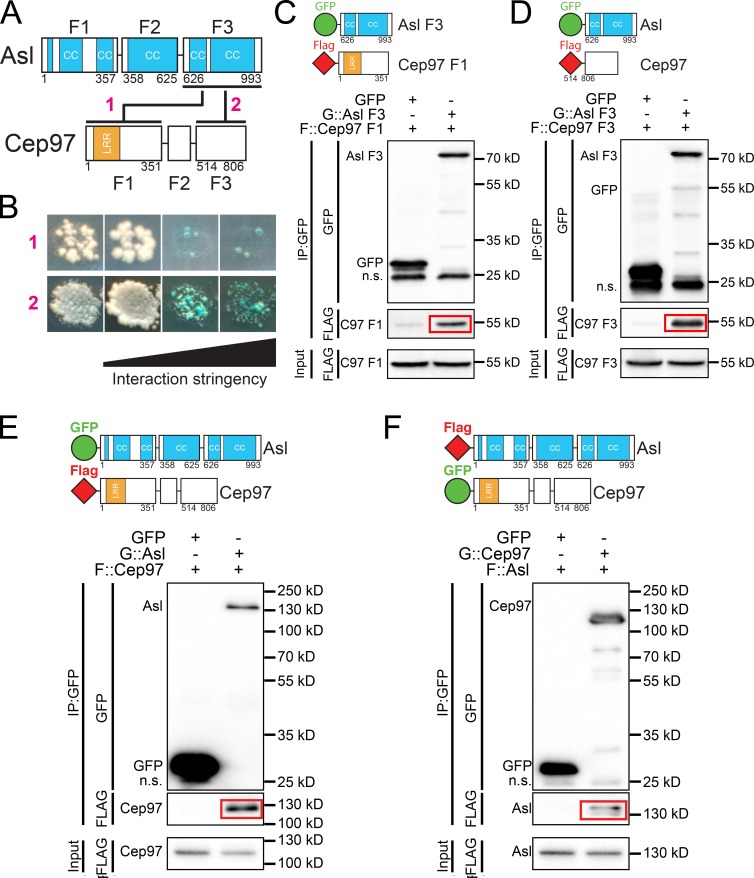
To better link Asl and Cep97, we tested if Asl and Cep97 physically interact. We divided both proteins into three subfragments, taking care to not disrupt any predicted motifs or secondary structures (Fig. 7 A). We then tested for direct interaction between these fragments by yeast-two hybrid (Y2H) screening, finding that the C-terminal region of Asl interacts with both the N- and C-terminal regions of Cep97 (Fig. 7, A and B; and Fig. S3 F). We confirmed these interactions by coimmunoprecipition (coIP) using fragments (Fig. 7, C and D) and full-length proteins (Fig. 7, E and F) overexpressed in S2 cells.
Figure 7.
Asterless and Cep97 interact. (A) Schematics of the subfragments (F1–F3) of Asl and Cep97 used for interaction assays. Red numbers refer to B. Numbers are amino acids. Lines, Y2H interactions; CC, coiled coil; LRR, leucine-rich repeat. (B) Asl F3 interacts with Cep97 F1 (1) and Cep97 F3 (2) by Y2H screening. The first column shows the presence of Y2H plasmids. The remaining columns show interaction under conditions of increasing stringency (see Materials and methods). Red numbers refer to A. (C) GFP::Asl F3 coIPs Flag::Cep97-F1 from S2 cells. (D) GFP::Asl F3 coIPs Flag::Cep97-F3 from S2 cells. (E) GFP::Asl full-length coIPs Flag::Cep97 full length from S2 cells. (F) GFP::Cep97 full-length coIPs Flag::Asl full length from S2 cells.
Cep97 functions downstream of Asl to regulate centriole length
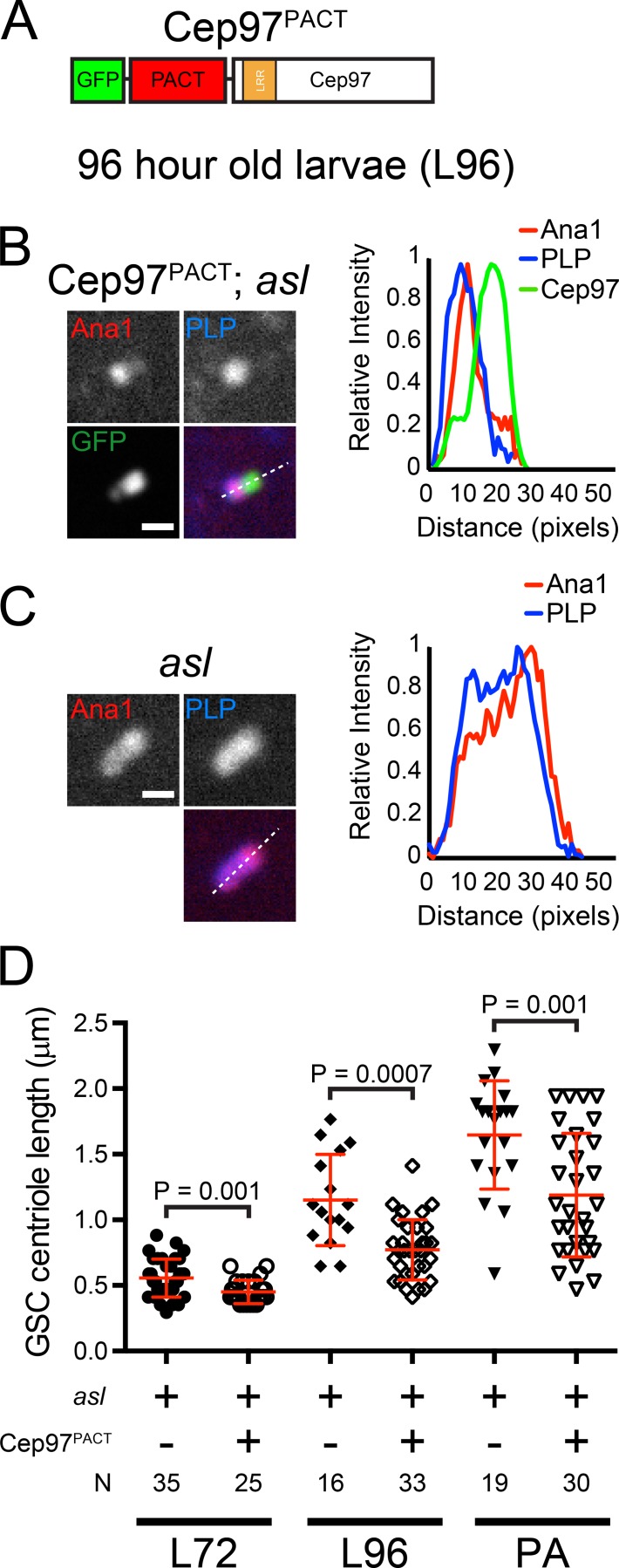
A simple model consistent with our findings is that Asl functions through Cep97 to regulate centriole length. In this case, we would predict that the long centriole phenotype in asl mGSCs might be overcome if large amounts of Cep97 was forced to the centriole. To accomplish this, we generated a transgenic fly ubiquitously expressing Cep97 fused to GFP and the PACT domain of PLP, a commonly used centrosome-targeting domain (Gillingham and Munro, 2000). We refer to this construct as Cep97PACT (Fig. 8 A). Cep97PACT localized to asl mutant mGSC centrioles, and these centrioles were significantly shorter than those in asl mGSCs not expressing Cep97PACT (Fig. 8, B–D; and Fig. S4, A and B). Expression of this construct in wt did not prevent the elongation of the normally giant centrioles in SCs (Fig. S4 C). Importantly, neither expression of Cep97 (Fig. S4 D) nor expression of PACT (Fig. S4 E) could suppress the long centriole phenotype of asl mutants, even when expressed at higher levels than Cep97PACT (Fig. S4, F and G). This strongly support a model where Cep97 is a downstream effector of Asl in centriole length control.
Figure 8.
Cep97 functions downstream of Asl in mGSC centriole length control. (A) Centriole targeted Cep97 construct. LRR, leucine-rich repeat; PACT, centriole targeting domain of PLP. (B) A representative centriole in asl mGSCs expressing Cep97PACT. PLP, blue; Cep97PACT, green; Ana1::tdTomato, red. Linescans (dashed line) along the long axis of the centriole, normalized to peak intensity. (C) A representative centriole in asl mutant mGSCs. PLP, blue; Ana1::tdTomato, red. Linescans as in B. (D) Length of Ana1::tdTomato in asl mutant mGSC centrioles at indicated developmental time from individuals with or without Cep97PACT expression. Additional examples are shown in Fig. S4. Error bars are ±SD. Comparison: unpaired t tests with Welch’s corrections when appropriate. N, centrioles measured. Bars, 1 µm.
We attempted to further link Asl and Cep97 function by generating a mutation in Asl specifically disrupting the Asl–Cep97 interaction. However, generating such a mutant proved to be extremely challenging. We produced >1,400 Asl mutant alleles using a reverse Y2H screen (see Materials and methods) and screened them for interaction with Cep97 and other proteins that interact with Asl (Fig. S5 A). We were unable to generate a mutant that specifically disrupted the interaction with both regions of Cep97 while maintaining the interaction with all other binding partners. This is consistent with the critical nature of the C terminus of Asl for centriole localization and Plk4-dependent centriole duplication (Klebba et al., 2015), and the large number of protein interactions made by this region (unpublished data). However, the direct interaction between Asl and Cep97, the similar mutant phenotypes, and the suppression of the asl phenotype by Cep97PACT all support a model where Asl and Cep97 function together to regulate centriole length.
Asl is required for basal body function
The progeny cells resulting from the asymmetric division of mGSCs undergo a differentiation program to become sperm, during which the centrioles undergo major changes. In SCs, centrioles elongate to form giant centrioles by extending the distal end and maintaining the cartwheel protein Sas-6 and PCM at the proximal end. Later, in spermatids (STs), centrioles act as BB that dock at the nuclear membrane and extend the flagellar axoneme from their distal end (Fig. 9 A; Fabian and Brill, 2012). As we have demonstrated in Fig. 3, unlike SC giant centrioles, the long centrioles in asl mGSCs adopt a proximal centriole character. Given this vastly different proximal/distal protein arrangement in asl mutant centrioles, we hypothesized that they would not be able to function as BBs. Because the SCs and STs are the progeny of the mGSCs, we reasoned that we could test this hypothesis if we could identify centrioles that grew long in the mGSC and were subsequently passed on to its progeny.
Figure 9.
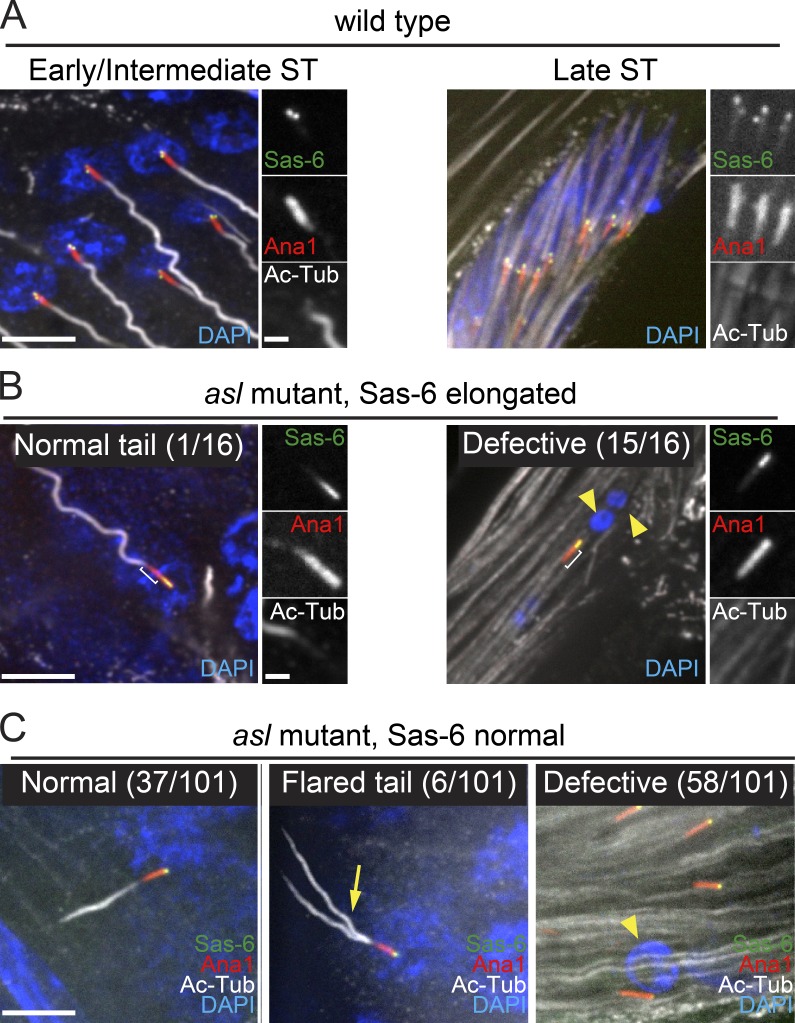
Asl is required for proper function of sperm basal bodies. (A–C) Sas-6 (green) on BBs (Ana1::tdTomato, red) in early/intermediate (left) or late (right) STs. Axonemes, Ac-tub (gray); DNA, DAPI (blue). (A) wt (asl/TM6) STs. In early/intermediate STs (left), basal bodies are docked at the nucleus and form axonemes. The nuclei of late STs are elongated and clustered. (B) Remnant centrioles in asl mutant STs with “elongated” region of Sas-6 (n = 16). ST at all stages were examined. Bracket, distal extension of Ana1 beyond Sas-6. (left) The only remnant centriole (early/intermediate ST) with elongated Sas-6 exhibiting normal nuclear attachment and axoneme nucleation. (right) Representative remnant ST centriole not attached to a nucleus or forming an axoneme. The nuclei of late stage asl STs are fragmented and dispersed (yellow arrowheads). (C) Remnant centrioles in asl mutants with a “normal” distribution of Sas-6 (restricted to the very proximal end; n = 101). (left) Representative remnant centriole in an early/intermediate ST exhibiting normal nuclear attachment and axoneme formation. (middle) Representative remnant centriole in an early/intermediate ST with a disrupted, flared axoneme (arrow). (right) Representative remnant centrioles in late STs not attached to nuclei or nucleating axonemes. Yellow arrowhead indicates nucleus that has not undergone reshaping. Additional examples are shown in Fig. S5. Bars: 5 µm; (insets) 1 µm. Defective, no axoneme or no nuclear attachment or both.
In asl mutant testes, we find remnant centrioles in all of the cell types derived from mGSCs (Fig. S1 D, zones 3 and 4). These cells could have received their centrioles via two mechanisms. (1) They could inherit centrioles from mGSCs before maternal Asl depletion. We predict these centrioles would have a normal proximal/distal character with Sas-6 restricted to a dot at the proximal end. We term these “Sas-6 normal” centrioles. (2) They could inherit a centriole that was in the mGSC when maternal Asl was depleted and grew long in the mGSC before being passed on. These centrioles would have a proximal/distal character similar to the centrioles in asl mutant mGSCs and have an extended Sas-6 localization. We term these centrioles “Sas-6 extended.”
We examined testes from pupae and PAs, and found remnant, Asl-free centrioles, in STs. The vast majority of centrioles were Sas-6 normal; however, we identified some Sas-6 elongated centrioles (n = 16 centrioles), identical to the centrioles in asl mGSCs. We found that 15 out of 16 (from 11 testis of ∼30 examined) of these centrioles were defective in nuclear attachment and axoneme formation (Figs. 9 B and S5 B). Interestingly, 14 of the 15 defective BBs formed an extension of Ana1 distal to the elongated Sas-6 signal. This suggests that they underwent the normal elongation of the distal end that takes place in SCs and that defects in BB function might not be a consequence of extended proximal character, but rather a consequence of an independent and as of yet undescribed function of Asl. If this is the case, we would predict that the Sas-6 normal centrioles would show similar BB defects.
When we examined Sas-6 normal remnant centrioles in spermatids, only 37 out of 101 (20 in early and 17 in late spermatids) BBs were attached to a nucleus and had built a normal-looking axoneme. The remaining 64 had defects in nuclear attachment, improper axoneme formation, or both (Figs. 9 C and S5 C). Most striking, we observed BBs in early spermatids that had forked axonemes as indicated by Ac-tub (Figs. 9 C and S5 C, arrows). Given these defects were seen in asl mutant BBs with normal proximal/distal character (Sas-6 normal), they are most likely a consequence of losing yet another unknown function of Asl specific to the BB. Consistent with this idea, Asl protein localization is dramatically rearranged during the final stages of BB formation from coating the centriole wall to a ring-like structure at the base of the axoneme, approximately where the BB to axoneme transition occurs (Fig. S5 D). Thus, we have identified a novel role for Asl in proper BB function during the final stages of spermatogenesis, and this role is independent of its role in centriole length control.
Discussion
Loss-of-function studies have identified proteins critical for the function of centrosomes (Goshima et al., 2007; Dobbelaere et al., 2008; Gönczy, 2012; Balestra et al., 2013). The loss of many of these proteins results in the loss of centrosomes from the cell, precluding analysis of other roles they might play beyond centrosome duplication or maintenance. Here, we investigate roles for Asl beyond its well-established function in centriole duplication. We take advantage of Drosophila zygotic mutant animals that build centrioles in early development using maternally provided Asl. We then identify these centrioles in larval and adult tissues where they now reside in an environment genetically null for Asl. These remnant centrioles have proven to be a critical tool to investigate Asl, but similar analysis can be performed on remnant centrioles generated using zygotic mutants of other centriole duplication factors.
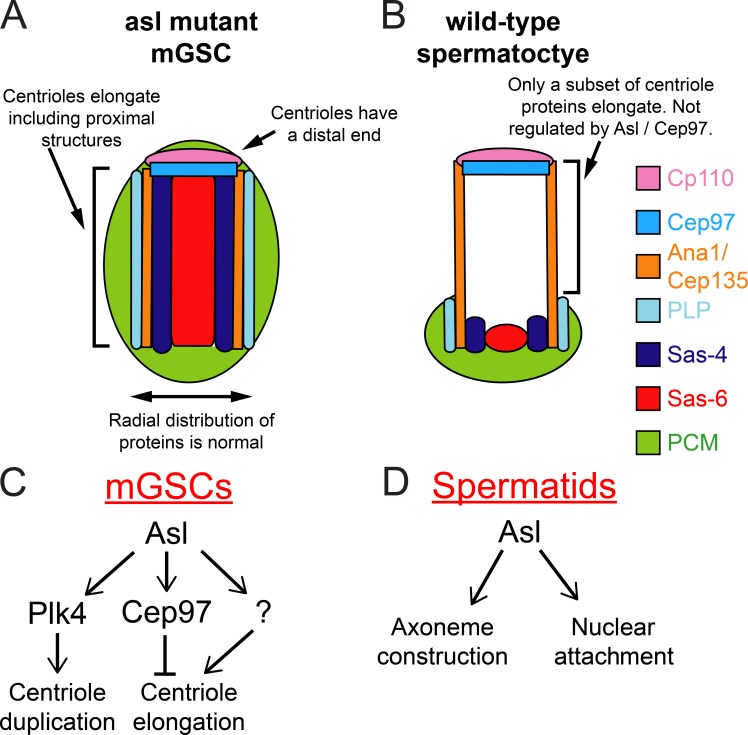
The most striking finding was that the absence of Asl resulted in the massive (up to 30-fold) elongation of centrioles in both germline and somatic cells. Although long, these centrioles maintain normal radial distributions of all the tested proteins and a characteristic distal centriole tip (Fig. 10 A). These long asl mutant centrioles are formed in a manner that results in proximal centriole proteins along the entire centriole (Fig. 10 A). This is different than the elongation seen in SC centrioles, where these proteins are restricted to the proximal end. Restricting the growth of the centriole proximal domain is a mechanism that has not been previously documented.
Figure 10.
Model for Asl function in centriole organization and length control. (A and B) Not all long centrioles are alike. Centrioles in asl mutant mGSCs (A) are organized in a manner distinct from the giant centrioles normally found in SCs (B). This indicates that these two structures, although of similar scale, arise via distinct mechanisms. (C and D) Expanded roles for Asterless at the centriole. In the male germline, Asl plays a critical role in regulating centrosome duplication via Plk4, and controlling centriole length upstream of, and likely directly through, Cep97. (D) In developing STs, Asl is required for normal nuclear attachment and axoneme formation.
Unlike in the other cells examined, remnant centrioles in asl hub cells do not grow long. This supports the model that centriole elongation requires cells to be actively proceeding through the cell cycle. Our data suggest that Asl’s role in controlling centriole length is most important in highly proliferative cells where a given centriole has the highest chance of being maintained in the same cell over multiple cell cycles. This is a circumstance common to stem cells. Thus, our identification of significant length control defects in two distinct stem cells suggests that length control by this mechanism might be especially critical in stem cells.
Direct protein binding and genetic analysis revealed that Cep97 functions downstream of Asl to exert a significant portion of its centriole length control (Fig. 10 C). The presence of overexpressed Cep97 at the distal end of Asl-free centrioles suggests that the mechanism by which Asl controls length is not simply to recruit Cep97. It remains a formal possibility that at endogenous levels Cep97 localization might be affected by loss of Asl. An attractive alternative model is that Cep97 localizes to centrioles via binding to a different protein but that binding to Asl enhances Cep97’s centriole length control activity. The ability of centriole targeted Cep97 to partially compensate for the loss of Asl is consistent with this model. Our data also suggest that Asl does not function exclusively through Cep97 because the long centrioles in cep97 mutants are shorter than those in similarly aged asl mutants.
Cep97 and CP110 have been implicated together in the control of centriole length. However, we found that a hypomorphic cep97 allele had a stronger effect on centriole length than a null cp110 mutant, suggesting a role for Cep97, independent of CP110. Additionally, the centrioles in both asl and cep97 mGSCs are morphologically different than the elongated centrioles reported in Drosophila cells lacking CP110. These centrioles were only ∼1.1 times (10%) longer than controls and had a significant MT protrusion (marked by Ac-tub) at one end (Franz et al., 2013); similar results have been reported in mammalian cells (Schmidt et al., 2009). Interestingly, loss of Klp10A in Drosophila cells results in both significantly longer centrioles (approximately two times longer [200%]) with additional MT extensions from its distal end, and its function does not require CP110 (Delgehyr et al., 2012; Franz et al., 2013). This supports the conclusion that centriole protein proximal-distal distribution, the length of the centriole as a whole, and the length of centriole wall MTs might all be regulated by distinct, possibly cell type–specific, mechanisms.
One interesting question is why cells have evolved a mechanism to control centriole length. It is possible that evolution of the centriole has selected for a minimum centriole “functional unit” that can efficiently accomplish its task, in this case, serving as an MTOC. Any unessential centriole length might require additional cellular resources, which could provide selective pressure to evolve a centriole length control mechanism. Another possibility is the presence of limited amounts of centriole proteins in each cell. In this case, limiting the length of the mother centriole would preserve materials needed for growth of the daughter. Yet another possibility is that regulating MTOC size is critical in cells that require movement and positioning of the centrioles, such as in cells dividing asymmetrically or forming cilia.
The effect of Asl loss is not limited to centriole structure; it also influences the distribution of PCM proteins (Fig. 10 A), but not in the predicted way. Based on its position on the outside of the centriole and antibody interference experiments in Drosophila embryos, Asl was suggested to be required for PCM recruitment (Conduit et al., 2010, 2014; Mennella et al., 2012). We found that PCM proteins were recruited to Asl-free centrioles in mitotic mGSCs and meiotic SCs, which can organize MTs and build a functional spindle. Interestingly, the location of PCM assembly corresponded to the position of proximal centriole proteins, even in cases where the proximalized asl mutant centrioles were >2 µm long. An intriguing hypothesis, with broad implications in centrosome assembly, is that the organization of the internal centriole proteins helps establish the position of the PCM through an unknown inside-out communication mechanism. There is precedence for such a mechanism. Previous work suggested that Sas-4/CPAP might help tether PCM proteins located on the outside of the centriole to the centriole via its interactions with Cep135 and Ana2, located inside the centriole (Zheng et al., 2014). Testing this model will be an exciting future direction that will involve the identification and analysis of the physical links between the centriole core and the PCM, a link that must span the MTs of the centriole wall.
We also identified a role for Asl in sperm BB function. In the absence of Asl in STs, we found significant defects in the attachment of the BB to the nucleus as well as in the assembly and/or maintenance of the flagellar axoneme (Fig. 10 D). Our initial hypothesis was that these defects were a result of the longer, proximalized asl mutant centrioles, but our data proved otherwise. We believe these defects uncover a unique and independent role for Asl during the final stages of sperm development. Asl is associated with the outer surface of BB throughout spermatogenesis, and we show it undergoes a dramatic change in localization during the final stages of ST development (Fig. S5 D). The localization of Asl during spermatogenesis is reminiscent of the “centriolar adjunct,” which is wrapped around the length of the basal body in early STs and becomes a ring structure during elongation (Tates, 1971). The defects in flagella assembly seen in asl could be the result of defects in the assembly, organization or maintenance of the centriolar adjunct and, thus, the axoneme. The scarcity of BBs in asl testis makes a detailed understanding of the role of Asl in flagella assembly especially challenging. Future identification of interactions of Asl with proteins involved in flagella assembly and the generation of separation of function mutations in Asl that retain centriole duplication function, but lose BB functions, will be required to further our understanding of this novel role for Asl.
Collectively, we have shown that Asl has roles additional to recruiting Plk4 for centriole duplication. Asl functions to control centriole length and the distribution of proximal centriole characteristics and to ensure proper BB functions during spermatogenesis. Insight into additional roles beyond duplication could help shed light on how specific lesions within centriole proteins manifest human diseases. For example, how does the missense mutation Q265P in Cep152 (the human Asl orthologue) lead to the development of human microcephaly (Guernsey et al., 2010)? What exactly does this lesion do to protein function? Our work expands the known roles of Asl, thus allowing one to hypothesize and test which functions are disrupted in specific disease mutations.
Materials and methods
Fly stocks
The following Drosophila lines were generous gifts: UAS-Ana1::tdTomato and UAS-Cep135::GFP are transgenic lines with ∼2 kb of upstream regulatory elements fused with the cDNA and sequence encoding fluorescent proteins all downstream of the yeast UAS sequence (T. Avidor-Reiss, University of Toledo, Toledo, OH; Blachon et al., 2008). Unc::GFP is under the control of its upstream regulatory elements (Baker et al., 2004). GFP::α-tubulin (T. Avidor-Reiss) and Sas-4::GFP (J. Raff, Univeristy of Oxford, Oxford, England, UK; Peel et al., 2007) contain cDNAs, fused to sequences encoding GFP under the control of the ubiquitin promoter. Act5C-Gal4 drives expression of yeast Gal4 under the control of the Act5C promoter (stock #3954; Bloomington Drosophila Stock Center). aslmecD (T. Avidor-Reiss; Blachon et al., 2008) and asl2 (M. Gatti, Sapienza Univerisity of Rome, Rome, Italy; Bonaccorsi et al., 1998) are loss-of-function alleles of asl. Df(3R)ED5177 is a deletion that removes the region surrounding asl (stock #8103; Bloomington Drosophila Stock Center). GFP::Sas-6 BAC carries an insertion from a BAC containing 20 kb of genomic sequence surrounding Sas-6 with GFP sequence recombineered in between the start codon of Sas-6 and the rest of its sequence (Lerit and Rusan, 2013). cep97LL01167 (cep97LL; Drosophila Genetic Resource Center at Kyoto Institute of Technology) is a piggyBAC insertion in the cep97 locus (Schuldiner et al., 2008). This transposable element has strong splice acceptors followed by stop codons in all three reading frames in both orientations. Because the first exon of cep97LL01167 only contains two complete codons, this insertion should lead to a significant reduction in the longest isoform of the protein, Cep97-PB. Cep97LL/Df(2L)A1 has long centrioles more frequently than Cep97LL/Cep97LL, indicating that cep97LL01167 is not a genetic null but rather a strong hypomorphic mutant. For plk4 mutant experiments, plk4c06612 (a piggyback insertion in the plk4 locus)/Df(3L)Pc-2q (a deletion removing the region surrounding plk4) was used as previously described (Bettencourt-Dias et al., 2005).
Transgenic lines were generated by BestGene, Inc. GFP::Cep97, CP110::GFP, and GFP::PACT are under the control of the ubiquitin promoter and constructed in pUGW or pUWG (Drosophila Genomics Resource Center). Cep97PACT was made by introducing the sequence encoding aa 2,687–2,796 of PLP-PF onto the 5′ end of cep97 cDNA, which was then introduced into pUGW to generate a GFP fusion.
Df(2L)A1 is a Flippase recognition target–mediated deletion (Parks et al., 2004) generated using P[XP]capud05064 and PBac[WH]f02748 (Thibault et al., 2004; the Exelixis Collection at the Harvard Medical School). The removal of the sequence in between the insertions was confirmed by PCR of genomic DNA from Df(2L)A1 followed by DNA sequencing. Both cep97LL/cep97LL and cep97LL/Df(2L)A1 individuals develop to adulthood. In the case of both heterozygotes and hemizygotes, we found long centrioles less frequently when cep97LL heterozygous mothers were used then with cep97LL homozygous mothers, suggesting a significant maternal effect. All experiments with Cep97LL were performed using individuals derived from homozygous Cep97LL00167/Cep97LL00167 mothers.
Immunofluorescence methods
Testes, ovaries, and wing discs from the indicate life stage were dissected in Drosophila S2 media and fixed in 9% formaldehyde in PBS for 20 min. Samples were washed three times in PBS + 0.3% Triton X-100 (PBST) and blocked for at least 1 h in PBST + 5% normal goat serum. Primary antibodies were incubated with testes overnight at 4°C followed by three 10-min washes in PBST. Secondary antibodies were incubated 4–8 h at room temperature followed by three 10-min washes in PBST. Staining with anti–Ac-tub antibody on pupal testis was performed, with the following modification. Before fixation, testes were treated with 100 μM colchicine in Drosophila Schneider’s medium for 30 min. Testes from pharate adults were prepared as described previously (Franz et al., 2013) with the following modification. Incubation in 45% and 60% acetic acid was omitted after fixation and testes were immediately transferred to coverslip for squashing. Samples were mounted under a No. 1.5 coverslip in Aquapolymount (Polysciences, Inc.) for conventional microscopy or Vectashield (Vector Laboratories) for SIM.
The following primary antibodies were used: guinea pig anti-Asl (gift of G. Rogers, University of Arizona, Tucson, AZ; 1:3,000), mouse anti–Ac-tub (1:500; T6793; Sigma-Aldrich), affinity-purified rabbit anti-PLP (1:4,000; gift of G. Rogers; University of Arizona, Tucson, AZ), rabbit anti-Cnn (1:3,000; this study, raised against aa 698–1,148), rabbit anti-Spd2 (1:2,000, gift of M. Gatti; Giansanti et al., 2008), mouse anti-γ-tubulin (1:500, T6556, GTU88; Sigma-Aldrich), mouse anti–β-tubulin (1:1,000, MAB3408; EMD Millipore). Secondary antibodies were Alexa Fluor 488, 568, or 647 conjugated (Thermo Fisher Scientific) 1:1500 in PBST plus 5% normal goat serum. DAPI (1:1,000) and Alexa Fluor 488– or 568–conjugated phalloidin (Thermo Fisher Scientific) 1:1,000 were added to some samples.
Microscopy and image analysis
Confocal microscopy was performed on an Eclipse Ti (Nikon) with a 100×/1.4 NA objective, a spinning disc confocal head (Visitech International), an interline-transfer cooled charge-coupled device camera (CoolSNAP HQ2; Photometrics), and illumination lasers at 405, 491, 561, and 642 nm. The microscope was controlled by and images were acquired using MetaMorph (Molecular Devices). SIM images were collected using an OMX4 (GE Healthcare). All data analysis was performed using ImageJ (National Institutes of Health). Length and width measurements were performed using line scans across centrosomes and the half maximal width of the line scan is presented. Centrioles were considered long when the measurement exceeded the mean measurement of the control plus 2 × SD. For intensity measurements, images were collected to avoid saturation and ensure that all pixel intensities fell within the linear response range of the camera. Confocal slices encompassing the volume of the centrosome were summed. The intensities presented are the pixel intensity of a region encompassing the centrosome minus the pixel intensity of an adjacent region of the cell of the same area. Data are normalized to mean intensity of centrosomes in control cells of a single day of imaging.
For live imaging of testes, larval testes were dissected in Drosophila S2 media. Whole testes were mounted in a drop of the same media, surrounded by Halocarbon oil 700 (Sigma-Aldrich) on a 50-mm lummox dish (Sarstedt), and covered with a #1.5 coverslip. Live images were acquired on a similar microscope as fixed images equipped with a stage incubator (Binomic System BC-110 Controller; 20/20 Technologies) set to 25°C, a 100×/1.49 NA objective, and an ORCA-Flash 4.0 CMOS camera (Hamamatsu Photonics). Stacks, over a 10–15-µm range, were collected at 1-µm intervals at 60- or 120-s time intervals. Images in supplemental videos are projections of multiple confocal stacks, displayed at 2 min intervals at 7 frames/s. Mitosis in mGSCs with remnant centrioles is a rare event. mGSCs are in mitosis, which lasts ∼10 min, about once a day. As such, ∼1% of mGSCs in a population are in mitosis. Each testis only has ∼10 mGSCs, and often only three or four are in a good position for live-cell imaging when the testis is mounted. As reported above, only ∼40% of mGSCs in larvae have a remnant centriole. Imaging dozens of testis for periods of 10–18 h was required to obtain only a few examples. We present representative examples of this small sample.
IP
For IP experiments, fusion proteins were expressed from pUGW and pFAHW plasmids. One well of a six-well plate of S2 cells at ∼50% confluence was transfected with 0.5 µg DNA using Effectene (QIAGEN) and allowed to recover for 48 h at 25°C. All subsequent steps were performed at 4°C. 50 µl of Protein A–conjugated Dynabeads (Thermo Fisher Scientific) was used per IP and resuspended in 1 ml PBS plus 0.01% Tween 20. 1 µl rabbit anti-GFP antibody (AB290; Abcam) was added and incubated with mixing for 1 h. Cells were harvested and lysed by resuspending in 1 ml RIPA buffer with 1 mM DTT, 1 µg/ml pepstatin A, 1 µg/ml leupeptin, and 1 mM PMSF. Lysate was cleared by a 5-min spin in a microfuge at full speed, then incubated with antibody-conjugated beads for 1 h. Beads were washed three times for 5 min each in RIPA buffer. Before the last wash, the beads were moved to a fresh tube. Beads were eluted by boiling in 25 µl of 2× Laemmli buffer for 5 min, and 10-µl samples were then analyzed by SDS-PAGE followed by Western blot.
Western blotting
For analysis of Asl protein levels in testes, whole testes dissected from young adult flies of the appropriate genotype were lysed in 2× Laemmli buffer (2 µl/testis) using a microfuge pestle. Samples were boiled for 5 min and SDS-PAGE performed followed by wet transfer to nitrocellulose or polyvinylidene fluoride membrane. After transfer, membranes were blocked with 5% milk in TBS plus 0.05% Tween 20. The following primary antibodies were used: guinea pig anti-Asl (1:5,000; gift from G. Rogers), mouse anti-GFP (JL8, Takara Bio Inc.; 1:10,000), mouse anti-FLAG M2 (1:1,000; Sigma-Aldrich), mouse anti–Αψ-tub (1:5,000, DM1A; Sigma-Aldrich), and chicken anti-Cep97 (raised against the entire protein, this study, 1:2,000). HRP-linked secondary antibodies (1:5,000) were used and SuperSignal West Dura Extended Duration Substrate (1:5,000; Thermo Fisher Scientific) was used for detection.
Y2H screening
Y2H screening to test the interaction between Asl and Cep97 fragments was performed exactly as described previously (Galletta et al., 2014). In brief, the indicated protein fragments were introduced into pDEST-PGADT7 (Rossignol et al., 2007) and pDEST-pGBKT7, modified to carry ampicillin resistance (Galletta et al., 2014) using the Gateway cloning system (Thermo Fisher Scientific). Clones in pGADT7 and pGBKT7 were transformed into Y187 and Y2HGold, respectively (Takara Bio Inc.). Liquid cultures of yeast carrying these plasmids were grown overnight at 30°C in SD −Leu or SD −Trp media, as appropriate, to maintain plasmid selection to an OD600 ∼0.5. Yeast strains of opposite mating types were mated by mixing 20 µl of a culture of a Y187 strain with 20 µl of a culture of a Y2HGold strain in 100 µl 2× YPAD media in a well of a 96-well plate. Mating was performed overnight at 30°C with shaking. Mating cultures were transferred to SD −Leu −Trp (DDO) plates using a Multi-Blot Replicator (V&P Scientific). DDO plates were grown for 5 d at 30°C. DDO plates were replica plated to DDO plates, QDO plates (SD −Ade −His −Leu −Trp), DDOXA plates (SD −Leu −Trp with Aurobasidin A [Takara Bio Inc.] and X-α-Gal [Gold Biotechnology]) and QDOXA plates (SD −Ade −His −Leu −Trp with Aurobasidin A and X-α-Gal). Plates were grown for 5 d at 30°C and scored for growth and development of color as appropriate. All Y2H constructs were tested for their ability to activate Y2H reporters in the absence of any binding partner (autoactivation).
The reverse two-hybrid screen to identify mutations in Asl (aa 626–994) that abrogate the Cep97 interaction was performed as described previously (Bennett et al., 2004) with significant modifications described elsewhere (Galletta and Rusan, 2015; Klebba et al., 2015). In brief, Asl (aa 626–994) in pGBKT7 was amplified using low-fidelity PCR conditions (Taq polymerase, in the supplied buffer with 0.05 mM MnCl2, 0.25 mM dCTP, 0.25 mM dGTP, 0.25 mM dTTP, and 0.06 mM dATP; New England Biolabs, Inc.) and primers designed to complement sequence in the vector (5′-TAATACGACTCACTATAGGGCG-3′ and 5′-CGGAATTAGCTTGGCTGC-3′). Amplified PCR products and pGBKT7, linearized with EcoRI and PstI, were gel purified using DEAE cellulose and cotransformed into Y2HGold yeast using standard methods. Yeast carrying the reconstituted plasmid were selected on SD −Trp plates. Approximately 1,400 independent clones containing randomly mutagenized Asl (aa 626–994) were obtained and then individually crossed to Y187 strains carrying Cep97 (aa 1–351) and Cep97 (514–806) in pGADT7. Diploids carrying bait and prey plasmids were selected by growth on DDO plates after 5 d at 30°C. Diploids were replica plated to DDO and QDOXA, as above, and clones that grew on DDO but failed to grow on QDOXA were selected. These clones were then counterscreened for their interaction with the following Asl interacting fragments (Fig. S5 A): Asl (aa 358–625), Asl (aa 626–994), CP110 (aa 1–325), Plk4 (aa 382–602), Sas-4 (aa 1–347), Sas-4 (aa 348–707), and Spd-2 (aa 664–1,146). No clones disrupted both of the Cep97 interactions without significantly disrupting other interactions.
Statistical analysis
Unpaired t tests, with Welch’s correction when appropriate, one-way analysis of variance followed by Tukey’s multiple comparisons tests, or Fisher’s exact tests were used for all statistical comparisons as indicated in figure legends. All statistical analysis was performed with Excel (Microsoft) or Prism (GraphPad Software).
Online supplemental material
Fig. S1 shows an immunoblot demonstrating loss of full-length Asl in aslmecD, images and schematics of the centrioles in whole testis from wt and aslmecD and demonstration that centrioles in aslmecD do not have Asl by immunofluorescence. Fig. S2 shows long centrioles in other allelic combinations of asl mutants, the localization of PCM proteins to centrioles during interphase in asl mutants, and stills from Video 3. Fig. S3 shows a western blot demonstrating the loss of Cep97 protein in the mutants used in this study, micrographs showing long centrioles in additional allelic combinations of cep97 mutants and the complete Y2H data demonstrating that Asl and Cep97 directly interact. Fig. S4 shows data that Cep97PACT also rescues centriole length in pharate adult asl testis and includes controls for these experiments and those in Fig. 8. Fig. S5 shows examples of reverse Y2H mutants in Asl that failed to disrupt both Asl–Cep97 interactions and not any other Asl interactions. This figure shows additional examples of defects in remnant centrioles/basal body function in asl spermatids like those show in Fig. 9. Video 1 shows mitosis in a wt mGSC. Video 2 shows mitosis in an asl mGSC with a single, elongated centriole. Video 3 shows mitosis in an asl mGSC with no centrioles. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201501120/DC1.
Supplementary Material
Acknowledgments
We thank X. Wu (National Heart, Lung, and Blood Institute Light Microscopy Core) for assistance with SIM; J. Ortega for assistance in reverse Y2H screening; and D.A. Lerit, T.A. Schoborg, and A.L. Zajac for critical reading of the manuscript.
N.M. Rusan is supported by the Division of Intramural Research at National Institutes of Health, National Heart, Lung, and Blood Institute (1ZIAHL006104).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Ac-tub
- acetylated tubulin
- Asl
- Asterless
- BB
- basal body
- Cnn
- Centrosomin
- fGSC
- female germline stem cell
- γ-tub
- γ-tubulin
- IP
- immunoprecipitation
- mGSC
- male germ line stem cell
- MT
- microtubule
- MTOC
- microtubule organizing center
- PA
- pharate adult
- PCM
- pericentriolar material
- PLP
- Pericentrin-like protein
- SC
- spermatocyte
- SIM
- structured illumination microscopy
- ST
- spermatid
- Unc
- Uncoordinated
- wt
- wild type
- Y2H
- yeast two hybrid
References
- Angus K.L., and Griffiths G.M.. 2013. Cell polarisation and the immunological synapse. Curr. Opin. Cell Biol. 25:85–91. 10.1016/j.ceb.2012.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J.D., Adhikarakunnathu S., and Kernan M.J.. 2004. Mechanosensory-defective, male-sterile unc mutants identify a novel basal body protein required for ciliogenesis in Drosophila. Development. 131:3411–3422. 10.1242/dev.01229 [DOI] [PubMed] [Google Scholar]
- Balestra F.R., Strnad P., Flückiger I., and Gönczy P.. 2013. Discovering regulators of centriole biogenesis through siRNA-based functional genomics in human cells. Dev. Cell. 25:555–571. (published erratum appears in Dev. Cell. 2013. 26:220) 10.1016/j.devcel.2013.05.016 [DOI] [PubMed] [Google Scholar]
- Bennett M.A., Shern J.F., and Kahn R.A.. 2004. Reverse two-hybrid techniques in the yeast Saccharomyces cerevisiae. Methods Mol. Biol. 261:313–326. 10.1385/1-59259-762-9:313 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M.K., Carmo N., Balloux F., Callaini G., and Glover D.M.. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15:2199–2207. 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Hildebrandt F., Pellman D., Woods G., and Godinho S.A.. 2011. Centrosomes and cilia in human disease. Trends Genet. 27:307–315. 10.1016/j.tig.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachon S., Gopalakrishnan J., Omori Y., Polyanovsky A., Church A., Nicastro D., Malicki J., and Avidor-Reiss T.. 2008. Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics. 180:2081–2094. 10.1534/genetics.108.095141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi S., Giansanti M.G., and Gatti M.. 1998. Spindle self-organization and cytokinesis during male meiosis in asterless mutants of Drosophila melanogaster. J. Cell Biol. 142:751–761. 10.1083/jcb.142.3.751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. 2012. The centrosome in cells and organisms. Science. 335:422–426. 10.1126/science.1209037 [DOI] [PubMed] [Google Scholar]
- Cizmecioglu O., Arnold M., Bahtz R., Settele F., Ehret L., Haselmann-Weiss U., Antony C., and Hoffmann I.. 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 191:731–739. 10.1083/jcb.201007107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit P.T., Brunk K., Dobbelaere J., Dix C.I., Lucas E.P., and Raff J.W.. 2010. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr. Biol. 20:2178–2186. 10.1016/j.cub.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Conduit P.T., Richens J.H., Wainman A., Holder J., Vicente C.C., Pratt M.B., Dix C.I., Novak Z.A., Dobbie I.M., Schermelleh L., and Raff J.W.. 2014. A molecular mechanism of mitotic centrosome assembly in Drosophila. eLife. 3:e03399 10.7554/eLife.03399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N., Rangone H., Fu J., Mao G., Tom B., Riparbelli M.G., Callaini G., and Glover D.M.. 2012. Klp10A, a microtubule-depolymerizing kinesin-13, cooperates with CP110 to control Drosophila centriole length. Curr. Biol. 22:502–509. 10.1016/j.cub.2012.01.046 [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Josué F., Suijkerbuijk S., Baum B., Tapon N., and Raff J.. 2008. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6:e224 10.1371/journal.pbio.0060224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Yu Q.D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., and Glover D.M.. 2010. Asterless is a scaffold for the onset of centriole assembly. Nature. 467:714–718. 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Tzolovsky G., Lipinszki Z., Schneider S., Lattao R., Fu J., Debski J., Dadlez M., and Glover D.M.. 2014. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr. Biol. 24:2526–2532. 10.1016/j.cub.2014.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elric J., and Etienne-Manneville S.. 2014. Centrosome positioning in polarized cells: common themes and variations. Exp. Cell Res. 328:240–248. 10.1016/j.yexcr.2014.09.004 [DOI] [PubMed] [Google Scholar]
- Fabian L., and Brill J.A.. 2012. Drosophila spermiogenesis: big things come from little packages. Spermatogenesis. 2:197–212. 10.4161/spmg.21798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong K.W., Choi Y.K., Rattner J.B., and Qi R.Z.. 2008. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol. Biol. Cell. 19:115–125. 10.1091/mbc.E07-04-0371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz A., Roque H., Saurya S., Dobbelaere J., and Raff J.W.. 2013. CP110 exhibits novel regulatory activities during centriole assembly in Drosophila. J. Cell Biol. 203:785–799. 10.1083/jcb.201305109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., and Glover D.M.. 2012. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2:120104 10.1098/rsob.120104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta B.J., and Rusan N.M.. 2015. A yeast two-hybrid approach for probing protein-protein interactions at the centrosome. Methods Cell Biol. 129:251–277. 10.1016/bs.mcb.2015.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta B.J., Guillen R.X., Fagerstrom C.J., Brownlee C.W., Lerit D.A., Megraw T.L., Rogers G.C., and Rusan N.M.. 2014. Drosophila pericentrin requires interaction with calmodulin for its function at centrosomes and neuronal basal bodies but not at sperm basal bodies. Mol. Biol. Cell. 25:2682–2694. 10.1091/mbc.E13-10-0617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti M.G., Bucciarelli E., Bonaccorsi S., and Gatti M.. 2008. Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr. Biol. 18:303–309. 10.1016/j.cub.2008.01.058 [DOI] [PubMed] [Google Scholar]
- Gillingham A.K., and Munro S.. 2000. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1:524–529. 10.1093/embo-reports/kvd105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P. 2012. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 13:425–435. 10.1038/nrm3373 [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan J., Guichard P., Smith A.H., Schwarz H., Agard D.A., Marco S., and Avidor-Reiss T.. 2010. Self-assembling SAS-6 multimer is a core centriole building block. J. Biol. Chem. 285:8759–8770. 10.1074/jbc.M109.092627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan J., Mennella V., Blachon S., Zhai B., Smith A.H., Megraw T.L., Nicastro D., Gygi S.P., Agard D.A., and Avidor-Reiss T.. 2011. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat. Commun. 2:359 10.1038/ncomms1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S.S., Zhang N., Scholey J.M., Vale R.D., and Stuurman N.. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 316:417–421. 10.1126/science.1141314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardo M., Callaini G., and Riparbelli M.G.. 2015. Structural characterization of procentrioles in Drosophila spermatids. Cytoskeleton, 72:576–584. 10.1002/cm.21260 [DOI] [PubMed] [Google Scholar]
- Guernsey D.L., Jiang H., Hussin J., Arnold M., Bouyakdan K., Perry S., Babineau-Sturk T., Beis J., Dumas N., Evans S.C., et al. 2010. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am. J. Hum. Genet. 87:40–51. 10.1016/j.ajhg.2010.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch E.M., Kulukian A., Holland A.J., Cleveland D.W., and Stearns T.. 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191:721–729. 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmke K.J., Heald R., and Wilbur J.D.. 2013. Interplay between spindle architecture and function. Int. Rev. Cell Mol. Biol. 306:83–125. 10.1016/B978-0-12-407694-5.00003-1 [DOI] [PubMed] [Google Scholar]
- Kalay E., Yigit G., Aslan Y., Brown K.E., Pohl E., Bicknell L.S., Kayserili H., Li Y., Tüysüz B., Nürnberg G., et al. 2011. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat. Genet. 43:23–26. 10.1038/ng.725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khire A., Vizuet A.A., Davila E., and Avidor-Reiss T.. 2015. Asterless Reduction during spermiogenesis is regulated by Plk4 and is essential for zygote development in Drosophila. Curr. Biol. 25:2956–2963. 10.1016/j.cub.2015.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., Park J.E., Shukla A., Choi S., Murugan R.N., Lee J.H., Ahn M., Rhee K., Bang J.K., Kim B.Y., et al. 2013. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proc. Natl. Acad. Sci. USA. 110:E4849–E4857. 10.1073/pnas.1319656110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D., Vakonakis I., Olieric N., Hilbert M., Keller D., Olieric V., Bortfeld M., Erat M.C., Flückiger I., Gönczy P., and Steinmetz M.O.. 2011. Structural basis of the 9-fold symmetry of centrioles. Cell. 144:364–375. 10.1016/j.cell.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba J.E., Galletta B.J., Nye J., Plevock K.M., Buster D.W., Hollingsworth N.A., Slep K.C., Rusan N.M., and Rogers G.C.. 2015. Two Polo-like kinase 4 binding domains in Asterless perform distinct roles in regulating kinase stability. J. Cell Biol. 208:401–414. 10.1083/jcb.201410105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier G., Loncarek J., Meng X., McEwen B.F., Mogensen M.M., Spektor A., Dynlacht B.D., Khodjakov A., and Gönczy P.. 2009. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 19:1012–1018. 10.1016/j.cub.2009.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniewski N., Cuevas R., Duensing A., and Duensing S.. 2010. Daughter centriole elongation is controlled by proteolysis. Mol. Biol. Cell. 21:3942–3951. 10.1091/mbc.E09-12-1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniewski N., Hohenfellner M., and Duensing S.. 2013. The centrosome as potential target for cancer therapy and prevention. Expert Opin. Ther. Targets. 17:43–52. 10.1517/14728222.2013.731396 [DOI] [PubMed] [Google Scholar]
- Lerit D.A., and Rusan N.M.. 2013. PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J. Cell Biol. 202:1013–1022. 10.1083/jcb.201303141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.N., Wu C.T., Lin Y.C., Hsu W.B., Tang C.J., Chang C.W., and Tang T.K.. 2013. CEP120 interacts with CPAP and positively regulates centriole elongation. J. Cell Biol. 202:211–219. 10.1083/jcb.201212060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V., Keszthelyi B., McDonald K.L., Chhun B., Kan F., Rogers G.C., Huang B., and Agard D.A.. 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14:1159–1168. 10.1038/ncb2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milán M., Campuzano S., and García-Bellido A.. 1996. Cell cycling and patterned cell proliferation in the wing primordium of Drosophila. Proc. Natl. Acad. Sci. USA. 93:640–645. 10.1073/pnas.93.2.640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., Čajánek L., and Arquint C.. 2014. The centrosome duplication cycle in health and disease. FEBS Lett. 588:2366–2372. 10.1016/j.febslet.2014.06.030 [DOI] [PubMed] [Google Scholar]
- Noatynska A., Gotta M., and Meraldi P.. 2012. Mitotic spindle (DIS)orientation and DISease: cause or consequence? J. Cell Biol. 199:1025–1035. 10.1083/jcb.201209015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak Z.A., Conduit P.T., Wainman A., and Raff J.W.. 2014. Asterless licenses daughter centrioles to duplicate for the first time in Drosophila embryos. Curr. Biol. 24:1276–1282. 10.1016/j.cub.2014.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Ashikawa T., Nozaki Y., Kozuka-Hata H., Goto H., Inagaki M., Oyama M., and Kitagawa D.. 2014. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 5:5267 10.1038/ncomms6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks A.L., Cook K.R., Belvin M., Dompe N.A., Fawcett R., Huppert K., Tan L.R., Winter C.G., Bogart K.P., Deal J.E., et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36:288–292. 10.1038/ng1312 [DOI] [PubMed] [Google Scholar]
- Peel N., Stevens N.R., Basto R., and Raff J.W.. 2007. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 17:834–843. 10.1016/j.cub.2007.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L., Ozlü N., Hannak E., Cowan C., Habermann B., Ruer M., Müller-Reichert T., and Hyman A.A.. 2004. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 14:863–873. 10.1016/j.cub.2004.04.012 [DOI] [PubMed] [Google Scholar]
- Pelletier L., O’Toole E., Schwager A., Hyman A.A., and Müller-Reichert T.. 2006. Centriole assembly in Caenorhabditis elegans. Nature. 444:619–623. 10.1038/nature05318 [DOI] [PubMed] [Google Scholar]
- Poulton J.S., Cuningham J.C., and Peifer M.. 2014. Acentrosomal Drosophila epithelial cells exhibit abnormal cell division, leading to cell death and compensatory proliferation. Dev. Cell. 30:731–745. 10.1016/j.devcel.2014.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli M.G., and Callaini G.. 2011. Male gametogenesis without centrioles. Dev. Biol. 349:427–439. 10.1016/j.ydbio.2010.10.021 [DOI] [PubMed] [Google Scholar]
- Riparbelli M.G., Callaini G., and Megraw T.L.. 2012. Assembly and persistence of primary cilia in dividing Drosophila spermatocytes. Dev. Cell. 23:425–432. 10.1016/j.devcel.2012.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol P., Collier S., Bush M., Shaw P., and Doonan J.H.. 2007. Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J. Cell Sci. 120:3678–3687. 10.1242/jcs.004119 [DOI] [PubMed] [Google Scholar]
- Sakakibara A., Ando R., Sapir T., and Tanaka T.. 2013. Microtubule dynamics in neuronal morphogenesis. Open Biol. 3:130061 10.1098/rsob.130061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.I., Kleylein-Sohn J., Westendorf J., Le Clech M., Lavoie S.B., Stierhof Y.D., and Nigg E.A.. 2009. Control of centriole length by CPAP and CP110. Curr. Biol. 19:1005–1011. 10.1016/j.cub.2009.05.016 [DOI] [PubMed] [Google Scholar]
- Schuldiner O., Berdnik D., Levy J.M., Wu J.S., Luginbuhl D., Gontang A.C., and Luo L.. 2008. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev. Cell. 14:227–238. 10.1016/j.devcel.2007.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen K.F., Schermelleh L., Leonhardt H., and Nigg E.A.. 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open. 1:965–976. 10.1242/bio.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen K.F., Gabryjonczyk A.M., Anselm E., Stierhof Y.D., and Nigg E.A.. 2013. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J. Cell Sci. 126:3223–3233. 10.1242/jcs.129502 [DOI] [PubMed] [Google Scholar]
- Spektor A., Tsang W.Y., Khoo D., and Dynlacht B.D.. 2007. Cep97 and CP110 suppress a cilia assembly program. Cell. 130:678–690. 10.1016/j.cell.2007.06.027 [DOI] [PubMed] [Google Scholar]
- Stevens N.R., Roque H., and Raff J.W.. 2010. DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Dev. Cell. 19:913–919. 10.1016/j.devcel.2010.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.J., Fu R.H., Wu K.S., Hsu W.B., and Tang T.K.. 2009. CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 11:825–831. 10.1038/ncb1889 [DOI] [PubMed] [Google Scholar]
- Tates A.D. 1971. Cytodifferentiation during spermatogenesis in Drosophila melanogaster: an electron microscope study. PhD thesis. Rijkuniversiteit, Leiden. 162 pp. [Google Scholar]
- Thibault S.T., Singer M.A., Miyazaki W.Y., Milash B., Dompe N.A., Singh C.M., Buchholz R., Demsky M., Fawcett R., Francis-Lang H.L., et al. 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36:283–287. 10.1038/ng1314 [DOI] [PubMed] [Google Scholar]
- van Breugel M., Hirono M., Andreeva A., Yanagisawa H.A., Yamaguchi S., Nakazawa Y., Morgner N., Petrovich M., Ebong I.-O., Robinson C.V., et al. 2011. Structures of SAS-6 suggest its organization in centrioles. Science. 331:1196–1199. 10.1126/science.1199325 [DOI] [PubMed] [Google Scholar]
- Varmark H., Llamazares S., Rebollo E., Lange B., Reina J., Schwarz H., and Gonzalez C.. 2007. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr. Biol. 17:1735–1745. 10.1016/j.cub.2007.09.031 [DOI] [PubMed] [Google Scholar]
- Vitre B.D., and Cleveland D.W.. 2012. Centrosomes, chromosome instability (CIN) and aneuploidy. Curr. Opin. Cell Biol. 24:809–815. 10.1016/j.ceb.2012.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y.M., Mahowald A.P., Perlin J.R., and Fuller M.T.. 2007. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 315:518–521. 10.1126/science.1134910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Gooi L.M., Wason A., Gabriel E., Mehrjardi N.Z., Yang Q., Zhang X., Debec A., Basiri M.L., Avidor-Reiss T., et al. 2014. Conserved TCP domain of Sas-4/CPAP is essential for pericentriolar material tethering during centrosome biogenesis. Proc. Natl. Acad. Sci. USA. 111:E354–E363. 10.1073/pnas.1317535111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., Lawo S., Bird A., Pinchev D., Ralph A., Richter C., Müller-Reichert T., Kittler R., Hyman A.A., and Pelletier L.. 2008. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr. Biol. 18:136–141. 10.1016/j.cub.2007.12.055 [DOI] [PubMed] [Google Scholar]
- Zimmerman W.C., Sillibourne J., Rosa J., and Doxsey S.J.. 2004. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell. 15:3642–3657. 10.1091/mbc.E03-11-0796 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.