The diaphanous formin mDia2, a protein involved in cytoskeletal control, is required for new CENP-A loading at centromeres during the cell cycle to maintain epigenetic markers.
Abstract
Centromeres of higher eukaryotes are epigenetically defined by centromere protein A (CENP-A), a centromere-specific histone H3 variant. The incorporation of new CENP-A into centromeres to maintain the epigenetic marker after genome replication in S phase occurs in G1 phase; however, how new CENP-A is loaded and stabilized remains poorly understood. Here, we identify the formin mDia2 as essential for stable replenishment of new CENP-A at centromeres. Quantitative imaging, pulse-chase analysis, and high-resolution ratiometric live-cell studies demonstrate that mDia2 and its nuclear localization are required to maintain CENP-A levels at centromeres. Depletion of mDia2 results in a prolonged centromere association of holiday junction recognition protein (HJURP), the chaperone required for CENP-A loading. A constitutively active form of mDia2 rescues the defect in new CENP-A loading caused by depletion of male germ cell Rac GTPase-activating protein (MgcRacGAP), a component of the small GTPase pathway essential for CENP-A maintenance. Thus, the formin mDia2 functions downstream of the MgcRacGAP-dependent pathway in regulating assembly of new CENP-A containing nucleosomes at centromeres.
Introduction
The epigenetic landscape of the chromosome is well inherited independent of underlying DNA sequences. In mammals, centromeres, the fundamental unit for chromosome segregation during mitosis, are defined epigenetically by nucleosomes containing the histone H3 variant centromere protein A (CENP-A; Cleveland et al., 2003; Nechemia-Arbely et al., 2012; Fukagawa and Earnshaw, 2014). To maintain centromere identity against CENP-A dilution as DNA replicates and cell divides, newly synthesized CENP-A proteins are deposited at centromeres during early G1 of each cell cycle (Jansen et al., 2007). This process is initiated by Plk1-mediated (McKinley and Cheeseman, 2014) centromeric recruitment of the Mis18 complex at anaphase onset (Hayashi et al., 2004; Fujita et al., 2007; Maddox et al., 2007) and involves the recruitment of holiday junction recognition protein (HJURP), the CENP-A chaperone (Dunleavy et al., 2009; Foltz et al., 2009).
The process to incorporate new CENP-A at centromeres remain poorly understood. A small GTPase molecular switch has been shown to stabilize newly loaded CENP-A. Depletion of Cdc42 or Rac1 in human cells leads to a decrease of CENP-A level at centromeres (Lagana et al., 2010). The downstream effectors of this small GTPase activity remain unidentified. Mammalian diaphanous-related (mDia) formins nucleate and assemble unbranched actin structures downstream of Rho family GTPase signaling (Xu et al., 2004). Recent studies have revealed potential nuclear roles for formins (Baarlink et al., 2013; Belin et al., 2015). Among mDia formin proteins (mDia1-3), only mDia2 shuttles between the cytoplasm and the nucleus (Miki et al., 2009; Baarlink et al., 2013). By affinity purification and mass spectrometry analysis, histones and topoisomerases have been identified as binding partners of mDia2, but neither mDia1 nor mDia3 (Daou et al., 2014).
Using quantitative imaging, we now provide direct evidence that the formin mDia2 is a novel cytoskeleton protein required for maintaining CENP-A levels at centromeres. As a constitutively active form of mDia2 rescues centromeric CENP-A levels caused by depletion of male germ cell Rac GTPase-activating protein (MgcRacGAP), a component of the small GTPase pathway essential for CENP-A maintenance, we additionally uncover mDia2 as the downstream effector of the GTPase activity for epigenetic centromere maintenance.
Results and discussion
Diaphanous formin mDia2 is essential to maintain CENP-A levels at centromeres
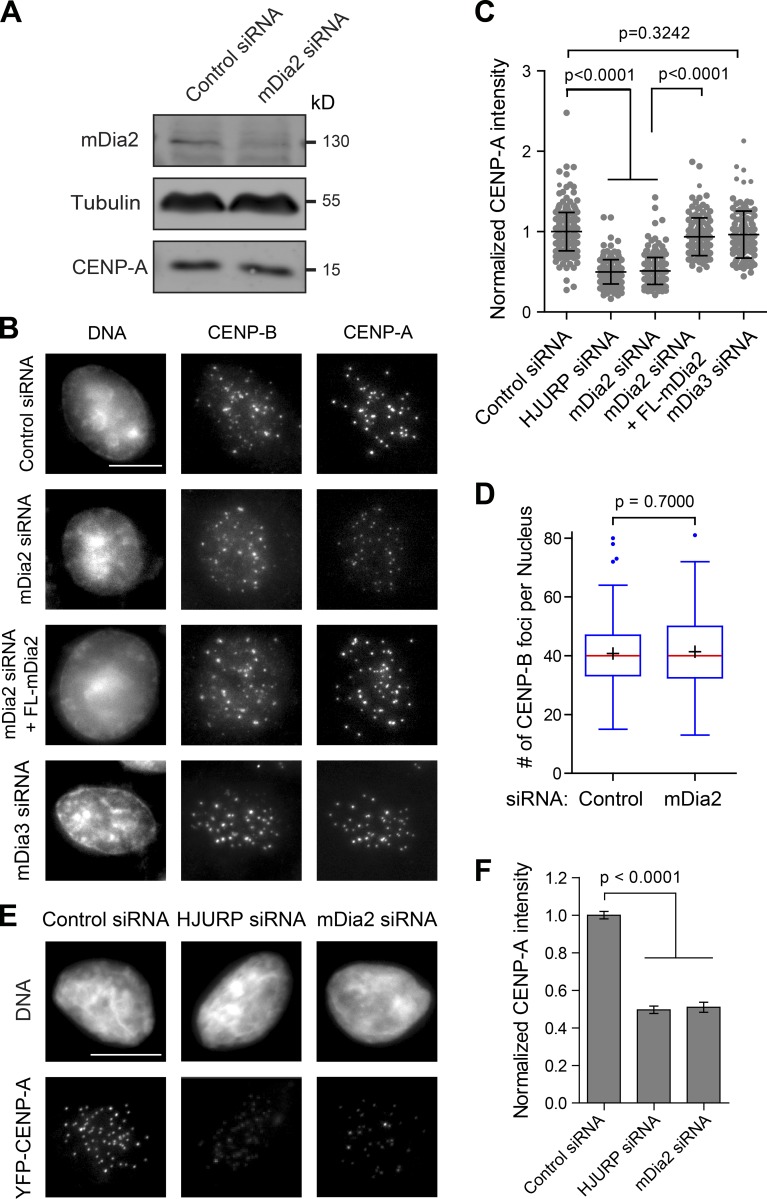
To test if the formin mDia2 is required for CENP-A level maintenance at centromeres, mDia2 protein levels were reduced in human cells (0.47 ± 0.11 relative to control, P < 0.0001) by the transfection of siRNA duplexes targeting mDia2 for 48 h (Fig. 1 A). The mDia2 depletion resulted in a decreased level, but not elimination, of CENP-A at centromeres (Fig. 1 B) without affecting total CENP-A protein levels (Fig. 1 A) compared with control cells (transfected with GAPDH siRNA). Significantly, the loss of CENP-A at centromeres could be rescued by the coexpression of the siRNA-resistant full-length mDia2 (Fig. 1 B), excluding the possibility of an off-target effect from mDia2 siRNA. CENP-A levels at centromeres from large numbers of cells were quantified using an automatic image-analysis algorithm (Fig. S1), designed in this study, without human bias. This confirmed the partial reduction in CENP-A levels at centromeres in mDia2-depeleted cells (Fig. 1 C). The decrease of CENP-A level was not caused by loss of centromere numbers in individual cells, judging by counting the immunostaining of CENP-B (Fig. 1 D), which localizes to centromeres independently of CENP-A (Masumoto et al., 1989). In contrast to mDia2, knockdown of mDia3, a formin protein that has been shown to associate with kinetochores and to be important for kinetochore-microtubule attachment (Yasuda et al., 2004; Cheng et al., 2011), did not result in loss of CENP-A at centromeres (Fig. 1, B and C). These results support a role for the formin mDia2 in CENP-A level maintenance at centromeres.
Figure 1.
The formin mDia2 is required to maintain CENP-A levels at centromeres. (A) Depletion of mDia2 does not affect CENP-A protein level. Immunoblotting analysis of HeLa cell lysates 48 h after transfection with control (GAPDH) and mDia2 siRNAs. (B) Immunofluorescence detection of CENP-A and CENP-B 48 h after transfection with the indicated siRNAs and the full-length mEmerald-tagged mDia2 (FL-mDia2) expression vector (DNA - DAPI). Transfected cells were identified by cotransfected fluorescence markers. Bar, 10 µm. (C) Scatterplot showing the distribution of normalized CENP-A integrated intensity per nucleus (mean ± SD overlaid with scatterplot) with the INCA method (Fig. S1). Control: n = 292; HJURP siRNA: n = 196; mDia2 siRNA: n = 260; mDia2 siRNA + FL-mDia2: n = 136; and mDia3 siRNA: n = 117 from three independent experiments. The p-value was computed using a two-tailed t test. (D) Depletion of mDia2 does not affect centromere numbers. Whisker-Tukey boxplots show the number of CENP-B foci. The boxes span the 25th to 75th percentile of the data, whereas the center bar denotes the median and + marks the mean. Control: n = 168, and mDia2 siRNA: n = 125, from three independent experiments. The p-value was computed using a two-tailed t test. (E) Depletion of mDia2 results in reduced levels of YFP-CENP-A at centromeres. Immunofluorescence images showing YFP-CENP-A in HeLa cells stably expressing YFP-CENP-A 48 h after transfection with indicated siRNAs (DNA - DAPI). Bar, 10 µm. (F) Quantification showing the mean levels of normalized integrated YFP-CENP-A intensity per nucleus (means ± 95% confidence intervals). Control (GAPDH siRNA): n = 456; HJURP siRNA: n = 250; and mDia2 siRNA: n = 158 cells from three independent experiments. The p-value was computed using a two-tailed t test.
To exclude the possibility that the CENP-A loss in mDia2-depleted cells could be caused by cell cycle–dependent transcriptional regulation of CENP-A, YFP-CENP-A levels were measured in fixed cells stably expressing a YFP-tagged CENP-A. Despite that the YFP-CENP-A fusion is controlled by the 5′ long terminal repeat of the virus (Foltz et al., 2009), a similar CENP-A loss at centromeres was observed upon mDia2 depletion (Fig. 1, E and F). Reduced CENP-A levels in mDia2-depleted cells is reminiscent of the depletion of the CENP-A chaperon HJURP (Fig. 1, E and F). These results are consistent with a role for mDia2 in regulating CENP-A levels at centromeres.
The mDia2 protein is specifically required for loading of new CENP-A
To determine if mDia2 is required for cell cycle–dependent incorporation of new CENP-A into centromeres of duplicated sister chromatids after mitotic exit into G1, YFP-CENP-A levels at individual centromeres were followed by high-resolution ratiometric live-cell imaging designed in this study (Fig. S2). In control cells, the increase of centromeric YFP-CENP-A levels began shortly after anaphase onset and continued for several hours (Fig. 2, A and B; and Video 1), which is consistent with previous observations (Jansen et al., 2007; Lagana et al., 2010). In contrast, the increase of YFP-CENP-A levels at centromeres in mDia2 depleted cells could not be maintained despite an initial slight increase within 2 h after anaphase onset (Fig. 2, A and B; and Video 2). The defect of YFP-CENP-A loading in mDia2-depleted cells was also manifested with a significantly shorter apparent half-time (Fig. S2, B and C), consistent with attempted yet failed loading events. These results clearly demonstrate that mDia2 is essential for replenishing CENP-A levels during early G1 phase, when new CENP-A is loaded onto centromeres marked with preexisting CENP-A.
Figure 2.
The mDia2 protein is required for loading new CENP-A during G1. (A) High-resolution ratiometric live-cell imaging showing defective YFP-CENP-A loading upon mDia2 knockdown. Pseudocolored live imaging stills following cells through the 10-h time window after anaphase onset. Identical lookup table (LUT, linear and covering the full range of data) was used over time. Bar, 10 µm (insets are 3× magnified). (B) Quantification of centromeric YFP-CENP-A levels during G1 phase (plotted as mean ± SD). Control: n = 4,100 centromeres from 12 G1 pairs, and mDia2 siRNA: n = 4,310 centromeres from 13 G1-pairs were measured from three independent transfections (see Materials and methods and Fig. S2 for more details). (C) Scheme for the SNAP pulse chase labeling to distinguish existing centromeric CENP-A protein (old) from newly synthesized CENP-A loaded onto centromeres (new). (D) Immunofluorescence analysis showing old CENP-A labeled by TMR-Star and total CENP-A stained with anti-HA antibody. In merge: red, TMR-Star; green, HA-tag. Bars: (main) 10 µm; (insets) 1 µm. (E) Quantification of SNAP-tag-labeled CENP-A (old, red) and total CENP-A (total, green; means ± 95% confidence intervals). The p-value was computed using a two-tailed t test. (F) Quantification showing the normalized ratio between total CENP-A and old CENP-A (mean ± SD overlaid with scatterplot). Control: n = 215, and mDia2 siRNA: n = 217 cells from three independent experiments. The p-value was computed using a two-tailed t test.
Whereas cell cycle distribution was not significantly altered upon depletion of mDia2 (Fig. S2 G), live-cell imaging analysis showed reduced levels of CENP-A recruitment at centromeres upon mDia2 depletion. Cells with fully loaded CENP-A at centromeres inherits 50% “old” CENP-A during S phase and recruit 50% “new” CENP-A at early G1. To test which population of CENP-A was affected by mDia2 depletion, old and new CENP-A levels were measured by the SNAP-tag pulse-chase method (Jansen et al., 2007; Lagana et al., 2010). The old inherited CENP-A was pulse-labeled (15 min) with a fluorescent mark, whereas new CENP-A was chased with a nonfluorescent label (Fig. 2 C). This analysis revealed that mDia2 depletion resulted in 37% reduction of total CENP-A levels, but unchanged levels of old CENP-A compared with control cells (Fig. 2, D–F). Thus, the formin mDia2 functions in the recruitment of new CENP-A onto centromeres in G1 cells.
The formin mDia2 is a downstream effector of the MgcRacGAP-dependent small GTPase pathway required to maintain CENP-A levels at centromeres
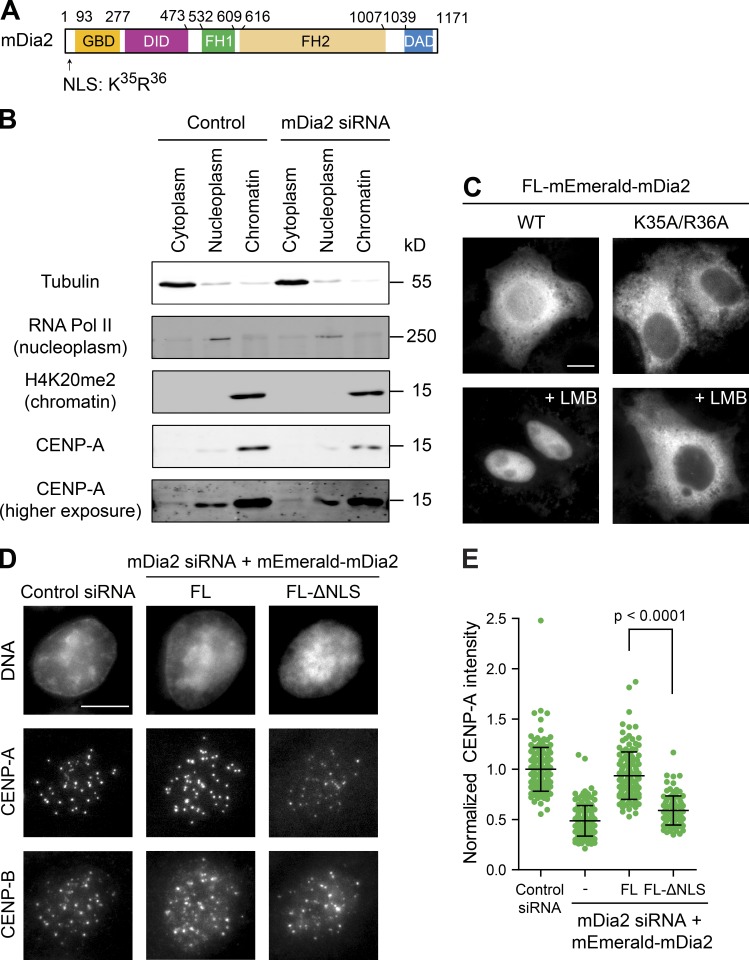
The mDia2 protein contains functional nuclear localization (Fig. 3 A) and nuclear export signals, and shuttles between the nucleus and the cytoplasm through importin-α/β– and CRM1-mediated nuclear transport mechanisms (Miki et al., 2009; Shao et al., 2015). Depletion of mDia2 did not change CENP-A distributions between the cytoplasm and the nucleus, though resulted in less CENP-A bound to chromatins, as expected (Figs. 3 B and S3 A). Full-length mEmerald-mDia2 proteins were accumulated in the nucleus upon treatment with leptomycin B (LMB) to block CRM1. In contrast, the K35A/R36A mutation within the nuclear localization signal (NLS) abolished nuclear accumulation of mEmerald-mDia2 upon LMB treatment (Fig. 3 C). Replacing endogenous mDia2 with this mutant resulted in significantly reduced levels of CENP-A at centromeres (Fig. 3, D and E), demonstrating that the nuclear function of mDia2 is required for its role in CENP-A maintenance.
Figure 3.
Nuclear mDia2 is required for CENP-A levels at centromeres. (A) The mDia2 protein structure showing the relative positions of GBD, DID, FH1, FH2, and DAD domains as well as the position of the NLS. (B) Depletion of mDia2 does not affect CENP-A distribution in the cytoplasm and the nucleus. Cellular fractionation and immunoblotting analysis showing the distribution of CENP-A proteins in the cytoplasm, nucleoplasm, and insoluble materials associated with the chromatin marked by tubulin, RNA polymerase II, and H4K20me2, respectively. A higher exposure of CENP-A blot was shown for its cytoplasmic distribution. (C) Cells expressing wild-type mDia2 or the mDia2K35A/R36A mutant (with a defective NLS) fused with mEmerald were treated with or without LMB (20 nM for 60 min) before fixation and imaging mEmerald. Bar, 10 µm. (D) Immunofluorescence detection of CENP-A and CENP-B 48 h after transfection with the indicated siRNAs and the full-length mDia2 (FL mDia2) or mDia2 ΔNLS mutant expression vectors (DNA - DAPI). Bar, 10 µm. (E) Quantification showing the normalized CENP-A integrated intensity per nucleus (mean ± SD overlaid with scatterplot). The p-value was computed using a two-tailed t test. Control: n = 174, mDia2 siRNA: n = 149, mDia2 siRNA + FL-mDia2: n = 136, and mDia2 siRNA + FL-ΔNLS: n = 103.
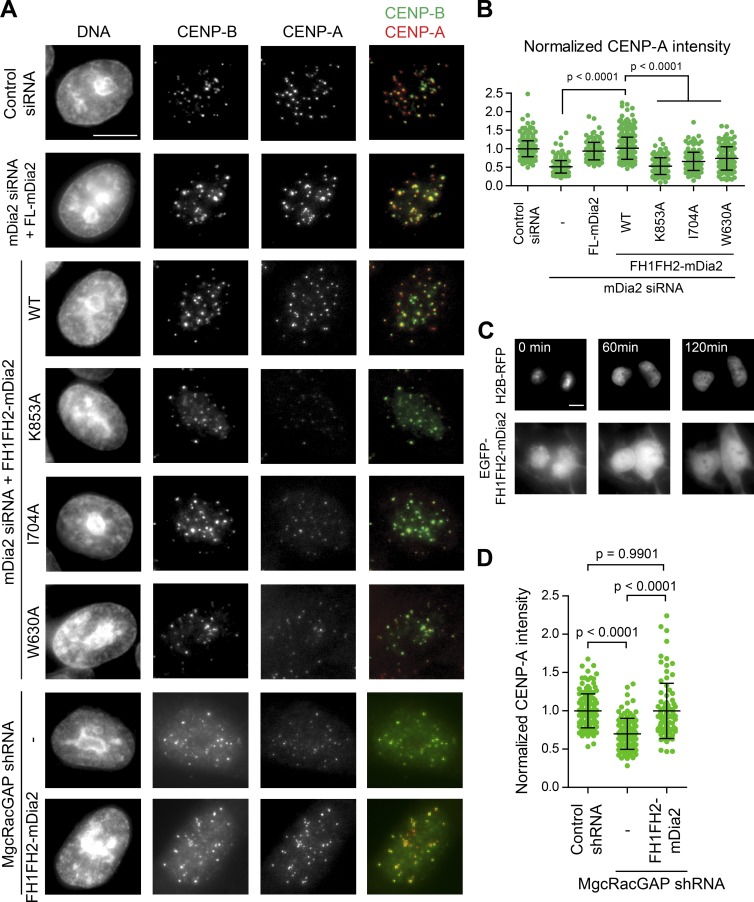
A small GTPase switch including MgcRacGAP, a Rho family GTPase activating protein, is involved in CENP-A maintenance at centromeres (Lagana et al., 2010). The mDia formin proteins are autoinhibited through an intramolecular interaction between N-terminal GBD-DID domains and C-terminal DAD domain (Alberts et al., 1998). The autoinhibition is relieved upon the binding of the small GTPase at the GBD domain (Watanabe et al., 1999). The mDia2 construct lacking the regulatory domains (FH1FH2-mDia2), but not FH1FH2-mDia2 constructs with actin assembly defective point mutations (K853A, I704A, and W630A; Bartolini et al., 2008), was able to restore CENP-A levels at centromeres as well as the full-length mDia2 in cells depleted of endogenous mDia2 (Fig. 4, A and B). Although lacking the known NLS, the EGFP-fused FH1FH2-mDia2 localized to the nucleus in G1 cells (Fig. 4 C). Further, the constitutively active form of mDia2 was very efficient in restoring CENP-A levels at centromeres in MgcRacGAP-depleted cells (Fig. 4 D). This is consistent with the formin mDia2 as a downstream effector of the MgcRacGAP-dependent GTPase pathway to play a role in centromeric CENP-A maintenance.
Figure 4.
The formin mDia2 is a downstream effector of the MgcRacGAP-dependent GTPase pathway to regulate epigenetic centromere maintenance. (A) HeLa cells 48 h after transfection of indicated siRNAs along with expression vectors were fixed and stained with DAPI (DNA), CENP-B, and CENP-A. Transfected cells were identified by fluorescence markers. Bar, 10 µm. (B and D) Quantifications of normalized CENP-A integrated intensity per nucleus plotted as means ± SD overlaid with scatterplot. The p-value was computed using a two-tailed t test. Control siRNA: n = 425, mDia2 siRNA: n = 260, mDia2 siRNA + FL-mDia2: n = 136, mDia2 siRNA + WT-FH1FH2-mDia2: n = 383, mDia2 siRNA + K853A-FH1FH2-mDia2: n = 151, mDia2 siRNA + I704A-FH1FH2-mDia2: n = 158, and mDia2 siRNA + W630A-FH1FH2-mDia2: n = 128. Control shRNA: n = 135, MgcRacGAP shRNA: n = 117, and MgcRacGAP shRNA + WT-FH1FH2-mDia2: n = 88 from at least three independent experiments. (C) Live-cell imaging stills showing EGFP-FH1FH2-mDia2 nuclear localization during G1 phase upon anaphase onset (0 min). Bar, 10 µm.
Depletion of mDia2 results in a prolonged centromere association of HJURP
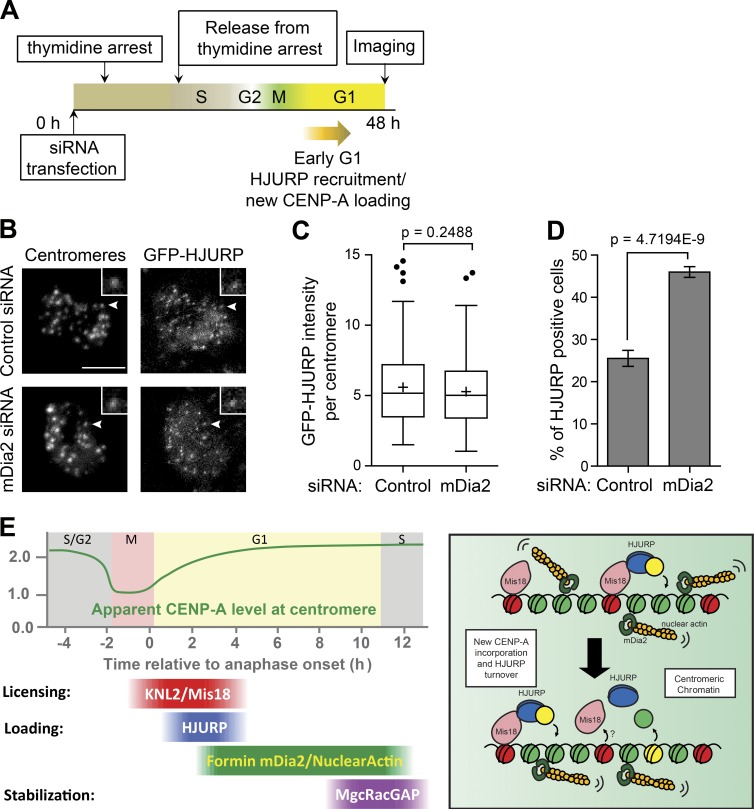
CENP-A loading at G1 centromeres requires the CENP-A histone chaperone HJURP, which also has the CENP-A nucleosome assembly activity (Barnhart et al., 2011). A subset of early G1 cells has HJURP colocalized to centromeres (Foltz et al., 2009). To test whether depletion of mDia2 affects the dynamics of HJURP at centromeres, we examined the HJURP centromere localization in early G1 cells expressing the EGFP-HJURP fusion upon releasing from thymidine arrest (Fig. 5 A). This revealed that mDia2 depletion did not affect the intensity of HJURP at centromeres (Fig. 5, B and C), but the percentage of HJURP positive cells was almost doubled compared with control cells (Fig. 5 D). These results indicate a prolonged attempting of new CENP-A nucleosome assembly by HJURP in the absence of mDia2. Stochastic simulation demonstrated that prolonged HJURP dwelling (i.e., reduced HJRUP turnover) not only suffices to cause a higher percentage of HJURP positive cells as observed experimentally, but also contributes to the inability of CENP-A accumulation on centromeres, which is in good quantitative agreement with ratiometric live cell measurement over time (Fig. S3, B–E). These results are consistent with a role of mDia2 in regulating CENP-A levels at centromeres (Fig. 5 E).
Figure 5.
Depletion of mDia2 produces a prolonged HJURP localization at centromeres. (A) Scheme for the siRNA transfection and the thymidine arrest to examine centromere localization of HJURP in early G1 cells. (B) Immunofluorescence images of GFP-HJURP. Centromeres were identified using ACA antibodies. The arrowheads point to centromeres shown in insets. Bars, 5 µm (insets are 2× magnified). (C) Whisker-Tukey boxplots show the relative intensity of GFP-HJURP foci at centromeres. The boxes span the 25th to 75th percentile of the samples, whereas the center bar denotes the median and the + marks the mean. Control: n = 174 centromeres from 26 cells, and mDia2 siRNA: n = 183 centromeres from 20 cells. The p-value was computed using a two-tailed t test. (D) Mean percentage of GFP-HJURP centromere-positive cells with error bars showing standard deviations from three experiments (control: n = 594 cells, and mDia2 siRNA: n = 277 cells). The p-value was computed using a two-tailed z-test. (E) Model of mDia2 regulating CENP-A loading. (left) Timeline of the epigenetic inheritance of CENP-A over cell cycle. (right) Schematic model showing mDia2 to be important for HJURP-mediated CENP-A chromatin assembly and timely HJURP turnover. Green, histone H3; red, old CENP-A; yellow, new CENP-A.
A novel nuclear function of formin mDia2 in centromere epigenetic regulation
Centromeres are epigenetically marked by the conserved histone H3 variant CENP-A. Each sister centromere inherits one half the number of CENP-A molecules upon DNA duplication. To maintain centromere identity, new CENP-A is added to double the number of CENP-A molecules at centromeres during G1. Loading of new CENP-A into centromeres includes the following steps (Fig. 5 E, left): (1) the assembly of licensing factors, the Mis18 complex, at anaphase onset (Hayashi et al., 2004; Fujita et al., 2007; Maddox et al., 2007), (2) new CENP-A deposition and nucleosome assembly by the CENP-A chaperone HJURP at early G1 (Barnhart et al., 2011), and (3) a maintenance step involving a small GTPase molecular switch (Lagana et al., 2010). Here, our results reveal a critical role for the formin mDia2 in regulating epigenetic maintenance of centromere identity. Quantitative imaging and high resolution ratiometric live cell studies have demonstrated that knockdown of mDia2 results in reduced levels of CENP-A at centromeres. In contrast, depletion of mDia2 does not affect the recruitment of H2A.Z, another histone variant in human cells (Fig. S3, F and G).
The formin mDia2 is not likely to play a role in centromere licensing or the recruitment of centromere components, as we have not been able to detect mDia2 centromere localization at any point in the cell cycle. This is further supported by the normal level of HJURP associated with centromeres in mDia2-depleted cells. In contrast, the increased percentage of HJURP positive cells upon mDia2 depletion indicates a role of mDia2 in regulating nucleosome assembly for new CENP-A incorporation (Fig. 5 E, right). Expressing a constitutively active form of mDia2 is able to rescue the CENP-A deposition defect caused by knockdown of MgcRacGAP, a phenotype that is consistent with the formin mDia2 being the downstream effector of the MgcRacGAP-dependent GTPase pathway during the maintenance step to stabilize newly incorporated CENP-A. The formin mDia2 could be involved in this process by, at least, two different mechanisms: (1) assisting chromatin-remodeling for new CENP-A incorporation, as the FH2 region of mDia formins has been shown to interact with CENP-A using a yeast two-hybrid assay (Yasuda et al., 2004); and (2) altering mobility or organization of chromatin. The latter possibility will require mDia2-mediated nuclear actin activity. Recent studies have demonstrated nuclear actin network assembly mediated by formin proteins in regulating the MAL/SRF (megakaryocytic acute leukemia/serum response factor) transcription function (Baarlink et al., 2013) and in DNA damage response (Belin et al., 2015). Although it has been shown that latrunculin A or cytochalasin D treatment does not affect CENP-A levels at centromeres (Lagana et al., 2010), actin in nucleus could form short oligomers or other forms of structures that are less sensitive to drug treatment than actin polymers (Gonsior et al., 1999; Schoenenberger et al., 2005; McDonald et al., 2006; Belin et al., 2013). It will be important to understand whether actin dynamics is important for epigenetic centromere maintenance in future studies.
Materials and methods
siRNA sequences, constructs, and antibodies
The siRNAs used in this study include GAPDH (NM_002046.4, 5′-UGGUUUACAUGUUCCAAUA-3′), DIAPH3/mDia2 (AB244756.1, 5′-CUCCGGCACAAUUCAGUUCAA-3′), DIAPH2/mDia3 (BC117414, 5′-CACCGTCTCAATGACATTCGA-3′), and HJURP (NM_018410.4, 5′-CUACUGGGCUCAACUGCAAU-3′).
The constructs used in this study include H2B-YFP, H2B-RFP (Foltz et al., 2009), FL-mEmerald-mDia2, FL-ΔNLS-mEmerald-mDia2, pEGFP-FH1FH2mDia2-WT, pEGFP-FH1FH2mDia2-K853A, pEGFP-FH1FH2mDia2-I704A, pEGFP-FH1FH2mDia2-W630A, pmCherry-C1 (catalog number 632524; Takara Bio Inc.), and MgcRacGAP MISSION shRNA plasmid (NM_013277.3-2165s21c1 and NM_013277.2-456s1c1; Sigma-Aldrich). The pEGFP-FH1FH2mDia2–based constructs were gifts from F. Bartolini (Columbia University Medical Center, New York, NY). The construct of mEmerald-mDia2-C-14 was a gift from M. Davidson (National High Magnetic Field Laboratory, Tallahassee, FL; plasmid 54158; Addgene). Site-directed mutagenesis was performed to generate the K35AR36A mutant, using QuikChange Lightning following the manufacturer’s instructions.
Primary antibodies used in this study include rabbit anti-mDia2 (Watanabe et al., 2008), rabbit anti-mDia3 (LS-C19007; Lifespan), mouse anti–CENP-A (Ab13939; Abcam), mouse antitubulin (T6199; Sigma-Aldrich), rabbit anti-HJURP (Ab100800; Abcam; Foltz et al., 2009), chicken anti-GFP (Ab16901; EMD Millipore), rabbit anti–CENP-B (Ab25734; Abcam), mouse anti-HA (MMS-101P; Covance), rabbit anti-MgcRacGAP (Ab61192; Abcam), and rabbit anti-H2A.Z (Ab4174; Abcam).
Cell culture, transfection, and drug treatment
HeLa cells were used for most of the quantitative imaging experiments in this study. A HeLa cell line stably expressing YFP-CENP-A was used for high-resolution ratiometric live-cell imaging. A HeLa cell line stably expressing GFP-HJURP (gift from D. Foltz, Northwestern University, Chicago, IL) was used for fixed cell imaging in Fig. 5. Cells were maintained in DME medium supplemented with 10% FBS (complete growth medium) at 37°C in 5% CO2. The transfection of siRNAs was performed with Hiperfect (QIAGEN) following the manufacturer’s protocol. For cotransfection experiments, siRNAs were transfected with at least 20-fold molar excess to marker plasmids encoding fluorescent proteins. Control cells were transfected with GAPDH siRNA. All knockdowns were confirmed by immunoblotting analysis. Cells were fixed for indirect immunofluorescence analysis or live cell imaging 48 h after transfection. The thymidine synchronization (also detailed in the section describing SNAP-tag pulse chase assay) was performed with 2 mM thymidine in complete growth medium for at least 17 h, washed twice in prewarmed PBS, and released into complete medium supplemented with 24 µM deoxycytidine (for 9 h if followed by another round of thymidine arrest). Nocodazole was used at 100 ng/ml. LMB (provided in methanol/water = 7:3) was used at 20 nM (final concentration) for 1 h. All drugs were purchased form Sigma-Aldrich.
Quantitative fixed-cell imaging and data analysis using the INCA method
For indirect immunofluorescence and fixed-cell imaging, cells grown on poly-d-lysine–coated coverslips were washed in PBS (prewarmed at 37°C), fixed in cold MeOH at −20°C for 5 min, and then permeablized with 0.1% Triton X-100 in PBS for 1 min. After being rehydrated in PBS briefly, fixed cells were blocked in 5% BSA in PBS at 4°C overnight. Coverslips were subjected to primary antibodies diluted in PBS and Alexa Fluor 488–, DyLight 594–, or CyTM5-conjugated secondary antibodies, both at room temperature for 1 h. DAPI (16.67 ng/ml) was used to stain DNA. Coverslips were mounted using antifade reagent (ProLong Gold; Molecular Probes). Images were acquired at room temperature using an inverted microscope (IX81; Olympus) with a 60× NA 1.42 Plan Apochromat oil immersion objective (Olympus), a monochrome charge-coupled device camera (Sensicam QE; Cooke Corporation) at 1 × 1 binning, which are all controlled by the SlideBook software (3i and Olympus). 10 optical sections 0.5 µm apart spanning 5 µm were acquired for each field. All images in each experiment along with appropriate controls were collected on the same day with identical exposure time. Representative images presented in figures are scaled identically across groups. Cells stably expressing YFP-CENP-A were fixed following a slightly different protocol to preserve YFP’s fluorescence: cells were washed in PBS (prewarmed at 37°C), fixed in freshly prepared EM-grade PFA (4% diluted in PBS; 16% stock; catalog number 15710-S; Electron Microscopy Sciences) at room temperature for 10 min, washed in PBS twice (5 min each), and then permeablized with 0.1% Triton X-100 in PBS for 1 min. The subsequent block and staining steps are the same as the general protocol.
Inspired by a previous study (Lagana et al., 2010), we developed an image analysis technique by measuring the integrated CENP-A intensity per nucleus: the integrated nuclear CENP-A (INCA) measurement, a custom-written software using MATLAB (R2013a; MathWorks). All 16-bit uncompressed raw images processed only by maximum Z-projection were fed into MATLAB for an automatic and unbiased measurement. In brief, raw images were subjected to band-pass filtering (Fig. S1 A), which eliminates noise by applying a narrow kernel and removes background with a wide kernel (kernel sizes were fixed throughout the study). Resultant DAPI images were subjected to Otsu thresholding (Otsu, 1979) and watershed algorithm (Lindeberg, 1993) to generate binary nuclear masks, which were subsequently applied to the cognate CENP-A images. The integrated fluorescence intensities on CENP-A images within the nuclear masks were eventually measured to reflect the “loading capacity” of all centromeric CENP-A in that particular nucleus or cell. To confirm quantification obtained using the INCA method, a more labor-intensive traditional method measuring the integrated intensity of single centromeres (with regions of interest slightly larger than each single centromere) was performed using ImageJ (National Institutes of Health) and yielded the same results as the INCA method (Fig. S1 B). The INCA method returned the integrated intensity value for every single nucleus. To compare the intensity values of nuclei between experiments, the measurements of each experiment were normalized against that of the control group in that dataset, therefore allowing cross-dataset comparison (Lagana et al., 2010). Imaging experiment with or without cotransfection markers were both performed and analyzed using the INCA method with the same conclusions.
High-resolution ratiometric live-cell imaging
HeLa cells stably expressing YFP-CENP-A (Foltz et al., 2009) were plated onto poly-d-lysine–coated 35-mm glass-bottom dishes (MatTek Corporation) and maintained in CO2 independent medium supplemented with 4 mM l-glutamine and 10% FBS, with an environmentally controlled chamber at 37°C during imaging. Images were acquired every 20 min for a total duration of ∼13 h, with 11 z-sections spanning 10 µm (1 µm apart each optical section) being acquired at each time point. Exposure time was kept constant throughout the duration of live imaging (200 ms for YFP after being optimized against photobleaching). All live imaging was performed using a motorized inverted microscope (IX81; Olympus) with a 60× NA 1.42 Plan Apochromat oil-immersion objective (Olympus) and a interline transfer cooled charge-coupled device camera (ORCA-R2 C10600-10B; Hamamatsu Photonics) at 1 × 1 binning, which were controlled by the MetaMorph software (Molecular Devices).
For live imaging data, because of a much lower signal-to-noise ratio than fixed samples, a different quantification method was developed based on a previous study (Lagana et al., 2010). Essentially, intensity measurement was performed for individual cells upon mitotic exit and during G1. Only cells staying in focus were subjected to measurement (mCherry-C1 was used as a cotransfection marker for identifying siRNA transfected cells). Measurements were performed on maximum z-projections with all 16-bit depth preserved. Ratiometric analysis was performed by normalizing the time course of each cell’s centromeric CENP-A intensities with a “reference frame,” which represents the averaged centromeric CENP-A level for that particular cell before CENP-A loading occurs. Because of resolution constraints (i.e., more than one centromeres are inevitably overlapped during anaphase/telophase), to ensure it is the single centromere’s intensity that was measured, the reference frame (against which the “loading curve” was normalized) was selected to be late prometaphase or early metaphase, where a single sister centromere can be identified and measured to obtain an internal reference intensity before any new CENP-A loading has occurred. During the course of telophase/G1 phase, only well-separated single centromeres, whose intensity distributions are circular-symmetric on z-projection images, were randomly selected and measured using ImageJ (National Institutes of Health). Because pixel intensities on each centromere approximately follow 2D-Gaussian distribution, local maxima were considered as a reasonable measurement of CENP-A levels per centromere. The measurements were followed by normalization: each data point throughout the time course was normalized against the host cell’s mean measurement in the “reference frame.” After normalization, data were plotted as mean ± SD. For visual comparison, representative maximum z-projection images were demonstrated, with linear intensity transformation function being applied (Inoué, 1986) and identical dynamic range/LUT being used for all images of each cell throughout the time course.
Regarding photobleaching, our protocol for long term live cell imaging has been optimized such that only minimal photobleach occurred by 10 h after imaging started (Fig. S2, D–F; and Video 3).
SNAP-tag pulse chase assay
The SNAP-tag pulse chase assay was performed based on published protocols (Jansen et al., 2007; Lagana et al., 2010). Essentially, HeLa cells stably expressing CENP-A-SNAP-3×HA were synchronized using double thymidine block (2 mM thymidine, 17 h each) with 9-h release in between (in medium supplemented with 24 µM deoxycytidine). During the second round of thymidine arrest, siRNAs were transfected to allow for ∼48-h knockdown time before fixation whereas only one round of CENP-A deposition during the immediate next G1 was affected. Upon releasing from the second round of thymidine block, cells were pulse-labeled with TMR-Star, a fluorescent SNAP substrate (3 µM), for 15 min, followed with complete washes and block. Cells were fixed and stained for total CENP-A (anti-HA tag). Quantitative imaging and image processing were performed using the INCA method.
Cell fractionation
Cell fractionation analysis was performed using an adapted protocol based on established methods (Szentirmay and Sawadogo, 2000; Baarlink et al., 2013; Bodor et al., 2014). In brief, cells (5 × 106) were harvested and washed with cold PBS. Cell pellet was resuspended in 500 µl 1× hypotonic buffer (20 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1 mM PMSF, and 1× protease inhibitor cocktail from Roche) by pipetting several times, followed by 15-min incubation on ice to allow for swelling. Then, 25 µl of 40% NP-40 was added into the system before vortexing for 10 s. The homogenate was then centrifuged (3,000 rpm for 10 min at 4°C) to separate the cytoplasmic fraction (supernatant) from the nuclear pellet. Nuclear pellet was rewashed with hypotonic buffer (without NP-40) and centrifuged and was then resuspended in 50 µl of relatively low-salt extraction buffer (10 mM Tris, pH 7.4, 2 mM Na3VO4, 100 mM NaCl, 1% Triton X-100, 1 mM EDTA, 10% glycerol, 1 mM EGTA, 0.1% SDS, 1 mM NaF, 0.5% deoxycholate, 20 mM Na4P2O7, 1 mM PMSF, and 1× protease inhibitor cocktail) and incubated on ice for 30 min, with vortexing every 10 min. The mixture was then centrifuged for 30 min at 14,000 g at 4°C. Supernatant is nucleoplasm and pellet is chromatin-associated materials. For immunoblotting, tubulin was used as the cytoplasmic marker (Baarlink et al., 2013), RNA polymerase II as the nucleoplasmic marker (Szentirmay and Sawadogo, 2000), and histone H4K20me2 as the chromatin-associated marker (Bodor et al., 2014).
Immunoblotting analysis
Immunoblotting was performed as previously described (Liu et al., 2015). In brief, cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.1% SDS, and 0.5% Na-deoxycholate acid) and then denatured using SDS sample buffer. Cell lysates were subsequently subjected to 10% SDS-PAGE followed by membrane transfer (Immobilon-P and Towbin transfer buffer, pH 8.3; EMD Millipore). Immunoblots on the membrane were blocked with 5% nonfat milk dissolved in Tris-buffered saline with Tween (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.05% Tween) and then probed with primary antibodies diluted in Tris-buffered saline with Tween. Primary antibodies were visualized using Alexa Fluor 680–conjugated secondary antibodies (Thermo Fisher Scientific) together with the LI-COR imaging system (LI-COR Biosciences).
Cell cycle analysis
Control HeLa cells or mDia2 knockdown cells (confluent) were trypsinized from six-well plates, fixed with MeOH (−20°C), and stained with DAPI. The BD LSRII Cell Analyzer was used for FACS experiments. The FlowJo was used for FACS data analysis with Gaussian fitting based on univariate cell cycle model (Watson et al., 1987).
Statistical analysis and plotting
All statistical analyses were performed with GraphPad Prism 5 (GraphPad Software) using an unpaired, two-tailed t test between groups unless noted otherwise (e.g., z-test with MATLAB to compare two sample proportions). All plots were prepared in MATLAB (R2013a; MathWorks), Prism (GraphPad Software), Origin 8.6 (OriginLab), or Excel (Microsoft). Control groups and mDia2-depleted groups in all experiments were pooled together after normalization and presented.
Stochastic simulation of HJURP turnover at centromeres
To test if extended dwelling time of HJURP molecules on centromere can contribute to the change in the observed percentage of HJURP-positive cells, we applied the Gillespie next reaction algorithm to numerically simulate the stochastic association/dissociation events of HJURP molecules on the centromere (Gillespie, 1977). Parameters were chosen based on reported numbers (kon, koff, etc.; Table S1) or realistic assumptions when no parameters are available (numbers of docking site per centromere). In brief, a realistic number of docking sites (30, which is smaller than the total number of CENP-A nucleosomes per centromere) were assigned for each single centromere and there is no HJURP associated on any docking site at the beginning. Each round of simulation starts with generating a series of random numbers (random probability [Prand]) for each time step (ti) and compare the Prand(ti) with the actual probability of either association (Pon) event or dissociation event (Poff) given the current docking site status is either unoccupied or occupied, respectively, at time ti. Pon and Poff are calculated using equations Pon = 1 − e−(kon*)*ti and Poff = 1 − e−(koff)*ti, where koff (dissociation rate constant, min−1) and kon* (pseudoassociation rate constant, min−1) are derived from parameters listed in Table S1.
If Pon(ti) > Prand(ti), it suggests that compared with random probability, the association event is more likely to happen given an unoccupied docking site. One molecule of HJURP will jump on the docking site. If Poff(ti) > Prand(ti), it suggests that compared with random probability, the dissociation event is more likely to happen given an occupied docking site. One molecule of HJURP will jump off and leave the docking site available for the next round of possible association event.
In the case of incomplete or failed incorporation, the increased level of CENP-A as a result of HJURP association will be removed after HJURP dissociation, giving rise to unsuccessful association and the inability of new CENP-A nucleosomes to build up. Meanwhile, koff is lowered by half compared with ordinary conditions to manifest on altered HJURP dwelling time and thus turnover rate (Fig. S3 A).
Extended dwelling time shouldn’t affect the number of dwelling events per docking site per centromere. To estimate the total numbers of time steps in case of lowered koff and failed incorporation to get similar numbers of dwelling events, we plotted the number of dwelling event per docking site with empirical increment of time steps. It turns out that 3,500 time steps under this circumstance is the minimum requirement to achieve similar numbers of total dwelling event per docking site (P = 0.0923). Total dwelling time per docking site therefore has the mean value of 333.384 min as compared with 185.06 min under ordinary conditions. Δt is therefore 2.4721 h longer under the condition of “failed incorporation + lower koff.”
Next, to assess the influence of temporal changes on the percentage of observable HJURP-positive cells, we initiated another matrix to simulate thousands of cells’ collective behavior. It is assumed that a cell shares the same temporal property of the docking site regarding HJURP’s presence on its centromeres. Instructed by experimental observations and practical experiences (Table S1), here we assumed 50% cells are synchronized around the G1/S boundary upon being released from single round of thymidine arrest. The exact position of each cell’s time line at the start of simulation (upon thymidine release) is stochastically distributed within a normal time window (µ = 0 and SD = 2). As a reference, a cell starts right at G1/S boundary (0 h) will proceed for 9 h to start having HJURP on its centromeres. After 3 h for this cell to be an HJURP-positive cell, HJURP will stop being associated with centromeres, therefore making 12 h the last time point for it to be an HJURP-positive cell (Fig. S3 C). “Green point” and “red point” will be used hereafter to name the start and end of HJURP association, respectively. Despite the intrinsic uncertainties associated with cell synchronization, all cells are fixed 11 h after thymidine release for imaging (invariant observational point). Consequently, all cells with their red point before the observational point, and all cells with their green point after the observational point, will not be documented as HJURP-positive cells. The percentage of HJURP-positive cells can then be calculated based on these criteria (Fig. S3 D).
To test if the number of HJURP molecules per centromere is altered at any given time during the period of being an HJURP-positive cell, 10 random position inside the HJURP-positive time window were chosen (per simulation) to count how many docking site are occupied at that particular time point. The number of occupied docking sites on that centromere reflects the number of HJURP molecules per centromere at that time. This process is repeated three times for plotting the simulated HJURP level per centromere (Fig. S3 D).
Finally, to visualize the time-dependent “loading” of CENP-A nucleosomes on the centromere, results from stochastic simulations were summed up over time to create loading curves of the accumulated number of CENP-A nucleosomes per centromere. Ordinary conditions and conditions with “failed incorporation + lower koff” were processed, respectively, and plotted to compare with experimental measurements (Fig. S3 E).
Online supplemental material
Fig. S1 shows the INCA measurement method developed in this study. Fig. S2 shows the method used for quantifying high-resolution ratiometric live cell imaging data and details of nonlinear regression. Fig. S3 shows the stochastic simulation of HJURP turnover at G1 centromeres. Video 1 shows live-cell imaging movies of a control cell and an mDia2 knockdown cell stably expressing YFP-CENP-A going through G1 phase. Video 2 shows the ratiometric live-cell imaging of YFP-CENP-A signals in a control cell and an mDia2 knockdown cell. Video 3 shows a representative cell expressing YFP-CENP-A arrested in G1/S phase (in the presence of thymidine) imaged every 20 min for 10 h for a photobleaching test. Table S1 shows parameters used for numerical simulation of HJURP turnover. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201512034/DC1.
Supplementary Material
Acknowledgments
We thank all members of the Mao laboratory for stimulating discussion. We thank G. Gundersen and F. Bartolini for insight on the project. We thank R. Vallee for inputs on the project and equipment for live cell imaging. We thank D. Foltz, M. Davidson, and D. Mullins for reagents. We are grateful to the training C. Liu received from the Physiology Course (2013) in the Marine Biological Laboratory (Woods Hole, MA), in particular computational programming from D. Odde and M. Gardner.
This work was supported by a grant from the National Institutes of Health (GM89768) to Y. Mao.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CENP-A
- centromere protein A
- FH
- formin-homology domain
- HJURP
- holiday junction recognition protein
- INCA
- integrated nuclear CENP-A
- LMB
- leptomycin B
- mDia
- mammalian diaphanous related
- MgcRacGAP
- male germ cell Rac GTPase-activating protein
References
- Alberts A.S., Bouquin N., Johnston L.H., and Treisman R.. 1998. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J. Biol. Chem. 273:8616–8622. 10.1074/jbc.273.15.8616 [DOI] [PubMed] [Google Scholar]
- Baarlink C., Wang H., and Grosse R.. 2013. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science. 340:864–867. 10.1126/science.1235038 [DOI] [PubMed] [Google Scholar]
- Barnhart M.C., Kuich P.H., Stellfox M.E., Ward J.A., Bassett E.A., Black B.E., and Foltz D.R.. 2011. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 194:229–243. 10.1083/jcb.201012017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F., Moseley J.B., Schmoranzer J., Cassimeris L., Goode B.L., and Gundersen G.G.. 2008. The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J. Cell Biol. 181:523–536. 10.1083/jcb.200709029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin B.J., Cimini B.A., Blackburn E.H., and Mullins R.D.. 2013. Visualization of actin filaments and monomers in somatic cell nuclei. Mol. Biol. Cell. 24:982–994. 10.1091/mbc.E12-09-0685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin B.J., Lee T., and Mullins R.D.. 2015. DNA damage induces nuclear actin filament assembly by Formin-2 and Spire-(1/2) that promotes efficient DNA repair. eLife. 4:e07735 10.7554/eLife.07735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor D.L., Mata J.F., Sergeev M., David A.F., Salimian K.J., Panchenko T., Cleveland D.W., Black B.E., Shah J.V., and Jansen L.E.. 2014. The quantitative architecture of centromeric chromatin. eLife. 3:e02137 10.7554/eLife.02137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Zhang J., Ahmad S., Rozier L., Yu H., Deng H., and Mao Y.. 2011. Aurora B regulates formin mDia3 in achieving metaphase chromosome alignment. Dev. Cell. 20:342–352. 10.1016/j.devcel.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D.W., Mao Y., and Sullivan K.F.. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 112:407–421. 10.1016/S0092-8674(03)00115-6 [DOI] [PubMed] [Google Scholar]
- Daou P., Hasan S., Breitsprecher D., Baudelet E., Camoin L., Audebert S., Goode B.L., and Badache A.. 2014. Essential and nonredundant roles for Diaphanous formins in cortical microtubule capture and directed cell migration. Mol. Biol. Cell. 25:658–668. 10.1091/mbc.E13-08-0482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., and Almouzni-Pettinotti G.. 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 137:485–497. 10.1016/j.cell.2009.02.040 [DOI] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Bailey A.O., Yates J.R. III, Bassett E.A., Wood S., Black B.E., and Cleveland D.W.. 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 137:472–484. 10.1016/j.cell.2009.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., and Yanagida M.. 2007. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell. 12:17–30. 10.1016/j.devcel.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Fukagawa T., and Earnshaw W.C.. 2014. The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell. 30:496–508. 10.1016/j.devcel.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie D.T. 1977. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81:2340–2361. 10.1021/j100540a008 [DOI] [Google Scholar]
- Gonsior S.M., Platz S., Buchmeier S., Scheer U., Jockusch B.M., and Hinssen H.. 1999. Conformational difference between nuclear and cytoplasmic actin as detected by a monoclonal antibody. J. Cell Sci. 112:797–809. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., and Yanagida M.. 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 118:715–729. 10.1016/j.cell.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Inoué , S. 1986. Digital image processing and analysis. In Video Microscopy. Chapter 10. Plenum Press, New York: 327–392. 10.1007/978-1-4757-6925-8_10 [DOI] [Google Scholar]
- Jansen L.E., Black B.E., Foltz D.R., and Cleveland D.W.. 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176:795–805. 10.1083/jcb.200701066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagana A., Dorn J.F., De Rop V., Ladouceur A.M., Maddox A.S., and Maddox P.S.. 2010. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat. Cell Biol. 12:1186–1193. 10.1038/ncb2129 [DOI] [PubMed] [Google Scholar]
- Lindeberg T. 1993. Detecting salient blob-like image structures and their scales with a scale-space primal sketch: A method for focus-of-attention. Int. J. Comput. Vis. 11:283–318. 10.1007/BF01469346 [DOI] [Google Scholar]
- Liu C., Chuang J.Z., Sung C.H., and Mao Y.. 2015. A dynein independent role of Tctex-1 at the kinetochore. Cell Cycle. 14:1379–1388. 10.1080/15384101.2014.1000217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P.S., Hyndman F., Monen J., Oegema K., and Desai A.. 2007. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 176:757–763. 10.1083/jcb.200701065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H., Masukata H., Muro Y., Nozaki N., and Okazaki T.. 1989. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 109:1963–1973. 10.1083/jcb.109.5.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D., Carrero G., Andrin C., de Vries G., and Hendzel M.J.. 2006. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J. Cell Biol. 172:541–552. 10.1083/jcb.200507101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley K.L., and Cheeseman I.M.. 2014. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell. 158:397–411. 10.1016/j.cell.2014.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Okawa K., Sekimoto T., Yoneda Y., Watanabe S., Ishizaki T., and Narumiya S.. 2009. mDia2 shuttles between the nucleus and the cytoplasm through the importin-alpha/beta- and CRM1-mediated nuclear transport mechanism. J. Biol. Chem. 284:5753–5762. 10.1074/jbc.M806191200 [DOI] [PubMed] [Google Scholar]
- Nechemia-Arbely Y., Fachinetti D., and Cleveland D.W.. 2012. Replicating centromeric chromatin: spatial and temporal control of CENP-A assembly. Exp. Cell Res. 318:1353–1360. 10.1016/j.yexcr.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu N. 1979. Threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9:62–66. 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- Schoenenberger C.A., Buchmeier S., Boerries M., Sütterlin R., Aebi U., and Jockusch B.M.. 2005. Conformation-specific antibodies reveal distinct actin structures in the nucleus and the cytoplasm. J. Struct. Biol. 152:157–168. 10.1016/j.jsb.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Shao X., Kawauchi K., Shivashankar G.V., and Bershadsky A.D.. 2015. Novel localization of formin mDia2: importin β-mediated delivery to and retention at the cytoplasmic side of the nuclear envelope. Biol. Open. 4:1569–1575. 10.1242/bio.013649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentirmay M.N., and Sawadogo M.. 2000. Spatial organization of RNA polymerase II transcription in the nucleus. Nucleic Acids Res. 28:2019–2025. 10.1093/nar/28.10.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N., Kato T., Fujita A., Ishizaki T., and Narumiya S.. 1999. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1:136–143. 10.1038/11056 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Ando Y., Yasuda S., Hosoya H., Watanabe N., Ishizaki T., and Narumiya S.. 2008. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol. Biol. Cell. 19:2328–2338. 10.1091/mbc.E07-10-1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J.V., Chambers S.H., and Smith P.J.. 1987. A pragmatic approach to the analysis of DNA histograms with a definable G1 peak. Cytometry. 8:1–8. 10.1002/cyto.990080101 [DOI] [PubMed] [Google Scholar]
- Xu Y., Moseley J.B., Sagot I., Poy F., Pellman D., Goode B.L., and Eck M.J.. 2004. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 116:711–723. 10.1016/S0092-8674(04)00210-7 [DOI] [PubMed] [Google Scholar]
- Yasuda S., Oceguera-Yanez F., Kato T., Okamoto M., Yonemura S., Terada Y., Ishizaki T., and Narumiya S.. 2004. Cdc42 and mDia3 regulate microtubule attachment to kinetochores. Nature. 428:767–771. 10.1038/nature02452 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.